LETTER
Oriental theileriosis is an economically important tick-borne disease of bovines, particularly in the Asia-Pacific region, and is caused by one or more genotypes of the Theileria orientalis complex (1, 2). Based on sequence analysis of the major piroplasm surface protein (MPSP) gene, at least 11 genotypes have been defined for T. orientalis (1); five of them (i.e., Buffeli, Chitose, Ikeda, and types 4 and 5) have been reported from Australia (3, 4). Previously, high infection intensities of the pathogenic genotypes Ikeda and Chitose have been linked to the clinical cases of oriental theileriosis in Australia and New Zealand (5–7); hence, it is important to estimate the burden of different genotypes of T. orientalis. To detect, differentiate, or quantitate these genotypes, three molecular diagnostic techniques, including conventional PCR (4), quantitative PCR using TaqMan probes (5, 6), and multiplexed tandem PCR (MT-PCR) (7), have been utilized.
Although a previously developed multiplexed tandem PCR (MT-PCR) for the detection, differentiation, and quantitation of four genotypes (Buffeli, Chitose, Ikeda, and type 5) of T. orientalis was sensitive, cost-effective, and time-effective (7), the direct comparison of the intensities of infection for different genotypes was challenging using the assay due to the use of multiple markers (i.e., the MPSP gene for Chitose and type 5, the p23 gene for Buffeli, and ITS-1 for Ikeda) with variable copy numbers per genome of T. orientalis (8). Furthermore, cross-reactivity was detected between genotypes Buffeli and Ikeda (using the p23 gene) when samples originating from other geographical regions such as Ethiopia and Pakistan were tested by MT-PCR (8). The objective of this study was to modify the existing MT-PCR assay to detect, differentiate, and quantitate the four common genotypes of T. orientalis (Buffeli, Chitose, Ikeda, and type 5) using the MPSP marker.
A total of 175 DNA samples (known test positives [n = 50] and known test negatives [n = 125] by conventional PCR and DNA sequencing) extracted from cattle blood samples were available from previous studies (9, 10). The 50 test-positive-control samples were positive for at least one of the four genotypes of T. orientalis (Buffeli, Chitose, Ikeda, and type 5) (9, 10).
The assay was conducted in the 24-well variant of the Easy-Plex platform, including the 96-well Easy-Plex Analyzer and PC with Easy-Plex software (catalog no. 9350; AusDiagnostics Pty. Ltd., Australia). The primary amplification (“target enrichment”) was conducted using genotype-specific primer pairs designed to the sequences of the MPSP gene for genotypes Buffeli, Chitose, Ikeda, and type 5 and the first internal transcribed spacer (ITS-1) for all Theileria spp. infecting bovines (Step 1 tubes for Theileria [8 well], catalog no. 78171S; AusDiagnostics). The secondary amplification for semiquantification employed nested primer pairs to internal regions of these loci (Theileria-EP96, 8 well, catalog no. 78171E; AusDiagnostics), and these internal primer pairs amplify a region of ∼90 to 110 bp from the MPSP and ITS-1 regions. Furthermore, another primer pair was included in each run as a reference for quantitation and to assess the efficiency of amplification from 10,000 copies of a synthetic oligonucleotide template (internal “spike control”).
For primary amplification (15 cycles of 10 s at 95°C, 20 s at 60°C, and 20 s at 72°C), 5 μl of genomic DNA representing each DNA sample or 5 μl of water (negative control) was dispensed into 0.2-ml PCR strips and placed into a 24-well thermocycling block in the Gene-Plex robotic platform (AusDiagnostics). Following the dispensing of each sample and the initiation of the assay, the following setup process and analysis were executed by the program Easy-Plex Assay Setup (AusDiagnostics), with the secondary amplification in MT-PCR and the melting curve analysis being semiautomated. Each sample was recorded as test positive using the autocall function of the Easy-Plex software (AusDiagnostics) if the amplicon produced a single melting curve which was within 1.5°C of the expected melting temperature, the height of the peak was higher than 0.2 dF/dT (where dF/dT is the derivative of fluorescence over temperature), and the peak width was ≤3.5°C; otherwise the sample was considered negative (AusDiagnostics). Additionally, cycle threshold (CT) values and DNA copy numbers for each genotype and/or pan-Theileria in each sample were determined by comparison with the data obtained from the internal spike control, which had a known DNA copy number of 10,000.
Based on the peak high-resolution melting (HRM) temperature analyses, genotypes Chitose, Buffeli, Ikeda, type 5, and pan-Theileria were assigned according to their mean HRM temperatures. DNA copy numbers (a measure of the intensity of infection) for the four genotypes of T. orientalis and pan-Theileria were log transformed (log10) and analyzed. Randomly selected amplicons representing each genotype were subjected to single-strand conformation polymorphism (SSCP) analysis and targeted sequencing (11).
