ABSTRACT
Zika virus (ZIKV) is a mosquito-borne flavivirus that is responsible for recent explosive epidemics in the Americas. Notably, ZIKV infection during pregnancy has been found to cause congenital birth defects, including microcephaly, and ZIKV has been associated with Guillain-Barré syndrome in adults. Diagnosis and surveillance of Zika in the Americas have been challenging due to similar clinical manifestations and extensive antibody cross-reactivity with endemic flaviviral diseases, such as dengue. We evaluated four serological and two reverse transcription-PCR (RT-PCR) methods in acute-phase (mean day, 1.8), early-convalescent-phase (mean day, 16.7), and late-convalescent-phase (mean, ~7 months) samples from the same individuals in a long-term pediatric cohort study in Nicaragua. Well-characterized samples from 301 cases of Zika, dengue, or non-Zika, nondengue febrile illnesses were tested. Compared to a composite reference, an in-house IgM antibody capture enzyme-linked immunosorbent assay (MAC-ELISA) and the NIAID-Biodefense and Emerging Infections (BEI) MAC-ELISA measuring IgM yielded sensitivities of 94.5% and 70.1% and specificities of 85.6% and 82.8%, respectively. The NS1 blockade-of-binding ELISA measuring anti-ZIKV NS1 antibody levels yielded sensitivities of 85.0% and 96.5% and specificities of 91.4% and 92.6% at early and late convalescence, respectively. An inhibition ELISA detecting total anti-ZIKV antibodies had sensitivity and specificity values of 68.3% and 58.3% for diagnosis and 94.0% and 98.6% for measuring annual infection incidence. Finally, the ZCD and Trioplex real-time RT-PCR assays detecting Zika, chikungunya, and dengue viruses both yielded a sensitivity of 96.1% and specificity of 100%. Together, these assays resolve the urgent need for diagnostic and surveillance tools for countries affected by Zika virus infections.
KEYWORDS: ELISA, RT-PCR, Zika virus, dengue virus, diagnosis, serological assay, surveillance
INTRODUCTION
Zika virus (ZIKV) is an emerging arbovirus belonging to the Flavivirus genus of the Flaviviridae family. ZIKV produces mild disease characterized by rash, moderate fever, conjunctivitis, and arthralgia; however, ZIKV has also been found to cause congenital birth defects, including microcephaly, as well as Guillain-Barré syndrome (1–7). The Zika epidemic in the Americas began in 2015 in Brazil and spread rapidly throughout the hemisphere (8, 9). The first laboratory-confirmed case in Nicaragua was detected in January 2016, with an explosive epidemic occurring from late June to mid-September.
The cocirculation of several arboviruses in the Americas that cause similar symptoms and display antibody cross-reactivity poses a challenge for diagnosis and surveillance (10–14). In particular, detection and differentiation of the antigenically related flaviviruses dengue virus (DENV) and ZIKV have relied to date mostly upon molecular methods (15–17). However, ZIKV viral loads are low, viremia is short-lived, and serological methods are cheaper and require less sample volume. Serological dengue and Zika assays are designed to detect antibodies to the envelope (E) and/or prM/M proteins on the virion surface or to nonstructural protein 1 (NS1) that is secreted from infected cells (18, 19). However, the standardization of serological methods that are sufficiently sensitive and that discriminate between anti-DENV and anti-ZIKV antibodies has been challenging.
Lack of access to well-characterized samples has been the fundamental problem for evaluation and standardization of new assays. In Nicaragua, a cohort of ~3,600 children has been followed since 2004; each febrile episode of suspected dengue, chikungunya, or Zika or of undifferentiated fever is well-documented and accompanied by blood samples drawn during the acute and early convalescent phases (20, 21). Additionally, an annual healthy blood sample is drawn from all participants during the dry season, when arboviral transmission is minimal. This study serves as a source of samples from DENV-immune individuals where the serotype and immune status are known and from ZIKV-infected participants, stratified by prior DENV exposure.
Here we evaluated the sensitivity and specificity of 2 real-time reverse transcription-PCR (rRT-PCR) assays, one designed by the CDC (15) and the other by Waggoner et al. (16), and of 4 serological methods (2 detecting IgM and 2 detecting total antibodies) using well-characterized samples from the cohort study. Two in-house serological assays, namely, an IgM antibody capture enzyme-linked immunosorbent assay (MAC-ELISA) and an inhibition ELISA method (IEM) detecting total anti-ZIKV antibodies, are new and were developed at the Centro Nacional de Diagnóstico y Referencia (CNDR) of the Nicaraguan Ministry of Health. Additionally, two existing assays were evaluated: an IgM capture technique developed by the CDC and supplied by the Biodefense and Emerging Infections (BEI) Research Resources Repository (NIAID-BEI MAC-ELISA) and a competition ELISA detecting anti-ZIKV NS1 antibodies (NS1 blockade-of-binding ELISA [BOB ELISA]) (22, 23).
MATERIALS AND METHODS
Ethics statement.
The Pediatric Dengue Cohort Study (PDCS) was approved by the Institutional Review Boards of the Nicaraguan Ministry of Health and the University of California, Berkeley. Parents or legal guardians of all subjects provided written informed consent, and subjects who were ≥6 years old provided assent.
Study population and sample selection.
The patient population of the PDCS is a community-based prospective cohort of children 2 to 14 years of age; the study has been ongoing since August 2004 in Managua, Nicaragua (20). In 2014, chikungunya was added to the PDCS, and in August 2015, ZIKV screening was initiated for participants meeting the clinical definition of dengue and/or chikungunya or presenting with undifferentiated fever. In February 2016, the case definition was expanded to include those presenting with signs and symptoms of Zika but without fever. Participants present at the first sign of illness to the Health Center Sócrates Flores Vivas, and study physicians use standardized forms to collect clinical data. Blood samples are collected during the acute phase and again during early convalescence for dengue, chikungunya, and Zika diagnostic testing. In addition, a healthy blood sample is collected annually from participants to determine inapparent infections (20).
Study samples and case definitions.
A total of 301 PDCS participants were included in the study, each providing an acute-phase sample (number of days post-onset of symptoms, 1.8 ± 0.9 [mean ± standard deviation {SD}]), an early-convalescent-phase sample (mean of 16.7 ± 2.9 days), and paired annual samples before and after the infection in question, although a few samples could not be tested by all methods due to insufficient volume (see Table S1 in the supplemental material). The first annual sample served as a baseline, and the second served as a late-convalescent-phase sample (∼7 months ± 29 days). The ZIKV-positive controls consisted of 127 children who tested positive for ZIKV infection by both rRT-PCR and at least one serological method (Zika MAC-ELISA, NS1 BOB ELISA, or IEM) (Table 1). Serologically positive cases were defined as (i) those with ZIKV seroconversion detected between acute-phase samples and either early or late convalescent-phase samples or between consecutive annual samples or (ii) those for which the titer at early convalescence was ≥4-fold higher than the acute-phase titer or for which a ≥4-fold increase in titer was observed between consecutive annual samples (IEM). Of the ZIKV-infected patients, 65 (51.2%) had been previously exposed to DENV (DENV-immune patients) and 62 (48.8%) were naïve (DENV-naïve patients). Confirmed ZIKV-positive cases were classified as DENV-naïve cases if they entered the cohort study with no detectable anti-DENV antibodies (as measured by DENV IEM) and had no documented DENV infections (symptomatic or inapparent) during their time in the cohort. Confirmed ZIKV-positive cases were classified as DENV-immune cases if they either entered the cohort study with detectable anti-DENV IEM antibodies or entered the cohort study with no detectable anti-DENV antibodies and had one or more documented DENV infections during their time in the cohort. A total of 97 children with previous RT-PCR-confirmed DENV serotype 1 (DENV1), DENV2, or DENV3 infections from 2007 to 2013, prior to the introduction of ZIKV in 2016, served as DENV-positive ZIKV-negative controls. In addition, 77 individuals who had never experienced ZIKV or DENV infection served as negative controls.
