Abstract
AIM
To detect the expression of miR-211 in age-related cataract tissue, explore the effects of miR-211 on lens epithelial cell proliferation and apoptosis, and identify its target gene.
METHODS
This study used real-time quantitative polymerase chain reaction (RT-qPCR) to measure the expression of miR-211 and its predicted target gene [silent mating-type information regulation 2 homolog 1 (SIRT1)] in 46 anterior lens capsules collected from age-related cataract patients. Human lens epithelial cell line (SRA01/04) cells were transfected with either miR-211 mimics, mimic controls, miR-211 inhibitors or inhibitor controls, 72h after transfection, miRNA and protein expression of SIRT1 were measured using RT-qPCR and Western blotting; then cells were exposed to 200 µmol/L H2O2 for 1h, whereupon cell viability was measured by MTS assay, caspase-3 assay was performed. Dual luciferase reporter assay was performed to verify the relationship between miR-211 of SIRT1.
RESULTS
Compared to the control group, expression of miR-211 was significantly increased (P<0.001), the miRNA and protein expression of SIRT1 were significantly decreased (P<0.001) in the anterior lens capsules of patients with age-related cataracts. Relative to the control group, SIRT1 miRNA and protein levels in the miR-211 mimic group were significantly reduced, cell proliferation activity significantly decreased, and caspase-3 activity was significantly increased (P<0.001). In the miR-211 inhibitor group, SIRT1 miRNA and protein expression were significantly increased, cell proliferation activity significantly increased, and caspase-3 activity was significantly decreased (P<0.001). A dual luciferase reporter assay confirmed that SIRT1 is a direct target of miR-211.
CONCLUSION
miR-211 is highly expressed in the anterior lens capsules of patients with age-related cataracts. By negatively regulating the expression of SIRT1, miR-211 promotes lens epithelial cell apoptosis and inhibits lens epithelial cell proliferation.
Keywords: miR-211, silent mating-type information regulation 2 homolog 1, cataract, apoptosis, proliferation
INTRODUCTION
Silent mating-type information regulation 2 homolog 1 (SIRT1) is an NAD+ dependent histone deacetylase, and is involved in the regulation of apoptosis and cell survival under stress[1]. SIRT1 is known to be increased by chronic stress[2], but inhibited by ultraviolet radiation B (UVB)[3]. Its activity is vital for the maintenance of chromosomal integrity and control of various cellular processes including cell metabolism and cellular stress response[4]. Albani et al[5] also showed that resveratrol, a putative SIRT1 activator, protects human neuroblastoma cells (SK-N-BE) from oxidative stress and α-synuclein-mediated toxicity.
MicroRNAs (miRNAs) are a class of tiny non-coding RNA, through binding to complementary sequences in the 3′-untranslated regions (3′-UTR) of their target miRNAs, miRNAs can induce miRNA degradation or translation suppression[6]. Prior studies have shown that miRNAs are involved in a variety of physiological and pathological processes[7]. It has also been shown that some miRNAs are associated with the onset of age-related cataracts, suggesting that miRNAs may become a new target for cataract diagnosis and treatment[8]. Bioinformatic investigation has predicted SIRT1 may be a target of miR-211 and this study was designed to detect miR-211 and SIRT1 expression levels in age-related cataract lens tissue. This was done to further clarify the role of miR-211 in regulating SIRT1 expression, human lens epithelial cell proliferation and apoptosis, and to reveal the mechanism of action of miR-211 in the progression of age-related cataracts.
SUBJECTS AND METHODS
Specimens
Of 46 fresh anterior lens capsules were collected at the Fourth Affiliated Hospital of China Medical University from age-related cataract patients undergoing phacoemulsification surgery (patients were excluded if they were affected by other eye diseases). Totally 21 of the samples were collected from males and 25 from females, aged 53-72 (61.23±9.41)y, and 24 anterior lens capsules of transparent lens (healthy) were obtained from the Fourth Affiliated Hospital of China Medical University Eye Bank, including 9 from males and 15 from females, aged 49-68 (59.12±6.17)y. All specimens were immediately stored in liquid nitrogen at the time of collection. This study was approved by the Ethics Committee of the Fourth Affiliated Hospital of China Medical University, and signed informed consent was obtained from each patient.
