Background and aim
Plasmid-mediated colistin resistance mechanisms have been identified worldwide in the past years. A multiplex polymerase chain reaction (PCR) protocol for detection of all currently known transferable colistin resistance genes (mcr-1 to mcr-5, and variants) in Enterobacteriaceae was developed for surveillance or research purposes. Methods: We designed four new primer pairs to amplify mcr-1, mcr-2, mcr-3 and mcr-4 gene products and used the originally described primers for mcr-5 to obtain a stepwise separation of ca 200 bp between amplicons. The primer pairs and amplification conditions allow for single or multiple detection of all currently described mcr genes and their variants present in Enterobacteriaceae. The protocol was validated testing 49 European Escherichia coli and Salmonella isolates of animal origin. Results: Multiplex PCR results in bovine and porcine isolates from Spain, Germany, France and Italy showed full concordance with whole genome sequence data. The method was able to detect mcr-1, mcr-3 and mcr-4 as singletons or in different combinations as they were present in the test isolates. One new mcr-4 variant, mcr-4.3, was also identified. Conclusions: This method allows rapid identification of mcr-positive bacteria and overcomes the challenges of phenotypic detection of colistin resistance. The multiplex PCR should be particularly interesting in settings or laboratories with limited resources for performing genetic analysis as it provides information on the mechanism of colistin resistance without requiring genome sequencing.
Keywords: polymerase chain reaction, multiplex, colistin, antimicrobial resistance, transferable resistance, polymyxin E, mcr, surveillance, mcr-4.3
Introduction
Colistin belongs to the antimicrobial class designated polymyxins which originates from the organism Paenibacillus polymyxa. This class consists of polymyxins A, B, C, D and E, of which only colistin (polymyxin E) and polymyxin B are used in clinical practice [1]. Colistin has been widely used in veterinary medicine in Asian, European and North American countries but human use was restricted due to its neuro- and nephrotoxicity [2-4]. In the past decade, emergence of multidrug-resistant Gram-negative bacteria (such as Enterobacteriaceae, Pseudomonas aeruginosa and Acinetobacter baumannii) led to an increase of colistin administration as a last resort antibiotic for human infections [5]. Prior to the detection of the plasmid-mediated resistance gene mcr-1, resistance to colistin in Enterobacteriaceae had only involved chromosomal mutations, including mutations in the pmrA/pmrB two-component system that regulate synthesis and structure of the lipopolysaccharide [6].
Global surveillance of plasmid-mediated colistin resistance is hindered by the technical difficulties of phenotypic tests owed to particular interactions of the drug with reagents or materials and resulting from its specific molecular structure. The main problems associated with phenotypic testing are the (i) irregular diffusion of colistin in agar media, (ii) interaction with cations in media and (iii) possible adsorption of the antibiotic to certain laboratory materials, which make the agreement of results between replicated testing and between laboratories difficult. Disk diffusion, agar dilution and Etest strips methods cannot be used to perform colistin susceptibility testing, and there is yet not enough information available regarding automated methods to reach a significant conclusion about their sensitivity [1,7]. Thus to date, only determination of minimum inhibitory concentration (MIC) by broth microdilution is recommended in technical guidance documents by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the Clinical and Laboratory Standards Institute (CLSI) [8,9].
The first plasmid-mediated colistin resistance gene was identified in an Escherichia coli strain from a pig in China and also found to be present in other Escherichia coli and Klebsiella pneumoniae isolates in samples from both animal and human origin, collected between 2011 and 2014 [4]. The mcr-1 gene (KP347127) encodes a phosphoetanolamine transferase that alters the lipid A in the lipopolysaccharide of the bacterial outer membrane. This reduces the attachment of colistin and therefore prevents cell lysis. Although this mechanism has only recently been discovered there are reports of isolates recovered in 1980 harbouring the mcr-1 gene, but most cases refer to isolates collected since 2009 [10]. After the first detection, additional four mcr genes were described: mcr-2 (LT598652) was found in E. coli from animal sources in Belgium [11], mcr-3 (KY924928) in E. coli from pigs in China [12], mcr-4 (MF543359) in E. coli and Salmonella enterica serovar Typhimurium from pigs in Italy, Spain and Belgium [13] and mcr-5 (KY807921) in Salmonella Paratyphi B dTa + from poultry in Germany [14]. Variants of mcr-1, mcr-3 and mcr-4 [15-18] as well as co-occurrence of mcr-1 and mcr-3 have been observed in Enterobacteriaceae [19,20].
Even though mcr genes have been more prevalent in animal and food isolates than in human strains, multiple cases of human infection by strains harbouring mcr-1, mcr-3 or mcr-4 have been observed [15,18,21]. Worldwide, over 100 Enterobacteriaceae strains harbouring mcr have been recovered from human hosts, mostly E. coli strains, in patients originating from, or who had recently travelled to, Asian countries [15]. This includes the first mcr-1 description by Liu et al. who identified 16 mcr-1-positive E. coli and K. pneumoniae clinical strains [4]. More recently, mcr-1 and mcr-1.2 were described in one E. coli and one K. pneumoniae strain from infected humans in Denmark [22] and Italy [23], respectively. Three additional E. coli strains carrying the mcr-1 gene were found in faecal samples of Dutch travellers [24] and a study of pilgrims travelling to Mecca revealed the gene in 10 E. coli and one K. pneumoniae strains [25]. These findings represent a snapshot of the cases so far described of mcr-1 in Enterobacteriaceae of human origin and illustrate the global spread of bacteria carrying mcr-1. In Denmark, one E. coli and 10 Salmonella clinical strains collected between 2009 and 2017 were found to harbour mcr-3, six of which were present in people reporting previous travel to Thailand or Vietnam [17,19]. In Italy, mcr-4-positive Salmonella strains have been isolated from two humans with gastroenteritis [18].
The discovery of plasmid-mediated colistin resistance has raised international concern as the spread of this resistance might hamper the efficacy of colistin in humans, although the full clinical relevance has not yet been determined. Plasmid-mediated resistance genes have the potential to be further transmitted and spread globally, including to and within regions with already high levels of antimicrobial resistance where multidrug-resistant bacteria represent a public health concern. This can lead to the emergence of colistin-resistance in already multidrug-resistant bacteria, for which colistin represents a last-resort antibiotic, and compromise antibiotic treatment. Thus, human infections for which therapeutic options have been depleted could occur in nosocomial settings.
