Abstract
Importance
There is a clear need for a molecular subtyping approach in prostate cancer to identify clinically distinct subgroups that benefit from specific therapies.
Objectives
To identify prostate cancer subtypes based on luminal and basal lineage and to determine associations with clinical outcomes and response to treatment.
Design, Setting, and Participants
The PAM50 classifier was used to subtype 1567 retrospectively collected (median follow-up, 10 years) and 2215 prospectively collected prostate cancer samples into luminal- and basal-like subtypes.
Main Outcomes and Measures
Metastasis, biochemical recurrence, overall survival, prostate cancer–specific survival, associations with biological pathways, and clinicopathologic variables were the main outcomes.
Results
Among the 3782 samples, the PAM50 classifier consistently segregated prostate cancer into 3 subtypes in both the retrospective and prospective cohorts: luminal A (retrospective, 538 [34.3%]; prospective, 737 [33.3%]), luminal B (retrospective, 447 [28.5%]; prospective, 723 [32.6%]), and basal (retrospective, 582 [37.1%]; prospective, 755 [34.1%]). Known luminal lineage markers, such as NKX3.1 and KRT18, were enriched in luminal-like cancers, and the basal lineage CD49f signature was enriched in basal-like cancers, demonstrating the connection between these subtypes and established prostate cancer biology. In the retrospective cohort, luminal B prostate cancers exhibited the poorest clinical prognoses on both univariable and multivariable analyses accounting for standard clinicopathologic prognostic factors (10-year biochemical recurrence-free survival [bRFS], 29%; distant metastasis-free survival [DMFS], 53%; prostate cancer-specific survival [PCSS], 78%; overall survival [OS], 69%), followed by basal prostate cancers (10-year bRFS, 39%; DMFS, 73%; PCSS, 86%; OS, 80%) and luminal A prostate cancers (10-year bRFS, 41%; DMFS, 73%; PCSS, 89%; OS, 82%). Although both luminal-like subtypes were associated with increased androgen receptor expression and signaling, only luminal B prostate cancers were significantly associated with postoperative response to androgen deprivation therapy (ADT) in a subset analysis in our retrospective cohorts (n = 315) matching patients based on clinicopathologic variables (luminal B 10-year metastasis: treated, 33% vs untreated, 55%; nonluminal B 10-year metastasis: treated, 37% vs untreated, 21%; P = .006 for interaction).
Conclusions and Relevance
Luminal- and basal-like prostate cancers demonstrate divergent clinical behavior, and patients with luminal B tumors respond better to postoperative ADT than do patients with non–luminal B tumors. These findings contribute novel insight into prostate cancer biology, providing a potential clinical tool to personalize ADT treatment for prostate cancer by predicting which men may benefit from ADT after surgery.
This study identifies prostate cancer subtypes based on luminal and basal lineage and assesses the association of those subtypes with clinical outcomes and response to treatment.
Key Points
Question
Is molecular subtyping by luminal and basal status clinically relevant in prostate cancer?
Findings
In this study of 3782 retrospectively and prospectively collected radical prostatectomy samples, molecular subtyping by the PAM50 classifier consistently segregates patients into luminal A, luminal B, and basal-like subtypes, which are associated with different lineage markers. Patients with luminal tumors exhibit increased androgen signaling, and those with luminal B tumors have poorer outcomes but potentially improved response to postoperative androgen deprivation therapy.
Meaning
Molecular subtyping by luminal and basal status in prostate cancer is prognostic for clinical outcomes and may be associated with response to postoperative androgen deprivation therapy.
Introduction
The lineage of prostate cancer is unknown. Prostate cancer was first thought to derive from glandular luminal cells; however, there is mounting evidence that basal cells may also play a role in prostate carcinogenesis. Mouse models have demonstrated that both luminal and basal cells include self-sustaining lineages that can give rise to prostate cancer. Recent work has sought to characterize luminal and basal cells that display characteristics similar to those of stem cells. However, this is an area of active research, and the similarities and differences of luminal and basal prostate cancer remain unresolved.
