This study examines the association between genomic metrics and the extent of immune infiltration in triple-negative breast cancers.
Key Points
Question
What are the genomic differences between triple-negative breast cancers with high lymphocytic infiltration and good prognosis and triple-negative breast cancers with less immune infiltration and worse prognosis?
Findings
In this study of genomic data sets, triple-negative breast cancers with high immune gene expression had lower clonal heterogeneity, fewer copy number alterations, lower somatic mutation, and lower neoantigen loads.
Meaning
This study suggests that antitumor immune surveillance in immune-rich triple-negative breast cancers may lead to elimination of clones, lower clonal heterogeneity, and “simpler” genomes; the surviving neoplastic cell population exists at a near equilibrium with immune surveillance, explaining the better prognosis, and immune-poor triple-negative breast cancers have greater genomic diversity attributable to lesser immune restraint.
Abstract
Importance
Why some triple-negative breast cancers (TNBCs) have high and others have low immune cell infiltration is unknown. Understanding how immune surveillance shapes the cancer genome could help in the selection of patients and the development of more effective immunotherapy strategies.
Objective
To examine the association between genomic metrics and the extent of immune infiltration in TNBCs.
Design, Setting, and Participants
This study, performed from June 1, 2015, through January 31, 2017, used DNA and RNA sequencing data and messenger RNA expression results from The Cancer Genome Atlas (TCGA) breast cancer data set (n = 1215) to calculate previously described immune metagene expression values and histologic lymphocyte counts to quantify immune infiltration and assign prognostic categories to TNBCs. It used the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) data set as an independent validation cohort. The study compared clonal heterogeneity, somatic total mutational load, neoantigen load, and somatic copy number alteration levels between immune-rich TNBC cohorts with good prognosis and immune-poor TNBC cohorts with poor prognosis. The study also compared the distribution of mutations in 119 canonical cancer genes.
Main Outcomes and Measures
Correlation between immune prognostic category and genomic metrics of the cancer.
Results
This study of 193 TNBC samples with patient survival information found an inverse association between clonal heterogeneity and immune metagene expression (ρ = −0.395, P = 2 × 10−8). The study also found an inverse association between immune metagene expression and somatic copy number alteration levels (ρ = −0.484, P = 2 × 10−10). Lymphocyte-rich TNBCs with good prognosis had significantly lower mutation and neoantigen counts than did lymphocyte-poor TNBCs with poor prognosis. The robustness of the study results was confirmed by using various immune metagenes in the same TCGA data set and in the independent METABRIC data set.
Conclusions and Relevance
This study suggests that immune-rich TNBCs may be under an immune surveillance that continuously eliminates many immunogenic clones, resulting in lower clonal heterogeneity. These cancers may also represent the subset of TNBCs that could derive benefit from immune checkpoint inhibitor therapy to tilt the balance in favor of the immune system.
Introduction
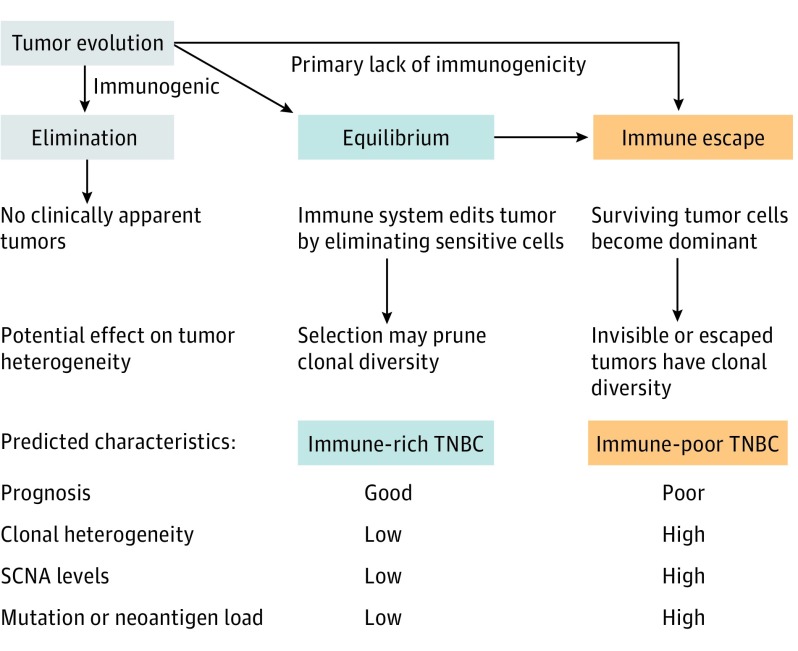
The importance of immune surveillance in determining the prognosis of various types of cancers is increasingly recognized. Understanding how the immune microenvironment influences the biology of cancer is important because it could lead to better patient selection strategies and more effective immunotherapies. More than 70% of breast cancers contain at least some tumor-infiltrating lymphocytes (TILs), and preclinical studies, as described by Schreiber et al, have found that antitumor immunity can eliminate some neoplastic cells, resulting in a precarious near equilibrium between the surviving clones and immune surveillance. Consistent with these observations, clinical studies also found that breast cancers with high immune