TABLE 1.
Characteristics of study population
| Characteristic | n of patients | % of patientsa |
|---|---|---|
| Total | 301 | |
| Sex | ||
| Female | 162 | 53.8 |
| Age | ||
| 2–4 yrs | 37 | 12.3 |
| 5–10 yrs | 146 | 48.5 |
| 11–14 yrs | 118 | 39.2 |
| ZIKV positive | 127 | 42.2a |
| DENV naïve | 62 | 48.8b |
| DENV immune | 65 | 51.2b |
| DENV positive | 97 | 32.2a |
| DENV1 positive | 24 | 24.7c |
| Primary | 12 | 50.0d |
| Secondary | 12 | 50.0d |
| DENV2 positive | 34 | 35.1c |
| Primary | 17 | 50.0e |
| Secondary | 17 | 50.0e |
| DENV3 positive | 39 | 40.2c |
| Primary | 19 | 48.7f |
| Secondary | 20 | 51.3f |
| Negative | 77 | 25.6a |
The population data for the subgroups denoted by roman superscript letters a through f add up to 100%.
Composite reference.
As there is no gold standard for Zika diagnosis, a composite reference standard was defined for the calculation of assay sensitivity and specificity (24, 25). To be considered Zika positive by the composite reference, the acute-phase serum sample of a case had to test positive by at least one molecular method and the patient had to seroconvert on the basis of at least one serological method. Sensitivity for each assay was defined as the percentage of cases that tested positive among the composite reference-positive cases. Specificity for each assay was defined as the percentage of cases that tested negative among the composite reference-negative cases.
NIAID-BEI MAC-ELISA.
A MAC-ELISA, developed on the basis of the initial CDC Emergency Use Authorization (March 2016), was manufactured and supplied by the NIAID-NIH BEI Resources Repository (catalog no. NR-50449) for the assessment of human IgM antibody to ZIKV in human serum (HS) by enzyme immunoassay (23). Briefly, 75 μl anti-human IgM antibody was coated on 8-well polystyrene Maxisorp strips (Nunc Thermo Scientific) and incubated overnight, followed by one wash with phosphate-buffered saline with 0.05% Tween 20 (PBS-T) and blocking with PBS-T containing 5% nonfat dry milk. After a 30-min incubation at room temperature, 50 μl of serum samples and negative control diluted 1:400 in PBS-T and positive control (anti-ZIKV human IgM positive control from the kit) diluted 1:125 in PBS-T were added to individual wells. After 1 h of incubation at 37°C, 50 μl of ZIKV antigen and negative-control antigen was added, and the reaction mixture was incubated overnight, followed by addition of 50 μl horseradish peroxidase (HRP)-conjugated monoclonal antibody (anti-flavivirus MAb clone 6B6C-1, which binds to a conserved domain of flavivirus E protein) diluted 1:1,000 in blocking solution (5% nonfat dry milk), and 75 μl of 3,3′-5,5′ tetramethylbenzidine (TMB; Sigma). The reaction was terminated with 50 μl 12.5% sulfuric acid. After each incubation period, wells were washed 4 times with PBS-T and 5 times after the addition of conjugated antibody. Optical absorbance was measured using a Microplate Photometer (Multiskan FC Thermo Scientific) at 450 nm. All samples with absorbance values of ≥0.2 were considered positive. A paired sample was considered positive when the acute-phase sample was negative and the convalescent-phase sample was positive (seroconversion).
CNDR MAC-ELISA.
ZIKV-specific IgM antibodies were detected by an in-house IgM-capture ELISA (CNDR MAC-ELISA) similar to that described for the detection of anti-DENV and anti-chikungunya virus (anti-CHIKV) IgM antibodies (26, 27). The assay was adapted to the detection of anti-ZIKV IgM antibodies using the strongly neutralizing ZIKV-specific monoclonal antibody (MAb) ZKA64 (Humabs Biomed SA) directed to ZIKV E domain III (14). Briefly, strips of polystyrene microwells were coated with 6.6 μg/ml goat anti-human IgM (Life Technologies) in sodium carbonate-bicarbonate buffer (pH 9.5) and incubated overnight at room temperature, followed by blocking in 150 μl of 1% bovine serum albumin (BSA) fraction V (Fisher) diluted in PBS-T. Each serum specimen was diluted 1:20 in PBS-T, and 50 μl was added to the wells and incubated for 1 h at 37°C; all samples were processed in duplicate. ZIKV antigen was produced from strain MR766 (obtained from Michael S. Diamond, Washington University in St. Louis) in suckling mouse brain (BALB/c strain) using the sucrose-acetone extraction method (28). A 50-μl volume of antigen diluted 1:20 in PBS-T was added to the wells, and the reaction mixture was incubated for 1 h at 37°C, followed by addition of 50 μl of MAb ZKA64 conjugated to horseradish peroxidase (HRP) diluted 1:1,000 in PBS-T with 2.5% normal human serum (NHS). After 1 h at 37°C, 50 μl of TMB substrate was added, and the strips were held at room temperature for 10 min until the reaction was terminated by the addition of 50 μl of 12.5% sulfuric acid. Optical absorbance was measured using a Microplate Photometer at 450/630 nm. After each incubation period, the wells were washed 4 times with PBS-T and 5 times after the addition of conjugated antibody. The cutoff value was determined so as to maximize specificity on the basis of testing negative controls and known DENV-positive samples collected during the period before the introduction of Zika and as such was defined as 6.6 times the average of the negative controls. All samples with absorbance values above the cutoff value were considered positive. As described above, seroconversion in paired samples indicated a positive result for the case.
NS1 blockade-of-binding ELISA (NS1 BOB).
ZIKV-specific anti-NS1 antibodies in samples were detected as described by Balmaseda et al. (22). Briefly, polystyrene Maxisorp plates were coated overnight with 1 μg/ml of ZIKV NS1 (MR766 strain; Native Antigen Company, Inc.) in coating buffer (PBS) at 4°C. Plates were blocked for 1 h with PBS containing 1% BSA and then washed two times with PBS-T. Serum (1:10 dilution in PBS–1% BSA) was added to NS1-coated ELISA plates. After 1 h, an equal volume of biotinylated anti-NS1 ZKA35 (Humabs Biomed) (20 ng/ml) was added to samples, and the mixture was incubated at room temperature for 15 min (final dilution of the plasma in the mixture, 1:20). Plates were washed with PBS-T four times, and alkaline phosphatase-conjugated streptavidin (Jackson Immuno Research) was added for 1 h. Plates were washed with PBS-T five times, and the substrate para-nitrophenyl phosphate was added for 20 min. Optical absorbance was then measured using a microplate photometer at 405 nm. BOB values were calculated as follows: {1 − [(OD sample − OD minimum value/delta value)]} × 100, where OD represents optical density. A BOB value of ≥50 was considered positive, and a BOB value of <50 was considered negative. As described above, seroconversion in paired samples indicated a positive result for the case.
Inhibition ELISA method (IEM).
