Abstract
We investigated influenza A(H5N6) viruses from migratory birds in Chungnam and Gyeonggi Provinces, South Korea following a reported die-off of poultry in nearby provinces in November 2017. Genetic analysis and virulence studies in chickens and ducks identified our isolate from December 2017 as a novel highly pathogenic avian influenza virus. It resulted from reassortment between the highly virulent H5N8 strain from Korea with the N6 gene from a low-pathogenic H3N6 virus from the Netherlands.
Keywords: Influenza A virus, H5N6, Reassortant, Highly pathogenic avian influenza (HPAI), Virulence, South Korea
In connection with an influenza A(H5N6) outbreak in poultry in November 2017 in JeollabukDo Province, South Korea [1], we collected influenza virus isolates investigated from wild migratory birds in the neighbouring Chungnam and Gyeonggi Provinces. Four novel reassortant highly pathogenic avian influenza (HPAI) H5N6 viruses were isolated on 13 December 2017 and continued to cause outbreaks in domestic poultry [personal communication: Dr Youn-Jeong Lee, Animal and Plant Quarantine Agency, South Korea, January 2018]. Genetic characterisation revealed that the haemagglutinin (HA) gene of the novel H5N6 viruses was closely associated with clade 2.3.4.4 influenza A(H5N8) viruses. Although genetically similar to H5N6 viruses that caused outbreaks in Japan (first reported on 10 November 2017) [2] and the Netherlands (7 December 2017) [3], there has not been a report about its pathogenic potential in poultry species. Therefore, we report here the genetic characterisation of the novel influenza A(H5N6) virus and the investigation of its pathogenic potential in chickens and ducks.
Genetic characterisation of novel influenza A(H5N6) viruses
Four influenza A(H5N6) viruses were isolated from faecal samples obtained from migratory bird habitats in Gyeonggi Province during a surveillance study conducted on 13 December 2017. Mitochondrial DNA sequence analysis of the faecal specimens revealed Anas platyrhynchos to be the viral host. In a full-length genomic sequence analysis, the viruses showed 99.9–100% nucleotide homology to one another but 97.2–99.4% homology with influenza A(H5N6) viruses from Japan and the Netherlands. Genetic and phylogenic analysis revealed that our strains clustered with the A/Tufted Duck/Germany/AR8459-L01988/2016(H5N8)-like viruses (clade 2.3.4.4 A/Brk/Korea/Gochang1/2014-like H5N8 lineage) (Figure 1), although their neuraminidase (NA) gene segments were derived from A/BG/Netherlands/2/2014(H3N6)-like viruses persisting in Europe during 2017 (Figure 2).
Figure 1.
Phylogenetic trees comparing the haemagglutinin nucleotide sequences of 2017/18 Korean influenza A(H5N6) viruses, South Korea, December 2017 (n = 4)
Bdk: broiler duck; Brk: breeder duck; BT: baikal teal; CK: chicken; CT: common teal; DK: duck; EHG: Eurasian herring gull; EM: environment; FP: ferruginous pochard; GBG: great black backed gull; GH: grey heron; GS: goose; HA: haemagglutinin; MD: mallard; Mdk: mallard duck; MS: mute swan; NS: northern shoveler; PC: peacock; PS: painted stork; RS: ruddy shelduck; SB: spoonbill; TD: tufted duck; WD: wild duck.
To investigate the origins of novel influenza A(H5N6) viruses (A/AP/Korea/W612/2017, A/AP/Korea/W613/2017, A/AP/Korea/W614/2017 and A/AP/Korea/W615/2017: marked with asterisks), full-length nucleotide sequences of HA genes were compared with available H5Nx and high blast-scoring virus sequences from GenBank and the Global Initiative on Sharing All Influenza Data (GISAID). The deposited GenBank accession numbers of HA genes are MG891804–MG891807; GISAID sequences are acknowledged in Supplement 1. Time-scaled phylogenies (dates shown on the horizontal axis) were inferred using strict-clock Bayesian Markov Chain Monte Carlo analysis. The month of isolation is indicated at the end of the virus name.
The yellow line indicates the 2016/17 influenza A(H5N6) subtypes while the blue line indicates the 2017/18 H5N6 subtype which was isolated from South Korea. The Group A of clade 2.3.4.4 viruses comprises influenza A(H5N8) viruses identified in South Korea in the 2013/14 and 2014/15 winter seasons. Group B comprises H5N8, H5N6 and H5N5 viruses identified in Asia and Europe during the 2013/14 to 2017/18 winter seasons. Group C of clade 2.3.4.4 viruses comprises H5N6 viruses identified in China and South Korea during the 2015/16 and 2017/18 winter seasons, respectively.