infiltration, particularly the triple-negative breast cancer (TNBC) and ERBB2 (formerly HER2 or HER2/neu)–positive subtypes, have better prognosis. According to the immunoediting hypothesis of cancer progression, some cancers may be eliminated by an antitumor immune response before diagnosis, whereas most clinically apparent cancers represent states of escape or partial control by immune surveillance. One hypothesis is that cancers with greater genomic instability will have higher mutational burden, greater clonal heterogeneity, and higher genomic diversity, resulting in more neoantigens and therefore greater immune infiltration. Indeed, a positive correlation between the overall mutation or neoantigen loads and immune infiltration has been observed across cancer types. Alternatively, another hypothesis is that extensive lymphocytic infiltration is a consequence of a strong antitumor immune response that results in pruning of the genomic heterogeneity of the cancer by eliminating many immunogenic cell clones, whereas cancers with low lymphocytic infiltration may represent immune escape that also allows tumor evolution toward greater clonal heterogeneity and genomic diversity (Figure 1). Several studies support an inverse association between immune cell infiltration and intratumor clonal heterogeneity and somatic copy number alterations (SCNAs). In some cancers, the neoantigen load is also lower than expected, suggesting selective elimination of immunogenic clones.
Figure 1. Influence of Immunoediting on Tumor Clonal Heterogeneity.
Most clinically apparent cancers represent states of partial immune control or escape. Immune cell–rich triple-negative breast cancers (TNBC) could represent the equilibrium phase, in which a strong immune response may result in pruning of cancer clonal and genomic heterogeneity by eliminating immunogenic sensitive cell clones. Cancers that have escaped immune surveillance have low lymphocytic infiltration and evolve toward greater clonal heterogeneity and genomic diversity. SCNA indicates somatic copy number alteration.
The goal of the present analysis was to assess the association between lymphocytic infiltration and genomic diversity in TNBCs. Specifically, we examine the association among immune infiltration measured by immune gene expression signatures, genomic complexity reflected by clonal heterogeneity, SCNAs, mutation load, neoantigen load, and patient prognosis.
Methods
In this study, performed from June 1, 2015, through January 31, 2017, previously reported prognostic immune gene expression signatures that were initially derived from DNA microarray data were transferred to RNA sequencing data of The Cancer Genome Atlas (TCGA) breast cancer cohort (n = 1215), as described in the eMethods and eFigure 1 in the Supplement. The RNA sequencing–based immune metagenes were highly correlated with the DNA microarray versions, successfully reproducing our previous immune clustering of TNBCs (n = 208) (eFigures 1-5 in the Supplement) and correlating well with histologic TIL quantification (eFigure 6 in the Supplement). Next, we classified the TNBC samples with survival information (n = 193) in the TCGA into good (n = 25) and poor prognosis (n = 168) categories. Good prognosis was defined as high immune infiltration (ie, major histocompatibility complex class II metagene expression in the top quartile) and low inflammation markers (ie, interleukin 8–vascular endothelial growth factor metagene expression below the median) (eFigures 7 and 20 in the Supplement). This classification was originally developed from an independent Affymetrix data set and remained strongly prognostic in the TCGA TNBC data (Figure 2A and eFigure 8 in the Supplement). We compared clonal heterogeneity measured by the mutant-allele tumor heterogeneity (MATH) score, which quantifies the dispersion of variant allele frequencies in each tumor, SCNAs as reported previously, mutational load, neoantigen load, and the distribution of mutations in 119 canonical cancer genes between the good and poor prognosis TNBC cohorts (eMethods, eTables 1-4, and eFigure 9 in the Supplement). All reported P values are 2-sided, and P < .05 was considered significant.