Cell culture and transfection human lens epithelial cell line (SRA01/04) was generously provided for experimental use by Dr. Yi-Sin Liu of the Doheny Eye Institute. SRA01/04 cells were cultured in Dulbecco's modified eagle medium (DMEM; Invitrogen, USA) supplemented with 10% fetal bovine serum (Gibco, USA), 100 U/mL penicillin and 100 mg/mL streptomycin (Thermo, USA), and were placed in a 37°C, 5% CO2 constant temperature incubator. SRA01/04 cells were seeded in 24-well cell culture plate for 24h. When 80%-85% confluence was reached, cells were divided into four groups, Lipofectamine RNAiMAX Transfection Reagent (Invitrogen, USA) was used according to the manufacturer instructions to transfect the cells with miR-211 mimics, mimic controls, miR-211 inhibitors, or inhibitor controls. The subsequent experiments were performed 72h after the completion of transfection.
Real-time Quantitative Polymerase Chain Reaction
Total RNA was extracted with Trizol reagent (Invitrogen, USA). For real-time quantitative polymerase chain reaction (RT-qPCR) analysis of miR-211, the total RNA isolated from cells was subsequently reverse transcribed to cDNA using a TaqMan miRNA reverse transcription kit (Applied Biosystems, USA). TaqMan miRNA assays (Applied Biosystems, USA) were used to detect miR-211 expression, and RNU6B was employed as an endogenous control. For RT-qPCR analysis of SIRT1, the total RNA was reverse transcribed using the PrimerScript RT reagent kit (Takara, China) and SIRT1 miRNA expression was detected using the TaqMan Universal Master Mix II kit (Applied Biosystems, USA), with β-actin designated as an endogenous control. The primers of miR-211 and RNU6B were purchased from ThermoFisher (USA) and their sequences can be found on their website. The SIRT1 primer sequences are as follows: forward: 5′-TCGGCAGGTCCCTTTGTCATCC-3′; reverse: 5′-TGCAGGTCAACTGGTGTCGT-3′; the β-actin primer sequences were: forward: 5′-CATCCGT AAAGACCTCTATGCCAAC-3′; reverse: 5′-ATGGAGCCACCGATCCACA-3′. PCR was performed on an ABI 7500 (Applied Biosystems, USA) RT-qPCR System. Three independent experiments were performed and 2−ΔΔCt quantitative analysis was performed to analyze relative expression levels.
Western Blotting
Total protein was extracted used radio-immunoprecipitation assay (RIPA) lysis buffer with protease inhibitor cocktail (Pierce, USA) and BCA kit (Thermo, USA) was employed to quantify protein concentration. Totally 40 µg of protein sample was added to each well of 10% NuPAGE Bis-Tris precast gels (Invitrogen, USA) for electrophoretic separation of proteins. Proteins were then transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were blocked with 5% nonfat milk for 1h at room temperature, then incubated with different primary antibodies: rabbit anti-SIRTl (1:1000, Abcam, USA) and rabbit anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:2000, Abcam, USA) overnight at 4°C. Horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (H+L) secondary antibody (1:2500, Promega, USA) was then added and samples were incubated at room temperature for 2h. The protein bands were visualized using an electrochemiluminescence (ECL) Western blotting substrate kit (Pierce, USA) after which analysis of protein bands was conducted using Image J software.
Dual Luciferase Reporter Assay
Using a human cDNA library as a template, the 3′-UTR region sequence of wild-type SIRT1 containing predicted miR-211 binding site was amplified and then cloned into the luciferase reporter vector (pGL3-Promoter vector; Promega, USA) to obtain the wild-type luciferase reporter gene vector (pGL3-SIRT1-wt). A point mutation was then induced in the predicted binding region of miR-211 on a separate sample of wild-type SIRT1 3′-UTR. This fragment was also cloned into a luciferase pGL3-promoter vector in the same site to obtain a mutant luciferase reporter gene vector (pGL3-SIRT1-mut). SRA01/04 cells were seeded in a 96-well cell culture plate for 24h then cotransfected with a luciferase reporter gene vector and miR-211 inhibitors or inhibitor control according to the specifications in the Lipofectamine RNAiMAX Transfection Reagent manual. The samples were divided into 8 groups: pGL3-SIRT1-wt+inhibitor controls, pGL3-SIRT1-wt+miR-211 inhibitors, pGL3-SIRT1-mut+inhibitor controls, pGL3-SIRT1-mut+miR-211 inhibitors, pGL3-SIRT1-wt+mimic controls, pGL3-SIRT1-wt+miR-211 mimics, pGL3-SIRT1-mut+mimic controls, and pGL3-SIRT1-mut+miR-211 mimics. Totally 72h following this transfection, a dual luciferase reporter assay system (Promega, USA) was used to detect the luciferase activity of each group. All experiments were performed in triplicates.