An in-house inhibition ELISA similar to that described by Galo et al. (26) was established and used to detect and determine the titer of total ZIKV-specific antibodies in serum. Briefly, 96-well polystyrene Maxisorp plates were coated with MAb ZKA64 (5 μg/ml). After three washes with PBS-T, 150 μl of 1% BSA diluted in PBS-T was added, and the reaction mixture was incubated for 30 min at 37°C. ZIKV antigen (100 μl) produced in infected suckling mouse brains (see above) and diluted 1:20 in PBS-T was added to each well, and plates were incubated for 1 h at 37°C. After four washes with PBS-T, 100 μl of samples, positive control (serially diluted in PBS-T–0.5% BSA from 1:10 to 1:100,000), and negative control (at a dilution of 1:10) were added, and the reaction mixture was incubated for 2 h at 37°C. After four washes with PBS-T, 100 μl of HRP-conjugated MAb ZKA64 diluted 1:1,000 in PBS–2.5% NHS was added to each well. The plates were incubated for 30 min at 37°C and washed five times with PBS-T, and then 50 μl of substrate TMB was added to each well. The plates were held at room temperature for 30 min, the reaction was stopped with 50 μl of 12.5% sulfuric acid, and the optical density was measured using a Microplate Photometer at 450 nm and 630 nm. The percentage of inhibition was calculated using the following formula:
| (1) |
The sample titer was then calculated using the Reed and Muench method [29] as follows:
| (2) |
where D≥50% is percent inhibition of the last serum dilution with ≥50% inhibition, T≥50% is the last serum dilution at which ≥50% inhibition was observed, and D<50% is percent inhibition of the first serum dilution with <50% inhibition. The cutoff value was determined based on previously published DENV and CHIKV IEM assays (18, 26, 30).
Molecular testing.
Viral RNA was extracted using a QIAamp viral RNA minikit (Qiagen) with 140 μl of serum and a 60-μl elution volume. All rRT-PCRs were performed on an ABI 7500 Fast instrument (Applied Biosystems) using 25-μl reaction mixtures, a SuperScript III Platinum One-Step qRT-PCR kit (Life Technologies), and 5 μl of RNA template.
The ZCD (Zika virus, chikungunya virus, and dengue virus) rRT-PCR assay detects and differentiates RNA from ZIKV, CHIKV, and DENV during the acute phase of illness, using target sequences in ZIKV NS4B (16). The assay was performed as previously described (16). Each run included a no-template control and positive controls for ZIKV, CHIKV, and DENV. A result was interpreted as positive if the reaction generated an exponential curve that crossed a threshold that was set manually, as described previously (31). The Trioplex real-time RT-PCR assay (Trioplex rRT-PCR) was developed by the CDC for the simultaneous qualitative detection of RNA from ZIKV, DENV, and CHIKV. The ZIKV primers and dually labeled hydrolysis probe target sequences in the E gene. The Trioplex assay was performed and interpreted according to the package insert instructions. The ZIKV result was interpreted as positive if the reaction generated an exponential curve that crossed the instrument-defined threshold within ≤38 cycles (15).
Statistical analysis.
Sensitivity and specificity values and 95% confidence intervals were calculated using Epi Info version 3.3.2 (CDC). Relative and absolute frequencies are reported for categorical variables, and means and standard deviations are reported for quantitative variables. We used RStudio Inc. software version 1.0.136 with data packet ggplot2 version 2.20 for generating graphs. Fisher's exact test was used to calculate P values.
RESULTS
Four serological and two molecular methods were evaluated for detection of ZIKV infection in 301 participants of the Pediatric Dengue Cohort Study (PDCS). Demographic data are shown in Table 1 (see also Table S1 in the supplemental material). Among the 301 individuals included, 97 had cases of RT-PCR-confirmed DENV infections (half primary and half secondary infections with each of serotypes DENV1, DENV2, and DENV3), 127 represented ZIKV-positive cases (62 DENV-naïve and 65 DENV-immune patients), and 77 were ZIKV negative and DENV negative (Table 1).
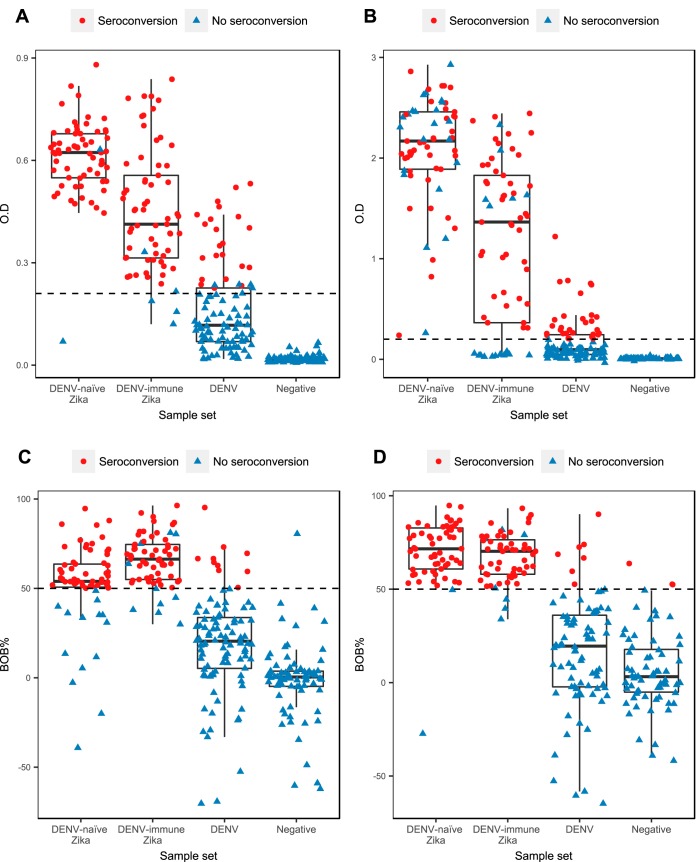
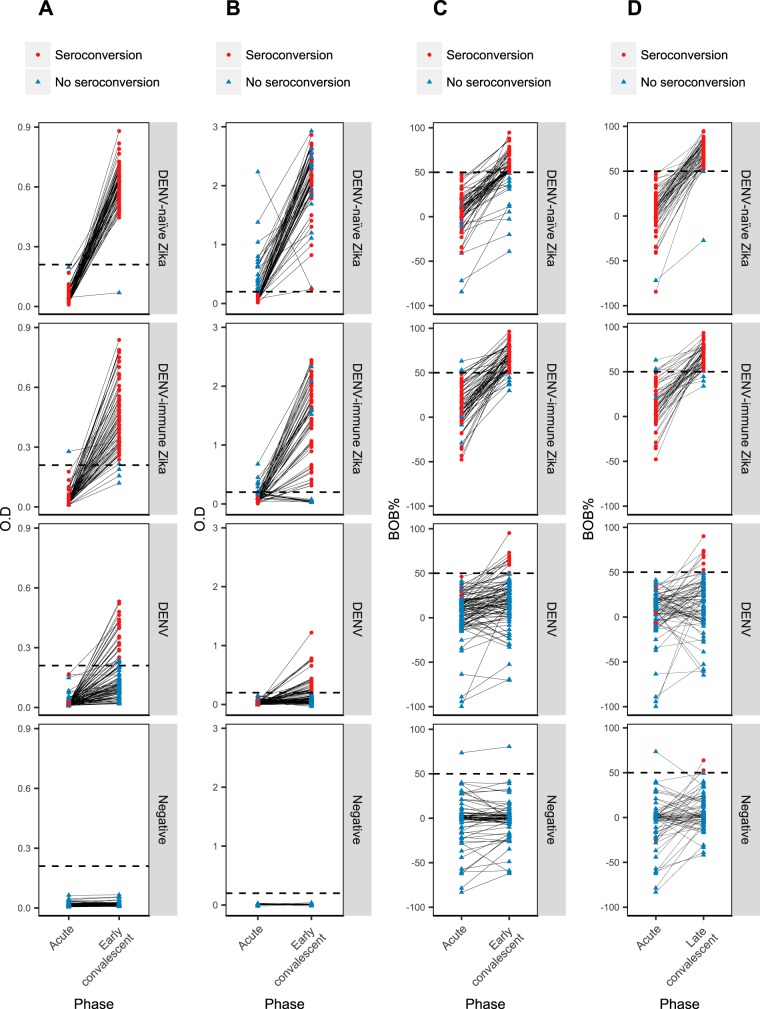
For detection of IgM seroconversion in acute-phase and early-convalescent-phase samples among Zika patients, the in-house CNDR MAC-ELISA had an overall sensitivity of 94.5% (95% confidence interval, 89.0 to 97.8), whereas the BEI MAC-ELISA yielded a sensitivity of 70.1% (61.3 to 77.9) (Table 2). Statistical analysis indicated that the CNDR MAC-ELISA was significantly more sensitive than the NIAID-BEI MAC-ELISA (P = 0.0399, Fisher's exact test). The respective sensitivities of the two assays for DENV-naïve and DENV-immune ZIKV infections were comparable. Panels A and B of Fig. 1 present the data obtained with early-convalescent-phase samples, and results from paired acute-phase and convalescent-phase samples are shown in Fig. 2A and B, with both figures indicating which individuals underwent seroconversion. The overall specificities were 85.6% (79.5% to 90.5%) and 82.8% (76.3% to 88.1%) for the CNDR MAC-ELISA and NIAID-BEI MAC-ELISA, respectively. Restricting the analysis to confirmed ZIKV-negative DENV-positive cases, the ZIKV specificity of the CNDR assay was higher (77.3% [67.7% to 85.2%]) than that of the BEI assay (69.1% [58.9% to 78.1%]). Interestingly, DENV1 showed the greatest serological cross-reactivity with ZIKV among the three DENV serotypes.