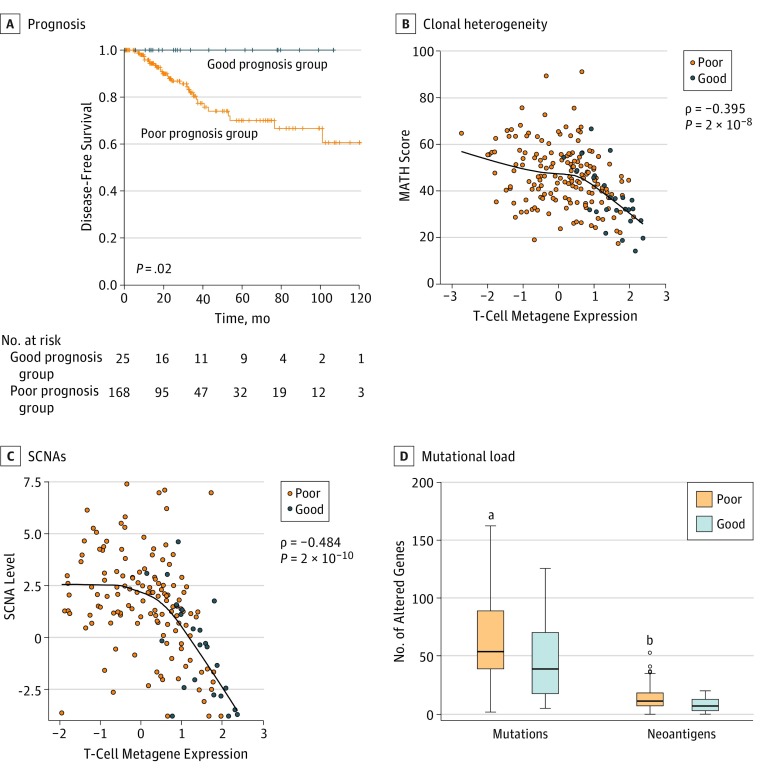
Figure 2. Association Between Clonal Heterogeneity and Prognostic Immune Signature.
A, A total of 193 triple-negative breast cancer (TNBC) samples with follow-up data from The Cancer Genome Atlas were classified according to a previously described prognostic immune signature based on metagenes for high lymphocyte infiltration (major histocompatibility complex class II gene signature) in combination with low interleukin 8–vascular endothelial growth factor signature expression. Kaplan-Meier analysis of disease-free survival of the good (n = 25) and poor (n = 168) prognosis groups is shown (P values are from the log-rank test). B, Inverse association between T-cell metagene expression and mutant-allele tumor heterogeneity (MATH) score (ie, clonal heterogeneity) in TNBCs (solid line is the locally weighted scatterplot smoothing [LOWESS] fit, Spearman rank correlation P value). TNBCs in the good prognosis group had significantly lower MATH scores (mean rank, 59.7 vs 98.8; Mann-Whitney test P = .001) (eFigure 22 in the Supplement). C, Inverse association between T-cell metagene expression and somatic copy number alteration (SCNA) levels in TNBCs (solid line is the LOWESS fit, Spearman rank correlation P value). SCNA levels were significantly lower in the good prognosis group (mean rank, 45.8 vs 84.2; Mann-Whitney test P < .001) (eFigure 22 in the Supplement). D, Differences in mutational load and predicted neoantigen load in good and poor prognosis TNBC groups; error bars indicate 95% CIs. The y-axis is cropped at 170 mutated genes per sample, which excludes individual hypermutated samples with 300 to 1200 mutations (P values from Mann-Whitney test).
aP = .02 compared with good prognosis.
bP = .04 compared with good prognosis.
This study uses only publicly freely available open access data from TCGA, which is not unique to an individual and therefore deidentified. Institutional review board approval was not required according to Exemption 45 CFR 46.101(b)(4) from the US Department of Health and Human Services and the local institutional review board.