Considering (i) the global concerns, (ii) the unreliability of most phenotypic methods for colistin susceptibility testing for surveillance purposes, and (iii) the limited availability of whole genome sequencing (WGS) technology in some settings or laboratories with a lack of adequate resources, we aimed to develop and validate a method that enables screening of relevant isolates in a cheap, rapid and efficient way to identify those possibly harbouring plasmid-mediated colistin resistance genes and meriting further characterisation. Here, we describe a multiplex PCR protocol that detects mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 genes rapidly and reliably for surveillance purposes and epidemiological research, particularly in food, animal and environmental samples.
Methods
The multiplex PCR method development
To allow fast and simultaneous detection of all the currently described mcr genes and their variants and improve visualisation by gel electrophoresis, we focused on conventional block multiplex PCR technique. It was chosen because of its simplicity that enables it to be used in all laboratories and settings with limited laboratory resources, limited or no access to WGS, and in situations where a rapid answer is desired.
The primers for mcr-5 were those described in the original study whereas the primers for mcr-1 (320 bp), mcr-2 (715 bp), mcr-3 (929 bp) and mcr-4 (1116 bp) were designed in this study (Table 1) in order to obtain sequential amplicons with a stepwise size separation of ca 200 bp and allow easy visualisation of bands on agarose gels.
Table 1. Primers and positive control strains used for multiplex PCR for detection of mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 genes.
| Primer name | Sequence (5’-3’) | Target gene | Size (bp) | Positive control strain | Reference |
|---|---|---|---|---|---|
| mcr1_320bp_fw | AGTCCGTTTGTTCTTGTGGC | mcr-1 | 320 | Escherichia coli 2012–60–1176–27 [22] | This study |
| mcr1_320bp_rev | AGATCCTTGGTCTCGGCTTG | ||||
| mcr2_700bp_fw | CAAGTGTGTTGGTCGCAGTT | mcr-2 | 715 | E. coli KP37 [11] | This study |
| mcr2_700bp_rev | TCTAGCCCGACAAGCATACC | ||||
| mcr3_900bp_fw | AAATAAAAATTGTTCCGCTTATG | mcr-3 | 929 | E. coli 2013-SQ352 | This study |
| mcr3_900bp_rev | AATGGAGATCCCCGTTTTT | ||||
| mcr4_1100bp_fw | TCACTTTCATCACTGCGTTG | mcr-4 | 1,116 | E. coli DH5α [13] | This study |
| mcr4_1100bp_rev | TTGGTCCATGACTACCAATG | ||||
| MCR5_fw | ATGCGGTTGTCTGCATTTATC | mcr-5 | 1,644 |
Salmonella 13-SA01718 [14] |
[14] |
| MCR5_rev | TCATTGTGGTTGTCCTTTTCTG |
As determined by the Basic Local Alignment Search Tool (BLAST) at the National Center for Biotechnology Information (NCBI), these primers amplify all mcr variants detected in Enterobacteriaceae to date: mcr-1.2 (KX236309), mcr-1.3 (KU934208), mcr-1.4 (KY041856), mcr-1.5 (KY283125), mcr-1.6 (KY352406), mcr-1.7 (KY488488), mcr-1.8 (KY683842), mcr-1.9 (KY964067.1), mcr-1.11 (KY853650.1), mcr-1.12 (LC337668.1), mcr-1.xx (a new mcr-1 variant, submitted for publication but not yet definitely named), mcr-3.2 (NPZH01000177.1), mcr-3.4 (FLXA01000011.1), mcr-3.5 (MF463699.1), mcr-3.10 (MG214533.1), mcr-3.11 (MG489958.1), mcr-4.2 (MG581979) and mcr-5.xx (a new mcr-5 variant, submitted for publication but not yet definitely named). The remaining variants known so far, mcr-1.10 (MF176238.1), mcr-2.2 (NG_055496.1), mcr-3.3 (MF495680.1), mcr-3.6 (MF598076.1), mcr-3.7 (MF598077.1), mcr-3.8 (MF598079.1) and mcr-3.9 (MF598080.1) have not been reported in Enterobacteriaceae, having only been found in Moraxella spp. and Aeromonas spp.. A full overview of mcr genes and their variants known to date can be found in the Supplement. BLAST analysis also revealed neither interaction between primer pairs nor unspecific binding to other genes.
The original reference strains for mcr-2 (E. coli KP37) and mcr-5 (Salmonella Paratyphi B dTa + 13-SA01718), and strains available at the Technical University of Denmark in Lyngby for mcr-1 (E. coli 2012–60–1176–27), mcr-3 (E. coli 2013-SQ352) and mcr-4 (E. coli DH5α with the entire mcr-4 gene cloned in pCR2 vector) were used as positive controls in the multiplex PCR (Table 1). DNA templates were obtained from overnight agar cultures by thermal cell lysis. Each PCR reaction consisted of 12.5 µL DreamTaq Green PCR Master Mix (Thermo Fisher Scientific, Waltham, Massachusetts, United States), 5.5 µL of nuclease-free water, 0.5 µL of each of the 10 primers solutions (10 µM), and 2 µL DNA lysate. Running conditions were: 1 cycle of denaturation at 94 °C for 15 min, followed by 25 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 90 s and elongation at 72 °C for 60 s, and a final cycle of elongation at 72 °C for 10 min. The amplification was visualised by electrophoresis using 1.5% agarose gel at 130V followed by staining in ethidium-bromide.
An internal amplification control was not used due to the incompatibility with DreamTaq Green PCR Master Mix, which contains a DNA polymerase synthesised in E. coli and thus yields amplicons when using 16S rRNA primers.