The concept of luminal- and basal-like cells and oncogenesis is not limited to prostate cancer. Luminal and basal features are thought to define key molecular subtypes in bladder cancer and, most notably, in breast cancer, in which the well-known PAM50 gene expression classifier identifies the major molecular subtypes of breast cancer. The PAM50 classifier categorizes breast cancer into luminal A, luminal B, HER2, and basal subtypes and is the basis for the commercially available Prosigna test (NanoString Technologies). Furthermore, these subtypes display significant differences in prognosis and response to treatment in both breast and bladder cancer. Luminal and basal subtypes of bladder cancer are correlated with the luminal and basal subtypes of breast cancer, suggesting that underlying biological differences that transcend the organ of origin can be identified using the PAM50 classifier.
Given that prostate and breast cancer are both hormonally driven tumors and share many oncogenic pathways, we hypothesized that the PAM50 algorithm could identify luminal- and basal-like subtypes in prostate cancer and that these subtypes would differ in clinical outcomes and treatment response. We used gene expression data from 3782 prostate cancer samples in 7 distinct cohorts on a clinical-grade microarray platform to investigate the prognostic and predictive utility of luminal and basal subtypes in prostate cancer.
Methods
Clinical Samples and Microarray Processing
Affymetrix Human Exon 1.0 ST microarray (Affymetrix) data from formalin-fixed, paraffin-embedded radical prostatectomy samples were obtained from 6 published retrospective patient cohorts (n = 1567) and 1 prospective cohort (n = 2215), for a total of 3782 samples. Retrospective cohorts were from the Mayo Clinic, Rochester, Minnesota (2 separate cohorts); Cleveland Clinic, Cleveland, Ohio; Johns Hopkins University, Baltimore, Maryland; Thomas Jefferson University, Philadelphia, Pennsylvania; and Durham Veterans Affairs, Durham, North Carolina. Additional cohort details can be found in the original articles. A total of 2215 deidentified, anonymized, and prospectively collected patients from clinical use of the Decipher test were obtained from Decipher GRID (clinicaltrials.gov Identifier: NCT02609269). Clinical outcomes were not available for Decipher GRID. Microarray processing was performed in a Clinical Laboratory Improvement Amendments–certified clinical operations laboratory (GenomeDx Biosciences, Inc). Microarrays were normalized using Single Channel Array Normalization. See the eAppendix in the Supplement for information regarding the androgen deprivation therapy (ADT)–matched analysis, microarray data, Gene Set Enrichment Analysis (GSEA), and statistical analyses. Data collection for the 6 retrospective cohorts was approved and supervised by the Mayo Clinic, Cleveland Clinic, Johns Hopkins University, Thomas Jefferson University, and Durham Veterans Affairs institutional review boards (IRBs). Patient consent for the 6 retrospective cohorts was waived by the IRB of each institution. Written informed patient consent and IRB approval for Decipher GRID were obtained through Quorum Review IRB (Seattle, Washington).
PAM50 Clustering
PAM50 clustering was performed based on the original algorithm from Parker et al. Source code was downloaded from the University of North Carolina Microarray Database (https://genome.unc.edu/pubsup/breastGEO/) and used without modification. Gene expression data were median centered in each cohort individually as required by the PAM50 algorithm. The normal-like subtype was excluded because the prostate cancer samples were macrodissected, limiting the amount of normal tissue present. The HER2 subtype was also excluded given the lack of ERBB2/HER2 (OMIM: 164870) amplification in prostate cancer. Assignment of subtype in the prostate cancer samples was thus assigned by greatest correlation with luminal A, luminal B, or basal.
Clinical End Points
All primary and secondary end points were preplanned. The primary clinical end point was distant metastasis–free survival (DMFS). Secondary clinical end points were biochemical recurrence-free survival (bRFS), prostate cancer–specific survival (PCSS), and overall survival (OS). All end points were defined from time of surgery until time of the event, death, or last follow-up. The primary analyses in the Decipher GRID cohort were to validate subtype gene expression patterns and associations with biological pathways and clinicopathologic markers in a contemporary cohort.