Results
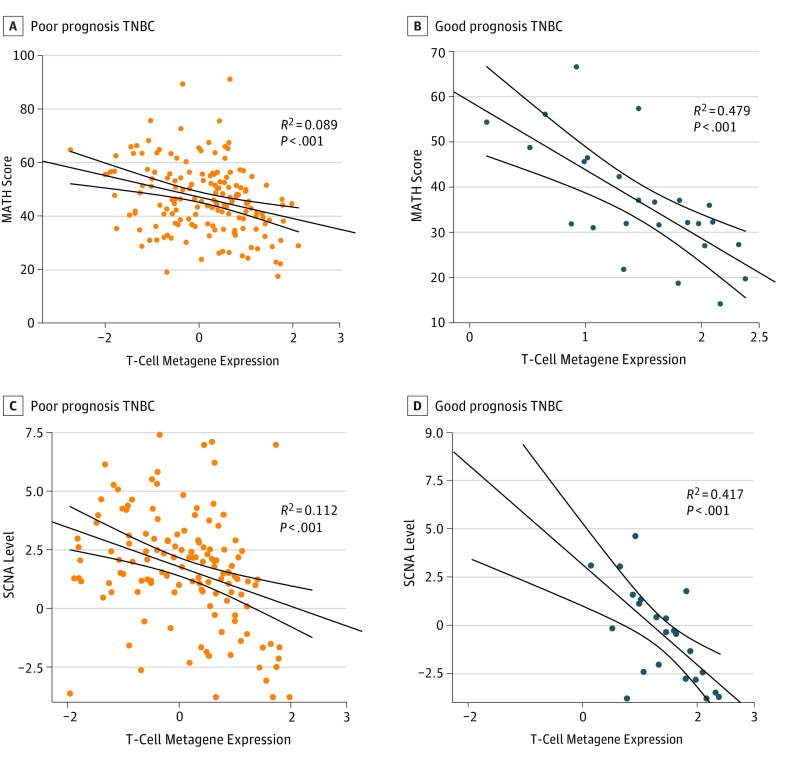
The immune-rich, good prognosis TNBC samples had significantly lower MATH scores, indicating lower clonal genomic heterogeneity (mean rank, 59.7 vs 98.8; Mann-Whitney test P = .001) (eFigure 22 in the Supplement). We observed a strong inverse association between MATH score and immune metagene expression across all TNBC samples (Figure 2B), which was particularly strong among the good prognosis samples (R2 = 0.479, P < .001) (Figure 3B). Levels of SCNAs were also significantly lower in the good prognosis group (mean rank, 45.8 vs 84.2; Mann-Whitney test P < .001) (eFigure 22 in the Supplement), with a significant inverse association between SCNAs and immune metagene expression across all samples (Figure 2C), which was again the strongest in the good prognosis group (R2 = 0.417) (Figure 3D). The SCNA levels and MATH scores showed only a weak positive correlation (R2 = 0.214) (eFigure 10 in the Supplement), suggesting that these metrics capture distinct genomic features, each separately associated with immune infiltration. The inverse association between immune infiltration and MATH score and SCNA levels was confirmed using different immune metagenes (the major histocompatibility complex class II metagene alone, B cell, and the cytolytic activity immune gene signature CYT) (eFigures 11 and 21 in the Supplement) and was also inversely correlated with histologic TIL counts (eFigures 12 and 13 in the Supplement). Good prognosis TNBCs also had significantly lower mutational load (mean rank, 70.4 vs 97.1; Mann-Whitney test P = .02) and neoantigen load (mean rank, 50.7 vs 70.1; Mann-Whitney test P = .04) (Figure 2D) compared with the poor prognosis samples. Lower overall mutation and neoantigen counts were also associated with high immune infiltration (eFigure 14 in the Supplement). Mutation load and neoantigen counts were highly correlated with one another (R2 = 0.68) (eFigure 15 in the Supplement) but not with MATH (R2 = 0.001) (eFigure 16 in the Supplement).
Figure 3. Association of Clonal Heterogeneity and Immune Signature in Prognostic Groups.
A and B, Association between T-cell metagene expression and mutant-allele tumor heterogeneity (MATH) score (ie, clonal heterogeneity) for triple-negative breast cancer (TNBC) in the poor (A) and good (B) prognosis groups (Figure 2A). C and D, Association between T-cell metagene expression and somatic copy number alteration (SCNA) levels for TNBC in the poor (C) and good (D) prognosis groups. Lines indicate 95 CIs. R2 and P values are from linear regression.
We also validated our results in the TNBC cohort (n = 283) of the independent METABRIC data set (eMethods in the Supplement). The cytolytic activity immune gene signature CYT showed a highly significant negative association with MATH (ρ = −0.286, P = 2 × 10−6) and a nonstatistically significant (ρ = −0.104, P = .14) association with chromosomal instability as a surrogate for SCNAs. The TIL-rich TNBC cluster also had a significantly lower MATH score compared with the TIL-poor cluster (eFigure 17 in the Supplement).
