ABSTRACT
Multistep algorithmic testing in which a sensitive nucleic acid amplification test (NAAT) is followed by a specific toxin A and toxin B enzyme immunoassay (EIA) is among the most accurate methods for Clostridium difficile infection (CDI) diagnosis. The obvious shortcoming of this approach is that multiple tests must be performed to establish a CDI diagnosis, which may delay treatment. Therefore, we sought to determine whether a preliminary diagnosis could be made on the basis of the quantitative results of the first test in algorithmic testing, which provide a measure of organism burden. To do so, we retrospectively analyzed two large collections of samples (n = 2,669 and n = 1,718) that were submitted to the laboratories of two Dutch hospitals for CDI testing. Both hospitals apply a two-step testing algorithm in which a NAAT is followed by a toxin A/B EIA. Of all samples, 208 and 113 samples, respectively, tested positive by NAAT. Among these NAAT-positive samples, significantly lower mean quantification cycle (Cq) values were found for patients whose stool eventually tested positive for toxin, compared with patients who tested negative for toxin (mean Cq values of 24.4 versus 30.4 and 26.8 versus 32.2; P < 0.001 for both cohorts). Receiver operating characteristic curve analysis was performed to investigate the ability of Cq values to predict toxin status and yielded areas under the curve of 0.826 and 0.854. Using the optimal Cq cutoff values, prediction of the eventual toxin A/B EIA results was accurate for 78.9% and 80.5% of samples, respectively. In conclusion, Cq values can serve as predictors of toxin status but, due to the suboptimal correlation between the two tests, additional toxin testing is still needed.
KEYWORDS: Clostridium difficile, diagnostic, NAAT quantitation
INTRODUCTION
Clostridium difficile (recently reclassified as Clostridioides difficile based on phenotypic, chemotaxonomic, and phylogenetic analyses [1]; for simplicity and consistency with previous literature, C. difficile is used in this paper) is an anaerobic, spore-producing bacterium that is responsible for C. difficile infection (CDI), the leading cause of nosocomial infectious diarrhea (2). Symptoms range from mild self-limiting diarrhea to potentially life-threatening fulminant colitis (3, 4). CDI occurs when alterations in the gut microbiome, particularly antibiotic-induced disruptions, create conditions favorable for C. difficile proliferation (5). Proliferation is followed by production of one or two enterotoxins, known as toxins A and B (toxin A/B), and in some strains a binary toxin, C. difficile transferase (CDT), whose inflammatory and necrotic effects on colonic tissue mediate the clinical symptoms of CDI (2).
CDI diagnostic methods continue to present problematic shortcomings. Establishing a CDI diagnosis is dependent on demonstrating the presence of toxin or toxigenic organism in stool samples (6). The two reference methods for doing so, namely, cell cytotoxicity neutralization assay (CCNA) and toxigenic culture, are lengthy laborious techniques whose clinical implementation is unrealistic. Therefore, rapid tests with the same aims have been developed. Enzyme immunoassays (EIAs) can be used to detect either toxin A/B or glutamate dehydrogenase (GDH), an abundant enzyme whose presence is indicative of C. difficile (both toxigenic and nontoxigenic strains). Similarly, nucleic acid amplification tests (NAATs) can detect the presence of toxin-producing genes. While these rapid tests are easily carried out in clinical settings, they too suffer from drawbacks. Toxin A/B EIA use was once widespread, given the etiological relationship between toxin and clinical symptoms, but recognition of the assay's low sensitivity (6) has changed this paradigm. Increasingly, NAATs have gained popularity, given their ease of use and high sensitivity. However, there is considerable debate regarding whether the presence of toxigenic organism alone warrants a diagnosis of CDI or should instead be considered C. difficile colonization (7–10). This has prompted the creation of multistep algorithms in which a first sensitive test (a NAAT or GDH EIA) is used to screen for the organism; in the event of a positive result, a highly specific second test for toxin detection (toxin A/B EIA) is used (6).
The algorithmic approach is currently recommended by common guidelines, such as those published by the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) (6). While algorithms do well to minimize false-positive and false-negative results, their obvious shortcoming is that, in the event of a positive first test, a second test must be performed to establish a diagnosis, potentially delaying treatment and isolation of true CDI patients or leading to premature treatment of non-CDI patients. In light of this shortcoming, we sought to determine whether the quantitative results of the first test (toxin A/B or toxin B NAAT) in a two-step algorithm could predict the eventual outcome of the second test (toxin A/B EIA).
MATERIALS AND METHODS
Study design and population.
This study was performed using CDI testing data from two Dutch hospitals, namely, the Leiden University Medical Center (LUMC) (a tertiary-care, university-affiliated hospital) and Amphia Hospital (a large general hospital). In both hospitals, CDI diagnoses are established using a two-step algorithm recommended by the ESCMID, in which a NAAT for the toxin A-producing gene (tcdA) and/or the toxin B-producing gene (tcdB) is, in the event of a positive result, followed by a toxin A/B EIA. All consecutive stool samples (from both inpatients and outpatients) that underwent CDI testing with this algorithm were considered. Samples from infants were included only if a specific request for CDI testing was made. For the LUMC, samples were included from January 2016 to March 2017; for Amphia Hospital, samples were included from January 2016 to January 2017. Additionally, LUMC data for adult asymptomatic patients who, upon admission for non-CDI-related reasons, agreed to have their stool tested for C. difficile and demonstrated positive culture results were included as controls. In the LUMC only, culture and ribotyping were performed for NAAT-positive samples.