Test isolates for validation of the multiplex PCR
To validate the multiplex PCR, 49 isolates from four European countries were used. These isolates were collected within the framework of the European Monitoring of Antimicrobial Resistance pertaining to the Commission Implementing Decision 2013/652/EU [26]. They were analysed at the European Union Reference Laboratory for Antimicrobial Resistance (EURL-AR) in the context of animal health and food safety, in accordance with the EURL-AR/European Food Safety Authority (EFSA) confirmatory testing, that consists of MIC determination by broth microdilution and WGS by Illumina Hi-Seq (Illumina, Inc., San Diego, California, United States) (European Nucleotide Archive project PRJEB21546). The confirmatory testing represents a random snapshot of submitted unusual phenotypes within each participating country based on selective criteria from EFSA.
Results
Isolates collected in 2016 from Spain (n = 19), Germany (n = 16), France (n = 10) and Italy (n = 4) were selected for this study due to the diverse colistin resistance profiles found by WGS in these countries. Forty-two isolates were E. coli from porcine (n = 20) and bovine (n = 22) sources (including caecum and meat samples) and seven isolates were Salmonella spp. from porcine (n = 5) and bovine (n = 2) sources.
The multiplex PCR protocol specifically amplified the fragments of the five mcr genes. Amplicon size ranged from 320 bp (mcr-1) to 1,644 bp (mcr-5) (Figure).
Figure.
Multiplex PCR for detection of mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5, European Union Reference Laboratory for Antimicrobial Resistance (EURL-AR) in the context of animal health and food safety, 2017
Visualisation of the bands on an agarose (1.5%) gel for the five positive control strains. GeneRuler 100 bp Plus DNA Ladder (Thermo Fisher Scientific, Waltham, Massachusetts, United States) was used as molecular size marker and the size of each amplicon is indicated at the side. Mix corresponds to results obtained with a 5 µL mixture of DNA from all five control strains. Neg. corresponds to the blank negative control.
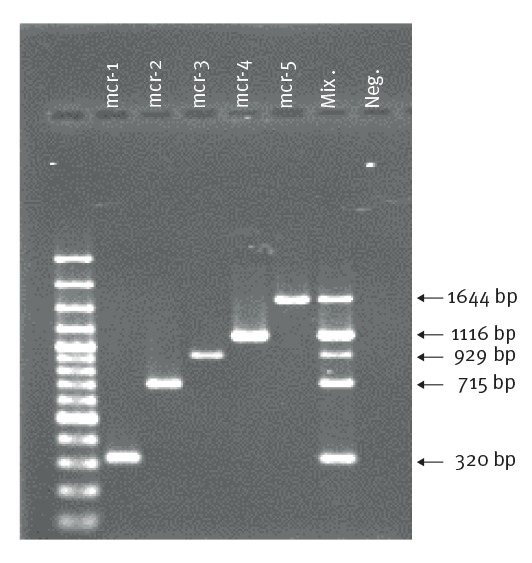
No interference between primer pairs and no unspecific products were observed. Analysing the validation set, the method detected mcr-1, mcr-3 and mcr-4 as singletons and in different combinations (Table 2).
Table 2. Multiplex PCR for detection of mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 compared with WGS results, applied to a collection of Escherichia coli and Salmonella spp. isolates from food animals and food, European Union Reference Laboratory for Antimicrobial Resistance in the context of animal health and food safety, Spain, Germany, France, Italy, 2016 (n = 49).
| Isolate ID | Species | Origin | Source | Colistin MIC | Colistin resistance by PCR | Colistin resistance by WGS | MLSTa | Plasmid contenta | Antimicrobial resistance genes and chromosomal mutationsa |
|---|---|---|---|---|---|---|---|---|---|
| Spain (n=19) | |||||||||
| 150721 | E. coli | Calf | Meat | ≤ 1 | - | - | ST-1488 | FII, FIB(AP001918), I1, Q1, Col(MG828) | bla CTX-M-32, bla TEM-1B, dfrA1, erm(B), mph(B), strA, strB, sul1, tet(A), tet(B), gyrA S83L, gyrA D87N, parC S80I |
| 151570 | Salmonella Kedougou | Pig | Carcass | 4 | mcr-4 | mcr-4.3 b (V236F) | ST-1543 | ColE10, ColpVC, ColRNAI | dfrA14, strB, sul2 |
| 151885 | E. coli | Pig | Meat | 4 | mcr-1 | mcr-1.xx | ST-533 | FIA(HI), HI1A, HI1B(R27) | aph(3')-Ic, bla CTX-M-32, dfrA1, floR, mph(E), msr(E), strA, strB, sul1, tet(B), gyrA S83L, gyrA D87N, parC S80I |
| 151916 | Salmonella Rissen | Pig | Carcass | ≤ 1 | - | - | ST-469 | ColRNAI, R | aadA1, aadA2, bla TEM-1B, dfrA12, mph(A), sul1, tet(A) |
| 152169 | E. coli | Pig | Meat | 4 | mcr-1 | mcr-1 | ST-58 | FII(pCoo), FIB(AP001918), I1, N, R, X4, Col156, Col8282 | aac(3)-IId, aadA2, bla CTX-M-1, bla TEM-1B, dfrA12, mph(A), qnrS1, sul1, tet(A) |
| ZTA15/00213–1EB1 | E. coli | Calf | Caecum | 8 | - | pmrB D283G/Y358N | ST-641 | FII, I1, X1, p0111, Col156, ColRNAI | aadA1, aadA2, bla SHV-12, bla TEM-1A, cmlA1, erm(B), sul1, sul3, tet(M), gyrA S83L, gyrA D87N, parC S80I |