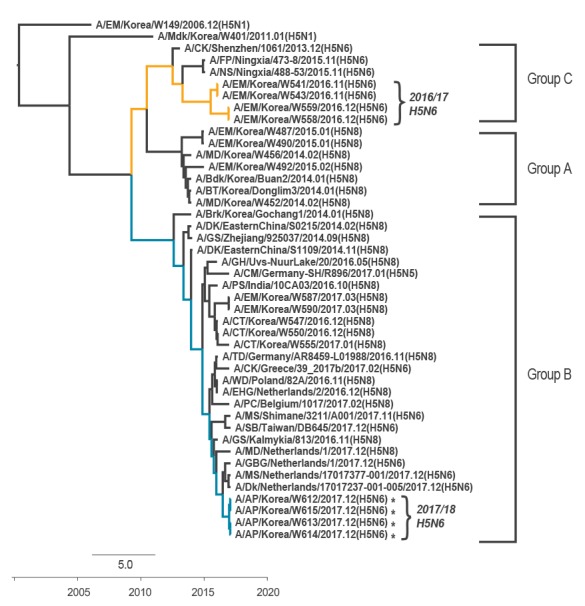
Figure 2.
Phylogenetic trees comparing the neuraminidase nucleotide sequences of novel influenza A(H5N6) viruses, South Korea, December 2017 (n = 4)
Bdk: broiler duck; BG: barnacle goose; Brk: breeder duck; BT: baikal teal; CK: chicken; CT: common teal; DK: duck; EHG: eurasian herring gull; EM: environment; FP: ferruginous pochard; GBG: great black backed gull; GH: grey heron; GS: goose; MD: mallard; Mdk: mallard duck; MS: mute swan; NA: neuraminidase; NS: northern shoveler; PC: peacock; PS: painted stork; RS: ruddy shelduck; SB: spoonbill; TD: tufted duck; WD: wild duck.
To investigate the origins of our novel influenza A(H5N6) viruses (A/AP/Korea/W612/2017, A/AP/Korea/W613/2017, A/AP/Korea/W614/2017 and A/AP/Korea/W615/2017: marked with asterisks), full-length nucleotide sequences of NA genes were compared with available H5Nx and high blast scoring virus sequences from the GenBank and Global Initiative on Sharing All Influenza Data (GISAID) databases; GISAID sequences are acknowledged in Supplement 1. The deposited GenBank accession numbers of NA genes are MG891812-MG891815.Time-scaled phylogenies (dates shown on the horizontal axis) were inferred using strict-clock Bayesian Markov Chain Monte Carlo analysis. The month of isolation is indicated at the end of the virus name.
The yellow line indicates the 2016/17 influenza A(H5N6) viruses while the blue line indicates the 2017/18 H5N6 viruses that were isolated from South Korea. The Group A of clade 2.3.4.4 viruses comprises influenza A(H5N8) viruses identified in South Korea in the 2013/14 and 2014/15 winter seasons. Group B comprises H5N8 and H5N6 viruses identified in Asia and Europe during the 2013/14 to 2017/18 winter seasons. Group C of clade 2.3.4.4 viruses comprises H5N6 viruses identified from China in the 2015/16 and South Korea in the 2016/17 winter seasons, respectively.