Diagnostic tests.
Both hospitals use an in-house NAAT targeting tcdB only (LUMC) or both tcdB and tcdA (Amphia Hospital). For both sites, we used the tcdB quantification cycle (Cq) value for all calculations. The LUMC NAAT was performed as described previously (11). For the in-house NAAT in Amphia Hospital, DNA extraction was performed using the Nuclisens EasyMag system (bioMérieux, Marcy l'Etoile, France). This in-house assay has been validated internally and complies with the quality criteria described in the requirements of the International Organization for Standardization (ISO) (12). In short, a feces sample approximately the size of a pinhead was suspended in 1 ml of stool transport and recovery (STAR) buffer (Roche Diagnostics, Almere, The Netherlands) and frozen before further processing. After thawing, samples were homogenized in a MagNA Lyser system (Roche Diagnostics, Almere, The Netherlands) (6,000 × g for 30 s) and then centrifuged (14,000 × g for 1 min). A total of 100 μl of supernatant was used for automated nucleic acid extraction using the EasyMag system. Amplification of the tcdA and tcdB genes was performed with an ABI TaqMan 7500 real-time PCR system (Applied Biosystems, Nieuwerkerk a/d Ijssel, The Netherlands). Primers and probes used for the Amphia Hospital NAAT were described previously (13). TaqMan Universal PCR master mix (Applied Biosystems) and PCR plates were prepared using a Piro pipetting robot (Dornier, Lindau, Germany), with 20 μl of master mix and 5 μl of extracted DNA. The amplification protocol included 5 min at 50°C, 10 min at 95°C, and 45 cycles of 10 s at 95°C and 32 s at 60°C. Both laboratories used phocine herpesvirus as an internal control to test for PCR inhibition. At both hospitals, NAAT results were quantitated by measuring the Cq, i.e., the cycle at which the fluorescence from amplification exceeded the background fluorescence, which served as an indirect measure of how many copies of DNA were present in the sample tested. At the LUMC, a Vidas C. difficile Toxin A&B test (bioMérieux) was performed; values of >0.37 were considered positive, according to the manufacturer's instructions. Amphia Hospital used an ImmunoCard Toxins A&B test (Meridian Bioscience, Cincinnati, OH, USA); results were not quantitative and instead are presented as positive or negative. Both assays were performed according to the manufacturer's instructions.
On working days, NAATs were performed on the day of receipt. For weekends and holidays, NAATs were performed on the next working day. In case of a positive NAAT result, a toxin A/B EIA was performed on the same day or the next day. Samples were stored at 4°C until they were tested. Culture and ribotyping of NAAT-positive samples from the LUMC cohort was performed as described previously (14).
Statistical analysis.
