ABSTRACT
The veterinary pathogens in the Staphylococcus intermedius group (SIG) are increasingly recognized as causes of human infection. Shared features between SIG and Staphylococcus aureus may result in the misidentification of SIG in human clinical cultures. This study examined the clinical and microbiological characteristics of isolates recovered at a tertiary-care academic medical center. From 2013 to 2015, 81 SIG isolates were recovered from 62 patients. Patients were commonly ≥50 years old, diabetic, and/or immunocompromised. Documentation of dog exposure in the electronic medical record was not common. Of the 81 SIG isolates, common sites of isolation included 37 (46%) isolates from wound cultures and 17 (21%) isolates from respiratory specimens. Although less common, 10 (12%) bloodstream infections were documented in 7 unique patients. The majority of SIG (65%) isolates were obtained from polymicrobial cultures. In comparison to S. aureus isolates from the same time period, significant differences were noted in proportion of SIG isolates that were susceptible to doxycycline (74% versus 97%, respectively; P < 0.001), trimethoprim-sulfamethoxazole (65% versus 97%, respectively; P < 0.001), and ciprofloxacin (78% versus 59%, respectively; P < 0.01). Methicillin resistance (MR) was detected in 12 (15%) of 81 SIG isolates. All MR isolates detected by an oxacillin disk diffusion test would have been misclassified as methicillin susceptible using a cefoxitin disk diffusion test. Thus, SIG is recovered from human clinical specimens, and distinction of SIG from S. aureus is critical for the accurate characterization of MR status in these isolates.
KEYWORDS: Staphylococcus intermedius group, Staphylococcus pseudintermedius, mecA, methicillin resistance
INTRODUCTION
Members of the Staphylococcus intermedius group (SIG) are commonly recovered from clinical specimens from animals. However, SIG strains are also associated with serious human infections. Recent advancements in bacterial identification methods, such as matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), have led to the observation that members of the SIG are increasingly isolated from human clinical specimens (1).
The SIG consists of S. pseudintermedius, S. intermedius, and S. delphini. These organisms are zoonotic pathogens that commonly colonize the mucosal surfaces of animals, particularly dogs (2). SIG strains are associated with skin and soft tissue infections, as well as invasive infections in animals. Previously, human infections caused by SIG strains were thought to be associated with animal contact, such as dog bites (3, 4). However, a recent investigation of SIG isolates obtained from 39 patients found an association with dogs in only 10% of cases (1). Prior reports have implicated SIG isolates in a variety of morbidities, including pneumonia, skin and soft tissue infections, hardware infections, and bacteremia (1, 4–6), but the epidemiology and clinical features of these infections have thus far not been extensively described.
In the past, phenotypic and biochemical profiling may have mistakenly identified SIG as Staphylococcus aureus, as both are coagulase-positive catalase-positive Gram-positive cocci that exhibit beta-hemolysis on blood agar plates (7, 8). Thus, the true incidence of the SIG as a cause of human infection may be underestimated. The performance of MALDI-TOF MS for the identification of SIG isolates has been evaluated, and while species-level identification within the complex is not well characterized, these studies support the idea that MALDI-TOF MS can accurately identify SIG isolates to a group level (9, 10).
Like S. aureus, SIG isolates can acquire methicillin resistance (MR) via expression of the mecA gene (11). The incidence of methicillin-resistant SIG isolates is increasing in veterinary clinical specimens (12, 13). Notably, prediction of MR in the SIG is most accurate using oxacillin as a surrogate for MR (14), and antimicrobial susceptibility testing (AST) standards have now been updated to reflect this detail (15, 16). For this reason, distinction from S. aureus is important, because cefoxitin is the recommended surrogate for resistance in S. aureus (15). In one recent study, the use of a cefoxitin disk diffusion test to predict MR in SIG resulted in very major error rates of 29.7% and 75.7% using coagulase-negative Staphylococcus (CNS) and S. aureus breakpoints, respectively (14). Conversely, there were no very major errors using an oxacillin disk diffusion test in combination with the S. pseudintermedius criteria from the CLSI VET01-S2 standards (14, 37), and standards for human AST are updated to reflect this (15). Thus, proper differentiation of the SIG from S. aureus is necessary to ensure accurate AST and facilitate appropriate therapy.
The objective of this study was to describe the epidemiology and clinical characteristics of SIG strains recovered from human clinical specimens. SIG isolates obtained as part of routine clinical care at a 1,250-bed tertiary-care academic medical center from 2013 to 2015 were retrospectively analyzed to define the salient characteristics surrounding each isolate. In addition, the antimicrobial susceptibility profiles were analyzed and compared to the institutional S. aureus antibiogram. To our knowledge, this is the largest study to date characterizing the epidemiology of human SIG infection.
(Preliminary results from this study were presented at the American Society for Microbiology Microbe Conference, 1 to 5 June 2017, New Orleans, LA, USA.)
MATERIALS AND METHODS
Isolate and subject characteristics.