Cell Proliferation Assay
The number of viable cells in proliferation was determined using the CellTiter96AQueous One Solution Cell Proliferation assay kit (Promega, China). The reagent contains a tetrazolium compound [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt; MTS]. Seventy-two hours after transfection, SRA01/04 cells were exposed to 200 µmol/L H2O2 for 1h. According to the manufacturer's instructions, 20 µL MTS solution was added to each well of the 96-well assay plate containing the cells in 100 µL of culture medium and the cells were then incubated for 4h at 37°C, 5% CO2. The absorbance of each group was read using an absorbance plate reader set to a 490 nm wavelength.
Caspase-3 Activity Assay
Caspase-3 activity was detected using a caspase-3 assay kit (Abcam, USA). Seventy-two hours after transfection, SRA01/04 cells were exposed to 200 µmol/L H2O2 for 1h. Then, in accordance with the manufacturer instructions, these cells were lysed in 50 µL of chilled cell lysis buffer and incubated on ice for 10min, centrifuged, the supernatant protein concentration was determined using the BCA method. Totally 50 µL of cell lysis buffer containing 100 µg protein were added to each well in a 96-well plate. Then, 50 µL 2× reaction buffer, 0.5 µL 10 mmol/L DL-dithiothreitol (DTT) and 5 µL caspase-3 catalytic substrate DEVD-pNA substrate were added to each well. The samples were incubated at 37°C for 2h. The optical density (OD) value was obtained using a microplate reader set at 405 nm wavelength. Each experiment was repeated three times. In the caspase-3 activity control group, OD was set to a value of 1 and the caspase-3 experimental group activity was standardized using the following calculation: (OD value of experimental group-blank well OD)/(OD value of control group-blank well OD)×100%.
Statistical Analysis
Each experiment was performed independently at least 3 times with similar results. Measurement data were presented as mean±standard deviation (SD). Differences between the groups were calculated using unpaired Student's t-tests. Statistical significant difference was considered at P<0.05. Statistical analysis was done using SPSS 16.0.
RESULTS
miR-211 Expression Increased in the Anterior Lens Capsules
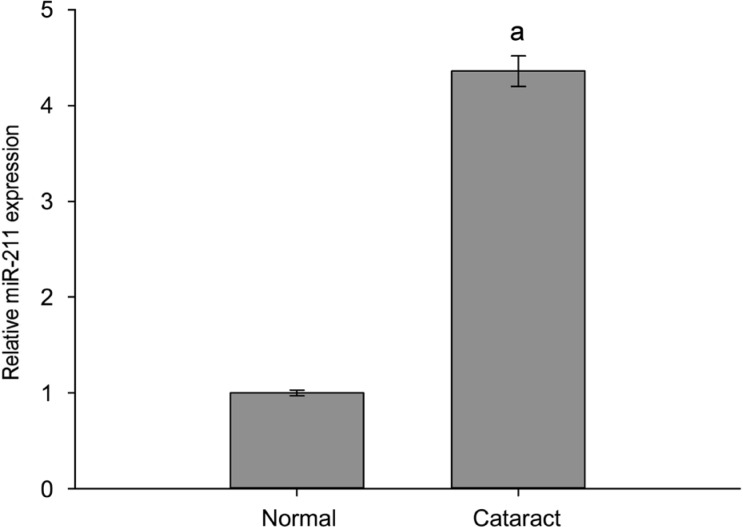
RT-qPCR detection of the expression level of miR-211 indicated that, as compared to the healthy transparent lens capsule group (normal group), the level of miR-211 expression in anterior capsules of patients with age-related cataracts (cataract group) was significantly higher (Figure 1).
Figure 1. miR-211 expression increased in the anterior lens capsules of age-related cataract patients.
RT-qPCR detection of the expression level of miR-211 in cataract group and normal group. The data were expressed as the mean±SD, three independent experiments were performed with pooled specimens. aP<0.001 compared with normal group.
Silent Mating-type Information Regulation 2 Homolog 1 Expression Decreased in the Anterior Lens Capsules
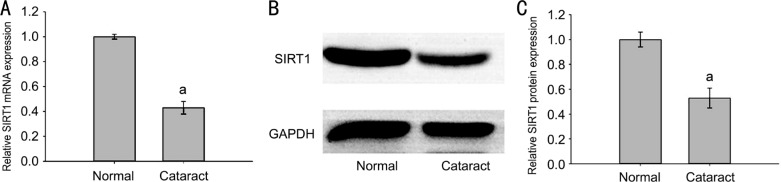
RT-qPCR and Western blotting assays showed a clear decrease in SIRT1 miRNA and protein expression in anterior capsules of age-related cataract patients (cataract group) as compared with the healthy transparent lens capsule group (normal group) (Figure 2).