Results
To subtype prostate cancers into luminal- and basal-like subtypes, we applied the PAM50 classifier to 1567 prostate cancer samples with a median clinical follow-up time of 10 years. A total of 538 samples (34.3%) are classified as luminal A, 447 (28.5%) as luminal B, and 582 (37.1%) as basal, with visually similar patterns of expression across all 6 independent cohorts (Figure 1A and eFigure 1 and eTable 1 in the Supplement). Expression patterns of PAM50 genes are also similar between breast and prostate cancer samples (eFigure 2 in the Supplement). Notably, the estrogen receptor, which is highest in luminal breast cancer tumors, and the progesterone receptor, which is highest in luminal A breast cancer tumors, do not demonstrate the same patterns in prostate cancer (eFigure 2 in the Supplement).
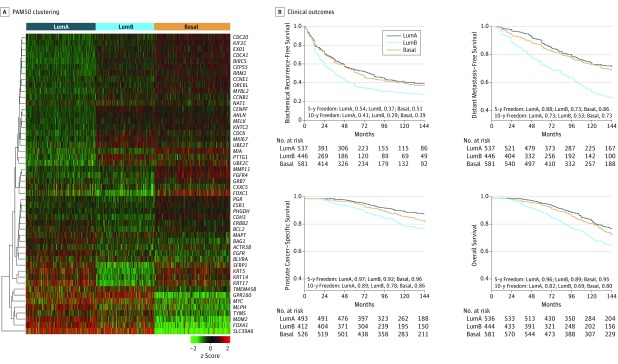
Figure 1. PAM50 Clustering and Clinical Outcomes in Prostate Cancer.
A, The PAM50 genes cluster prostate cancer samples into 3 subtypes, luminal A (LumA), luminal B (LumB), and basal, in the pooled prostate cancer cohorts (Mayo Clinic I and II, Cleveland Clinic, Thomas Jefferson University, Johns Hopkins University, and Durham Veterans Affairs) using hierarchical clustering of the genes. Each column represents a patient sample, and each row represents a gene. B, Kaplan-Meier curves showing that the PAM50 clusters risk stratify biochemical recurrence-free survival, distant metastasis–free survival, prostate cancer–specific survival, and overall survival.
We next examined associations of the luminal A, luminal B, and basal subtypes with clinical outcomes. Patients with luminal B tumors consistently have significantly poorer outcomes for all end points compared with those with luminal A and basal tumors (Figure 1B). The 10-year actuarial rates for bRFS are 29% for luminal B compared with 41% for luminal A and 39% for basal; for DMFS, 53% for luminal B compared with 73% for luminal A and basal subtypes; for PCSS, 78% for luminal B compared with 89% for luminal A and 86% for basal; and for OS, 69% for luminal B vs 82% for luminal A and 80% for basal.
On univariable Cox proportional hazards analysis (Table and eTable 2 in the Supplement), compared with the luminal B subtype, the basal and luminal A subtypes had improved bRFS (basal: hazard ratio [HR], 0.69; 95% CI, 0.59-0.81; P < .001; luminal A: HR, 0.66; 95% CI, 0.57-0.78; P < .001), DMFS (basal: HR, 0.50; 95% CI, 0.40-0.61; P < .001; luminal A: HR, 0.42; 95% CI, 0.34-0.53; P < .001), PCSS (basal: HR, 0.59; 95% CI, 0.44-0.79; P < .001; luminal A: HR, 0.38; 95% CI, 0.27-0.53; P < .001), and OS (basal: HR, 0.69; 95% CI, 0.56-0.85; P < .001; luminal A: HR, 0.56; 95% CI, 0.45-0.70; P < .001). However, the luminal A subtype does not exhibit significantly different bRFS (HR, 1.04; 95% CI, 0.89-1.22; P = .61) or DMFS (HR, 1.17; 95% CI, 0.93-1.49; P = .18) compared with the basal subtype. Luminal A does demonstrate poorer PCSS (HR, 1.54; 95% CI, 1.09-2.16; P = .01) and OS (HR, 1.25; 95% CI, 1.00-1.55; P = .05) compared with basal, although this finding is difficult to interpret in the setting of nonsignificant differences in metastasis and biochemical recurrence. Consistent with our data demonstrating that patients with luminal B tumors have the poorest clinical outcomes, those with luminal B tumors also have