| ZTA15/00420EB1 | E. coli | Pig | Caecum | 8 | mcr-1 and mcr-4 | mcr-1 and mcr-4 (441delT) | ST-10 | FII(pRSB107), HI2, HI2A, I1, Q1, X4, TrfA, Col(MG828), Col156, ColE10, ColRNAI | aac(3)-IVa, aadA1, aph(3')-Ia, aph(4)-Ia, bla CTX-M-14, bla TEM-1B, dfrA1, mph(B), strA, strB, sul1, sul2, tet(A), tet(M), gyrA S83L |
| ZTA15/00685EB1 | E. coli | Pig | Caecum | 2 | mcr-1 | mcr-1 | ST-457 | FIB(AP001918), FIC(FII), FII, I1, R, X4, Col(MG828), ColRNAI | aac(3)-IVa, aadA1, aadA2, aph(3')-Ia, aph(4)-Ia, bla CTX-M-27, catA2, cmlA1, dfrA12, erm(B), floR, mph(A), strA, strB, sul2, sul3, tet(M), gyrA S83L, gyrA D87N, parC S80I |
| ZTA15/01045EB1 | E. coli | Pig | Caecum | ≤ 1 | - | - | ST-156 | FIA, FIB(AP001918), FII, I1, X1, Col(MG828), ColRNAI | bla CTX-M-14, tet(A), gyrA S83L |
| ZTA15/01169–1EB1 | E. coli | Calf | Caecum | 8 | mcr-1 and mcr-3 | mcr-1 and mcr-3.2 | ST-533 | FIB(AP001918), FIC(FII), HI2, HI2A, I1, Y, TrfA, Col156, ColRNAI | aac(3)-IId, aadA2, bla CTX-M-55, bla TEM-1A, dfrA1, floR, mph(A), mph(B), strA, strB, sul1, sul3, tet(A), tet(M), gyrA S83L, gyrA D87N, parC S80I |
| ZTA15/00944–1EB1 | E. coli | Calf | Caecum | ≤ 1 | - | - | ST-1721 | FIB(pHCM2), R, X1 | aac(3)-IVa, aph(4)-Ia, bla CTX-M-1, bla TEM-1B, floR, mph(A), qnrS1, strA, sul2, tet(B) |
| ZTA15/00877EB1 | E. coli | Pig | Caecum | ≤ 1 | - | - | ST-654 | FIB(K), X1, Y | aadA1, aadA2, bla TEM-1B, bla SHV-12, cmlA1, floR, qnrS1, sul3, tet(A) |
| ZTA15/00421EB1 | E. coli | Pig | Caecum | ≤ 1 | - | - | ST-410 | FII, FIA, FIB(AP001918), ColRNAI | aac(6')-Ib-cr, aadA1, aadB, bla CTX-M-15, bla OXA-1, catB4, floR, sul1, sul2, tet(A), tet(M), gyrA S83L, gyrA D87N, parC S80I |
| ZTA15/00380EB1 | E. coli | Pig | Caecum | ≤ 1 | - | - | ST-533 | FII(p14), FIA, FIB(AP001918), I1, Y, ColRNAI | aac(3)-IVa, aadA1, aph(4)-Ia, bla CTX-M-27, dfrA1, floR, strA, strB, sul1, sul2, tet(A), gyrA S83L, gyrA D87N, parC S80I |
| ZTA15/00747EB1 | E. coli | Pig | Caecum | ≤ 1 | - | - | ST-75 | FII, FIA, FIB(AP001918), I1, Col8282, ColRNAI | bla CTX-M-1, bla TEM-1B, mph(A), tet(A) |
| ZTA15/00919EB1 | E. coli | Pig | Caecum | ≤ 1 | - | - | ST-453 | FII, FIB(AP001918), B/O/K/Z, X1, Q1 | aadA2, bla CTX-M-1, bla TEM-1B, dfrA5, qnrS1, strA, strB, sul2, gyrA S83L, gyrA D87N, parC S80I |
| ZTA15/01816–1EB1 | E. coli | Calf | Caecum | ≤ 1 | - | - | ST-58 | FII(pCoo), FIA(HI1), FIB(pB171), B/O/K/Z, ColRNAI | bla CTX-M-14, strA, strB, sul2, tet(B) |
| ZTA15/00170EB1 | E. coli | Pig | Caecum | ≤ 1 | - | - | ST-48 | FII, FIB(AP001918), I1, Q1, X1 | aadA1, bla CTX-M-14, bla TEM-1B, dfrA1, mph(B), qnrS1, strA, strB, sul1, sul2, tet(A) |
| ZTA15/02174–1EB1 | E. coli | Calf | Caecum | ≤ 1 | - | - | ST-10 | FIB(AP001918), FIC(FII), I1, Col156, ColRNAI | aadA1, aph(3')-Ic, bla CTX-M-1, bla OXA-1, bla TEM-1B, catA1, floR, strA, strB, sul1, sul2, tet(B), gyrA S83L, gyrA D87N, parC S80I, parE S458A |
| Germany (n=16) | |||||||||
| 15-AB01393_0 | E.coli | Calf | Meat | ≤ 1 | - | - | ST-58 | FII, FIB, Q1, X1 | bla CTX-M-1, mph(A), strA, strB, sul2 |
| 15-AB01299_0 | E.coli | Pig | Caecum | 4 | mcr-4 | mcr-4.2 | ST-410 | FII, FII(pRSB107), FIA, FIB(AP001918), X1, ColE10, ColRNAI | aadA1, aadB, bla CTX-M-15, floR, sul1, sul2, tet(A), gyrA S83L, gyrA D87N, parC S80I, parE S458A |
| 15-AB02002_0 | E.coli | Calf | Caecum | 8 | mcr-1 | mcr-1 | ST-10 | FII, FIB(AP001918), HI2, HI2A, I1, Q1, X1, TrfA, Col156, ColE10, ColRNAI | aadA1, aadA2, aac(3)-IIa, aph(3')-Ic, bla TEM-1A, catA1, cmlA1, dfrA1, strA, strB, sul1, sul2, sul3, tet(A), tet(B) |
| 15-AB01235_0 | E.coli | Pig | Meat | ≤ 1 | - | - | ST-1952 | FII, I1, Q1, Col(MG828) | aadA1, bla CTX-M-1, bla TEM-1B, dfrA1, floR, mph(A), strA, strB, sul1, sul2, tet(A), gyrA S83L |
| 15-AB01370_0 | E.coli | Pig | Caecum | ≤ 1 | - | - | ST-542 | I1 | aadA1, aadA5, bla CTX-M-1, dfrA17, sul2 |
| 15-AB01894_0 | E.coli | Pig | Caecum | ≤ 1 | - | - | ST-88 | FII(pCoo), FIB(AP001918), FIC(FII), I1, IncX1, Col(MG828), Col8282, ColRNAI | aadA1, aadA5, bla CTX-M-1, dfrA1, dfrA17, sul1, sul2 |
| 15-AB01509_0 | E.coli | Calf | Caecum | ≤ 1 | - | - | ST-694 | FII, FII(pRSB107), FIA, FIB(AP001918), Col156, ColRNAI, B/O/K/Z | aac(3)-IIa, aac(6')-Ib-cr, aadA5, bla CTX-M-15, dfrA17, mph(A), strA, strB, sul1, sul2, tet(B), gyrA S83L, gyrA D87N, parC S80I, parE S458A |