TABLE 2.
Results of CNDR MAC-ELISA and NIAID-BEI MAC-ELISA in acute-phase and early convalescent-phase samples for diagnosis of Zika casesa
| Patient group | CNDR MAC-ELISA |
NIAID-BEI MAC-ELISA |
||
|---|---|---|---|---|
| % Sens (95% CI) | % Spec (95% CI) | % Sens (95% CI) | % Spec (95% CI) | |
| Overall | 94.5 (89.0–97.8) | 85.6 (79.5–90.5) | 70.1 (61.3–77.9) | 82.8 (76.3–88.1) |
| ZIKV positive | 94.5 (89.0–97.8) | NA | 70.1 (61.3–77.9) | NA |
| DENV naïve | 96.8 (88.8–99.6) | NA | 69.4 (56.3–80.4) | NA |
| DENV immune | 92.3 (83.0–97.5) | NA | 70.8 (58.2–81.4) | NA |
| DENV positive | NA | 77.3 (67.7–85.2) | NA | 69.1 (58.9–78.1) |
| DENV1 | NA | 66.7 (44.7–84.4) | NA | 66.7 (44.7–84.4) |
| DENV2 | NA | 82.4 (65.5–93.2) | NA | 70.6 (52.5–84.9) |
| DENV3 | NA | 79.5 (63.5–90.7) | NA | 69.2 (52.4–83) |
| Primary | NA | 70.8 (55.9–83.0) | NA | 52.1 (37.2–66.7) |
| Secondary | NA | 83.7 (70.3–92.7) | NA | 85.7 (72.8–94.1) |
| Negative | NA | 96.1 (89.0–99.2) | NA | 100 (95.3–100.0) |
Sens, sensitivity. Sensitivity values were calculated for each assay based on ZIKV-positive samples, stratified by those who had been previously infected with DENV (DENV immune) and those who were DENV naïve. Spec, specificity. Specificity values were calculated for each assay based on two groups of ZIKV-negative samples: (i) ZIKV-negative DENV-positive samples, stratified by DENV subtype (DENV1 to DENV3) and primary and secondary infection, and (ii) ZIKV-negative DENV-negative controls. 95% CI, 95% confidence interval. NA, not applicable: data from ZIKV-positive cases served only for determination of sensitivity and were not used for the specificity calculation. Similarly, data from ZIKV-negative cases served only for determination of specificity and were not used for the sensitivity calculation.
FIG 1.
Box-and-whisker plots graphing results from convalescent-phase samples obtained using each serological method, stratified by sample set. (A) CNDR MAC-ELISA, early-convalescent-phase samples. (B) NIAID-BEI MAC-ELISA, early-convalescent-phase samples. (C) ZIKV NS1 BOB ELISA, early-convalescent-phase samples. (D) ZIKV NS1 BOB ELISA, late-convalescent-phase samples. Red dots represent results from convalescent-phase samples that underwent seroconversion from the acute to convalescent phase in paired samples. Blue triangles represent results from convalescent-phase samples that did not undergo seroconversion. The dotted horizontal lines indicate the cutoff values for each method. For the CNDR MAC-ELISA, the mean cutoff value is depicted, while the other methods have a fixed cutoff value.
FIG 2.
Graphical representation of paired acute-phase and convalescent-phase samples for each serological method, stratified by sample set. (A) CNDR MAC-ELISA, acute-phase and early-convalescent-phase samples. (B) NIAID-BEI MAC-ELISA, acute-phase and early-convalescent-phase samples. (C) Zika NS1 BOB ELISA, acute-phase and early-convalescent-phase samples. (D) Zika NS1 BOB ELISA, acute-phase and late-convalescent-phase samples. Red dots represent the acute/convalescent pairs that underwent seroconversion. Blue triangles represent the acute/convalescent pairs that did not undergo seroconversion. The dotted horizontal lines represent the cutoff values for each method. For the CNDR MAC-ELISA, the mean cutoff value is depicted, while the other methods have a fixed cutoff value.
For the NS1 BOB ELISA, we analyzed acute-phase/early-convalescent-phase seroconversion and acute-phase/late-convalescent-phase seroconversion separately to evaluate its utility for diagnosis as well as for surveillance in samples collected several months postinfection. The NS1 BOB ELISA was significantly more sensitive at the late-convalescent-phase time point, with a sensitivity of 96.5% (91.2 to 99.0) compared to 85.0% (77.6 to 90.7) at the early-convalescent-phase time point (P = 0.0015; Table 3). This is clearly depicted in Fig. 1C and D and in plots of paired acute-phase and convalescent-phase samples (Fig. 2C and D), where the individuals who underwent seroconversion are indicated. At early convalescence, the NS1 BOB ELISA displayed higher sensitivity in DENV-immune patients (92.3% [83.0 to 97.5]) than in DENV-naïve patients (77.4% [65.0 to 87.1]): however, this difference was not apparent at late convalescence. High specificity was observed at both the early (91.4% [86.2 to 95.1]) and late (92.6% [87.2 to 96.3]) time points.
TABLE 3.