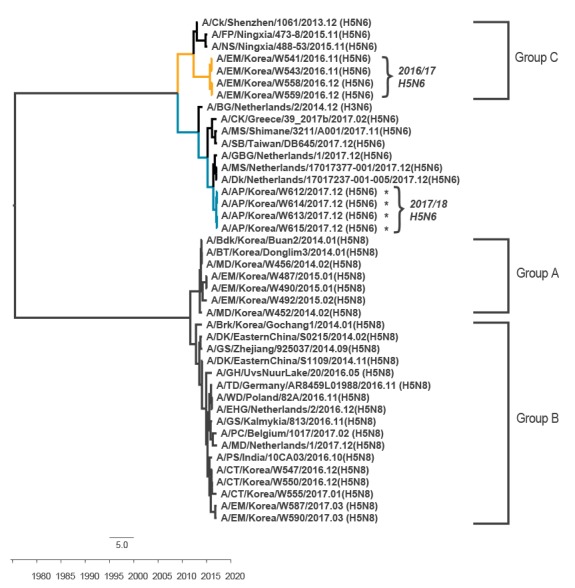
In contrast to the previous 2016/17 Korean influenza A(H5N6) viruses [4], the HA gene of 2017/18 Korean influenza A(H5N6) viruses did not belong to Group C of the clade 2.3.4.4 HPAI H5 viruses. Instead, they were closely associated with Group B of the clade 2.3.4.4 H5N8 viruses that have mainly been circulating in Eurasia [5]. Further, the PB2, NP, M and NS genes of 2017/18 Korean influenza A(H5N6) viruses were closely related to A/EM/Korea/W437/2012(H7N7)-like influenza strains, the PB1 gene was closely related to the A/DK/Mongolia/709/2015(H10N7)-like virus, while the PA genes were closely related to the A/EM/Korea/W401/2011(H5N1)-like virus and clustered together with the European influenza A(H5N8) viruses (A/TD/Germany/AR8459-L01988/2016-like viruses) (Figure 3).
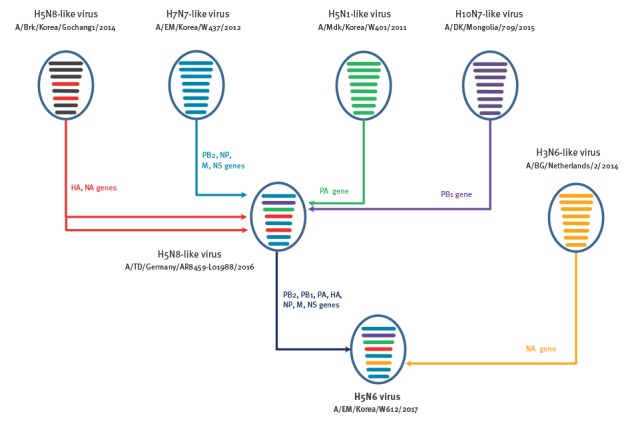
Figure 3.
Genotype illustration of the reassortment process of 2017/18 Korean influenza A(H5N6) viruses, South Korea
BG: barnacle goose; Brk: breeder duck; DK: duck; EM: environment; HA: haemagglutinin; M: matrix; Mdk: mallad duck; NA: neuraminidase; NP: nucleoprotein; NS: nonstructural; PA: polymerase acidic; PB: polymerase basic; TD: tufted duck.
The AP/W612/H5N6 virus is a reassortant of at least five different subtypes (H5N8, H7N7, H5N1, H10N7, and H3N6) from the natural gene pool in Eurasian avian influenza viruses. The eight gene segments in each illustrative virus particle represent (from top to bottom) the PB2, PB1, PA, HA, NP, NA, M and NS genes. Each colour represents a virus lineage. Black: origin from A/Brk/Korea/Gochang1/2014-like; blue: A/EM/Korea/W437/2012-like; purple: A/DK/Mongolia/709/2015-like; light green: A/Mdk/Korea/W401/2011-like; red: Clade 2.3.4.4 H5 HA and NA genes.
Molecular analysis demonstrated that the HA cleavage site of the 2017/18 Korean influenza A(H5N6) viruses contained polybasic residues (REKRRK/G), which denotes a high-pathogenicity phenotype in chickens. All four 2017/18 Korean influenza A(H5N6) viruses maintained the glutamine residue at position 226 (H3 numbering) and a glycine residue at position 228. While all influenza A(H5N6) strains isolated in Korea in 2016 had one amino acid deletion in their HA1 133 site, a mutation commonly found in the 2.3.4.4 HA genes of human infectious influenza A(H5N6) viruses [4], the 2017/18 Korean influenza A(H5N6) viruses do not contain this deletion (Table 1). In addition, the 2017/18 Korean influenza A(H5N6) viruses did not contain the characteristic amino acid deletion (position 49–68 bases) in the NA gene that is present in the 2016 influenza A(H5N6) virus. With the exception of the NS gene, the internal genes of 2017/18 Korean influenza A(H5N6) viruses displayed the same characteristics as the 2016/17 Korean influenza A(H5N8) viruses (Table 1).