The analytical sensitivity (using a 10-fold serial dilution) was determined using known positive samples, and the analytical specificity of the assay was determined by testing known positive samples for each of the targeted genotypes of T. orientalis (Chitose, Buffeli, type 5, and Ikeda) and other Theileria spp. (T. parva, T. annulata, T. mutans, and T. velifera) as well as other pathogens, including Babesia bovis, Anaplasma marginale, and Anaplasma centrale. The repeatability of the assay for the detection of the expected genotype(s) and the DNA copy numbers were also assessed using known positive samples which were tested three times in different runs and on different days. The epiR package (12) in R (13) was used to calculate the proportion (and its 95% confidence interval [CI]) of test-positive samples using both conventional PCR and MT-PCR.
Using the MT-PCR, each of the four primer sets designed to the MPSP gene amplified products exclusively from the expected genotypes (Fig. 1). The identity of individual products was confirmed by SSCP analysis and sequencing, and no products were amplified from other pathogens tested (i.e., A. marginale, A. centrale, B. bovis, T. parva, T. annulata, T. mutans, and T. velifera). Furthermore, the pan-Theileria primer pair successfully amplified the products for the tested Theileria species (T. parva, T. annulata, T. mutans, and T. velifera). Based on the peak HRM temperature analyses, each genotype of T. orientalis and the pan-Theileria were produced as a single and distinct melt curve, and genotypes Chitose, Buffeli, Ikeda, and type 5 and pan-Theileria had mean HRM temperatures of 82.3 ± 1.5°C, 82.8 ± 1.5°C, 83.7 ± 1.5°C, 81.4 ± 1.5°C, and 84.2 ± 1.5°C, respectively (Fig. 1).
FIG 1.
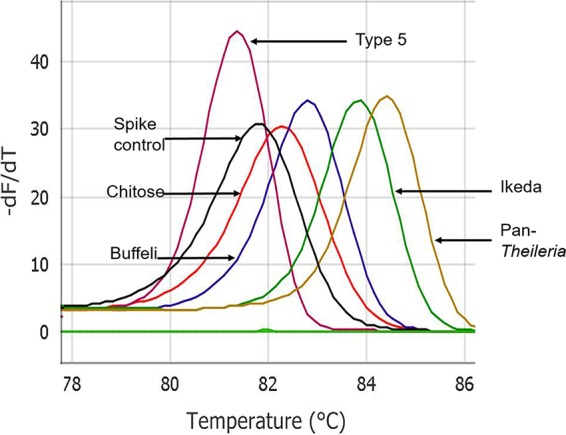
High-resolution melting (HRM) curve analysis of four genotypes of Theileria orientalis, pan-Theileria, and spike control using the MT-PCR assay.
The MT-PCR assay was then validated using the known positive-control (n = 50) and negative-control (n = 125) samples. All 50 positive and seven of the negative-control samples were test positive using the MT-PCR (32.6%; 57/175) (Table 1). Furthermore, all of the four genotypes (i.e., Chitose, Buffeli, type 5, and Ikeda) were detected, and genotype Ikeda was detected in 22% (38/175) of samples, followed by Buffeli (21%; 37/175), Chitose (19%; 33/175), and type 5 (7%; 13/175) (Table 1). Mixed infections with two or more genotypes of T. orientalis were more common (19%; 33/175) than monogenotypic infections (14%; 24/175). The relative intensity of infection (average DNA copies) with genotype Ikeda was higher (average number of DNA copies, 3,654), followed by Buffeli (2,572 copies), Chitose (2,173 copies), and type 5 (2,028 copies) (Table 1). The proportion of samples positive by conventional PCR that were also positive by MT-PCR and the proportion of samples negative by conventional PCR that were also negative by MT-PCR were 1.0 (95% CI, 0.93 to 1.0) and 0.94 (95% CI, 0.89 to 0.97), respectively.
TABLE 1.
Genotypes of Theileria orientalis and their infection intensities detected by the modified MT-PCR assayb
| Genotype/pathogen detected | No. of test positives for each genotype/pathogen (%) | Avg no. of DNA copies | Median no. of DNA copies (min, max)a |
|---|---|---|---|
| Chitose | 33 (18.9) | 2,173 | 1,082 (2, 11,102) |
| Buffeli | 37 (21.1) | 2,572 | 2,098 (1, 9,111) |
| Type 5 | 13 (7.4) | 2,028 | 3,29 (2, 13,433) |
| Ikeda | 38 (21.7) | 3,654 | 1,011 (1, 24,584) |
| Pan-Theileria | 57 (32.6) | 8,517 | 5,060 (1, 47,443) |
min, minimum; max, maximum.
Overall number of test-positive samples was 57/175 (32.6%).
The modified MT-PCR assay utilizes only one marker (i.e., MPSP), which is genotype specific and sensitive for the detection, differentiation, and quantitation of the four genotypes Chitose, Buffeli, type 5, and Ikeda of T. orientalis. The cross-amplification observed using the previous MT-PCR assay (8) established by Perera et al. (7) using multiple markers is thus resolved here, indicating that the MPSP gene is a suitable marker when amplifying even short segments of the gene compared to the p23 gene and other molecular markers to detect genotypes of T. orientalis. Furthermore, the modified MT-PCR assay includes pan-Theileria primers to detect all the known Theileria spp. infecting bovines, thereby increasing the usefulness of the assay globally.