Figure 2. SIRT1 expression decreased in the anterior lens capsules of age-related cataract patients.
A: miRNA expression of SIRT1 in cataract group and normal group; B: SIRT1 protein expression in cataract group and normal group; C: Strip chart of SIRT1 protein. The data were expressed as the mean±SD, three independent experiments were performed with pooled specimens. aP<0.001 compared with normal group.
miR-211 Regulated Silent Mating-type Information Regulation 2 Homolog 1 Expression in Human Lens Epithelial Cells
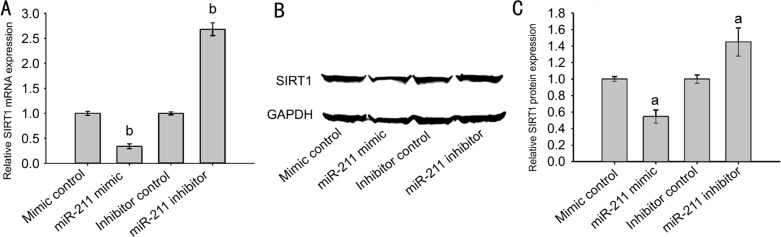
SRA01/04 cells were transfected with miR-211 mimics, mimic controls, miR-211 inhibitors or inhibitor controls. Seventy-two hours after transfection, RT-qPCR was used to measure SIRT1 miRNA expression and Western blotting was used to measure SIRT1 protein levels. As compared to the mimic control group, SIRT1 miRNA (Figure 3A) and protein (Figure 3B, 3C) expression levels in the miR-211 mimic group were significantly decreased (P<0.05). In addition, SIRT1 miRNA (Figure 3A) and protein (Figure 3B, 3C) expression levels in the miR-211 inhibitor group were significantly increased (P<0.05) when compared with the inhibitor control group.
Figure 3. miR-211 regulated SIRT1 expression in human lens epithelial cells.
A: miRNA expression of SIRT1 in each group; B: Protein expression of SIRT1 in each group; C: Strip chart of SIRT1 protein. The data were expressed as the mean±SD, all experiments were performed in triplicates. aP<0.05, bP<0.001 compared with control group.
Silent Mating-type Information Regulation 2 Homolog 1 is the Target of miR-211
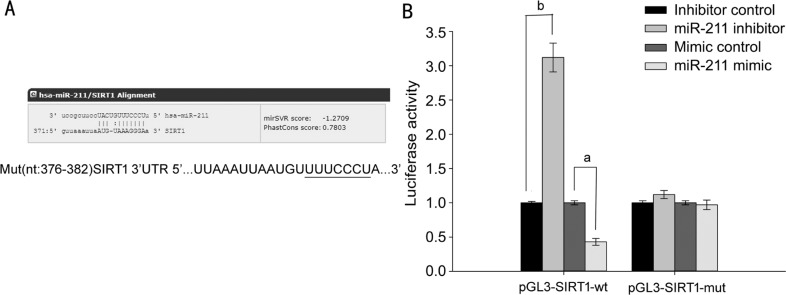
The online software miRanda predicted SIRT1 may be the target gene of miR-211(Figure 4A). This prediction was confirmed using a dual-luciferase reporter assay. SRA01/04 cells were co-transfected with a miR-211 inhibitor or miR-211 mimics and either the pGL3-SIRT1-wt luciferase reporter gene (3′-UTR sequence containing the wild-type SIRT1 miR-211 binding region) or pGL3-SIRT1-mut (containing a mutated 3′-UTR SIRT1 sequence in the miR-211 binding region). Seventy-two hours after transfection, relative luciferase activity was measured. The results showed that, compared with the miR-211 inhibitor control group, cells co-transfected with pGL3-SIRT1-wt+miR-211 inhibitors had significantly increased luciferase activity (P<0.001; Figure 4B). Cells co-transfected with pGL3-SIRT1-mut+miR-211 inhibitors had no significant change in their luciferase activity (P>0.05; Figure 4B), otherwise compared with the miR-211 mimic control group, cells co-transfected with pGL3-SIRT1-wt+miR-211 mimics had significantly decreased luciferase activity (P<0.05; Figure 4B). Cells co-transfected with pGL3-SIRT1-mut+miR-211 mimics had no significant change in their luciferase activity (P>0.05; Figure 4B), demonstrating that SIRT1 is a direct target of miR-211.