Table 1. Molecular comparison of H5 avian influenza viruses and similar isolates, South Korea, December 2017 (n = 8).
| Virusesa | HA clade | HA sequence (aa) | HA deletion |
NA stalk deletion | NS1 | PB2 sequence at aa | Expression of PB1-F2 protein | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cleavage site | Receptor binding sites | Deletion of aa 80–84 | aa residue at | |||||||||||||||
| 335–348b | 158 | 193 | 222 | 224 | 226 | 227 | 228 | 318 | 133 | 49–68 | 92 | C-term | 627 | 701 | ||||
| AP/Korea/W612/17 | 2.3.4.4 | REKRRK__ /G | N | N | Q | N | Q | R | G | T | No | No | No | D | GSEV | E | D | Yes |
| PC/Belgium/1017/17 | 2.3.4.4 | REKRRK__ /G | N | N | Q | N | Q | R | G | T | No | No | No | D | GSEV | E | D | Yes |
| TD/Germany/AR8459-L01988/16 | 2.3.4.4 | REKRRK__ /G | N | N | Q | N | Q | R | G | T | No | No | No | D | GSEV | E | D | Yes |
| EM/Korea/W541/16 | 2.3.4.4 | RERRRK__/G | N | N | Q | N | Q | Q | G | T | Yes | Yes | Yes | E | ESEV | E | D | Yes |
| CT/Korea/W555/17 | 2.3.4.4 | REKRRK__ /G | N | N | Q | N | Q | R | G | T | No | No | No | D | GSEV | E | D | Yes |
| MD/Korea/W452/14 | 2.3.4.4 | RERRRK__/G | N | N | Q | N | Q | R | G | T | No | No | No | D | ESEVRG | E | D | Yes |
| BDk/Korea/Gochang1/14 | 2.3.4.6 | REKRRK __/G | N | N | Q | N | Q | R | G | T | No | No | No | D | ESEV | E | D | Yes |
| EM/Korea/W149/06 | 2.2 | GERRRKKR/G | N | K | K | N | Q | S | G | T | No | Yes | Yes | D | ESKV | K | D | Yes |
aa: amino acid; AP: Anas platyrhynchos; BDk: breeder duck; CT: common teal; C-term: 4 aa sequence at the C-terminal end; EM: environment; HA: haemaggutinin; MD: mallard duck; NA: neuraminidase; PC: peacock; RBS: receptor binding site; TD: tufted duck.
a The isolate in bold was the 2017 Korean highly pathogenic avian influenza H5N6 viruses examined in this study.
b H3 numbering.
Virulence in chickens and ducks
One representative virus, A/AP/Korea/W612/2017(H5N6), was selected for further study. The intravenous pathogenicity index (IVPI) was measured in accordance with World Organisation for Animal Health (OIE) standards. In chickens, the IVPI score was 2.76, resulting in classification of the A/AP/Korea/W612/2017(H5N6) virus as HPAI [6]. To measure the chicken and duck lethal dose 50% (CLD50 and DLD50), specific pathogen-free chickens and ducks were infected with 107–102 egg infectious dose (EID)50/mL by intranasal inoculation. The CLD50 was 2.83 log10EID50/mL, with which the chickens died within 3 to 5 days after infection. In contrast, none of the infected ducks died during the experimental period (14 days) suggesting a DLD50 of more than 107 EID50/mL.
To investigate the pathogenicity and horizontal transmission ability of the A/AP/Korea/W612/2017(H5N6) virus in chickens and ducks, we proceeded with a group of intranasally inoculated (106 EID50/mL) animals (n = 14) and added a direct contact group (n = 3) one day after infection. Oropharyngeal and cloacal swab samples were collected for virus titration for 14 days post infection (dpi). Chickens in the infection and direct contact groups all succumbed by 4 dpi and 5 dpi, respectively. Moreover, virus was detected at all time points of swab collection and peaked at 2 dpi at 4.8 log10 EID50/mL in oropharyngeal swabs (Table 2). In the direct contact group, the virus was detected from the second day after contact and the highest cloacal swab viral titre (5.6 log10 EID50/mL) was measured on the fourth day. To determine virus tissue distribution of the A/AP/Korea/W612/2017(H5N6) virus in infected chickens, we collected the lung, brain, kidney, spleen, heart, liver and colon from three birds each at 3 and 5 dpi using individual sterile equipment to avoid cross-contamination between them. The virus was detected in all organs collected from the inoculated chicken group at 3 dpi, the last point at which samples could be collected due to the lethality of the virus (Table 2).