| 15-AB01308_0 | E.coli | Calf | Caecum | ≤ 1 | - | - | ST-224 | Y | bla CTX-M-15, bla TEM-1B, dfrA14, qnrS1, strA, strB, sul2, tet(A), gyrA S83L, gyrA D87N, parC S80I, parE S458A |
| 15-AB01312_0 | E.coli | Calf | Caecum | ≤ 1 | - | - | ST-641 | FII, Q1 | aac(3)-IVa, aph(4)-Ia, bla CTX-M-1, bla TEM-1B, dfrA7, mph(A), strA, strB, sul2, tet(A), gyrA S83L |
| 15-AB01045_0 | E.coli | Calf | Caecum | ≤ 1 | - | - | ST-410 | FII, FIB(AP001918), FIC(FII), I1, Y, Col(BS512), ColRNAI | aadA1, aph(3')-Ia, bla TEM-1, dfrA1, floR, strA, strB, sul2, tet(B), ampC T32A, gyrA S83L, gyrA D87N, parC S80I, parE S458A |
| 16-AB00129_0 | E.coli | Pig | Caecum | ≤ 1 | - | - | ST-10 | FII, FIA, FIB(AP001918), I1, Col(BS512), Col8282 | aadA1, aadA5, bla CTX-M-1, dfrA17, mph(A), strA, strB, sul1, sul2, tet(A) |
| 16-AB00148_0 | E.coli | Calf | Caecum | ≤ 1 | - | - | ST-349 | FII(pCoo), Y | aac(3)-IVa, aph(4)-Ia, bla CTX-M-1, mph(A), strA, strB |
| 16-AB00307_0 | E.coli | Calf | Caecum | ≤ 1 | - | - | ST-224 | FII, FII(pRSB107), FIA, FIB(AP001918), X1, p0111 | aac(3)-IIa, aac(6')-Ib-cr, aadA5, bla CTX-M-15, bla OXA-1, bla TEM-1B, catB4, dfrA17, mph(A), qnrS1, sul1, tet(A), gyrA S83L, gyrA D87N, parC S80I, parE S458A |
| 16-AB00409_0 | E.coli | Calf | Caecum | ≤ 1 | - | - | ST-118 | FII(pSE11), FIB(AP001918), B/O/K/Z, I1, Q1 | bla CTX-M-1, mph(A), strA, strB, sul2, tet(A) |
| 16-AB00430_0 | E.coli | Calf | Caecum | 8 | mcr-1 | mcr-1 | ST-950 | FII, FIB(AP001918), B/O/K/Z, HI2, HI2A, I2, TrfA | aac(3)-IVa, aadA1, aph(4)-Ia, bla CTX-M-1, bla OXA-1, floR, mph(A), strA, strB, sul1, sul2, tet(Y), gyrA S83L, gyrA D87Y, parC S80I |
| 15-SA02327_0 | Salmonella 1,4 [4],12:i:- | Pig | Carcass | ≤ 1 | - | - | ST-34 | FIB(AP001918), FIC(FII) | aadA1, aadA2, bla TEM-1B, cmlA1, dfrA12, mef(B), sul3, tet(B) |
| France (n=10) | |||||||||
| 15F001188 | E. coli | Pig | Animal | ≤ 1 | - | - | ST-744 | FII, FII(pHN7A8), FIB(AP001918), Q1, Col(MG828), Col156 | aadA5, aph(3')-Ia, bla CTX-M-55, bla TEM-1B, dfrA17, fosA3, mph(A), strA, strB, sul1, sul2, tet(B), gyrA S83L, gyrA D87N, parC A56T, parC S80I |
| 15F001211 | E. coli | Calf | Animal | 4 | mcr-3 | mcr-3.2 | ST-744 | FIB(AP001918), FIC(FII), Q1 | aac(3)-IId, aadA5, bla CTX-M-55, bla TEM-1B, catA1, dfrA17, floR, mph(A), strA, strB, sul1, sul2, tet(B), gyrA S83L, gyrA D87N, parC A56T, parC S80I |
| 15F001279 | E. coli | Calf | Animal | 4 | mcr-1 | mcr-1 | ST-648 | FIB(AP001918), FIC(FII), HI2, HI2A, Q1, X1, TrfA, ColRNAI | aac(3)-IVa, aadA1, aadA2, aph(3')-Ia, aph(4)-Ia, blaTEM-1A, dfrA1, dfrA12, erm(B), mph(B), strA, strB, sul2, tet(A), gyrA S83L, gyrA D87N, parC S80I, parE L416F |
| 15F002387 | E. coli | Calf | Animal | 4 | mcr-1 | mcr-1 | ST-744 | FIB(AP001918), FIC(FII), HI2, HI2A, Q1, TrfA | aadA1, aadA2, aadA5, bla CTX-M-1, bla TEM-1B, cmlA1, dfrA1, dfrA12, dfrA17, floR, mph(A), strA, strB, sul1, sul2, sul3, tet(A), tet(B), tet(M), gyrA S83L, gyrA D87N, parC A56T, parC S80I |
| 15Q003557 | Salmonella Rissen | Calf | Carcass | ≤ 1 | - | - | ST-469 | Col(MG828), ColRNAI | tet(A) |
| 15Q003582 | Salmonella Rissen | Pig | Carcass | ≤ 1 | - | - | ST-469 | I1, ColRNAI | aadA1, aadA2, cmlA1, dfrA12, tet(A), sul3 |
| 15Q003631 | Salmonella 4,12:i:- | Pig | Carcass | 8 | mcr-1 | mcr-1 | ST-34 | HI2, HI2A, I1, Q1, TrfA, Col156 | aac(3)-IVa, aph(3')-Ia, aph(4)-Ia, bla TEM-1B, catA1, strA, strB, sul2, tet(A), tet(B) |
| 15Q004074 | Salmonella 4,12:i:- | Calf | Carcass | 4 | mcr-4 | mcr-4.2 | ST-34 | Q1, Col156, ColE10, ColRNAI, | blaTEM-1B, strA, strB, sul2, tet(B) |
| 15F001226 | E. coli | Calf | Animal | ≤ 1 | - | - | ST-1011 | Q1 | aac(3)-IId, aadA2, bla CTX-M-32, bla TEM-1B, catA1, dfrA12, mph(A), strA, strB, sul1, sul2, tet(A), gyrA S83L, gyrA D87N, parC S80I |
| 16F000284 | E. coli | Pig | Animal | ≤ 1 | - | - | ST-871 | Col(BS512), ColRNAI, Y | - |
| Italy (n=4) | |||||||||
| 15051805COYF25 | E. coli | Calf | Animal | 8 | mcr-1 | mcr-1 | ST-4096 | FII, HI2, HI2A, TrfA, ColE10 | aac(3)-IIa, aadA1, aph(3')-Ic, bla CTX-M-15, dfrA1, floR, strA, strB, sul3, tet(A), tet(M), gyrA S83L |
| 150542127ARP05 | E. coli | Calf | Animal | 4 | mcr-1 and mcr-3 | mcr-1 and mcr-3.2 | ST-744 | FIB(AP001918), FIC(FII), R, X1, X4, Col156 | aac(3)-IId, aadA2, aadA5, bla CTX-M-55, bla TEM-1B, dfrA12, floR, mph(A), sul1, sul3, tet(B), tet(M), gyrA S83L, gyrA D87N, parC S80I, parC A56T |