Figure 4. SIRT1 is the target of miR-211.
A: Bioinformatic analysis of the wild-type and mutant miR-211 target sequences with the 3′UTR of SIRT1 cDNA; B: The relative luciferase activity was measured by a dual-luciferase reporter assay. The data were expressed as the mean±SD, all experiments were performed in triplicates. aP<0.05, bP<0.001 compared with control group.
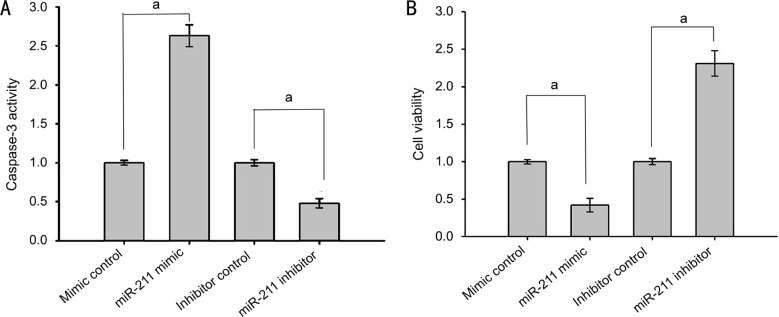
miR-211 Regulated Human Lens Epithelial Cell Proliferation and Apoptosis
Considering the discovery that SIRT1 inhibited apoptosis, further investigation was conducted into the effect of miR-211 on apoptosis and proliferation. SRA01/04 cells were transfected with either miR-211 mimics, mimic controls, miR-211 inhibitors, or inhibitor controls. Seventy-two hours after transfection, the cells were exposed to 200 µmol/L H2O2 for 1h, after which caspase-3 activity was assessed and cell viability was determined using a MTS assay. Compared with the control group, the miR-211 mimic group had significantly elevated caspase-3 activity (P<0.001), while the caspase-3 activity of the miR-211 inhibitor group was markedly decreased (P<0.001; Figure 5A). These results suggest that miR-211 promotes apoptosis in human lens epithelial cells. The MTS assay results showed that compared with the control group, there were significantly fewer viable cells in the miR-211 mimic group cells (P<0.001), while the number of viable cells was significantly increased in the miR-211 inhibitor group (P<0.001; Figure 5B). These results further indicate that miR-211 inhibits the proliferation of human lens epithelial cells.
Figure 5. miR-211 regulated human lens epithelial cell proliferation and apoptosis.
A: Caspase-3 activity in each group; B: Cell viability measured by the MTS assay in each group. The data were expressed as the mean±SD, all experiments were performed in triplicates. aP<0.001 compared with control group.
DISCUSSION
miRNAs are a class of endogenous noncoding RNA ranging from 21 to 25 nucleotides[9]. They often regulate gene expression by complementary pairing with the 3′-UTR of a target gene and mediating the degradation of target gene miRNA[10]–[11]. miRNAs are involved in a variety of physiological and pathological processes including cell growth and apoptosis, hormone secretion, aging, organ development, immune response and other pathologies[7],[12]–[16]. Other studies have shown that miRNAs play an important role not only in ocular development but also the progression of a variety of ocular diseases such as cataracts, glaucoma, age-related macular degeneration and choroidal neovascularization[17]–[25]. miRNAs represent a relatively new and intriguing potential target for the diagnosis, prevention and treatment of ocular disease[26]–[27]. This study highlights the mechanism of miRNA-211 in silencing SIRT1 expression and elucidates its impact on the development of age-related cataracts.
Abnormal expression of miRNAs in cataract tissue has been reported previously. For instance, Qin et al[28] found low expression of miR-125b in the anterior lens capsules of patients with age-related cataracts and further discovered that miR-125b inhibited human lens epithelial cells apoptosis. Other studies showed that expression of miR-421 and miR-133b were decreased in the lens epithelial cells of age-related cataract patients while miR-15a and miR-16-1 expression was increased[8],[29]–[30].