Table 2. Viral titres in chickens and ducks experimentally inoculated with influenza A/AP/Korea/W612/2017(H5N6), South Korea, December 2017 (n = 17).
| Experimental setup | Chicken | Duck | Seroconversion (HI GMT) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Infected group: swaba |
Time point | 1 dpi | 2 dpi | 3 dpi | 4 dpic | 1 dpi | 3 dpi | 5 dpi | 7 dpi | 9 dpi | 640 | ||||
| Oropharyngeal | 2.7 ± 0.4 | 4.8 ± 0.5 | 4.1 ± 0.4 | NC | 3.0 ± 0.3 | 5.1 ± 0.2 | 4.9 ± 0.4 | 1.9 ± 0.4 | ND | ||||||
| Cloacal | 1.1 ± 0.4 | 3.1 ± 0.4 | 3.2 ± 0.2 | NC | 1.0 ± 0.4 | 1.8 ± 0.5 | 1.3 ± 0.3 | ND | ND | ||||||
| Contact group: swaba |
Time point | 1 dpc | 2 dpc | 3 dpc | 4 dpc | 5 dpcc | 1 dpc | 2 dpc | 3 dpc | 4 dpc | 5 dpc | 6 dpc | 7 dpc | 320 | |
| Oropharyngeal | ND | 1.3 ± 0.1 | 3.1 ± 0.5 | 4.2 ± 0.2 | NC | 1.1 ± 0.2 | 1.2 ± 0.3 | 4.7 ± 0.4 | 4.5 ± 0.4 | 3.8 ± 0.5 | 2.8 ± 0.5 | ND | |||
| Cloacal | ND | 1.1 ± 0.2 | 4.0 ± 0.5 | 5.6 ± 0.4 | NC | ND | 0.8 ± 0.5 | 1.9 ± 0.5 | 2.4 ± 0.4 | 2.2 ± 0.3 | 1.0 ± 0.2 | ND | |||
| Infected group: tissueb | Time point | 3 dpi | 4 dpic | 3 dpi | 5 dpi | ||||||||||
| Lung | 4.3 ± 0.1 | NC | 4.8 ± 0.5 | 4.0 ± 0.2 | |||||||||||
| Brain | 1.3 ± 0.1 | NC | ND | ND | |||||||||||
| Kidney | 3.8 ± 0.4 | NC | 5.1 ± 0.3 | 4.3 ± 0.1 | |||||||||||
| Spleen | 4.4 ± 0.4 | NC | 3.9 ± 0.5 | 2.5 ± 0.4 | |||||||||||
| Heart | 3.5 ± 0.4 | NC | 5.0 ± 0.5 | 3.0 ± 0.5 | |||||||||||
| Liver | 4.2 ± 0.3 | NC | 4.2 ± 0.3 | 1.3 ± 0.1 | |||||||||||
| Colon | 3.2 ± 0.3 | NC | 5.1 ± 0.2 | 2.0 ± 0.5 | |||||||||||
dpc: days post contact; dpi: days post infection; GMT: geometric mean titre; HI: haemagglutination inhibition; NC: samples not collected; ND: none detected.
a Viral titres are expressed as mean log10EID50/mL.
b Viral titres are expressed as mean log10EID50/g.
c Samples were not collected because chickens were all dead.
Titres are indicated with standard deviation. Swabs from chickens were collected daily for the infected and contact group. Swabs from ducks were collected every 2 days for the infected group and daily for the contact group.
In ducks, we measured the viral replication and transmission efficacy although no mortalities were observed. However, high viral titres were detected at all time points in oropharyngeal swabs of both groups, peaking at 3 dpi (5.1 log10 EID50/mL) for the infection group (Table 2). In contrast, low titres were observed in the cloacal swabs, which peaked at only 2.4 log10 EID50/mL at 4 dpi in the direct contact group. Furthermore, the virus was not detectable in cloacal swabs at 7 dpi or in cloacal or oropharyngeal swabs at 9 dpi for the inoculated group. For the contact group, the virus was not detected in oropharyngeal or cloacal swabs at 7 days post contact. Seroconversion was observed in all ducks inoculated with the A/AP/Korea/W612/2017(H5N6) virus and in their contact ducks, and haemagglutination inhibition geometric mean titres were 640 and 320 in the inoculated and direct contact group, respectively (Table 2). To investigate the tissue distribution of A/AP/Korea/W612/2017(H5N6) virus in ducks, we collected the lung, brain, kidney, spleen, heart, liver and colon from three birds each at 3 and 5 dpi. The A/AP/Korea/W612/2017(H5N6) virus was detected in all organs harvested from ducks in this experiment, except for the brain (Table 2).