Average Cq values were compared by t test and analysis of variance (ANOVA). The ability of Cq values to predict toxin presence was assessed with receiver operating characteristic (ROC) curves. Positive predictive values and negative predictive values were calculated for different Cq cutoff points. Results were considered significant below the 0.05 level. Analyses of data were performed using SPSS 23.0 statistical software (IBM, Armonk, NY, USA) and Stata SE 12.1 statistical software (StataCorp, College Station, TX, USA).
RESULTS
LUMC.
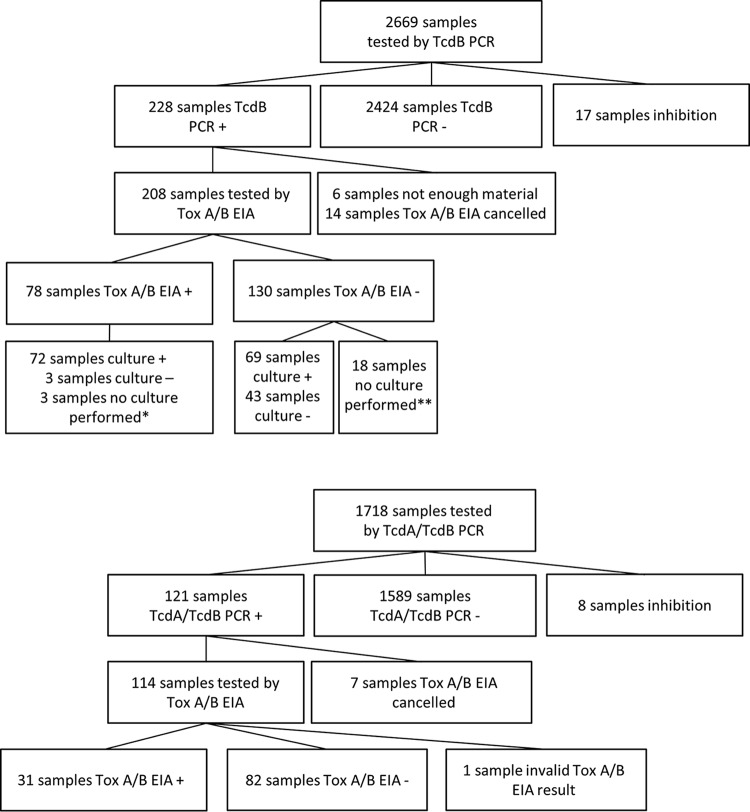
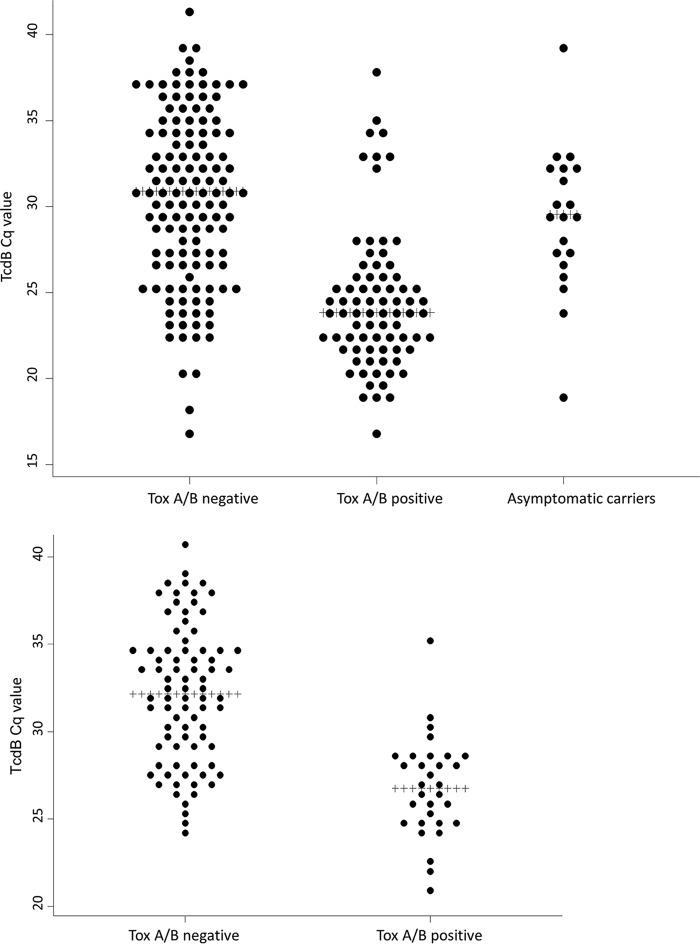
In total, 2,669 unformed stool samples from patients suspected of having CDI were tested with an in-house NAAT. Of those, 2,424 had negative results and 17 showed inhibition with the NAAT and were excluded from further analysis. Of the remaining 228 NAAT-positive samples, 20 were excluded from further analysis because the toxin A/B EIA was not performed (either because there was an insufficient amount of feces to perform the assay or because the assay was stopped for another reason). The remaining samples underwent toxin A/B EIA testing, yielding a final sample size of 208 (Fig. 1, top). Cq values for patients with positive (n = 78) or negative (n = 130) toxin A/B EIA results and for asymptomatic individuals who were found, via culture, to be asymptomatically colonized by C. difficile upon hospital admission are shown in Fig. 2, top. Comparable mean Cq values were observed for symptomatic patients with negative toxin results (mean Cq, 30.4 [95% confidence interval [CI], 29.5 to 31.3]) and asymptomatic carriers (mean Cq, 29.2 [95% CI, 27.3 to 31.2]), while symptomatic patients with positive toxin results had significantly lower mean Cq values, according to ANOVA (mean Cq, 24.4 [95% CI, 23.5 to 25.3]; P < 0.001). Seventeen outliers that were positive by toxin A/B EIA and had high Cq values were retested with a TcdB NAAT; the mean Cq value for those samples did not decrease after retesting. Samples were evaluated for PCR inhibition or irregular amplification curves, but neither was found to be a cause of these anomalies. Clinical data showed that only 1 of these samples was submitted for CDI testing during metronidazole treatment, 14 samples were submitted while no CDI antibiotics were being used, and antibiotic use was not clear for 2 samples. All except 1 of these 17 samples exhibited positive culture results, yielding 11 different ribotypes. The only culture-negative sample was from a patient with clinical suspicion of a CDI recurrence 4 months after a previous episode. After the positive CDI test result, the patient received oral metronidazole treatment.
FIG 1.
Flowchart of included samples. (Top) LUMC cohort. (Bottom) Amphia Hospital cohort. *, No culture was performed because a culture with positive results had been performed within the previous week. **, No culture was performed because a culture with positive results had been performed within the previous week (n = 1) or because positive TcdB results were obtained retrospectively during the implementation phase of the tcdB NAAT, when samples were routinely tested by toxin (Tox) A/B EIA only.
FIG 2.
Dot plots of observed Cq values. (Top) LUMC cohort and asymptomatic carriers. (Bottom) Amphia Hospital cohort.