Following approval from the Washington University in St. Louis institutional review board, a retrospective analysis was performed on all SIG strains isolated (n = 81) from clinical specimens from adult patients (≥18 years of age) submitted to the microbiology laboratory of a 1,250-bed tertiary-care hospital between 2013 and 2015. All isolates were identified and reported according to standard-of-care procedures at the time of isolation. The laboratory information system was queried for the microbiologic characteristics of each specimen, including source, colony morphology, biochemical testing results, Gram stain description, method of identification, number and type of microorganisms in polymicrobial cultures, and antimicrobial susceptibility testing results. Demographic and clinical data were obtained through chart reviews of 62 individual subjects included in the study. To compare the seasonality of SIG infections, the average temperature of the St. Louis area was compared to the number of SIG strains isolated in each quarter of 2013 to 2015. Average temperatures were pulled from data archived online (see https://www.weather.gov/media/lsx/climate/stl/temp/temp_stl_monthly_seasonal_averages.pdf).
Identification methods.
SIG isolates were identified either by phenotypic testing or MALDI-TOF MS using the Bruker Biotyper (Bruker Daltonics, Billerica, MA), according to the standard operating procedures of the laboratory at the time an isolate was recovered. The Bruker Biotyper software versions in use over the course of the study included 3.1, 4.0, and 5.0. Phenotypic testing included coagulase, catalase, pyrrolidonyl arylamidase (PYR; Thermo Scientific/Oxoid, Waltham, MA), and Staphylococcus latex agglutination (StaphTEX; Hardy Diagnostics, Santa Maria, CA) testing. Alternatively, isolates were phenotypically identified using the Vitek 2 system with Gram-positive ID (GPID) cards (bioMérieux, St. Louis, MO).
Antimicrobial susceptibility testing.
All SIG isolates were routinely tested for antimicrobial susceptibility using the Vitek 2 system or Kirby-Bauer disk diffusion test performed on Mueller-Hinton agar (BD, Franklin Lakes, NJ). AST on the Vitek 2 system was performed using the GP70 card, according to the manufacturer's recommendations. Testing was performed and interpreted according to CLSI standards (15, 18). Prior to the recommendation that oxacillin be used as a surrogate for methicillin resistance in SIG isolates, cefoxitin was used as a surrogate and interpreted according to the coagulase-negative Staphylococcus breakpoints (19). The susceptibility testing results were compared to the 2015 Staphylococcus aureus antibiogram that was produced in accordance with CLSI guidelines (20). Penicillin-binding protein 2a (PBP2a) was tested using a rapid immunochromatographic assay according to the manufacturer's instructions (PBP2a culture colony test; Alere, Waltham, MA).
Statistical analysis.
Fisher's exact test and linear regression analyses were performed in R (version 3.3.1).
RESULTS
Epidemiology of human SIG infections.
A total of 81 SIG isolates from 62 unique patients recovered from 2013 to 2015 were included in the analysis. A total of 32 (52%) patients were male, and the overall median age was 58 years (interquartile range [IQR], 45.3 to 63.8 years) (Table 1). In comparison, 55% of patients with either S. aureus (n = 2,655) or CNS (n = 1,537) isolated in culture from our laboratory during the same time period were male (P = 0.83) (21). The majority of patients (n = 32 [52%]) with SIG infection were between the ages of 50 and 69 years old, in comparison to 46% of patients with S. aureus and 42% with CNS (21). Common comorbidities for SIG infection included diabetes (n = 18 [29%]) and immunosuppression (n = 17 [27%]), and medical hardware was reported to be present in 18 cases (29%) (Table 1). A review of medical records found evidence of any contact with a dog in only 4 patients (6%). Patients with SIG infection were most commonly managed by dermatology (n = 14 [23%]) or otolaryngology (n = 11 [18%]). The primary source of specimens was wounds (n = 37 [46%]), in comparison to 17% of S. aureus and 12% of CNS isolates reported in wound cultures during the same time period (P < 0.001). Respiratory specimens were the other major source (n = 17 [21%]) of SIG isolates, compared to 17% of S. aureus and 0.2% of CNS isolates (P < 0.001) (21). The remainder of the SIG isolates were obtained from sterile specimens, including tissues (n = 13 [16%]), bone and synovial fluid (n = 4 [5%]), and blood (n = 10 [12%]). Notably, SIG isolates were obtained from the blood of 7 (9%) unique patients.
TABLE 1.