Discussion
In contrast to influenza A(H5N8) viruses, the clade 2.3.4.4 H5N6 virus first emerged in China in 2013 and became established in Asian countries such as Laos, Vietnam and mainland China [7-9] before causing large outbreaks in 2016 [4,10]. During the 2016/17 winter season, South Korea experienced large outbreaks of two different HPAI strains (H5N6 and H5N8) and as a result destroyed 640 million poultry (almost 30% of poultry in South Korea) [10-12].
In this study, we report the identification of a novel reassortant HPAI H5N6 virus that caused major outbreaks in domestic poultry during the 2017/18 winter season. This H5N6 virus is a reassortment of multiple subtypes (H5N8, H7N7, H5N1, H10N7 and H3N6) in the Eurasian gene pool of avian influenza viruses. Animal studies revealed that this novel influenza A(H5N6) virus is highly pathogenic in chickens but has attenuated virulence in ducks, which may result in their function as a virus reservoir. With the high susceptibility but attenuated virulence of HPAI infections in ducks, the duck can also be used as a sentinel for avian influenza virus surveillances [13]. Since the first avian influenza A(H5N8) (clade 2.3.4.4) virus was reported in Korean poultry in 2014 [14,15] and has continued to spread in South Korea, migratory birds have spread the virus to wild birds worldwide, including in Europe [16,17] and North America [18]. This rapid and widespread proliferation and reassortment underscores the need for continuous monitoring of avian influenza viruses in wild migratory birds, including virulence and pathogenicity studies using laboratory animals.
Acknowledgements
This work was partially supported by grants from the Korea Health Technology R&D Project, funded by the Ministry of Health and Welfare, Republic of Korea (Grant number: HI16C1032 ) and by the research fund of Animal and Plant Quarantine Agency (grant number: B-1543418-2018-20-01). Animal experiments were progressed in an enhanced animal biosafety level 3 facility in Chungbuk National University permitted by the K-CDC (permit number KCDC-14-3-07). All animal experiment protocols were approved by the Medical Research Institute and Laboratory Animal Research Center (LARC) (approval number CBNUA-1084-18-01). The authors would like to thank the eight originating and submitting laboratories who contributed sequences used in phylogenetic analysis to GISAID (listed in Supplement 1). All submitters of data may be contacted directly via the GISAID website www.gisaid.org
Conflict of interest: None declared.
Authors’ contributions: Wrote the manuscript: YK Choi, MH Lee, YI Kim, YJ Si. Performed laboratory investigations: YI Kim, YJ Si, EH Kim, SJ Park, NJ Robles, HD Nguyen, MA Yu, KM Yu. Performed phylogenetic analyses: YJ Si, YK Choi, YI Kim, HI Kwon, YJ Lee; Animal experiment: HI Kwon, YI Kim, YJ Si. All authors reviewed the manuscript.