Results of Zika NS1 BOB ELISA in early and late convalescent-phase samples for diagnosis and surveillance of ZIKV infectionsa
| Patient population | NS1 BOB ELISA |
|||
|---|---|---|---|---|
| Early convalescence |
Late convalescence |
|||
| Sens (95% CI) | Spec (95% CI) | Sens (95% CI) | Spec (95% CI) | |
| Overall | 85.0 (77.6–90.7) | 91.4 (86.2–95.1) | 96.5 (91.2–99.0) | 92.6 (87.2–96.3) |
| ZIKV positive | 85.0 (77.6–90.7) | NA | 96.5 (91.2–99.0) | NA |
| DENV naïve | 77.4 (65.0–87.1) | NA | 98.3 (90.8–100.0) | NA |
| DENV immune | 92.3 (83.0–97.5) | NA | 94.5 (84.9–98.9) | NA |
| DENV positive | NA | 88.7 (80.6–94.2) | NA | 90.0 (81.2–95.6) |
| DENV1 | NA | 95.8 (78.9–99.9) | NA | 85.0 (62.1–96.8) |
| DENV2 | NA | 85.3 (68.9–95.0) | NA | 89.7 (72.2–97.8) |
| DENV3 | NA | 87.2 (72.6–95.7) | NA | 93.5 (78.6–99.2) |
| Primary | NA | 100.0 (92.6–100.0) | NA | 89.5 (75.2–97.1) |
| Secondary | NA | 77.6 (63.4–88.2) | NA | 90.5 (77.4–97.3) |
| Negative | NA | 94.8 (87.2–98.6) | NA | 95.7 (87.8–98.1) |
Sens, sensitivity. Sensitivity values were calculated for each assay based on ZIKV-positive samples, stratified by those who had been previously infected with DENV (DENV immune) and those who were DENV naïve. Spec, specificity. Specificity values were calculated for each assay based on two groups of ZIKV-negative samples: (i) ZIKV-negative DENV-positive samples, stratified by DENV subtype (DENV1 to DENV3) and primary and secondary infection, and (ii) ZIKV-negative DENV-negative controls. 95% CI, 95% confidence interval. NA, not applicable: data from ZIKV-positive cases served only for determination of sensitivity and were not used for the specificity calculation. Similarly, data from ZIKV-negative cases served only for determination of specificity and were not used for the sensitivity calculation.
With the IEM, a case was considered positive when the titer at early convalescence was ≥4-fold higher than the acute-phase titer or when seroconversion was observed. In this diagnostic scenario, the sensitivity was 68.3% (59.4% to 76.3%) and the specificity was 58.8% (51.0% to 66.3%) (Table 4; see also Fig. S1 in the supplemental material). Another application of this method is comparing titers in paired annual samples in cohort studies to determine the incidence of ZIKV infection (20). When paired annual samples were evaluated with the IEM, positivity was defined as seroconversion or a ≥4× increase in titer (Fig. S1), yielding a sensitivity of 94.0% (88.0% to 97.5%) and a specificity of 98.6% (94.9% to 98.8%) (Table 4). The IEM was significantly more sensitive and specific (P < 0.0001 for both) in paired annual samples than in samples from the acute-phase and early-convalescent-phase time points. Comparing the NS1 BOB ELISA and IEM techniques for paired acute-phase and early-convalescent-phase samples, the NS1 BOB ELISA was significantly more sensitive and specific than the IEM (P = 0.0027 and <0.0001, respectively).
TABLE 4.
Results of IEM for diagnosis of Zika cases and determination of ZIKV infection incidencea
| Patient population | Inhibition ELISA |
|||
|---|---|---|---|---|
| Diagnosis |
Infection incidence |
|||
| Sens (95% CI) | Spec (95% CI) | Sens (95% CI) | Spec (95% CI) | |
| Overall | 68.3 (59.4–76.3) | 58.8 (51.0–66.3) | 94.0 (88.0–97.5) | 98.6 (94.9–98.8) |
| Zika virus positive | 68.3 (59.4–76.3) | NA | 94.0 (88.0–97.5) | NA |
| DENV naïve | 55.7 (42.4–68.5) | NA | 96.6 (88.1–99.6) | NA |
| DENV immune | 80.0 (68.2–88.9) | NA | 90.9 (80.0–97.0) | NA |
| Dengue virus positive | NA | 40.6 (30.7–51.1) | NA | 97.2 (90.3–99.7) |
| DENV1 | NA | 41.7 (22.1–63.4) | NA | 100.0 (82.4–100.0) |
| DENV2 | NA | 38.2 (22.2–56.4) | NA | 100.0 (86.3–100.0) |
| DENV3 | NA | 42.1 (26.3–59.2) | NA | 92.3 (76.5–99.1) |
| Primary | NA | 60.4 (45.3–74.2) | NA | 100.0 (89.4–100.0) |
| Secondary | NA | 20.8 (10.5–35.0) | NA | 94.9 (82.7–99.4) |
| Negative | NA | 82.4 (71.8–90.3) | NA | 100.0 (94.7–100.0) |
Sens, sensitivity. Sensitivity values were calculated for each assay based on ZIKV-positive samples, stratified by those who had been previously infected with DENV (DENV immune) and those who were DENV naïve. Spec, specificity. Specificity values were calculated for each assay based on two groups of ZIKV-negative samples: (i) ZIKV-negative DENV-positive samples, stratified by DENV subtype (DENV1 to DENV3) and primary and secondary infection, and (ii) ZIKV-negative DENV-negative controls. 95% CI, 95% confidence interval. NA, not applicable: data from ZIKV-positive cases served only for determination of sensitivity and were not used for the specificity calculation. Similarly, data from ZIKV-negative cases served only for determination of specificity and were not used for the sensitivity calculation.
The two molecular methods performed similarly and demonstrated very good agreement (kappa = 0.93) for ZIKV detection in acute-phase samples. The overall sensitivity and specificity of each assay were 96.1% and 100%, respectively (Table 5). There were 10 samples with discordant rRT-PCR results, with 5 samples testing positive in only one of the two assays. All discordant samples had late threshold cycle (CT) values in the respective assays of >38 for samples positive only in the ZCD assay and >35 for samples positive only in the Trioplex assay. However, all rRT-PCR-positive samples also tested positive by at least one serological method.
TABLE 5.
Evaluation of real-time RT-PCR methodsa
| Patient population | RT-PCR |
|||
|---|---|---|---|---|
| ZCD |
Trioplex |
|||
| Sens (95% CI) | Spec (95% CI) | Sens (95% CI) | Spec (95% CI) | |
| Overall | 96.1 (91.1–98.7) | 100 (97.9–100) | 96.1 (91.1–98.7) | 100 (97.9–100) |
| Zika virus positive | 96.1 (91.1–98.7) | NA | 96.1 (91.1–98.7) | NA |
| DENV naïve | 93.5 (84.3–98.2) | NA | 95.2 (86.5–99) | NA |
| DENV immune | 98.5 (91.7–100) | NA | 96.9 (89.3–99.6) | NA |
| Dengue virus positive | NA | 100 (96.3–100) | NA | 100 (96.3–100) |
| DENV1 | NA | 100 (85.8–100) | NA | 100 (85.8–100) |
| DENV2 | NA | 100 (89.7–100) | NA | 100 (89.7–100) |
| DENV3 | NA | 100 (91–100) | NA | 100 (91–100) |
| Negative | NA | 100 (95.3–100) | NA | 100 (95.3–100) |
Sens, sensitivity. Sensitivity values were calculated for each assay based on ZIKV-positive samples, stratified by those who had been previously infected with DENV (DENV immune) and those who were DENV naïve. Spec, specificity. Specificity values were calculated for each assay based on two groups of ZIKV-negative samples: (i) ZIKV-negative DENV-positive samples, stratified by DENV subtype (DENV1 to DENV3) and primary and secondary infection, and (ii) ZIKV-negative DENV-negative controls. 95% CI, 95% confidence interval. NA, not applicable: data from ZIKV-positive cases served only for determination of sensitivity and were not used for the specificity calculation. Similarly, data from ZIKV-negative cases served only for determination of specificity and were not used for the sensitivity calculation.