Patient characteristics
| Patient characteristic | No. (%) |
|---|---|
| Sex | |
| Male | 32 (52) |
| Female | 30 (48) |
| Age (yr) | |
| <20 | 2 (3) |
| 20–29 | 2 (3) |
| 30–39 | 9 (15) |
| 40–49 | 7 (11) |
| 50–59 | 13 (21) |
| 60–69 | 19 (31) |
| 70–79 | 9 (15) |
| 80–89 | 1 (2) |
| Comorbidities | |
| Diabetes | 18 (29) |
| Immunosuppression | 17 (27) |
| Transplant | 7 (41) |
| Steroids | 3 (18) |
| Chemotherapy | 6 (35) |
| Cystic fibrosis | 1 (6) |
| Hardware | 18 (29) |
| Central catheter | 11 (61) |
| Orthopedic hardware | 3 (17) |
| Othera | 4 (22) |
| Evidence of any contact with dog in medical record | 4 (6) |
| Medical services | |
| Bone marrow transplant/oncology | 5 (8) |
| Dermatology | 14 (23) |
| Orthopedics | 5 (8) |
| Otolaryngology | 11 (18) |
| Medicine | 5 (8) |
| Emergency department | 5 (8) |
| Otherb | 17 (27) |
| Infection source | |
| Wound | 37 (46) |
| Respiratory/oticc | 17 (21) |
| Tissue | 13 (16) |
| Blood | 10 (12) |
| Other, steriled | 4 (5) |
Peripheral catheter, left ventricular assist device, insulin pump, and tracheostomy tube.
Cardiology, gynecology, infectious disease, neurology, podiatry, pulmonary, and surgery.
Lower respiratory (n = 9), sinus drainage (n = 7), and ear drainage (n = 1).
Bone and synovial fluid.
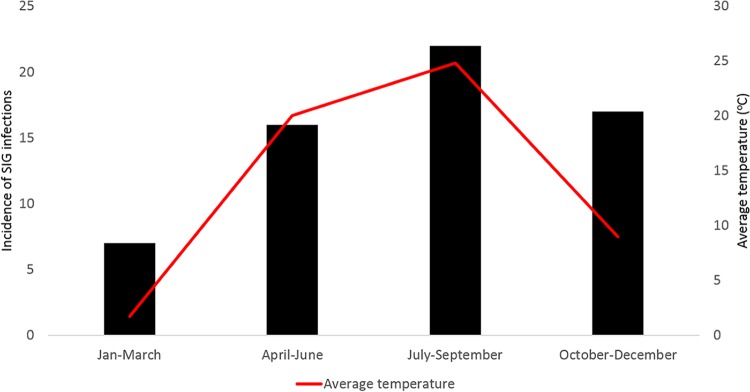
The incidence of SIG infections was compared to the months in which SIG isolates were obtained (Fig. 1). The majority of SIG isolates were reported between the months of July and September, while the incidence was lowest in the months of January to March. This trend tracked with the average temperature for the area (Fig. 1). Linear regression analysis showed a statistically significant (P < 0.01) although relatively minor increase in the monthly incidence of SIG infection with increasing average monthly temperature (0.05 with each increase of 1°C).
FIG 1.
Comparison of average seasonal temperature (degrees Celsius) and incidence of SIG infection. The numbers of SIG isolates obtained quarterly from 2013 to 2015 (gray bars) are compared to the average temperature for the area (red line) in the same time period.
Microbiologic characteristics of human clinical isolates of SIG.
Specimen Gram stains were performed and reported on 70 of 81 specimens. Of these, 34 specimens (49%) had no polymorphonuclear leukocytes (PMNs) present, 21 specimens (30%) had rare to few PMNs (≤10 present per low-power field), and 15 specimens (21%) had moderate to abundant PMNs (>10 present per low-power field) (Table 2). Gram-positive cocci were noted on 42 (60%) of the Gram stains and were associated with PMNs in 22 (31%) Gram-stained specimens (Table 2).
TABLE 2.
Culture characteristics
| Culture characteristics | No. (%) |
|---|---|
| Specimen Gram stain | |
| Polymorphonuclear leukocytes | |
| None | 34 (49) |
| Rare/few | 21 (30) |
| Moderate/abundant | 15 (21) |
| Gram-positive cocci present | 42 (60) |
| PMNs and Gram-positive cocci present | 22 (31) |
| Βeta-hemolysis on blood agar | 78 (96) |
| Method of SIG identification | |
| Phenotypic | 29 (36) |
| Errors in initial identification | 6 (21) |
| MALDI-TOF MSa | 52 (64) |
| Errors in initial identification | 0 (0) |
| PYRb | |
| Positive | 26 (96) |
| Negative | 1 (4) |
| Staphylococcal latex agglutination | |
| Positive | 6 (15) |
| Negative | 19 (47.5) |
| Questionable | 15 (37.5) |
Matrix-assisted laser desorption ionization–time of flight mass spectrometry with Bruker Biotyper.
Pyrrolidonyl arylamidase.
The majority of SIG isolates exhibited beta-hemolysis on blood agar plates (78/81 [96%]). Isolates were identified using phenotypic methods (29/81 [36%]) prior to the implementation of MALDI-TOF MS, which was used for identification of the majority of isolates (52/81 [64%]). Notably, errors in the initial identification of SIG isolates were reported in patient charts in 6 out of 29 (21%) instances using phenotypic methods, while no errors in identification occurred with the use of MALDI-TOF MS. Biochemical testing using the PYR test was used to differentiate SIG isolates from other Staphylococcus species. The result was positive in 26 of 27 isolates (96%) tested. Staphylococcus latex agglutination testing was performed on 39 isolates, with results recorded as positive, negative, or questionable for 6 (15%), 19 (47.5%), and 15 (37.5%) isolates, respectively.