In conclusion, the modified MT-PCR assay utilizing the MPSP gene is a rapid and effective molecular diagnostic tool to detect, differentiate and quantitate genotypes Chitose, Buffeli, type 5, and Ikeda of T. orientalis. Additionally, this assay will be a useful tool for the surveillance of oriental theileriosis and for monitoring the spread of disease during cattle exportation/importation, particularly in the Asia-Pacific region (14).
ACKNOWLEDGMENTS
This project was supported by Collaborative Research and Early Career Researchers grants (the University of Melbourne) (A.J.). H.G. is a grateful recipient of the Melbourne International Research Scholarships (MIRS) and Melbourne International Fee Remission Scholarships (MIFRS) through The University of Melbourne.
We extend our appreciation to the farmers and veterinarians who contributed their time and expertise to this study.
REFERENCES
- 1.Sivakumar T, Hayashida K, Sugimoto C, Yokoyama N. 2014. Evolution and genetic diversity of Theileria. Infect Genet Evol 27:250–263. doi: 10.1016/j.meegid.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 2.Perera PK, Gasser RB, Firestone SM, Anderson GA, Malmo J, Davis G, Beggs DS, Jabbar A. 2014. Oriental theileriosis in dairy cows causes a significant milk production loss. Parasit Vectors 7:73. doi: 10.1186/1756-3305-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamau J, de Vos AJ, Playford M, Salim B, Kinyanjui P, Sugimoto C. 2011. Emergence of new types of Theileria orientalis in Australian cattle and possible cause of theileriosis outbreaks. Parasit Vectors 4:22. doi: 10.1186/1756-3305-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perera PK, Gasser RB, Anderson GA, Jeffers M, Bell CM, Jabbar A. 2013. Epidemiological survey following oriental theileriosis outbreaks in Victoria, Australia, on selected cattle farms. Vet Parasitol 197:509–521. doi: 10.1016/j.vetpar.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 5.Bogema D, Deutscher A, Fell S, Collins D, Eamens G, Jenkins C. 2015. Development and validation of a quantitative PCR assay using multiplexed hydrolysis probes for detection and quantification of Theileria orientalis isolates and differentiation of clinically relevant subtypes. J Clin Microbiol 53:941–950. doi: 10.1128/JCM.03387-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pulford DJ, Gias E, Bueno IM, McFadden AMJ. 2016. Developing high throughput quantitative PCR assays for diagnosing ikeda and other Theileria orientalis types common to New Zealand in bovine blood samples. N Z Vet J 64:29–37. doi: 10.1080/00480169.2015.1089798. [DOI] [PubMed] [Google Scholar]
- 7.Perera PK, Gasser RB, Firestone SM, Smith L, Roeber F, Jabbar A. 2015. Semi-quantitative multiplexed tandem PCR for detection and differentiation of four Theileria orientalis genotypes in cattle. J Clin Microbiol 53:79–87. doi: 10.1128/JCM.02536-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gebrekidan H, Gasser RB, Jabbar A. 2017. Inadequate differentiation of Theileria orientalis genotypes buffeli and ikeda in a multiplexed tandem PCR (MT-PCR) assay using the p23 gene as a marker. J Clin Microbiol 55:641–644. doi: 10.1128/JCM.02017-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gebrekidan A, Gasser RB, Baneth G, Yasur-Landau D, Nachum-Biala Y, Hailu A, Jabbar A. 2016. Molecular characterization of Theileria orientalis from cattle in Ethiopia. Ticks Tick Borne Dis 7:742–747. doi: 10.1016/j.ttbdis.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Gebrekidan H, Abbas T, Wajid M, Ali A, Gasser RB, Jabbar A. 2017. Molecular characterisation of Theileria orientalis from bovines in Pakistan. Infect Genet Evol 47:19–25. doi: 10.1016/j.meegid.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Cufos N, Jabbar A, de Carvalho LM, Gasser RB. 2012. Mutation scanning-based analysis of Theileria orientalis populations in cattle following an outbreak. Electrophoresis 33:2036–2040. doi: 10.1002/elps.201200082. [DOI] [PubMed] [Google Scholar]
- 12.Stevenson M, Nunes T, Heuer C, Marshall J, Sanchez J, Thornton R, Reiczigel J, Robison-Cox J, Sebastiani P, Solymos P, Yoshida K, Jones G, Pirikahu S, Firestone S, Kyle R. 2017. epiR: tools for the analysis of epidemiological data. R package version 0.9-90. http://fvas.unimelb.edu.au/veam.
- 13.R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- 14.Gebrekidan H, Nelson L, Smith G, Gasser RB, Jabbar A. 2017. An outbreak of oriental theileriosis in dairy cattle imported to Vietnam from Australia. Parasitology 144:738–746. doi: 10.1017/S0031182016002328. [DOI] [PubMed] [Google Scholar]