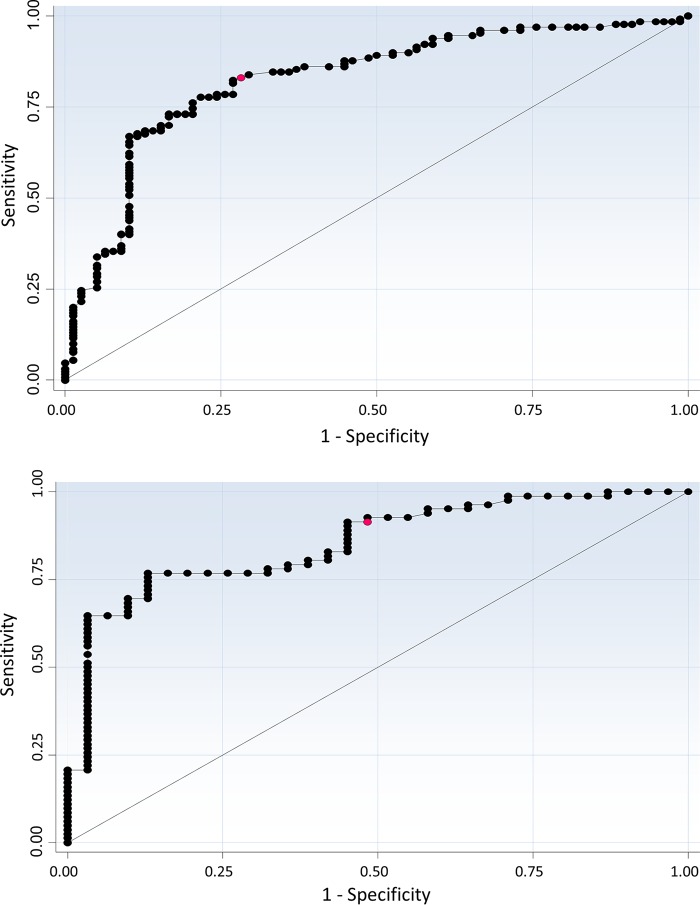
Based on the significantly lower Cq values observed for toxin-positive samples, a ROC curve was generated to calculate the ability of Cq values to predict toxin A/B EIA outcomes (Fig. 3, top). The area under the curve (AUC) was found to be 0.826 (P < 0.001), with an ideal cutoff value of 25.3 cycles (the value best able to discriminate between outcomes); 78.9% of samples would be correctly classified as toxin A/B EIA positive or negative using this Cq cutoff value. Measures of accuracy for the ideal cutoff value and others are shown in Table 1.
FIG 3.
ROC curves assessing the ability of Cq values to predict the presence of toxin. Optimal cutoff values are shown in red. (Top) LUMC cohort. (Bottom) Amphia Hospital cohort.
TABLE 1.
Accuracy of the ability of NAAT Cq cutoff values to predict toxin A/B EIA outcomesa
| Cohort and Cq cutoff value | Sensitivity (95% CI) (%) | Specificity (95% CI) (%) | Accuracy (95% CI) (%) | PPV (95% CI) (%) | NPV (95% CI) (%) |
|---|---|---|---|---|---|
| LUMC cohort | |||||
| 22.0 | 24.36 (15.35–35.40) | 96.92 (92.31–99.16) | 69.71 (62.98–75.87) | 82.61 (62.65–93.08) | 68.11 (65.23–70.86) |
| 23.0 | 41.03 (30.01–52.75) | 93.85 (88.23–97.31) | 74.04 (67.52–79.86) | 80.0 (66.02–89.17) | 72.62 (68.68–76.24) |
| 24.0 | 51.28 (39.69–62.77) | 88.46 (81.68–93.4) | 74.52 (68.03–80.29) | 72.73 (61.25–81.81) | 75.16 (70.50–79.30) |
| 25.3 | 71.79 (60.47–81.41) | 83.08 (75.51–89.08) | 78.85 (72.66–84.19) | 71.79 (62.92–79.25) | 83.08 (77.36–87.58) |
| 26.0 | 75.64 (64.6–84.65) | 77.69 (69.56–84.52) | 76.92 (70.59–82.47) | 67.05 (59.04–74.17) | 84.17 (78.06–88.82) |
| 27.0 | 82.05 (71.72–89.83) | 73.08 (64.60–80.48) | 76.44 (70.08–82.03) | 64.65 (57.49–71.20) | 87.16 (80.67–91.69) |
| 28.0 | 87.18 (77.68–93.68) | 68.46 (59.73–76.33) | 75.48 (69.05–81.17) | 62.39 (55.94–68.42) | 89.90 (83.14–94.14) |
| 29.0 | 89.74 (80.79–95.47) | 64.62 (55.75–72.80) | 74.04 (67.52–79.86) | 60.34 (54.38–66.02) | 91.30 (84.33–95.35) |
| Amphia Hospital cohort | |||||
| 24.0 | 9.68 (2.04–25.75) | 100 (95.60–100) | 75.22 (66.22–82.86) | 100 | 74.55 (72.3–76.67) |
| 25.0 | 29.03 (14.22–48.04) | 97.56 (91.47–99.7) | 78.76 (70.07–85.89 | 81.82 (50.72–95.16) | 78.43 (74.33–82.04) |
| 26.0 | 38.71 (21.85–57.81) | 95.12 (87.98–98.66) | 79.65 (71.04–86.64) | 75.0 (51.13–89.59) | 80.41 (75.55–84.51) |
| 27.0 | 51.61 (33.06–69.85) | 91.46 (83.2–96.5) | 80.53 (72.02–87.38) | 69.57 (51.01–83.38) | 83.33 (77.55–87.86) |
| 28.0 | 64.52 (45.37–80.77) | 78.05 (67.54–86.44) | 74.34 (65.26–82.09) | 52.63 (40.63–64.33) | 85.33 (78.12–90.46) |
| 29.0 | 87.1 (70.17–96.37) | 74.39 (63.56–83.4) | 77.88 (69.10–85.14) | 56.25 (46.46–65.57) | 93.85 (85.83–97.46) |
| 30.0 | 90.32 (74.25–97.96) | 68.29 (57.08–78.13) | 74.34 (65.26–82.09) | 51.85 (43.44–60.16) | 94.92 (86.31–98.22) |
| 31.0 | 96.77 (83.30–99.92) | 64.63 (53.30–74.88) | 73.45 (64.32–81.32) | 50.85 (43.40–58.26) | 98.15 (88.45–99.73) |
| 32.0 | 96.77 (83.30–99.92) | 56.10 (44.7–67.04) | 67.26 (57.79–75.79) | 45.45 (39.29–51.77) | 97.87 (86.89–99.69) |