| 15056414J9PUD1 | E. coli | Pig | Animal | 4 | mcr-1 | mcr-1.xx | ST-5995 | FIB(AP001918), FIC(FII) | catA1 |
| 15058525J9PUD5 | E. coli | Pig | Animal | ≤ 1 | - | - | ST-1684 | FIA, FIB(AP001918), FIC(FII), I1, I2, Col(MG828), Col8282, ColRNAI | aadA1, bla CMY-2, erm(C), tet(B) |
MIC: minimum inhibitory concentration; MLST: multilocus sequence typing; ST: sequence type; WGS: whole genome sequencing.
-: Not detected.
Grey: phenotypic resistance according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) epidemiological cut-off value [29].
a As determined by using online tools at the Center for Genomic Epidemiology (http://www.genomicepidemiology.org/).
b New variant detected in this study.
Validation test and comparison with WGS data
The validation test set revealed eight colistin-resistant E. coli isolates (MIC 4 – 8 mg/L), five from bovine and three from porcine origin, harbouring the mcr-1 gene or one of its variants. An additional E. coli isolate from a pig phenotypically classified as borderline susceptible (colistin MIC of 2 mg/L), on repeated testing at two laboratories also harboured the mcr-1 gene. The multiplex PCR also yielded mcr-1 in one porcine Salmonella spp. isolate with colistin MIC of 8 mg/L. One E. coli isolate of bovine origin with MIC of 4 mg/L harboured the mcr-3 gene. Three isolates, with colistin MIC of 4 mg/L, harboured the mcr-4 gene, corresponding to two Salmonella spp. (one isolated from pig and the other from calf) and one porcine E. coli. Co-occurrence of mcr-1 and mcr-3 was observed in two bovine E. coli isolates with colistin MICs of 4 and 8 mg/L, respectively. One porcine E. coli isolate with colistin MIC of 8 mg/L harboured both mcr-1 and mcr-4 genes.
The results were 100% concordant with WGS data and notably, the multiplex PCR was able to detect a novel variant described in this study for the first time, mcr-4.3, that varies in one amino-acid from the original mcr-4 encoded protein (Table 2). The genome of the isolate harbouring mcr-4.3 (Salmonella enterica serovar Kedougou 151570) is deposited in ENA with the accession number ERS1801979. Additionally, the multiplex PCR also detected a mcr-4 gene with a deletion in nt 441 that alters the reading frame and introduces a premature stop codon. There was one E. coli isolate presenting phenotypic resistance to colistin (MIC = 8 mg/L) not due to the presence of mcr genes, but instead attributed to the mutations pmrB D283G/Y358N in the pmrA/pmrB two-component system as determined by analysis of WGS data (Table 2).
Discussion
The multiplex PCR reaction showed 100% specificity (no unpredicted amplification products) and 100% sensitivity (all of the predicted products were visualised in agarose gel) for the validation set, according to WGS data.
One notable finding was the detection of mcr-1 in one E. coli isolate classified as wildtype (susceptible) (colistin MIC of 2 mg/L). Preliminary results from other studies revealed similar findings of E. coli isolates, particularly from poultry, with MIC of 2 mg/L and carrying the mcr-1 gene, collected in Denmark (data not shown) and Poland (Dariusz Wasyl, National Veterinary Research Institute, Puławy, Poland, personal communication, January 2018). These findings seem to indicate that there is a need to review of the established epidemiological cut-off value for colistin. Also based on the discovery of a new mcr-4 variant (mcr-4.3), it becomes clear that colistin resistance is not yet fully uncovered although our knowledge is evolving rapidly, filling the gaps that compromise the prevention and control of multidrug-resistant bacteria.
Another observation of interest was the detection of two pmrA/pmrB point mutations in one colistin-resistant isolate, which reveals a need to further investigate and characterise this two-component system in order to accurately predict which mutations lead to phenotypic colistin resistance. Taking into account that the mutations found in this isolate are not consistently associated with a resistant phenotype we cannot confirm, without further analysis, if they represent the underlying mechanism of colistin resistance for this isolate [27,28].