Discussion
Our findings may appear to contradict an earlier publication that reported a weak positive association between neoantigen load and the cytolytic activity immune gene signature CYT when all breast cancers subtypes were examined together. We also observed this overall association but noticed that it may be in part attributable to the higher somatic mutation burden and higher immune infiltration in TNBCs compared with luminal cancers (eFigure 18 in the Supplement), as well as a small positive correlation in luminal B subtype (eFigure 19 in the Supplement), which was recently reported. When TNBC cancers are examined separately, the positive correlations between immune infiltration and genomic heterogeneity and mutation load are no longer seen; in fact, the opposite is observed, which is consistent with an immune pruning effect in TNBCs. Two other reports also support our observations. An earlier report noted that TNBCs with low clonal heterogeneity but high clonal mutational burden (ie, mutation burden adjusted for tumor clonality) have higher neoantigens per neoplastic clone and higher immune gene expression that is associated with greater chemotherapy sensitivity. Davoli et al independently observed a negative correlation between tumor aneuploidy and immune gene expression in a pan-cancer study.
Limitations
A limitation of our study is our inability to determine a cause-and-effect relationship because our observations are correlative in nature. It is therefore possible that genomic alterations are also sculpting the immune system, and we observed a result of the interactive effect of each other. In addition, tumor purity may affect mutation calling and confound the analysis. Further discussion of both these issues can be found in the eMethods.
Conclusions
We demonstrate that high immune infiltration is mostly seen in primary TNBCs with low clonal heterogeneity, fewer SCNAs, and lower somatic mutation and neoantigen loads. We suggest that these findings may be a consequence of effective immune surveillance that continuously eliminates immunogenic clones, resulting in lower clonal heterogeneity. The better prognosis of these cancers is consistent with strong immune surveillance and precarious equilibrium between the cancer and the immune system. Surgical resection of the primary tumor and adjuvant chemotherapy may assist the immune system. These cancers may also represent the subset of TNBCs that could derive further benefit from immune checkpoint inhibitor therapy.
eMethods. Supplementary Methods
eFigure 1. Strategy of RNA-Seq and Whole-Exome-Seq Analyses for TNBC Classification
eFigure 2. ER, PR, and HER2 expression assessed by RNA-Seq and Agilent arrays
eFigure 3. Dependency of platform correlation on gene expression level
eFigure 4. Correlation between RNA-Seq and Affymetrix for Metagene clusters
eFigure 5. Classification of TNBC (n = 208) based on RNA-Seq data
eFigure 6. Correlation of MHC2 metagene expression and histological quantification of TILs in TCGA samples
eFigure 7. Classification algorithm of the prognostic immune signature
eFigure 8. Validation of improved prognosis of TNBC patients with “Good prognosis” signature in RNA-Seq data
eFigure 9. Mutational count distribution in 186 TNBC
eFigure 10. Relationship between SCNA levels and MATH in TNBC from TCGA
eFigure 11. Validation of Inverse Relationships Between Measures of Genomic Complexity and Immune Cell Infiltration in TNBC Using Different Immune Metagenes
eFigure 12. Prognostic value of histologically quantified TILs in the TCGA TNBC data Set
eFigure 13. Validation of Inverse Relationship of genomic heterogeneity and Immune Cell Infiltration Using Histologically Quantified TILs in the TCGA TNBC Data Set
eFigure 14. Differences in Mutation Count by Immune Cell Infiltration Metagenes and IL8/VEGF metagene expression categories
eFigure 15. Correlation of the Number of Predicted Neoantigens and Mutational Load
eFigure 16. Independence of MATH Score and Total Mutation Counts
eFigure 17. Validation Analyses in METABRIC Data Set
eFigure 18. Differences in Mutation Count, Neoantigen Count, and CYT by Molecular Subtype in Breast Cancer
eFigure 19. Confounding of Molecular Breast Cancer Subtypes on Predicted Neoantigen Count and CYT
eFigure 20. High intercorrelation of TIL metagenes
eFigure 21. Association between clonal heterogeneity and immune metagene expression
eFigure 22. Association of MATH and SCNA with prognostic groups in TNBC
eTable 1. Annotated Cancer Genes Mutated in ≥3 samples
eTable 2. TCGA samples included in the study
eTable 3. “Cancer genes” curated by Vogelstein and colleagues
eTable 4. Individual Genes Constituting TNBC Metagenes and Their Correlation With Affymetrix Microarray
References