Values were calculated as follows: sensitivity = samples with Cq values beneath the cutoff value/all toxin-positive samples; specificity = samples with Cq values above the cutoff value/all toxin-negative samples; accuracy = all correctly classified specimens; positive predictive value (PPV) = chance of positive toxin A/B EIA result among samples with Cq values beneath the cutoff value; negative predictive value (NPV) = chance of negative toxin A/B EIA result among samples with Cq values above the cutoff value.
Because LUMC data included PCR ribotypes, we investigated whether ribotype had an effect on our findings. Ribotype distributions were comparable for toxin A/B EIA-positive and -negative patients, by the chi-squared test (P = 0.26), and we did not find any differences in the mean Cq values for different ribotype categories (P = 0.55 for toxin-negative samples and P = 0.11 for toxin-positive samples) (Table 2).
TABLE 2.
Ribotype distribution and mean Cq values for toxin-positive and toxin-negative samples (LUMC cohort)
| Ribotype | Mean Cq value (no. of samples) |
|
|---|---|---|
| Toxin A/B EIA-negative samples | Toxin A/B EIA-positive samples | |
| 001 | 35.55 (2) | 26.38 (4) |
| 002 | 32.23 (4) | 21.15 (2) |
| 003 | 22.35 (2) | |
| 005 | 30.93 (3) | 22.83 (3) |
| 012 | 26.6 (2) | 22.35 (2) |
| 013 | 22.75 (2) | |
| 014/020 | 28.46 (14) | 22.56 (15) |
| 015 | 22.96 (5) | |
| 017 | 24.7 (1) | |
| 019 | 32.0 (1) | |
| 023 | 30.55 (2) | |
| 026 | 22.2 (1) | |
| 031 | 23.9 (1) | |
| 034 | 30.3 (1) | |
| 037 | 32.65 (2) | |
| 050 | 34.4 (1) | 21.8 (1) |
| 053 | 26.3 (1) | |
| 057 | 33.4 (1) | 27.3 (1) |
| 070 | 30.9 (1) | 21.9 (2) |
| 076 | 27.7 (1) | |
| 078/126 | 28.3 (15) | 25.79 (15) |
| 081 | 24.2 (2) | |
| 104 | 32.7 (1) | |
| 123 | 30.9 (1) | |
| 127 | 24.1 (1) | |
| 154 | 23.9 (1) | |
| 156 | 18.6 (1) | |
| 168 | 30.2 (1) | |
| 198 | 23.6 (1) | |
| 258 | 22.8 (1) | |
| 262 | 25.3 (1) | |
| 265 | 31.0 (3) | 29.55 (2) |
| 293 | 23.95 (2) | |
| 328 | 27.78 (4) | |
| 356 | 23.4 (1) | |
| Unknown | 26.92 (5) | 25.06 (5) |
Amphia Hospital.
A total of 1,718 unformed stool samples from patients suspected of having CDI were tested with an in-house NAAT (different from the LUMC NAAT). Of those, 1,589 had negative results and 8 showed inhibition and were excluded from further analysis. Seven of the 121 NAAT-positive samples were not tested with the toxin A/B EIA (2 were repeat samples from the same patient on the same day, 1 sample was from a gut biopsy, and 4 other samples were not tested otherwise). The remaining 114 samples underwent toxin A/B EIA testing. One sample had an invalid result from the second test (no detectable color in the reaction port) and was also excluded from further analysis, yielding a final sample size of 113 (Fig. 1, bottom). Cq values for toxin-positive (n = 31) and toxin-negative (n = 82) samples are shown in Fig. 2, bottom. Significantly lower mean Cq values were found for toxin-positive patients than for toxin-negative patients (mean Cq values of 26.8 [95% CI, 25.8 to 27.9] versus 32.2 [95% CI, 31.3 to 33.0]; P < 0.001). Evaluation of the 1 outlier that was positive by toxin A/B EIA and had a high Cq value revealed a normal shape for the amplification curve but no diarrhea at the time of the results (without treatment).