The first novel finding in this study is that miR-211 expression was significantly increased in the anterior lens capsules of patients with age-related cataracts, suggesting that miR-211 may be involved in the development of age-related cataracts. Other studies have reported low expression of miR-211 in various tumor tissues, and have investigated the effects of miR-211 in suppressing tumor cell proliferation, migration, invasion and induce apoptosis[31]–[33]. However, the expression and functions of known miRNAs are highly tissue-specific, and thus the precise mechanism and effects of miR-211 in human lens epithelial cells requires further study.
This is the first study to report that levels of SIRT1 expression were significantly decreased in the anterior lens capsules of age-related cataract patients, suggesting that SIRT1 activity could be linked to miR-211 expression in mediating the development of age-related cataracts. The protein encoded by the SIRT1 gene is an NAD+ dependent histone deacetylase, it belongs to the human Sirtuin protein family, which is highly conserved in various species and that is present in almost all cell types of the body[34]. It has been suggested that SIRT1 may facilitate p53 acetylation, thereby decreasing its ability to induce apoptosis[35]. Calapre et al[36] demonstrated that heat-induced SIRT1 activation mediates survival of DNA damaged keratinocytes through deacetylation of p53 after exposure to UVB plus heat. Furthermore, SIRT1 overexpression in osteoblasts suppressed hypoxia-induced apoptosis[1]. SIRT1 seems to play a protective role in a variety of pathological conditions; however, the mechanism of SIRT1 in cataract development remains unclear.
The online software miRanda predicted that SIRT1 may be the target gene of miR-211. We verified that prediction in this study by transient transfecting lens epithelial cells with miR-211 mimics or miR-211 inhibitors and monitoring resultant SIRT1 levels. This experiment found a striking inverse relationship between miR-211 and SIRT1 expression. Finally, dual-luciferase reporter assay confirmed SIRT1 as a direct target of miR-211.
We then transfected lens epithelial cells with miR-211 mimics or miR-211 inhibitors exposed them to 200 µmol/L H2O2 for 1h to induce apoptosis, and found that overexpression of miR-211 group resulted in significantly higher caspase-3 activity as well as significantly fewer viable cell. The miR-211 inhibitor group, by contrast, showed a significant decrease in caspase-3 activity while cell activity was significantly increased, indicating that miR-211 plays a role in inducing lens epithelial cell apoptosis and inhibiting the proliferation of lens epithelial cells. These results indicate that miR-211 induces lens epithelial cell apoptosis by SIRT1 expression, suggesting that it may play a key role in the progression of age-related cataracts.
In summary, miR-211 expression greatly elevated in the anterior lens capsules of patients with age-related cataracts. miR-211 induces apoptosis and decreasing the viability of lens epithelial cells by directly targeting SIRT1, miR-211 may play a key role in the development of age-related cataracts. The findings of this study provide strong evidence that miR-211 could be a potential new target for the diagnosis and treatment of cataracts.
Acknowledgments
Foundations: Supported by the National Natural Science Foundation of China (No.81170836; No.81570838); the Natural Science Foundation of Liaoning Province, China (No.2015020474); the Liaoning Provincial Hospital Program for Building Treatment Capacity in Key Clinical Departments (No.LNCCC-D15-2015).
Conflicts of Interest: Lu B, None; Christensen IT, None; Ma LW, None; Wang XL, None; Jiang LF, None; Wang CX, None; Feng L, None; Zhang JS, None; Yan QC, None.