DISCUSSION
Since the introduction of ZIKV into the Americas in 2015, accurately differentiating ZIKV and DENV infections has been a challenge. Serological cross-reactivity among flaviviruses (8, 10, 11, 13) and the low ZIKV viral load and short period of detectable viremia (32, 33) have complicated serological diagnosis and molecular diagnosis, respectively. Hence, it is necessary to thoroughly evaluate multiple diagnostic assays. However, the evaluation of different assays has been hindered by limited access to adequate numbers of well-characterized samples. Over the past 14 years, the Nicaraguan PDCS, with an average size of ~3,600 active participants each year, has accumulated a large collection of serum samples, including paired acute- and early-convalescent-phase samples from episodes of febrile illness together with annual healthy samples from subjects with well-documented histories of arbovirus infections (20). Hence, these samples are well-suited for diagnostic assay evaluation. In the present study, we assessed four serological and two molecular methods for the diagnosis and surveillance of ZIKV infection using sample sets from a total of 301 patients, including flavivirus-immune and flavivirus-naïve individuals with Zika, dengue, or other febrile illnesses.
Among the serological methods evaluated, two detect anti-ZIKV IgM antibodies; one detects total antibodies against the ZIKV NS1 protein, and the other measures total antibodies against ZIKV. The CNDR MAC-ELISA was significantly more sensitive and more specific than the NIAID-BEI MAC-ELISA. The use of a conjugated MAb, ZKA64, which is highly neutralizing and is specific for domain III of the ZIKV E protein (14), in the CNDR MAC-ELISA, compared to the NIAID-BEI MAC-ELISA, which uses a less-specific anti-flavivirus MAb, could explain the differences between the assays. Importantly, there are substantial differences in processing time between the two IgM ELISAs. The CNDR MAC-ELISA takes 3 to 4 h, whereas the NIAID-BEI MAC-ELISA BEI takes 2 to 3 days. The shorter processing time is very important for epidemiological surveillance during epidemics, as evidenced in Nicaragua during the 2016 Zika virus disease epidemic.
Overall, the CNDR and NIAID-BEI MAC-ELISAs performed well. In another report, Safronetz et al. (34) evaluated 5 commercial Zika immunoassays and concluded that most of them had adequate (higher than 90%) specificity in returned Canadian travelers but low sensitivity (37% to 65%), except for the InBios ELISA, which had 100% sensitivity but low specificity in samples from dengue cases. Another study reported sensitivity for Zika IgM detection by the Euroimmun assay of 39.5%, which improved to 83% when the assay was combined with IgG detection (19). The specificity was 92%; however, only samples from flavivirus-naïve or likely primary DENV infections were evaluated, whereas our study measured specificity by using samples from both primary and secondary DENV infections as well as from flavivirus-naïve individuals.
In contrast to the MAC-ELISA, the Zika NS1 BOB ELISA can be used for both diagnosis and seroprevalence studies as it likely primarily measures levels of IgG antibodies. However, as it shows higher sensitivity in later convalescence, the optimal use is for surveillance and seroprevalence studies. The Zika NS1 BOB method was initially evaluated in a multicenter study involving laboratories from 5 countries, yielding sensitivity and specificity values of 92% and 96%, respectively (22). In the present evaluation, similar results were obtained, with higher sensitivity in the late-convalescent-phase samples (mean of 223 days postinfection) than in the early-convalescent phase samples (mean of 16.7 days). This is consistent with the previous results, where the sensitivity of the NS1 BOB ELISA was increased at >20 days post-onset of symptoms (22). We also noted that the sensitivity in early convalescence was greater in DENV-immune patients than in DENV-naïve Zika patients, suggesting more rapid kinetics of the antibody response in secondary flavivirus infections, as expected. Thus, unlike DENV or CHIKV infections, where antibody titers rise sufficiently by days 14 to 17 postinfection, it appears that anti-NS1 antibodies rise more slowly in ZIKV infections, with even slower kinetics in DENV-naïve Zika patients. One possible explanation is the lower viremia of ZIKV than of DENV and CHIKV, which could result in lower antibody titers and/or slower antibody kinetics.
Regarding specificity, Balmaseda et al. (22) reported a specificity of 89% for the NS1 BOB ELISA in evaluation of dengue cases, coinciding with the current study, which yielded 88.7% specificity for dengue cases. The reported specificity increased to 96% when samples from patients infected with other flaviviruses or other viruses and healthy donors were included (22); in the present study, the specificity increased to 91.4 to 92.6% when samples from flavivirus-negative patients were included. In general, these results indicate that the NS1 BOB ELISA can be useful for seroprevalence studies of anti-ZIKV antibodies at the population level, differentiating individuals infected by ZIKV from those infected by other flaviviruses.
Two applications of the Zika IEM were evaluated, based on our experience with the dengue IEM (35–37). To diagnose febrile episodes, a ≥4-fold increase in titer or seroconversion between the acute-phase samples and early-convalescent-phase samples was used; however, the Zika IEM performed poorly in this setting. Again, this was potentially due to low ZIKV viremia resulting in delayed and lower antibody responses. The other scenario is to detect incidence of ZIKV infections by comparing titers in paired annual samples bounding the intervening year. Here, the IEM performed better, with a sensitivity value of 94% and specificity of 98.6%. The IEM has been used successfully for 14 years for anti-DENV antibody detection in the PDCS and other Nicaraguan studies (18, 30, 35–37). A similar method was adapted for chikungunya diagnosis and surveillance, with sensitivity and specificity greater than 95% (26).
For Zika diagnosis, using paired acute-phase and early-convalescent-phase samples, the IgM-based CNDR MAC-ELISA technique is recommended, since it is based on a domain III ZIKV E monoclonal antibody and yielded 94.5% sensitivity and 85.6% specificity, performing better than the NIAID-BEI MAC ELISA, the NS1 BOB ELISA, and the IEM. For paired acute-phase and late-convalescent-phase samples, as well as seroprevalence studies, the NS1 BOB ELISA is recommended, as it was highly sensitive (96.5%) and specific (92.6%) and has a shorter processing time than the IEM. If paired annual samples are available (as in a cohort study), the IEM technique is recommended for determination of infection incidence.
Regarding molecular methods, two multiplex rRT-PCR assays were evaluated, the ZCD method targeting the ZIKV NS4B gene and the Trioplex assay directed to the E gene of the ZIKV genome. Both rRT-PCR assays demonstrated high sensitivity and specificity for ZIKV (96.1% and 100%, respectively), which is consistent with previous results for the ZCD assay evaluated against a monoplex rRT-PCR (16). Thus, the multiplex design of the two assays evaluated here does not appear to affect the sensitivity of ZIKV detection. As there is no gold standard for Zika diagnosis, the current study utilized a composite reference standard that required at least one positive molecular result and one positive serological result for a case to be considered positive. This, coupled with the use of samples from confirmed dengue cases and from cases of nondengue, non-Zika febrile illnesses, provides strong evidence that these rRT-PCR assays provide sensitive and specific ZIKV detection in the acute setting.
Overall, the strengths of this study were the sample size and the use of well-characterized cases. Importantly, equal numbers of DENV-immune and DENV-naïve Zika cases were evaluated, as well as equal numbers of primary and secondary infections with DENV1, DENV2, and DENV3. Data corresponding to prior DENV exposure were not determined by preinfection antibody titers, which can potentially be misleading, but rather by inclusion of children who had entered the cohort study in a DENV-naïve state and who had had no DENV infections (DENV-naïve children) or had experienced a documented DENV infection or had entered the PDCS with serological evidence of prior DENV infection (DENV-immune children). Of note, all the evaluations were performed in Nicaragua. One limitation was that there is very little circulation of DENV4 in Nicaragua and that clinical illness due to DENV4 is very rare; therefore, this analysis focused on DENV serotypes 1, 2, and 3.