As performed for the other cohort, a ROC curve was generated to determine the ability of Cq values to predict toxin A/B EIA outcomes (Fig. 3, bottom). The AUC was 0.854 (P < 0.001), with an ideal cutoff value of 27.0; 80.5% of samples would be correctly classified. Measures of accuracy for the ideal cutoff value and others are shown in Table 1.
DISCUSSION
This study sought to determine whether quantitation of NAAT results could predict the presence or absence of toxin in subsequent testing. Significantly lower Cq values were found in stool samples that tested positive for toxin in two large cohorts from different hospitals. Concomitant ROC curves for both cohorts showed that, using the optimal Cq cutoff value, the toxin result could be predicted for at least 78% of the samples. With the recent emergence of NAATs as standalone tests or as the first step in an algorithm, there has been increasing interest in the use or nonuse of quantitation of NAAT results. There is a growing body of work showing an association between Cq values and toxin presence; toxin-positive samples are associated with lower Cq values or greater bacterial loads (15–22). Toxin presence is generally thought to be associated with CDI severity and outcomes (7, 8). Some studies indeed found Cq values to be predictors of clinical severity or outcomes, probably mediated through the presence or absence of toxins (22–24), although this was not confirmed in all studies (25, 26). A very recent paper was the first to describe the performance characteristics of NAAT Cq cutoff values for discriminating toxin-positive and toxin-negative stool samples (27). Our study adds to the literature by confirming that Cq values can indeed be used to predict toxin status. In our two cohorts, the optimal Cq cutoff values detected toxin-positive samples with positive and negative predictive values of 71.8% and 83.1% and 69.6% and 83.3%, respectively. Our study also indicates that local assessment of NAAT performance is warranted to determine cutoff values that can be adopted for clinical use, as Cq values are semiquantitative and depend on many factors involving the sample material, materials used, and assay conditions.
Clinical implementation of these findings may be beneficial. Algorithmic testing requires more time to establish a CDI diagnosis than standalone tests, which has been shown to negatively affect patient care (28). One way of addressing this problem would be to use Cq values to establish a preliminary diagnosis; this can be done by using the optimal Cq cutoff values to consider samples either likely toxin positive or likely toxin negative. Using this approach, as many samples as possible will be classified correctly. It might be argued, however, that toxin-positive samples should not be missed, as delayed treatment or delayed isolation measures may negatively affect patient care and C. difficile transmission. In that case, a Cq cutoff value with a high negative predictive value should be chosen, to classify samples with Cq values above the cutoff value as probably toxin negative. As an example, Cq cutoff values of 29.0 and 32.0 for the LUMC and Amphia Hospital cohorts, respectively, would correctly classify 91.3% and 97.9% of samples with Cq values above the cutoff values as negative. A preliminary diagnosis based on one of these two approaches might be used, for instance, when a clinician considers CDI treatment for a patient before the results of toxin testing are available. We do recognize, however, that the correlation between Cq values and toxin positivity and the positive and negative predictive values for the diverse cutoff values are far from perfect. Therefore, we think that Cq values may be helpful in questionable cases but NAAT quantitation should not be seen as a surrogate for free toxin testing or clinical judgment. It would be interesting to investigate whether the incorporation of Cq values into an algorithm improves patient outcomes, compared to testing in which diagnoses, and consequent treatment, are dependent exclusively on demonstrating the presence of toxin.
Our study had some limitations. First, we used EIAs to detect toxin, tests that are known to suffer from low sensitivities. Automated toxin A/B EIAs such as the Vidas C. difficile Toxin A&B test used by the LUMC have reported sensitivities ranging from 53 to 98%, compared to CCNA; membrane-type toxin A/B EIAs such as the ImmunoCard Toxins A&B test have sensitivities ranging from 85 to 96%, compared to CCNA (6). It is possible that some of the outliers we observed, with low Cq values but no toxin present, were actually false-negative results from the toxin A/B EIA. CCNA, the gold standard of toxin detection, ideally should have been used but, because these analyses were conducted retrospectively, using clinical data with toxin testing performed with toxin A/B EIAs, this was not possible. In the study by Senchyna and colleagues, a membrane-type EIA that detected both GDH and toxin A/B, CCNA, and a well-type toxin A/B EIA were combined to detect toxin-positive samples (27). Using these combined tests as the reference standard, the optimal threshold cycle (CT) cutoff value detected toxin-positive samples with a positive predictive value of 81.7%, somewhat greater than those found for our cohorts, which may be explained by the more sensitive reference standard those authors used. A second limitation of our study is that we analyzed all samples that were tested for CDI and did not exclude samples from the same patient, samples from children, samples obtained during the same diarrheal episode, or samples submitted during or after treatment. The heterogeneity of the cohorts may have obscured some associations, such as higher Cq values for certain ribotypes, as reported previously for ribotype 014 (21), or an aberrant association between Cq values and toxin positivity in children. However, the inclusion of all submitted samples yielded cohorts that are representative of the actual situations. Information on repeat samples and CDI treatment is often lacking, and the eventual ribotype (if CDI is confirmed) is not available when the samples arrive at the laboratory. Therefore, we think that this study demonstrated the usefulness of NAAT quantitation in two unbiased cohorts that are highly representative of samples that are submitted for CDI testing, both in a universal hospital and in a general hospital.