REFERENCES
- 1.Zhou L, Wang SI, Moon YJ, Kim KM, Lee KB, Park BH, Jang KY, Kim JR. Overexpression of SIRT1 prevents hypoxiainduced apoptosis in osteoblast cells. Mol Med Rep. 2017;16(3):2969–2975. doi: 10.3892/mmr.2017.6917. [DOI] [PubMed] [Google Scholar]
- 2.Wang B, Lian YJ, Su WJ, Peng W, Dong X, Liu LL, Gong H, Zhang T, Jiang CL, Wang YX. HMGB1 mediates depressive behavior induced by chronic stress through activating the kynurenine pathway. Brain Behav Immun. 2017;S0889-1591(17):30514–30518. doi: 10.1016/j.bbi.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 3.Chou WW, Chen KC, Wang YS, Wang JY, Liang CL, Juo SH. The role of SIRT1/AKT/ERK pathway in ultraviolet B induced damage on human retinal pigment epithelial cells. Toxicol In Vitro. 2013;27(6):1728–1736. doi: 10.1016/j.tiv.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Wilking MJ, Singh C, Nihal M, Zhong W, Ahmad N. SIRT1 deacetylase is overexpressed in human melanoma and its small molecule inhibition imparts anti-proliferative response via p53 activation. Arch Biochem Biophys. 2014;563:94–100. doi: 10.1016/j.abb.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albani D, Polito L, Batelli S, De Mauro S, Fracasso C, Martelli G, Colombo L, Manzoni C, Salmona M, Caccia S, Negro A, Forloni G. The SIRT1 activator resveratrol protects SK-N-BE cells from oxidative stress and against toxicity caused by alpha-synuclein or amyloid-beta (1-42) peptide. J Neurochem. 2009;110(5):1445–1456. doi: 10.1111/j.1471-4159.2009.06228.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Xiao W, Chen W, Liu X, Wu M, Bo Q, Luo Y, Ye S, Cao Y, Liu Y. MicroRNA-26a and -26b inhibit lens fibrosis and cataract by negatively regulating Jagged-1/Notch signaling pathway. Cell Death Differ. 2017;24(8):1431–1442. doi: 10.1038/cdd.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou X, Jiao Z, Ji J, Li S, Huang X, Lu X, Zhao H, Peng J, Chen X, Ji Q, Ji Y. Characterization of mouse serum exosomal small RNA content: The origins and their roles in modulating inflammatory response. Oncotarget. 2017;8(26):42712–42727. doi: 10.18632/oncotarget.17448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F, Meng W, Tong B. Down-regulation of microRNA-133b suppresses apoptosis of lens epithelial cell by up-regulating BCL2L2 in age-related cataracts. Med Sci Monit. 2016;22:4139–4145. doi: 10.12659/MSM.896975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113(6):673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 10.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 11.Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhuang L, Wang X, Wang Z, Ma X, Han B, Zou H, Wu Z, Dong S, Qu Z, Zang Y, Wu L. MicroRNA-23b functions as an oncogene and activates AKT/GSK3beta/beta-catenin signaling by targeting ST7L in hepatocellular carcinoma. Cell Death Dis. 2017;8(5):e2804. doi: 10.1038/cddis.2017.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marta GN, Garicochea B, Carvalho AL, Real JM, Kowalski LP. MicroRNAs, cancer and ionizing radiation: where are we? Rev Assoc Med Bras (1992) 2015;61(3):275–281. doi: 10.1590/1806-9282.61.03.275. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Tan S, Kooger R, Zhang C, Zhang Y. MicroRNAs as novel biological targets for detection and regulation. Chem Soc Rev. 2014;43(2):506–517. doi: 10.1039/c3cs60312a. [DOI] [PubMed] [Google Scholar]
- 15.Sellitti DF, Doi SQ. MicroRNAs in renal cell carcinoma. Microrna. 2015;4(1):26–35. doi: 10.2174/2211536604666150713105247. [DOI] [PubMed] [Google Scholar]
- 16.Bhaskaran M, Mohan M. MicroRNAs: history, biogenesis, and their evolving role in animal development and disease. Vet Pathol. 2014;51(4):759–774. doi: 10.1177/0300985813502820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berber P, Grassmann F, Kiel C, Weber BH. An eye on age-related macular degeneration: the role of micrornas in disease pathology. Mol Diagn Ther. 2017;21(1):31–43. doi: 10.1007/s40291-016-0234-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunmire JJ, Lagouros E, Bouhenni RA, Jones M, Edward DP. MicroRNA in aqueous humor from patients with cataract. Exp Eye Res. 2013;108:68–71. doi: 10.1016/j.exer.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Zhao F, Xin M, Li G, Luna C, Li G, Zhou Q, He Y, Yu B, Olson E, Gonzalez P, Wang S. Regulation of intraocular pressure by microRNA cluster miR-143/145. Sci Rep. 2017;7(1):915. doi: 10.1038/s41598-017-01003-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghanbari M, Erkeland SJ, Xu L, Colijn JM, Franco OH, Dehghan A, Klaver CCW, Meester-Smoor MA. Genetic variants in microRNAs and their binding sites within gene 3′UTRs associate with susceptibility to age-related macular degeneration. Hum Mutat. 2017;38(7):827–838. doi: 10.1002/humu.23226. [DOI] [PubMed] [Google Scholar]