the highest preoperative prostate-specific antigen (PSA) levels, Gleason score, and rates of extracapsular extension (ECE) and lymph node invasion (LNI), followed by those with basal and then luminal A tumors (eTable 1 in the Supplement). On multivariable analysis (Table and eTable 2 in the Supplement), adjusting for clinicopathologic variables (age, PSA, Gleason score, positive surgical margin status, ECE, seminal vesicle invasion [SVI], and LNI), patients with basal and luminal A tumors have significantly better independent prognosis than those with luminal B tumors for bRFS (basal: HR, 0.81; 955 CI, 0.69-0.96; P = .01; luminal A: HR, 0.79; 95% CI, 0.66-0.93; P = .005) and DMFS (basal: HR, 0.66; 95% CI, 0.53-0.82; P < .001; luminal A: HR, 0.55; 95% CI, 0.43-0.69; P < .001). Patients with luminal A tumors also have significantly improved outcomes compared with those with luminal B tumors for PCSS (HR, 0.50; 95% CI, 0.35-0.71; P < .001) and OS (HR, 0.69; 95% CI, 0.55-0.87; P = .002). To provide comparison with a composite clinical classifier, we similarly show that the basal and luminal A subtypes have significantly better prognosis than the luminal B subtype for all end points on multivariable analysis adjusting for age, LNI, and the assessment by risk using the classifier by D’Amico et al (eTable 3 in the Supplement).
Table. Univariable and Multivariable Analysis.
| Characteristic | Univariable | Multivariable | ||
|---|---|---|---|---|
| P Value | HR (95% CI) | P Value | HR (95% CI) | |
| Distant Metastasis–Free Survival | ||||
| Age, per 1-y increase | .88 | 1.00 (0.99-1.02) | .15 | 0.99 (0.98-1.00) |
| PSA 10-20 vs <10 | .64 | 1.05 (0.85-1.31) | .29 | 0.89 (0.71-1.11) |
| PSA >20 vs <10 | <.001 | 1.42 (1.12-1.79) | .16 | 0.83 (0.64-1.08) |
| Gleason score 7 vs <7 | <.001 | 4.57 (2.14-9.73) | <.001 | 3.49 (1.63-7.47) |
| Gleason score 8-10 vs <7 | <.001 | 14.32 (6.75-30.37) | <.001 | 8.8 (4.10-18.88) |
| SM | .08 | 1.18 (0.98-1.42) | .74 | 1.03 (0.85-1.25) |
| SVI | <.001 | 2.57 (2.14-3.08) | <.001 | 1.72 (1.39-2.11) |
| ECE | <.001 | 2.04 (1.67-2.50) | .07 | 1.23 (0.99-1.54) |
| LNI | <.001 | 2.56 (2.06-3.19) | .01 | 1.39 (1.09-1.78) |
| Basal vs luminal B | <.001 | 0.50 (0.40-0.61) | <.001 | 0.66 (0.53-0.82) |
| Luminal A vs luminal B | <.001 | 0.42 (0.34-0.53) | <.001 | 0.55 (0.43-0.69) |
| Prostate Cancer–Specific Survival | ||||
| Age, per 1-y increase | .86 | 1.00 (0.98-1.02) | .24 | 0.99 (0.97-1.01) |
| PSA 10-20 vs <10 | .79 | 1.04 (0.76-1.42) | .16 | 0.80 (0.58-1.09) |
| PSA >20 vs <10 | .07 | 1.35 (0.97-1.86) | .01 | 0.62 (0.43-0.89) |
| Gleason score 7 vs <7 | .002 | 3.35 (1.22-9.19) | .06 | 2.70 (0.98-7.46) |
| Gleason score 8-10 vs <7 | <.001 | 13.76 (5.08-37.23) | <.001 | 8.60 (3.12-23.68) |
| SM | <.001 | 1.56 (1.20-2.02) | .11 | 1.25 (0.95-1.64) |
| SVI | <.001 | 3.15 (2.43-4.08) | <.001 | 2.06 (1.53-2.78) |
| ECE | <.001 | 2.22 (1.67-2.96) | .29 | 1.19 (0.87-1.63) |
| LNI | <.001 | 3.19 (2.40-4.25) | <.001 | 1.60 (1.15-2.21) |
| Basal vs luminal B | <.001 | 0.59 (0.44-0.79) | .21 | 0.83 (0.61-1.12) |
| Luminal A vs luminal B | <.001 | 0.38 (0.27-0.53) | <.001 | 0.50 (0.35-0.71) |
Abbreviations: ECE, extracapsular extension; HR, hazard ratio; LNI, lymph node invasion; PSA, prostate-specific antigen; SM, positive surgical margin status; SVI, seminal vesicle invasion.