In conclusion, four serological methods were evaluated, confirming their usefulness for the diagnosis of ZIKV infections (e.g., both IgM MAC-ELISAs) and/or for surveillance and seroprevalence studies (e.g., NS1 BOB ELISA, Zika IEM), including two in-house serological assays and the NS1 BOB ELISA, which provide low-cost alternatives to commercial kits. Overall, together with the two molecular assays that performed well, we have established and evaluated both molecular and serological methods that allow discrimination of current and past ZIKV infection in regions where infections by other flaviviruses, such as DENV, are also endemic.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lionel Gresh for his advice and assistance. We thank past and present members of the study team based at the Centro de Salud Sóìcrates Flores Vivas, the National Virology Laboratory in the Centro Nacional de Diagnóstico y Referencia, and the Sustainable Sciences Institute in Nicaragua for their dedication and high-quality work, and we are grateful to the study participants and their families.
This research was funded by grants from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health R01AI099631 (A.B.) and P01AI106695 (E.H.); the cohort study was also supported by NIH grant U19AI118610 (E.H.), as well as by Pediatric Dengue Vaccine Initiative grant VE-1 (E.H.) and a FIRST grant (E.H.), both from the Bill and Melinda Gates Foundation. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We report no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01785-17.
REFERENCES
- 1.do Rosário MS, de Jesus PA, Vasilakis N, Farias DS, Novaes MA, Rodrigues SG, Martins LC, Vasconcelos PF, Ko AI, Alcântara LC, de Siqueira IC. 2016. Guillain-Barre syndrome after Zika virus infection in Brazil. Am J Trop Med Hyg 95:1157–1160. doi: 10.4269/ajtmh.16-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mlakar J, Korva M, Tul N, Popovic M, Poljsak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodusek V, Vizjak A, Pizem J, Petrovec M, Avsic Zupanc T. 2016. Zika virus associated with microcephaly. N Engl J Med 374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 3.Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. 2016. Zika virus and birth defects—reviewing the evidence for causality. N Engl J Med 374:1981–1987. doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- 4.Siu R, Bukhari W, Todd A, Gunn W, Huang QS, Timmings P. 2016. Acute Zika infection with concurrent onset of Guillain-Barre syndrome. Neurology 87:1623–1624. doi: 10.1212/WNL.0000000000003038. [DOI] [PubMed] [Google Scholar]
- 5.Cicuto Ferreira Rocha NA, de Campos AC, Cicuto Ferreira Rocha F, Pereira Dos Santos Silva F. 2017. Microcephaly and Zika virus: neuroradiological aspects, clinical findings and a proposed framework for early evaluation of child development. Infant Behav Dev 49:70–82. doi: 10.1016/j.infbeh.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Ventura CV, Maia M, Bravo-Filho V, Gois AL, Belfort R Jr. 2016. Zika virus in Brazil and macular atrophy in a child with microcephaly. Lancet 387:228. doi: 10.1016/S0140-6736(16)00006-4. [DOI] [PubMed] [Google Scholar]
- 7.Ventura CV, Maia M, Ventura BV, Linden VV, Araujo EB, Ramos RC, Rocha MA, Carvalho MD, Belfort R Jr, Ventura LO. 2016. Ophthalmological findings in infants with microcephaly and presumable intra-uterus Zika virus infection. Arq Bras Oftalmol 79:1–3. doi: 10.5935/0004-2749.20160002. [DOI] [PubMed] [Google Scholar]
- 8.Aziz H, Zia A, Anwer A, Aziz M, Fatima S, Faheem M. 2017. Zika virus: global health challenge, threat and current situation. J Med Virol 89:943–951. doi: 10.1002/jmv.24731. [DOI] [PubMed] [Google Scholar]
- 9.Faria NR, Azevedo R, Kraemer MUG, Souza R, Cunha MS, Hill SC, Theze J, Bonsall MB, Bowden TA, Rissanen I, Rocco IM, Nogueira JS, Maeda AY, Vasami F, Macedo FLL, Suzuki A, Rodrigues SG, Cruz ACR, Nunes BT, Medeiros DBA, Rodrigues DSG, Queiroz ALN, da Silva EVP, Henriques DF, da Rosa EST, de Oliveira CS, Martins LC, Vasconcelos HB, Casseb LMN, Simith DB, Messina JP, Abade L, Lourenco J, Alcantara LCJ, de Lima MM, Giovanetti M, Hay SI, de Oliveira RS, Lemos PDS, de Oliveira LF, de Lima CPS, da Silva SP, de Vasconcelos JM, Franco L, Cardoso JF, Vianez-Junior J, Mir D, Bello G, Delatorre E, Khan K, et al. . 2016. Zika virus in the Americas: early epidemiological and genetic findings. Science 352:345–349. doi: 10.1126/science.aaf5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T, Sakuntabhai A, Cao-Lormeau VM, Malasit P, Rey FA, Mongkolsapaya J, Screaton GR. 2016. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat Immunol 17:1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. 2008. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landry ML, St George K. 2017. Laboratory diagnosis of Zika virus infection. Arch Pathol Lab Med 141:60–67. doi: 10.5858/arpa.2016-0406-SA. [DOI] [PubMed] [Google Scholar]
- 13.Raboni SM, Bonfim C, Almeida BM, Zanluca C, Koishi AC, Rodrigues P, Kay CK, Ribeiro LL, Scola RH, Duarte Dos Santos CN. 2017. Flavivirus cross-reactivity in serological tests and Guillain-Barre syndrome in a hematopoietic stem cell transplant patient: a case report. Transpl Infect Dis 19:e12700. doi: 10.1111/tid.12700. [DOI] [PubMed] [Google Scholar]
- 14.Stettler K, Beltramello M, Espinosa DA, Graham V, Cassotta A, Bianchi S, Vanzetta F, Minola A, Jaconi S, Mele F, Foglierini M, Pedotti M, Simonelli L, Dowall S, Atkinson B, Percivalle E, Simmons CP, Varani L, Blum J, Baldanti F, Cameroni E, Hewson R, Harris E, Lanzavecchia A, Sallusto F, Corti D. 2016. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 353:823–826. doi: 10.1126/science.aaf8505. [DOI] [PubMed] [Google Scholar]
- 15.CDC. 2017. Trioplex real-time RT-PCR assay; instructions for use. CDC, Atlanta, GA. [Google Scholar]