The majority of SIG isolates were obtained from polymicrobial cultures (53/81 [65%]). A SIG isolate was accompanied by one other organism in 15 (19%) cultures, while 28 (35%) isolates were obtained from mixed cultures (>3 organisms) (Table 3). In polymicrobial cultures, the organisms most commonly isolated from SIG cultures included CNS species (9/53 [17%]), Streptococcus agalactiae (4/53 [8%]), and Escherichia coli (4/53 [8%]). Both S. aureus and Staphylococcus lugdunensis were recovered from 3 of 53 (6%) polymicrobial cultures (Table 3).
TABLE 3.
Composition of SIG cultures
| SIG culture characteristic | No. (%) |
|---|---|
| Monomicrobial cultures | 28 (35) |
| Polymicrobial cultures | 53 (65) |
| No. of other organisms in culture: | |
| 1 | 15 (19) |
| 2 | 8 (10) |
| 3 | 2 (2) |
| Mixed | 28 (35) |
| Organisms in polymicrobial cultures | |
| Gram-positive bacteria | |
| Staphylococcus aureus | 3 |
| Staphylococcus lugdunensis | 3 |
| Coagulase-negative Staphylococcus spp. | 9 |
| Enterococcus faecalis | 3 |
| Coryneform bacteria | 3 |
| Streptococcus agalactiae | 4 |
| Streptococcus anginosus group | 1 |
| Streptococcus pneumoniae | 1 |
| Other Streptococcus spp. | 4 |
| Non-anthracis Bacillus | 1 |
| Gram-negative bacteria | |
| Pseudomonas aeruginosa | 3 |
| Other Pseudomonas spp. | 2 |
| Escherichia coli | 4 |
| Enterobacter cloacae | 2 |
| Serratia marcescens | 1 |
| Acinetobacter calcoaceticus-A. baumannii complex | 2 |
| Stenotrophomonas maltophilia | 1 |
| Other Gram-negative bacilli | 2 |
| Anaerobes | |
| Bacteroides fragilis complex | 1 |
| Fusobacterium nucleatum | 1 |
| Fungi | |
| Yeast | 3 |
| Aspergillus fumigatus | 1 |
Antimicrobial susceptibility profiles of SIG.
The susceptibilities of SIG isolates to various antimicrobial agents were compared to the institutional S. aureus antibiogram from the same time period (Table 4). While there was no difference in susceptibility to clindamycin (70% susceptible for both SIG isolates and S. aureus, P = 1.0), there were significant differences in the percentages of SIG isolates that were susceptible compared to S. aureus for doxycycline (74% versus 97%, respectively; P < 0.001), trimethoprim-sulfamethoxazole (65% versus 97%, respectively; P < 0.001), and ciprofloxacin (78% versus 59%, respectively; P < 0.01). Of the SIG isolates, 12 (15%) isolates exhibited MR. Among the MR isolates, 25% and 8% tested were susceptible to clindamycin and trimethoprim-sulfamethoxazole, respectively. Ten MR isolates were tested for doxycycline and ciprofloxacin, of which 50% and 0% were susceptible, respectively.
TABLE 4.
Comparison of antibiotic susceptibility of SIG and S. aureus isolatesa
| Antibiotic | Susceptible (%) |
P value | |
|---|---|---|---|
| SIG | S. aureusb | ||
| Oxacillin (surrogate)c | 81 | NA | |
| Cefoxitin (surrogate) | NA | 50 | |
| Clindamycin | 70 | 70 | 1.0 |
| Doxycycline | 74 | 97 | <0.001 |
| Trimethoprim-sulfamethoxazole | 65 | 97 | <0.001 |
| Ciprofloxacin | 78 | 59 | <0.01 |
S. aureus represents data from a 2015 S. aureus antibiogram. NA, not available.
Interpretive criteria from CLSI, M100, 25th edition (38).
n = 54 (67%) isolates for which an oxacillin result was available.