Besides the representative cohorts that were used, there were some other strengths in our study. First, samples from the LUMC cohort underwent culture and PCR ribotyping and we were thus able to evaluate any differences in Cq levels among different ribotype categories. Culture and ribotyping results were also used to evaluate the outliers. Because 16/17 outlier samples had positive culture results and the 1 remaining sample had a clear clinical suspicion of CDI, false-positive toxin A/B EIA results were considered less likely. Laboratory and clinical evaluations, including retesting with a TcdB NAAT, were performed, but no clear explanation for the outliers with high Cq values but positive toxin A/B EIA results was found. Another strength of our study is the unique comparison with a group of asymptomatic carriers, which clearly demonstrated that Cq levels among asymptomatic carriers and symptomatic patients testing negative for toxins were comparable, suggesting that the latter group indeed represented CDI carriers with diarrhea not due to CDI.
In conclusion, we found Cq values to be predictors of toxin status in two large representative cohorts, although the suboptimal accuracy underscores the need for additional toxin A/B EIA testing. Additional studies are needed to determine whether the inclusion of Cq values in algorithmic testing may aid clinicians in reaching faster but still accurate preliminary CDI diagnoses while awaiting the results of free toxin testing.
ACKNOWLEDGMENTS
Sample collection from the asymptomatic carriers was supported by the Netherlands Organization for Health Research and Development (grant 50-52200-98-035).
We thank the molecular microbiologist Els Wessels and the technician Ingrid Sanders from the LUMC and the molecular microbiologist Tanja Geelen from the Amphia Hospital.
We have no conflicts of interest to declare.
REFERENCES
- 1.Lawson PA, Citron DM, Tyrrell KL, Finegold SM. 2016. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O'Toole 1935) Prevot 1938. Anaerobe 40:95–99. doi: 10.1016/j.anaerobe.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ. 2016. Clostridium difficile infection. Nat Rev Dis Primers 2:16020. doi: 10.1038/nrdp.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dobson G, Hickey C, Trinder J. 2003. Clostridium difficile colitis causing toxic megacolon, severe sepsis and multiple organ dysfunction syndrome. Intensive Care Med 29:1030. doi: 10.1007/s00134-003-1754-7. [DOI] [PubMed] [Google Scholar]
- 4.Leffler DA, Lamont JT. 2015. Clostridium difficile infection. N Engl J Med 373:287–288. [DOI] [PubMed] [Google Scholar]
- 5.Theriot CM, Bowman AA, Young VB. 2016. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere 1:e00045-15. doi: 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crobach MJ, Planche T, Eckert C, Barbut F, Terveer EM, Dekkers OM, Wilcox MH, Kuijper EJ. 2016. European Society of Clinical Microbiology and Infectious Diseases: update of the diagnostic guidance document for Clostridium difficile infection. Clin Microbiol Infect 22(Suppl 4):S63–S81. doi: 10.1016/j.cmi.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Planche TD, Davies KA, Coen PG, Finney JM, Monahan IM, Morris KA, O'Connor L, Oakley SJ, Pope CF, Wren MW, Shetty NP, Crook DW, Wilcox MH. 2013. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C. difficile infection. Lancet Infect Dis 13:936–945. doi: 10.1016/S1473-3099(13)70200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar S, Pollok R, Muscat I, Planche T. 2017. Diagnosis and outcome of Clostridium difficile infection by toxin enzyme immunoassay and polymerase chain reaction in an island population. J Gastroenterol Hepatol 32:415–419. doi: 10.1111/jgh.13504. [DOI] [PubMed] [Google Scholar]
- 9.Guerrero DM, Chou C, Jury LA, Nerandzic MM, Cadnum JC, Donskey CJ. 2011. Clinical and infection control implications of Clostridium difficile infection with negative enzyme immunoassay for toxin. Clin Infect Dis 53:287–290. doi: 10.1093/cid/cir361. [DOI] [PubMed] [Google Scholar]
- 10.Humphries RM, Uslan DZ, Rubin Z. 2013. Performance of Clostridium difficile toxin enzyme immunoassay and nucleic acid amplification tests stratified by patient disease severity. J Clin Microbiol 51:869–873. doi: 10.1128/JCM.02970-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terveer EM, Crobach MJ, Sanders IM, Vos MC, Verduin CM, Kuijper EJ. 2017. Detection of Clostridium difficile in feces of asymptomatic patients admitted to the hospital. J Clin Microbiol 55:403–411. doi: 10.1128/JCM.01858-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.International Organization for Standardization. 2012. Medical laboratories: requirements for quality and competence. ISO 15189:2012. International Organization for Standardization, Geneva, Switzerland. [Google Scholar]