- 16.Waggoner JJ, Gresh L, Mohamed-Hadley A, Ballesteros G, Davila MJ, Tellez Y, Sahoo MK, Balmaseda A, Harris E, Pinsky BA. 2016. Single-reaction multiplex reverse transcription PCR for detection of Zika, chikungunya, and dengue viruses. Emerg Infect Dis 22:1295–1297. doi: 10.3201/eid2207.160326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brasil P, Pereira JP Jr, Moreira ME, Ribeiro Nogueira RM, Damasceno L, Wakimoto M, Rabello RS, Valderramos SG, Halai UA, Salles TS, Zin AA, Horovitz D, Daltro P, Boechat M, Raja Gabaglia C, Carvalho de Sequeira P, Pilotto JH, Medialdea-Carrera R, Cotrim da Cunha D, Abreu de Carvalho LM, Pone M, Machado Siqueira A, Calvet GA, Rodrigues Baiao AE, Neves ES, Nassar de Carvalho PR, Hasue RH, Marschik PB, Einspieler C, Janzen C, Cherry JD, Bispo de Filippis AM, Nielsen-Saines K. 2016. Zika virus infection in pregnant women in Rio de Janeiro. N Engl J Med 375:2321–2334. doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balmaseda A, Saborio S, Tellez Y, Mercado JC, Perez L, Hammond SN, Rocha C, Kuan G, Harris E. 2008. Evaluation of immunological markers in serum, filter-paper blood spots, and saliva for dengue diagnosis and epidemiological studies. J Clin Virol 43:287–291. doi: 10.1016/j.jcv.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 19.L'Huillier AG, Hamid-Allie A, Kristjanson E, Papageorgiou L, Hung S, Wong CF, Stein DR, Olsha R, Goneau LW, Dimitrova K, Drebot M, Safronetz D, Gubbay JB. 2017. Evaluation of Euroimmun anti-Zika virus IgM and IgG enzyme-linked immunosorbent assays for Zika virus serologic testing. J Clin Microbiol 55:2462–2471. doi: 10.1128/JCM.00442-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuan G, Gordon A, Aviles W, Ortega O, Hammond SN, Elizondo D, Nunez A, Coloma J, Balmaseda A, Harris E. 2009. The Nicaraguan pediatric dengue cohort study: study design, methods, use of information technology, and extension to other infectious diseases. Am J Epidemiol 170:120–129. doi: 10.1093/aje/kwp092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balmaseda A, Gordon A, Gresh L, Ojeda S, Saborio S, Tellez Y, Sanchez N, Kuan G, Harris E. 2016. Clinical attack rate of Chikungunya in a cohort of Nicaraguan children. Am J Trop Med Hyg 94:397–399. doi: 10.4269/ajtmh.15-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balmaseda A, Stettler K, Medialdea-Carrera R, Collado D, Jin X, Zambrana JV, Jaconi S, Cameroni E, Saborio S, Rovida F, Percivalle E, Ijaz S, Dicks S, Ushiro-Lumb I, Barzon L, Siqueira P, Brown DWG, Baldanti F, Tedder R, Zambon M, de Filippis AMB, Harris E, Corti D. 2017. Antibody-based assay discriminates Zika virus infection from other flaviviruses. Proc Natl Acad Sci U S A 114:8384–8389. doi: 10.1073/pnas.1704984114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.BEI Resources. 2016. NIAID Zika virus (ZIKV) MAC-ELISA assay sheet. BEI Resources, Manassas, VA. [Google Scholar]
- 24.Waggoner JJ, Abeynayake J, Sahoo MK, Gresh L, Tellez Y, Gonzalez K, Ballesteros G, Balmaseda A, Karunaratne K, Harris E, Pinsky BA. 2013. Development of an internally controlled real-time reverse transcriptase PCR assay for pan-dengue virus detection and comparison of four molecular dengue virus detection assays. J Clin Microbiol 51:2172–2181. doi: 10.1128/JCM.00548-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waggoner JJ, Abeynayake J, Sahoo MK, Gresh L, Tellez Y, Gonzalez K, Ballesteros G, Guo FP, Balmaseda A, Karunaratne K, Harris E, Pinsky BA. 2013. Comparison of the FDA-approved CDC DENV-1-4 real-time reverse transcription-PCR with a laboratory-developed assay for dengue virus detection and serotyping. J Clin Microbiol 51:3418–3420. doi: 10.1128/JCM.01359-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galo SS, González K, Téllez Y, García N, Pérez L, Gresh L, Harris E, Balmaseda Á. 2017. Development of in-house serological methods for diagnosis and suveillance of chikungunya. Rev Panam Salud Publica 41:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balmaseda A, Guzman MG, Hammond S, Robleto G, Flores C, Tellez Y, Videa E, Saborio S, Perez L, Sandoval E, Rodriguez Y, Harris E. 2003. Diagnosis of dengue virus infection by detection of specific immunoglobulin M (IgM) and IgA antibodies in serum and saliva. Clin Diagn Lab Immunol 10:317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarke DH, Casals J. 1958. Techniques for hemagglutination and hemagglutination-inhibition with arthropod-borne viruses. Am J Trop Med Hyg 7:561–573. doi: 10.4269/ajtmh.1958.7.561. [DOI] [PubMed] [Google Scholar]
- 29.Reed LJ, Muench H. 1938. A simple method of estimating 50% endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 30.Balmaseda A, Hammond SN, Tellez Y, Imhoff L, Rodriguez Y, Saborio SI, Mercado JC, Perez L, Videa E, Almanza E, Kuan G, Reyes M, Saenz L, Amador JJ, Harris E. 2006. High seroprevalence of antibodies against dengue virus in a prospective study of schoolchildren in Managua, Nicaragua. Trop Med Int Health 11:935–942. doi: 10.1111/j.1365-3156.2006.01641.x. [DOI] [PubMed] [Google Scholar]
- 31.Waggoner J, Heath CJ, Ndenga B, Mutuku F, Sahoo MK, Mohamed-Hadley A, Vulule J, Mukoko D, Desiree LaBeaud A, Pinsky BA. 2017. Development of a real-time reverse transcription polymerase chain reaction for O'nyong-nyong virus and evaluation with clinical and mosquito specimens from Kenya. Am J Trop Med Hyg 97:121–124. doi: 10.4269/ajtmh.17-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fourcade C, Mansuy JM, Dutertre M, Delpech M, Marchou B, Delobel P, Izopet J, Martin-Blondel G. 2016. Viral load kinetics of Zika virus in plasma, urine and saliva in a couple returning from Martinique, French West Indies. J Clin Virol 82:1–4. doi: 10.1016/j.jcv.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 33.Pessoa R, Patriota JV, de Souza MDL, Abd El Wahed A, Sanabani SS. 2016. Detection of Zika virus in Brazilian patients during the first five days of infection-urine versus plasma. Euro Surveill 21(30):pii=30302 http://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2016.21.30.30302. [DOI] [PubMed] [Google Scholar]
- 34.Safronetz D, Sloan A, Stein DR, Mendoza E, Barairo N, Ranadheera C, Scharikow L, Holloway K, Robinson A, Traykova-Andonova M, Makowski K, Dimitrova K, Giles E, Hiebert J, Mogk R, Beddome S, Drebot M. 2017. Evaluation of 5 commercially available Zika virus immunoassays. Emerg Infect Dis 23:1577–1580. doi: 10.3201/eid2309.162043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balmaseda A, Standish K, Mercado JC, Matute JC, Tellez Y, Saborio S, Hammond SN, Nunez A, Aviles W, Henn MR, Holmes EC, Gordon A, Coloma J, Kuan G, Harris E. 2010. Trends in patterns of dengue transmission over 4 years in a pediatric cohort study in Nicaragua. J Infect Dis 201:5–14. doi: 10.1086/648592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon A, Kuan G, Mercado JC, Gresh L, Aviles W, Balmaseda A, Harris E. 2013. The Nicaraguan pediatric dengue cohort study: incidence of inapparent and symptomatic dengue virus infections, 2004–2010. PLoS Negl Trop Dis 7:e2462. doi: 10.1371/journal.pntd.0002462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montoya M, Gresh L, Mercado JC, Williams KL, Vargas MJ, Gutierrez G, Kuan G, Gordon A, Balmaseda A, Harris E. 2013. Symptomatic versus inapparent outcome in repeat dengue virus infections is influenced by the time interval between infections and study year. PLoS Negl Trop Dis 7:e2357. doi: 10.1371/journal.pntd.0002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.