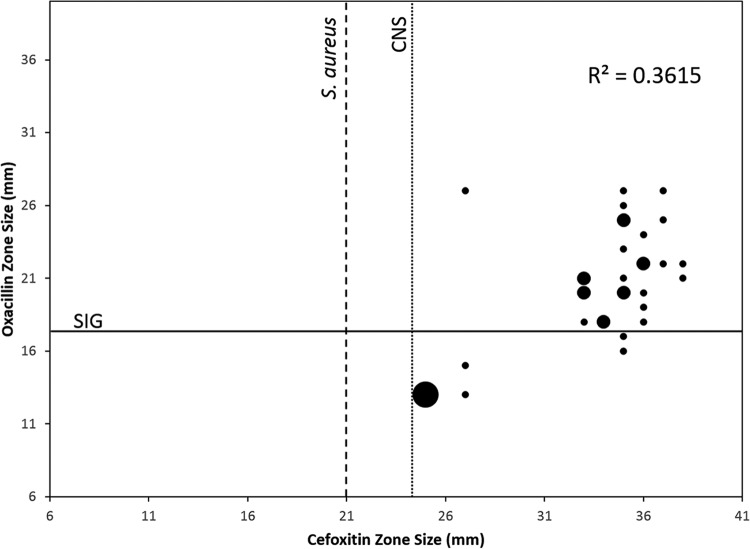
In isolates that were tested using oxacillin as a surrogate for MR prediction in SIG isolates (n = 54), 81% were methicillin susceptible. In comparison, S. aureus isolates tested within the same time period (n = 1,604) were 50% susceptible using cefoxitin as a surrogate for MR (Table 4). Both cefoxitin and oxacillin disk zone sizes were recorded for 34 SIG isolates. A comparison of the zones found no correlation (R2 value = 0.3615) between the cefoxitin and oxacillin test results (Fig. 2). Seven of 34 (21%) isolates tested by both cefoxitin and oxacillin disk diffusion tests exhibited MR using oxacillin as a surrogate. Using either the S. aureus or CNS breakpoints derived from the CLSI M100-S27 standards (15), all 7 of the MR SIG isolates would have been falsely predicted as susceptible if cefoxitin were used as a surrogate for methicillin susceptibility. Two of 10 SIG isolates that were tested for PBP2a by rapid immunoassay were positive. The PBP2a result correlated with 7 of 7 isolates tested by oxacillin disk diffusion and 3 of 3 tested by cefoxitin disk diffusion.
FIG 2.
Correlation of oxacillin and cefoxitin zone sizes (in millimeters) for a subset of SIG isolates (n = 34) tested by both cefoxitin and oxacillin disk diffusion. The lines represent susceptibility breakpoints for coagulase-negative Staphylococcus (CNS, dotted line), S. aureus (dashed line), and SIG (solid line), as described in CLSI standards (17). Seven MR isolates were mischaracterized as methicillin susceptible by cefoxitin (lower right section of graph). Small points indicate 1 isolate, medium-sized points represent 2 isolates, and the largest point represents 3 isolates.
DISCUSSION
SIG isolates are increasingly recognized as a cause of human infection (1, 4, 5, 7). Our retrospective review of all SIG isolates obtained in our clinical microbiology laboratory during a 3-year period found that SIG isolates are most commonly recovered from wounds and respiratory specimens. Risk factors associated with SIG infection included diabetes, immunosuppression, and the presence of hardware. A previous analysis of 24 human cases of SIG infection found that 92.1% of cases were related to contact with dogs at the time of infection (4). However, a review of the medical record in our study found an association with dogs in less than 7% of cases, with only one case of an injury from a dog bite. This is not to say that the patients in our cohort did not have contact with dogs; rather, animal exposure was not documented often in the medical record. Our findings are consistent with the results of a previous study in which animal contact was noted in less than 10% of the clinical documentation of SIG cases (1). In spite of this, dogs are potential sources for human infection, as both dog-to-dog and dog-to-human transmission have been described (22, 23). Interestingly, we noted a trend toward increased incidence of SIG infection reported in warmer months and less occurrence in the colder months. This is consistent with a higher likelihood of outdoor activities and possible animal exposure.
Given the polymicrobial makeup of the majority of cases in our study, the significance of SIG isolation may be questionable. However, the presence of PMNs and Gram-positive cocci was noted in the majority of isolates in which a Gram stain was available, supporting the idea that SIG isolates are important contributors to infection in many of these instances. In addition, SIG isolates were associated with 8 cases of invasive disease in our study, including joint infection and bacteremia. The majority of cases of bloodstream infections occurred in immunosuppressed patients with catheters. However, in one diabetic patient without hardware, SIG bacteria were isolated from neck abscess, joint fluid, and blood specimens. In this case, the presence of three dogs in the patient's household was detailed in the medical record.
The use of MALDI-TOF MS for bacterial species identification has led to the emergence of several microorganisms once thought to be uncommon human pathogens (24–27). Phenotypic characteristics of SIG, including the presence of beta-hemolysis on blood agar and a positive coagulase reaction, may lead to the mischaracterization of SIG as S. aureus in human clinical isolates (7, 8). This is supported by our data, in which six errors were made in the initial identification of SIG isolates using phenotypic methods, compared to no errors in reporting the identification after implementation of MALDI-TOF MS. Notably, no errors were the result of a false identification by the Vitek 2 system. A comparison of the local S. aureus antibiogram found that SIG isolates are significantly more likely to be resistant to doxycycline and trimethoprim-sulfamethoxazole, which could serve as an additional unique identifying characteristic of SIG isolates of which laboratories should be aware. While this pattern of multidrug resistance was most strongly associated with MR SIG isolates, this resistance profile was significantly different from S. aureus among methicillin-susceptible SIG isolates as well. Additionally, the PYR test is a useful biochemical test for the differentiation of SIG from S. aureus and most other Staphylococcus species (with S. lugdunensis being a notable exception) (28). One isolate in our study was negative by manual PYR testing. This could have been due to a technical error during PYR testing, as the isolate was identified as SIG by Vitek 2 and MALDI-TOF MS.