Although the presented multiplex PCR does not allow identification of the specific variants and detects non-functional genes, it still provides valuable information for surveillance purposes as well as for research. The results of the multiplex PCR also allow the selection of isolates for further studies based on genomic epidemiology, focusing on characterisation of mcr genes, their mobilisation mechanisms and the plasmids they are harboured on. While the multiplex PCR has not yet been validated on human strains, it is expected to provide the same sensitivity and specificity as for animal isolates. The vast majority of mcr genes are located on transferable plasmids that are not restricted to reservoirs or hosts. Similarly, we expect that this multiplex PCR design will maintain its usefulness should new gene variants among mcr-1 to mcr-5 be detected in Enterobacteriaceae, taking into account that the chance of variations in the primer annealing positions should be small and no such variations have been observed so far. A great advantage of this method is the ability to detect mcr genes in phenotypically susceptible bacteria, allowing it to be used as a screening technique for isolates recovered from human and animal sources. We believe future efforts should include the screening of large pools of borderline susceptible Enterobacteriaceae in order to understand if, and how, the epidemiological cut-off value for colistin should be reviewed.
Although an internal amplification control was not used in this study, it should be possible to use a different reaction mix containing another DNA polymerase in order to allow the addition of 16S rRNA primers to obtain an internal reaction control. We encourage researchers to adapt the PCR reaction to their specific needs.
This screening method can provide a useful and accurate description of colistin resistance in Enterobacteriaceae collections, particularly when used in connection with MIC susceptibility testing. The protocol and standard operating procedure are freely available at the EURL-AR website (https://www.eurl-ar.eu/). The intended use for this multiplex PCR is as part of the surveillance workflow, to screen colistin-resistant and borderline susceptible isolates after initial MIC determination by the standard broth dilution method. Colistin MIC determination allows classification of isolates as susceptible, borderline susceptible or resistant and, subsequently, the isolates determined borderline susceptible and resistant (MIC ≥ 2 mg/L) can be subjected to the multiplex PCR to provide further insight into the underlying resistance mechanism and potentially reveal the presence of the known mcr genes. For an initial surveillance analysis we do not recommend to perform the multiplex PCR before MIC determination, since it could lead to overlooking colistin resistant isolates with other underlying mechanisms of resistance. The method can, however, be used on its own if the research is focused, for example, on plasmid dissemination or mcr genes’ epidemiology. Advanced investigations could be conducted according to available procedures and surveillance/research goals, and would ideally involve WGS of the bacterial genome, even though it may not be easily accessible in some laboratories or countries, in particular, low and middle income countries (LMIC). Colistin resistant bacteria lacking any of the currently known mcr genes should be screened for other mechanisms of resistance, such as mutations in the pmrA/pmrB system, new mcr genes or variants, or even presently unknown mechanisms.
Future prospects include validation of the multiplex PCR for human clinical strains, to assure that the protocol follows the principles of the One Health perspective and can be used to directly screen human, veterinary and food bacteria. Other prospects are to clone mcr-1, mcr-2, mcr-3 and mcr-5 amplicons into TA-cloning vectors (Thermo Fisher Scientific) in order to obtain non-pathogenic and easy to handle control strains, and be able to confirm and update the protocol continuously as new variants are being detected.
In conclusion, we developed a multiplex PCR that rapidly detects all mcr genes described in Enterobacteriaceae to date. The multiplex PCR method is critical for epidemiological surveillance and antimicrobial resistance investigation, especially in settings where laboratory resources and access to WGS are limited, and where plasmid-mediated colistin resistance may threaten the effectiveness of available antimicrobials.
Acknowledgements
This work was supported by the European Union Reference Laboratory for Antimicrobial Resistance (EURL-AR) in the context of animal health and food safety, (http://www.eurl-ar.eu/) and the World Health Organization (WHO) Collaborating Centre for Antimicrobial Resistance in Foodborne Pathogens and Genomics including the WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR) (http://www.who.int/foodsafety/areas_work/antimicrobial-resistance/agisar/en/) and Global Antimicrobial Resistance Surveillance System (GLASS) (http://www.who.int/antimicrobial-resistance/publications/surveillance-system-manual/en/).
Disclaimer
Beatriz Guerra is employed with the European Food Safety Authority (EFSA) in its BIOCONTAM Unit that provides scientific and administrative support to EFSA’s scientific activities. The positions and opinions presented in this article are those of the authors alone and are not intended to represent the views or scientific works of EFSA.
* Erratum
Due to a technical error, the name of Cristina De Frutos Escobar was incorrectly displayed in the list of authors. The mistake was corrected on 12 Feb 2018.
Conflict of interest: None declared.
Authors’ contributions: BM, MB, JAH, AB, AF, PA, APG, SAG, CDFE, SMK, BG, LV and AC selected and/or provided either control strains or strains for the validation. ARR designed the multiplex PCR. VB, JSK, RSH, PL and IMH conducted and analysed the WGS data. VB, JSK, RSH, BG and SKP revised the concordance with phenotypical data. All participated in the coordination and concept of the manuscript. ARR and RSH drafted and coordinated the manuscript. All contributed writing the manuscript.