- 21.Romano GL, Platania CBM, Drago F, Salomone S, Raqusa M, Barbagallo C, Di Pietro C, Purrello M, Reibaldi M, Avitabile T, Longo A, Bucolo C. Retinal and circulating miRNAs in age-related macular degeneration: an in vivo animal and human study. Front Pharmacol. 2017;8:168. doi: 10.3389/fphar.2017.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szemraj M, Bielecka-Kowalska A, Oszajca K, Krajewska M, Gos R, Jurowski P, Kowalski M, Szemraj J. Serum microRNAs as potential biomarkers of AMD. Med Sci Monit. 2015;21:2734–2742. doi: 10.12659/MSM.893697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Lee AY, Wigg JP, Peshavariya H, Liu P, Zhang H. miRNA involvement in angiogenesis in age-related macular degeneration. J Physiol Biochem. 2016;72(4):583–592. doi: 10.1007/s13105-016-0496-2. [DOI] [PubMed] [Google Scholar]
- 24.Wang S, Koster KM, He Y, Zhou Q. miRNAs as potential therapeutic targets for age-related macular degeneration. Future Med Chem. 2012;4(3):277–287. doi: 10.4155/fmc.11.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romano GL, Platania CB, Forte S, Salomone S, Drago F, Bucolo C. MicroRNA target prediction in glaucoma. Prog Brain Res. 2015;220:217–240. doi: 10.1016/bs.pbr.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 26.Simonson B, Das S. MicroRNA therapeutics: the next magic bullet? Mini Rev Med Chem. 2015;15(6):467–474. doi: 10.2174/1389557515666150324123208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenberg DS, Soreq H. MicroRNA therapeutics in neurological disease. Curr Pharm Des. 2014;20(38):6022–6027. doi: 10.2174/1381612820666140314151924. [DOI] [PubMed] [Google Scholar]
- 28.Qin Y, Zhao J, Min X, Wang M, Luo W, Wu D, Yan Q, Li J, Wu X, Zhang J. MicroRNA-125b inhibits lens epithelial cell apoptosis by targeting p53 in age-related cataract. Biochim Biophys Acta. 2014;1842(12 Pt A):2439–2447. doi: 10.1016/j.bbadis.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Li G, Song H, Chen L, Yang W, Nan K, Lu P. TUG1 promotes lens epithelial cell apoptosis by regulating miR-421/caspase-3 axis in age-related cataract. Exp Cell Res. 2017;356(1):20–27. doi: 10.1016/j.yexcr.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Liu S, Zhang F, Jiang P, Wu X, Liang Y. Expression of the microRNAs hsa-miR-15a and hsa-miR-16-1 in lens epithelial cells of patients with age-related cataract. Int J Clin Exp Med. 2015;8(2):2405–2410. [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Shen YF, Shi ZM, Shang XJ, Jin DL, Xi F. Overexpression miR-211-5p hinders the proliferation, migration, and invasion of thyroid tumor cells by downregulating SOX11. J Clin Lab Anal. 2017 doi: 10.1002/jcla.22293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen LL, Zhang ZJ, Yi ZB, Li JJ. MicroRNA-211-5p suppresses tumour cell proliferation, invasion, migration and metastasis in triple-negative breast cancer by directly targeting SETBP1. Br J Cancer. 2017;117(1):78–88. doi: 10.1038/bjc.2017.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K, Jin W, Jin P, Fei X, Wang X, Chen X. miR-211-5p suppresses metastatic behavior by targeting SNAI1 in renal cancer. Mol Cancer Res. 2017;15(4):448–456. doi: 10.1158/1541-7786.MCR-16-0288. [DOI] [PubMed] [Google Scholar]
- 34.Yonamine CY, Pinheiro-Machado E, Michalani ML, Alves-Wagner AB, Esteves JV, Freitas HS, Machado UF. Resveratrol improves glycemic control in type 2 diabetic obese mice by regulating glucose transporter expression in skeletal muscle and liver. Molecules. 2017;22(7):pii:E1180. doi: 10.3390/molecules22071180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu X, Gu B, Lv X, Yu Z, Wang R, Zhou X, Qiao W, Mao Z, Zuo G, Li Q, Miao D, Jin J. 1, 25-dihydroxy-vitamin D3 with tumor necrosis factor-alpha protects against rheumatoid arthritis by promoting p53 acetylation-mediated apoptosis via Sirt1 in synoviocytes. Cell Death Dis. 2016;7(10):e2423. doi: 10.1038/cddis.2016.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calapre L, Gray ES, Kurdykowski S, David A, Descarques P, Ziman M. SIRT1 activation mediates heat-induced survival of UVB damaged Keratinocytes. BMC Dermatol. 2017;17(1):8. doi: 10.1186/s12895-017-0060-y. [DOI] [PMC free article] [PubMed] [Google Scholar]