Studies of both human and veterinary isolates suggest that globally, SIG isolates are genetically heterogeneous and have arisen from multiple recombination events (29, 30). The spread of MR in SIG isolates is postulated to occur via gene transfer rather than clonal spread, which is supported by the finding that MR occurred in diverse SIG populations (29). Several studies have noted that MR in veterinary isolates is associated with prior antimicrobial treatment (29, 31, 32).
With the rising incidence of MR, correct identification of SIG is essential to ensure accurate prediction of MR in SIG isolates. Like S. aureus, MR in SIG isolates is usually mediated by the production of PBP2a encoded by mecA (11). Unlike S. aureus, in which disk diffusion testing using cefoxitin is the surrogate for MR, disk diffusion testing with oxacillin is the indicator of MR for SIG isolates (15). This is supported by our data, which found no correlation between oxacillin and cefoxitin zone sizes for a proportion of SIG isolates that were tested using both agents. Twelve isolates were methicillin resistant in our study, and this was almost always accompanied by multidrug resistance to other antibiotics, including clindamycin, ciprofloxacin, and doxycycline. Methicillin resistance in our isolates may have been underreported, as 13 susceptible isolates were tested using only cefoxitin disk diffusion testing prior to the updated recommendation to use oxacillin. In the absence of oxacillin disk diffusion testing, PBP2a may be an acceptable substitute to predict methicillin resistance (11, 14). Two recent studies that evaluated the performance of a rapid immunochromatographic assay found that the method was able to detect methicillin resistance with 100% sensitivity and specificity, albeit some isolates required induction by cefoxitin (11, 14). Notably, PBP2a testing was performed for 3 of the 13 susceptible isolates in our study, all with a negative result.
In the past, the isolation of Staphylococcus species that were not S. aureus were often disregarded as contaminants, and species-level identification of many staphylococci required cumbersome or time-consuming methods. However, advances in bacterial identification methods have facilitated species-level identification of many Staphylococcus species, which in turn has revealed the contribution of many CNS species to clinically significant disease (33–35). Furthermore, the importance of species-level identification of Staphylococcus species may be necessary to accurately predict MR, as in the case of the SIG and Staphylococcus schleiferi (36). It is likely that as our understanding of the importance of many Staphylococcus species continues to improve, the necessity for additional species-specific breakpoints may be necessary.
To our knowledge, this is the largest and most comprehensive study of the clinical and microbiological characteristics of human clinical infection with the SIG to date. One limitation of this study is that multiple identification methods were used over the course of the study, and the methods used could reliably resolve SIG isolates only to the group level. While this is sufficient for clinical management, we were unable to determine if certain members of the group were more or less common in human infection than others. Additionally, it is possible that some SIG isolates were not detected by the laboratory in polymicrobial cultures or as a result of errors in the identification of SIG isolates using phenotypic methods. Due to the retrospective study design, the colony morphology, Gram stain descriptions, biochemical testing, and antimicrobial agents tested were limited to what was described in the standard-of-care culture record. Finally, this is a single-center study, and it is unknown how these findings might apply to other settings.
In conclusion, SIG bacteria are an underappreciated cause of human infection, especially skin and soft tissue infections, bacteremia, and respiratory infections. As dogs and other animals are potential reservoirs for MR SIG isolates, knowledge of animal exposure in patients with SIG isolates growing in culture is helpful to determine clinical significance and prevent further transmission. Correct species-level identification is essential for an optimal interpretation of antimicrobial susceptibility testing results and subsequent antimicrobial therapy. While SIG isolates can be reliably identified by MALDI-TOF MS, clinical microbiology laboratories that rely on phenotypic methods of identification should be aware of the microbiological characteristics of this organism that differentiate it from other Staphylococcus species.