References
- 1.World Organisation for Animal Health (OIE). H5N6 Update on avian influenza in animals (types H5 and H7). Paris: OIE; 19 Nov 2017. Available from: http://www.oie.int/wahis_2/temp/reports/en_imm_0000025260_20171120_171201.pdf
- 2.World Organisation for Animal Health (OIE). H5N6 Update on avian influenza in animals (types H5 and H7). Paris: OIE; 10 Nov 2017. Available from: http://www.oie.int/wahis_2/temp/reports/en_imm_0000025189_20171110_182530.pdf
- 3. Beerens N, Heutink R, Bergervoet SA, Harders F, Bossers A, Koch G. Multiple Reassorted Viruses as Cause of Highly Pathogenic Avian Influenza A(H5N8) Virus Epidemic, the Netherlands, 2016. Emerg Infect Dis. 2017;23(12):1974-81. 10.3201/eid2312.171062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Si Y-J, Lee IW, Kim E-H, Kim Y-I, Kwon H-I, Park S-J, et al. Genetic characterisation of novel, highly pathogenic avian influenza (HPAI) H5N6 viruses isolated in birds, South Korea, November 2016. Euro Surveill. 2017;22(1):30434. 10.2807/1560-7917.ES.2017.22.1.30434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pohlmann A, Starick E, Harder T, Grund C, Höper D, Globig A, et al. Outbreaks among wild birds and domestic poultry caused by reassorted influenza A (H5N8) clade 2.3. 4.4 viruses, Germany, 2016. Emerg Infect Dis. 2017;23(4):633-6. 10.3201/eid2304.161949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Organisation for Animal Health (OIE). Infection with avian influenza viruses. In: Terrestrial Animal Health Code. Paris: OIE; 2017 Available from: http://www.oie.int/index.php?id=169&L=0&htmfile=chapitre_avian_influenza_viruses.htm
- 7. Shen H, Wu B, Chen Y, Bi Y, Xie Q. Influenza A(H5N6) virus reassortant, southern China, 2014. Emerg Infect Dis. 2015;21(7):1261-2. 10.3201/eid2107.140838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wong FY, Phommachanh P, Kalpravidh W, Chanthavisouk C, Gilbert J, Bingham J, et al. Reassortant highly pathogenic influenza A(H5N6) virus in Laos. Emerg Infect Dis. 2015;21(3):511-6. 10.3201/eid2103.141488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu S, Zhao Z, Zhang J, Wang W, He X, Yu M, et al. Genetics, pathogenicity and transmissibility of novel reassortant H5N6 highly pathogenic avian influenza viruses first isolated from migratory birds in western China. Emerg Microbes Infect. 2018;7(1):6. 10.1038/s41426-017-0001-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Organisation for Animal Health (OIE). H5N6 Update on avian influenza in animals (types H5 and H7). Paris: OIE; 23 Nov 2017. Available from: http://www.oie.int/wahis_2/public%5C.%5Ctemp%5Creports/en_imm_0000021637_20161123_134128.pdf
- 11.World Organisation for Animal Health (OIE). H5N8 Update on avian influenza in animals (types H5 and H7). Paris: OIE; 20 Nov 2017. Available from: http://www.oie.int/wahis_2/temp/reports/en_fup_0000025168_20171120_170003.pdf
- 12. Kim Y-I, Park S-J, Kwon H-I, Kim E-H, Si Y-J, Jeong J-H, et al. Genetic and phylogenetic characterizations of a novel genotype of highly pathogenic avian influenza (HPAI) H5N8 viruses in 2016/2017 in South Korea. Infect Genet Evol. 2017;53:56-67. 10.1016/j.meegid.2017.05.001 [DOI] [PubMed] [Google Scholar]
- 13. Globig A, Baumer A, Revilla-Fernández S, Beer M, Wodak E, Fink M, et al. Ducks as sentinels for avian influenza in wild birds. Emerg Infect Dis. 2009;15(10):1633-6. 10.3201/eid1510.090439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim Y-I, Pascua PNQ, Kwon H-I, Lim G-J, Kim E-H, Yoon S-W, et al. Pathobiological features of a novel, highly pathogenic avian influenza A(H5N8) virus. Emerg Microbes Infect. 2014;3(10):e75. 10.1038/emi.2014.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee Y-J, Kang H-M, Lee E-K, Song B-M, Jeong J, Kwon Y-K, et al. Novel reassortant influenza A(H5N8) viruses, South Korea, 2014. Emerg Infect Dis. 2014;20(6):1087-9. 10.3201/eid2006.140233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Adlhoch C, Gossner C, Koch G, Brown I, Bouwstra R, Verdonck F, et al. Comparing introduction to Europe of highly pathogenic avian influenza viruses A(H5N8) in 2014 and A(H5N1) in 2005. Euro Surveill. 2014;19(50):20996. 10.2807/1560-7917.ES2014.19.50.20996 [DOI] [PubMed] [Google Scholar]
- 17. Poen MJ, Bestebroer TM, Vuong O, Scheuer RD, van der Jeugd HP, Kleyheeg E, et al. Local amplification of highly pathogenic avian influenza H5N8 viruses in wild birds in the Netherlands, 2016 to 2017. Euro Surveill. 2018;23(4):17-00449. 10.2807/1560-7917.ES.2018.23.4.17-00449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee D-H, Torchetti MK, Winker K, Ip HS, Song C-S, Swayne DE. Intercontinental spread of Asian-origin H5N8 to North America through Beringia by migratory birds. J Virol. 2015;89(12):6521-4. 10.1128/JVI.00728-15 [DOI] [PMC free article] [PubMed] [Google Scholar]