References
- 1. Poirel L, Jayol A, Nordmann P. Polymyxins: Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30(2):557-96. 10.1128/CMR.00064-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Catry B, Cavaleri M, Baptiste K, Grave K, Grein K, Holm A, et al. Use of colistin-containing products within the European Union and European Economic Area (EU/EEA): development of resistance in animals and possible impact on human and animal health. Int J Antimicrob Agents. 2015;46(3):297-306. 10.1016/j.ijantimicag.2015.06.005 [DOI] [PubMed] [Google Scholar]
- 3. Rhouma M, Beaudry F, Thériault W, Letellier A. Colistin in Pig Production: Chemistry, Mechanism of Antibacterial Action, Microbial Resistance Emergence, and One Health Perspectives. Front Microbiol. 2016;7:1789. . https://doi.org/10.3389/fmicb.2016.01789 10.3389/fmicb.2016.01789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161-8. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 5.European Medicines Agency (EMA). Updated advice on the use of colistin products in animals within the European Union: development of resistance and possible impact on human and animal health. London: EMA; Jul 2016. Available from: http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2016/07/WC500211080.pdf
- 6. McPhee JB, Lewenza S, Hancock RE. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol. 2003;50(1):205-17. 10.1046/j.1365-2958.2003.03673.x [DOI] [PubMed] [Google Scholar]
- 7.European Committee on Antimicrobial Susceptibility Testing (EUCAST). EUCAST warnings concerning antimicrobial susceptibility testing products or procedures. Växjö: EUCAST; Jun 2017. Available from: http://www.eucast.org/ast_of_bacteria/warnings/
- 8.European Committee on Antimicrobial Susceptibility Testing (EUCAST). Recommendations for MIC determination of colistin (polymyxin E). As recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. Växjö: EUCAST; Jun 2016. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf
- 9.Clinical and Laboratory Standards Institute (CLSI) Subcommittee on Antimicrobial Susceptibility Testing. CLSI AST News Update Volume 1, Issue 2. Philadelphia: CLSI; Dec 2016. Available from: https://clsi.org/media/1700/clsi-news-winter-2016.pdf
- 10. Shen Z, Wang Y, Shen Y, Shen J, Wu C. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect Dis. 2016;16(3):293. 10.1016/S1473-3099(16)00061-X [DOI] [PubMed] [Google Scholar]
- 11. Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016;21(27):30280. 10.2807/1560-7917.ES.2016.21.27.30280 [DOI] [PubMed] [Google Scholar]
- 12. Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, et al. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. MBio. 2017;8(3):e00543-17. 10.1128/mBio.00543-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, et al. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 2017;22(31):30589. 10.2807/1560-7917.ES.2017.22.31.30589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother. 2017;72(12):3317-24. 10.1093/jac/dkx327 [DOI] [PubMed] [Google Scholar]
- 15. Skov RL, Monnet DL. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill. 2016;21(9):30155. 10.2807/1560-7917.ES.2016.21.9.30155 [DOI] [PubMed] [Google Scholar]
- 16. Lu X, Hu Y, Luo M, Zhou H, Wang X, Du Y, et al. MCR-1.6, a New MCR variant carried by an IncP plasmid in a colistin-resistant Salmonella enterica serovar Typhimurium isolate from a healthy individual. Antimicrob Agents Chemother. 2017;61(5):e02632-16. 10.1128/AAC.02632-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roer L, Hansen F, Stegger M, Sönksen UW, Hasman H, Hammerum AM. Novel mcr-3 variant, encoding mobile colistin resistance, in an ST131 Escherichia coli isolate from bloodstream infection, Denmark, 2014. Euro Surveill. 2017;22(31):30584. 10.2807/1560-7917.ES.2017.22.31.30584 [DOI] [PubMed] [Google Scholar]
- 18.Carretto E, Brovarone F, Nardini P, Russello G, Barbarini D, Pongolini S, et al. Detection of mcr-4 positive Salmonella enterica serovar Typhimurium in clinical isolates of human origin, Italy, October to November 2016. Euro Surveill. 2018. 23(2):pii=17-00821. https://doi.org/10.2807/1560-7917.ES.2018.23.2.17-00821 [DOI] [PMC free article] [PubMed]
- 19. Litrup E, Kiil K, Hammerum AM, Roer L, Nielsen EM, Torpdahl M. Plasmid-borne colistin resistance gene mcr-3 in Salmonella isolates from human infections, Denmark, 2009-17. Euro Surveill. 2017;22(31):30587. 10.2807/1560-7917.ES.2017.22.31.30587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hernández M, Iglesias MR, Rodríguez-Lázaro D, Gallardo A, Quijada N, Miguela-Villoldo P, et al. Co-occurrence of colistin-resistance genes mcr-1 and mcr-3 among multidrug-resistant Escherichia coli isolated from cattle, Spain, September 2015. Euro Surveill. 2017;22(31):30586. 10.2807/1560-7917.ES.2017.22.31.30586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kluytmans J. Plasmid-encoded colistin resistance: mcr-one, two, three and counting. Euro Surveill. 2017;22(31):30588. 10.2807/1560-7917.ES.2017.22.31.30588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hasman H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agersø Y, et al. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill. 2015;20(49):30085. 10.2807/1560-7917.ES.2015.20.49.30085 [DOI] [PubMed] [Google Scholar]
- 23. Di Pilato V, Arena F, Tascini C, Cannatelli A, Henrici De Angelis L, Fortunato S, et al. mcr-1.2, a new mcr variant carried on a transferable plasmid from a colistin-resistant KPC carbapenemase-producing Klebsiella pneumoniae strain of Sequence Type 512. Antimicrob Agents Chemother. 2016;60(9):5612-5. 10.1128/AAC.01075-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. von Wintersdorff CJ, Wolffs PF, van Niekerk JM, Beuken E, van Alphen LB, Stobberingh EE, et al. Detection of the plasmid-mediated colistin-resistance gene mcr-1 in faecal metagenomes of Dutch travellers. J Antimicrob Chemother. 2016;71(12):3416-9. 10.1093/jac/dkw328 [DOI] [PubMed] [Google Scholar]
- 25. Leangapichart T, Gautret P, Brouqui P, Mimish Z, Raoult D, Rolain J-M. Acquisition of mcr-1 plasmid-mediated colistin resistance in Escherichia coli and Klebsiella pneumoniae during Hajj 2013 and 2014. Antimicrob Agents Chemother. 2016;60(11):6998-9. 10.1128/AAC.01486-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.European Commission. Commission implementing decision 2013/652/EU on the monitoring and reporting of antimicrobial resistance in zoonotic and commensal bacteria. Official Journal of the European Union. Luxembourg: Publications Office of the European Union. 14.11.2013:L 303. Available from: http://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=OJ:L:2013:303:FULL&from=EN
- 27. Brennan E, Martins M, McCusker MP, Wang J, Alves BM, Hurley D, et al. Multidrug-Resistant Escherichia coli in Bovine Animals, Europe. Emerg Infect Dis. 2016;22(9):1650-2. 10.3201/eid2209.160140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Quesada A, Porrero MC, Téllez S, Palomo G, García M, Domínguez L. Polymorphism of genes encoding PmrAB in colistin-resistant strains of Escherichia coli and Salmonella enterica isolated from poultry and swine. J Antimicrob Chemother. 2015;70(1):71-4. 10.1093/jac/dku320 [DOI] [PubMed] [Google Scholar]
- 29.The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Version 8.0. Växjö: EUCAST; 2018. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.0_Breakpoint_Tables.pdf