REFERENCES
- 1.Lee J, Murray A, Bendall R, Gaze W, Zhang L, Vos M. 2015. Improved detection of Staphylococcus intermedius group in a routine diagnostic laboratory. J Clin Microbiol 53:961–963. doi: 10.1128/JCM.02474-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weese JS. 2012. Staphylococcal infections, p 340–348. In Greene CE. (ed), Infectious diseases of the dog and cat, 4th ed Elsevier Saunders, St. Louis, MO. [Google Scholar]
- 3.Talan DA, Goldstein E, Staatz D, Overturf GD. 1989. Staphylococcus intermedius: clinical presentation of a new human dog bite pathogen. Ann Emerg Med 18:410–413. doi: 10.1016/S0196-0644(89)80582-7. [DOI] [PubMed] [Google Scholar]
- 4.Somayaji R, Priyantha MA, Rubin JE, Church D. 2016. Human infections due to Staphylococcus pseudintermedius, an emerging zoonosis of canine origin: report of 24 cases. Diagn Microbiol Infect Dis 85:471–476. doi: 10.1016/j.diagmicrobio.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Viau R, Hujer AM, Hujer KM, Bonomo RA, Jump RL. 2015. Are Staphylococcus intermedius infections in humans cases of mistaken identity? A case series and literature review. Open Forum Infect Dis 2: ofv110. doi: 10.1093/ofid/ofv110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerstadt K, Daly JS, Mitchell M, Wessolossky M, Cheeseman SH. 1999. Methicillin-resistant Staphylococcus intermedius pneumonia following coronary artery bypass grafting. Clin Infect Dis 29:218–219. doi: 10.1086/520168. [DOI] [PubMed] [Google Scholar]
- 7.Pottumarthy S, Schapiro JM, Prentice JL, Houze YB, Swanzy SR, Fang FC, Cookson BT. 2004. Clinical isolates of Staphylococcus intermedius masquerading as methicillin-resistant Staphylococcus aureus. J Clin Microbiol 42:5881–5884. doi: 10.1128/JCM.42.12.5881-5884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Börjesson S, Gomez-Sanz E, Ekstrom K, Torres C, Gronlund U. 2015. Staphylococcus pseudintermedius can be misdiagnosed as Staphylococcus aureus in humans with dog bite wounds. Eur J Clin Microbiol Infect Dis 34:839–844. doi: 10.1007/s10096-014-2300-y. [DOI] [PubMed] [Google Scholar]
- 9.Murugaiyan J, Walther B, Stamm I, Abou-Elnaga Y, Brueggemann-Schwarze S, Vincze S, Wieler LH, Lubke-Becker A, Semmler T, Roesler U. 2014. Species differentiation within the Staphylococcus intermedius group using a refined MALDI-TOF MS database. Clin Microbiol Infect 20:1007–1015. doi: 10.1111/1469-0691.12662. [DOI] [PubMed] [Google Scholar]
- 10.Dubois D, Grare M, Prere MF, Segonds C, Marty N, Oswald E. 2012. Performances of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for rapid identification of bacteria in routine clinical microbiology. J Clin Microbiol 50:2568–2576. doi: 10.1128/JCM.00343-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold AR, Burnham CA, Ford BA, Lawhon SD, McAllister SK, Lonsway D, Albrecht V, Jerris RC, Rasheed JK, Limbago B, Burd EM, Westblade LF. 2016. Evaluation of an immunochromatographic assay for rapid detection of penicillin-binding protein 2a in human and animal Staphylococcus intermedius group, Staphylococcus lugdunensis, and Staphylococcus schleiferi clinical isolates. J Clin Microbiol 54:745–748. doi: 10.1128/JCM.02869-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bemis DA, Jones RD, Videla R, Kania SA. 2012. Evaluation of cefoxitin disk diffusion breakpoint for detection of methicillin resistance in Staphylococcus pseudintermedius isolates from dogs. J Vet Diagn Invest 24:964–967. doi: 10.1177/1040638712452112. [DOI] [PubMed] [Google Scholar]
- 13.Humphries RM, Wu MT, Westblade LF, Robertson AE, Burnham CA, Wallace MA, Burd EM, Lawhon S, Hindler JA. 2016. In vitro antimicrobial susceptibility of Staphylococcus pseudintermedius isolates of human and animal origin. J Clin Microbiol 54:1391–1394. doi: 10.1128/JCM.00270-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu MT, Burnham CA, Westblade LF, Dien Bard J, Lawhon SD, Wallace MA, Stanley T, Burd E, Hindler J, Humphries RM. 2016. Evaluation of oxacillin and cefoxitin disk and MIC breakpoints for prediction of methicillin resistance in human and veterinary isolates of Staphylococcus intermedius group. J Clin Microbiol 54:535–542. doi: 10.1128/JCM.02864-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CLSI. 2017. Performance standards for antimicrobial susceptibility testing; 27th informational supplement. CLSI document M100-S27. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 16.European Committee on Antimicrobial Susceptibility Testing. 2017. Breakpoint tables for interpretation of MICs and zone diameters. Version 7.1. European Committee on Antimicrobial Susceptibility Testing, Växjö, Sweden: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_7.1_Breakpoint_Tables.pdf. [Google Scholar]
- 17.Reference deleted.