- 13.Knetsch CW, Bakker D, de Boer RF, Sanders I, Hofs S, Kooistra-Smid AM, Corver J, Eastwood K, Wilcox MH, Kuijper EJ. 2011. Comparison of real-time PCR techniques to cytotoxigenic culture methods for diagnosing Clostridium difficile infection. J Clin Microbiol 49:227–231. doi: 10.1128/JCM.01743-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crobach MJT, van Dorp SM, Terveer EM, Harmanus C, Sanders IMJG, Kuijper EJ, Notermans DW, de Greeff SC, Albas J, van Dissel JT. 2017. Eleventh annual report of the National Reference Laboratory for Clostridium difficile and results of the sentinel surveillance, May 2016–May 2017. Leiden University Medical Center, Leiden, Netherlands: http://www.rivm.nl/dsresource?objectid=9eceaf93-6a82-42c5-b724-d12d7b7cbce1&type=PDF. [Google Scholar]
- 15.Beaulieu C, Dionne LL, Julien AS, Longtin Y. 2014. Clinical characteristics and outcome of patients with Clostridium difficile infection diagnosed by PCR versus a three-step algorithm. Clin Microbiol Infect 20:1067–1073. doi: 10.1111/1469-0691.12676. [DOI] [PubMed] [Google Scholar]
- 16.Polage CR, Gyorke CE, Kennedy MA, Leslie JL, Chin DL, Wang S, Nguyen HH, Huang B, Tang YW, Lee LW, Kim K, Taylor S, Romano PS, Panacek EA, Goodell PB, Solnick JV, Cohen SH. 2015. Overdiagnosis of Clostridium difficile infection in the molecular test era. JAMA Intern Med 175:1792–1801. doi: 10.1001/jamainternmed.2015.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker I, Leeming JP, Reynolds R, Ibrahim I, Darley E. 2013. Clinical relevance of a positive molecular test in the diagnosis of Clostridium difficile infection. J Hosp Infect 84:311–315. doi: 10.1016/j.jhin.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Kaltsas A, Simon M, Unruh LH, Son C, Wroblewski D, Musser KA, Sepkowitz K, Babady NE, Kamboj M. 2012. Clinical and laboratory characteristics of Clostridium difficile infection in patients with discordant diagnostic test results. J Clin Microbiol 50:1303–1307. doi: 10.1128/JCM.05711-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dionne LL, Raymond F, Corbeil J, Longtin J, Gervais P, Longtin Y. 2013. Correlation between Clostridium difficile bacterial load, commercial real-time PCR cycle thresholds, and results of diagnostic tests based on enzyme immunoassay and cell culture cytotoxicity assay. J Clin Microbiol 51:3624–3630. doi: 10.1128/JCM.01444-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung HS, Lee M. 2017. Evaluation of the performance of C. DIFF QUIK CHEK COMPLETE and its usefulness in a hospital setting with a high prevalence of Clostridium difficile infection. J Investig Med 65:88–92. doi: 10.1136/jim-2016-000231. [DOI] [PubMed] [Google Scholar]
- 21.Leslie JL, Cohen SH, Solnick JV, Polage CR. 2012. Role of fecal Clostridium difficile load in discrepancies between toxin tests and PCR: is quantitation the next step in C. difficile testing? Eur J Clin Microbiol Infect Dis 31:3295–3299. doi: 10.1007/s10096-012-1695-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies KA, Planche T, Shetty N, Wren M, Crook D, Wilcox M. 2015. Toxin gene nucleic acid amplification test cycle threshold result is associated with severity of C. difficile infection and poor patient outcomes. Open Forum Infect Dis 2(Suppl 1):S235. doi: 10.1093/ofid/ofv133.667. [DOI] [Google Scholar]
- 23.Jazmati N, Hellmich M, Licanin B, Plum G, Kaasch AJ. 2016. PCR cycle threshold value predicts the course of Clostridium difficile infection. Clin Microbiol Infect 22:e7–e8. doi: 10.1016/j.cmi.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 24.Reigadas E, Alcala L, Valerio M, Marin M, Martin A, Bouza E. 2016. Toxin B PCR cycle threshold as a predictor of poor outcome of Clostridium difficile infection: a derivation and validation cohort study. J Antimicrob Chemother 71:1380–1385. doi: 10.1093/jac/dkv497. [DOI] [PubMed] [Google Scholar]
- 25.El Feghaly RE, Stauber JL, Deych E, Gonzalez C, Tarr PI, Haslam DB. 2013. Markers of intestinal inflammation, not bacterial burden, correlate with clinical outcomes in Clostridium difficile infection. Clin Infect Dis 56:1713–1721. doi: 10.1093/cid/cit147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anikst VE, Gaur RL, Schroeder LF, Banaei N. 2016. Organism burden, toxin concentration, and lactoferrin concentration do not distinguish between clinically significant and nonsignificant diarrhea in patients with Clostridium difficile. Diagn Microbiol Infect Dis 84:343–346. doi: 10.1016/j.diagmicrobio.2015.11.022. [DOI] [PubMed] [Google Scholar]
- 27.Senchyna F, Gaur RL, Gombar S, Truong CY, Schroeder LF, Banaei N. 2017. Clostridium difficile PCR cycle threshold predicts free toxin. J Clin Microbiol 55:2651–2660. doi: 10.1128/JCM.00563-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbut F, Surgers L, Eckert C, Visseaux B, Cuingnet M, Mesquita C, Pradier N, Thiriez A, Ait-Ammar N, Aifaoui A, Grandsire E, Lalande V. 2014. Does a rapid diagnosis of Clostridium difficile infection impact on quality of patient management? Clin Microbiol Infect 20:136–144. doi: 10.1111/1469-0691.12221. [DOI] [PubMed] [Google Scholar]