- 1.Pusztai L, Karn T, Safonov A, Abu-Khalaf MM, Bianchini G. New strategies in breast cancer: immunotherapy. Clin Cancer Res. 2016;22(9):2105-2110. doi: 10.1158/1078-0432.CCR-15-1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331(6024):1565-1570. doi: 10.1126/science.1203486 [DOI] [PubMed] [Google Scholar]
- 3.Bianchini G, Qi Y, Alvarez RH, et al. Molecular anatomy of breast cancer stroma and its prognostic value in estrogen receptor-positive and -negative cancers. J Clin Oncol. 2010;28(28):4316-4323. doi: 10.1200/JCO.2009.27.2419 [DOI] [PubMed] [Google Scholar]
- 4.Karn T, Pusztai L, Rody A, Holtrich U, Becker S. The influence of host factors on the prognosis of breast cancer: stroma and immune cell components as cancer biomarkers. Curr Cancer Drug Targets. 2015;15(8):652-664. [DOI] [PubMed] [Google Scholar]
- 5.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1-2):48-61. doi: 10.1016/j.cell.2014.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown SD, Warren RL, Gibb EA, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014;24(5):743-750. doi: 10.1101/gr.165985.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris LGT, Riaz N, Desrichard A, et al. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget. 2016;7(9):10051-10063. doi: 10.18632/oncotarget.7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355(6322):eaaf8399. doi: 10.1126/science.aaf8399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rody A, Karn T, Liedtke C, et al. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 2011;13(5):R97. doi: 10.1186/bcr3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karn T, Pusztai L, Holtrich U, et al. Homogeneous datasets of triple negative breast cancers enable the identification of novel prognostic and predictive signatures. PLoS One. 2011;6(12):e28403. doi: 10.1371/journal.pone.0028403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol. 2013;49(3):211-215. doi: 10.1016/j.oraloncology.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333-339. doi: 10.1038/nature12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546-1558. doi: 10.1126/science.1235122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Safonov A, Jiang T, Bianchini G, et al. Immune gene expression is associated with genomic aberrations in breast cancer [published online April 20, 2017]. Cancer Res. doi: 10.1158/0008-5472.CAN-16-3478 [DOI] [PubMed] [Google Scholar]
- 15.Jiang T, Shi W, Wali VB, et al. Predictors of chemosensitivity in triple negative breast cancer: an integrated genomic analysis. PLoS Med. 2016;13(12):e1002193. doi: 10.1371/journal.pmed.1002193 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Supplementary Methods
eFigure 1. Strategy of RNA-Seq and Whole-Exome-Seq Analyses for TNBC Classification
eFigure 2. ER, PR, and HER2 expression assessed by RNA-Seq and Agilent arrays
eFigure 3. Dependency of platform correlation on gene expression level
eFigure 4. Correlation between RNA-Seq and Affymetrix for Metagene clusters
eFigure 5. Classification of TNBC (n = 208) based on RNA-Seq data
eFigure 6. Correlation of MHC2 metagene expression and histological quantification of TILs in TCGA samples
eFigure 7. Classification algorithm of the prognostic immune signature
eFigure 8. Validation of improved prognosis of TNBC patients with “Good prognosis” signature in RNA-Seq data
eFigure 9. Mutational count distribution in 186 TNBC
eFigure 10. Relationship between SCNA levels and MATH in TNBC from TCGA
eFigure 11. Validation of Inverse Relationships Between Measures of Genomic Complexity and Immune Cell Infiltration in TNBC Using Different Immune Metagenes
eFigure 12. Prognostic value of histologically quantified TILs in the TCGA TNBC data Set
eFigure 13. Validation of Inverse Relationship of genomic heterogeneity and Immune Cell Infiltration Using Histologically Quantified TILs in the TCGA TNBC Data Set
eFigure 14. Differences in Mutation Count by Immune Cell Infiltration Metagenes and IL8/VEGF metagene expression categories
eFigure 15. Correlation of the Number of Predicted Neoantigens and Mutational Load
eFigure 16. Independence of MATH Score and Total Mutation Counts
eFigure 17. Validation Analyses in METABRIC Data Set
eFigure 18. Differences in Mutation Count, Neoantigen Count, and CYT by Molecular Subtype in Breast Cancer
eFigure 19. Confounding of Molecular Breast Cancer Subtypes on Predicted Neoantigen Count and CYT
eFigure 20. High intercorrelation of TIL metagenes
eFigure 21. Association between clonal heterogeneity and immune metagene expression
eFigure 22. Association of MATH and SCNA with prognostic groups in TNBC
eTable 1. Annotated Cancer Genes Mutated in ≥3 samples
eTable 2. TCGA samples included in the study
eTable 3. “Cancer genes” curated by Vogelstein and colleagues
eTable 4. Individual Genes Constituting TNBC Metagenes and Their Correlation With Affymetrix Microarray