- 18.CLSI. 2015. Performance standards for antimicrobial disk susceptibility tests; approved standard, 12th ed CLSI document M02-A12. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 19.CLSI. 2013. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. CLSI document M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 20.CLSI. 2014. Analysis and presentation of cumulative antimicrobial susceptibility test data; approved guideline, 4th ed CLSI document M39-A4. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 21.McMullen AR, Anderson NW, Wallace MA, Shupe A, Burnham CA. 2017. When good bugs go bad: epidemiology and antimicrobial resistance profiles of Corynebacterium striatum, an emerging multidrug resistant, opportunistic pathogen. Antimicrob Agents Chemother 62:e01111-17. doi: 10.1128/AAC.01111-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lozano C, Rezusta A, Ferrer I, Perez-Laguna V, Zarazaga M, Ruiz-Ripa L, Revillo MJ, Torres C. 2017. Staphylococcus pseudintermedius human infection cases in Spain: dog-to-human transmission. Vector Borne Zoonotic Dis 17:268–270. doi: 10.1089/vbz.2016.2048. [DOI] [PubMed] [Google Scholar]
- 23.Windahl U, Gren J, Holst BS, Borjesson S. 2016. Colonization with methicillin-resistant Staphylococcus pseudintermedius in multi-dog households: a longitudinal study using whole genome sequencing. Vet Microbiol 189:8–14. doi: 10.1016/j.vetmic.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 24.Le Brun C, Robert S, Bruyere F, Lanotte P. 2015. Emerging uropathogens: point for urologists and biologists. Prog Urol 25:363–369. (In French.) doi: 10.1016/j.purol.2015.01.021. [DOI] [PubMed] [Google Scholar]
- 25.Leal SM Jr, Jones M, Gilligan PH. 2016. Clinical significance of commensal Gram-positive rods routinely isolated from patient samples. J Clin Microbiol 54:2928–2936. doi: 10.1128/JCM.01393-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Non LR, Nazinitsky A, Gonzalez MD, Burnham CA, Patel R. 2015. Actinobaculum schaalii bacteremia: a report of two cases. Anaerobe 34:84–85. doi: 10.1016/j.anaerobe.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 27.Opota O, Prod'hom G, Andreutti-Zaugg C, Dessauges M, Merz L, Greub G, Chave JP, Jaton K. 2016. Diagnosis of Aerococcus urinae infections: importance of matrix-assisted laser desorption ionization time-of-flight mass spectrometry and broad-range 16S rDNA PCR. Clin Microbiol Infect 22:e1-2. doi: 10.1016/j.cmi.2015.08.026. [DOI] [PubMed] [Google Scholar]
- 28.Becker K, Skov RL, Von Eiff C. 2015. Staphylococcus, Micrococcus, and other catalase-positive cocci, p 354–382. In Jorgensen JH, Pfaller MA, Carroll KC, Funke G, Landry ML, Richter SS, Warnock DW (ed), Manual of clinical microbiology, 11th ed, vol 1 ASM Press, Washington, DC. [Google Scholar]
- 29.Grönthal T, Eklund M, Thomson K, Piiparinen H, Sironen T, Rantala M. 2017. Antimicrobial resistance in Staphylococcus pseudintermedius and the molecular epidemiology of methicillin-resistant S. pseudintermedius in small animals in Finland. J Antimicrob Chemother 72:1021–1030. doi: 10.1093/jac/dkx086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pires Dos Santos T, Damborg P, Moodley A, Guardabassi L. 2016. Systematic review on global epidemiology of methicillin-resistant Staphylococcus pseudintermedius: inference of population structure from multilocus sequence typing data. Front Microbiol 7:1599. doi: 10.3389/fmicb.2016.01599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eckholm NG, Outerbridge CA, White SD, Sykes JE. 2013. Prevalence of and risk factors for isolation of meticillin-resistant Staphylococcus spp. from dogs with pyoderma in northern California, USA. Vet Dermatol 24: 154–161. doi: 10.1111/j.1365-3164.2012.01051.x. [DOI] [PubMed] [Google Scholar]
- 32.Nienhoff U, Kadlec K, Chaberny IF, Verspohl J, Gerlach GF, Kreienbrock L, Schwarz S, Simon D, Nolte I. 2011. Methicillin-resistant Staphylococcus pseudintermedius among dogs admitted to a small animal hospital. Vet Microbiol 150:191–197. doi: 10.1016/j.vetmic.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 33.Argemi X, Riegel P, Lavigne T, Lefebvre N, Grandpre N, Hansmann Y, Jaulhac B, Prevost G, Schramm F. 2015. Implementation of matrix-assisted laser desorption ionization–time of flight mass spectrometry in routine clinical laboratories improves identification of coagulase-negative staphylococci and reveals the pathogenic role of Staphylococcus lugdunensis. J Clin Microbiol 53:2030–2036. doi: 10.1128/JCM.00177-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elamin WF, Ball D, Millar M. 2015. Unbiased species-level identification of clinical isolates of coagulase-negative staphylococci: does it change the perspective on Staphylococcus lugdunensis? J Clin Microbiol 53:292–294. doi: 10.1128/JCM.02932-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Becker K, Heilmann C, Peters G. 2014. Coagulase-negative staphylococci. Clin Microbiol Rev 27:870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bemis DA, Jones RD, Hiatt LE, Ofori ED, Rohrbach BW, Frank LA, Kania SA. 2006. Comparison of tests to detect oxacillin resistance in Staphylococcus intermedius, Staphylococcus schleiferi, and Staphylococcus aureus isolates from canine hosts. J Clin Microbiol 44:3374–3376. doi: 10.1128/JCM.01336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.CLSI 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; 2nd informational supplement. CLSI document VET01-S2 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 38.CLSI 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI document M100-S25 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]