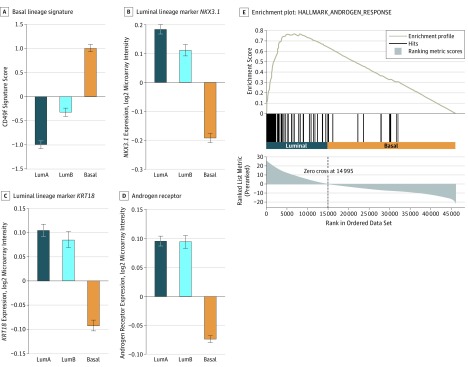
We then investigated the relationship between these subtypes and luminal and basal prostate cancer lineage markers. The basal lineage CD49f signature is increased in basal-like samples (Figure 2A). Concordantly, the luminal markers NKX3.1 (OMIM: 602041), KRT18 (OMIM: 148070), and AR (OMIM: 313700) are increased in luminal-like samples (Figure 2B-C). Consistent with our findings for AR, the androgen activity pathway is enriched in the luminal subtypes compared with the basal subtype (GSEA normalized enrichment score, 3.93; P < .001; Figure 2C). Examining the top GSEA hallmark concepts comparing luminal with basal subtypes (eAppendix in the Supplement) reveals that the MYC pathway is the top enriched pathway in luminal-like samples, and genes downregulated by KRAS are the top positive pathway in basal-like samples (negatively enriched in luminal samples). These results are concordant with MYC (OMIM: 190080) and KRAS (OMIM: 190070) expression, which are both increased in luminal-like samples (eFigure 3 in the Supplement). On observing that proliferation genes, such as MKI67 (OMIM: 176741), are low in luminal A (Figure 1A), we formally examined the subtypes using the PAM50 proliferation score. The luminal A subtype has a lower proliferation score than the luminal B and basal subtypes (eFigure 4 in the Supplement), which may explain the divergent clinical outcomes despite the biological similarities between the luminal A and B subtypes.
Figure 2. Association of Basal and Luminal Subtypes With Basal and Luminal Lineage Markers.
A, Examination of the prostate basal lineage 91-gene CD49f signature shows higher scores in the basal subtype (P < .001). B and C, Conversely, prostate luminal lineage has been characterized by high expression of NKX3.1 (P < .001) and KRT18 (P < .001), and the expression of these genes is higher in the luminal subtypes. D, Androgen receptor expression (also a luminal lineage marker) is increased in the luminal subtypes (P < .001). E, On Gene Set Enrichment Analysis, androgen response targets are positively enriched in the luminal samples (P < .001). Bar graphs show the mean (SE) of median-centered gene expression, and P values are determined via analyses of variance. LumA indicates luminal A subtype; LumB, luminal B subtype.
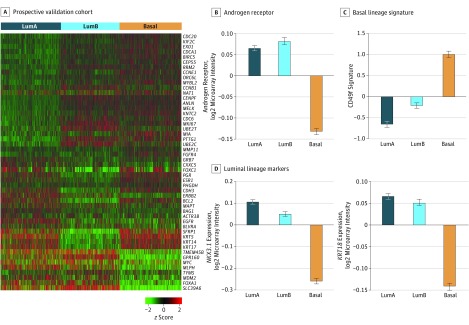
We next independently validated the associations of these subtypes with biological and clinicopathologic factors in Decipher GRID, a prospectively collected cohort of 2215 expression profiles of patients who underwent prostatectomy. The PAM50 gene expression patterns are similar to those in the pooled retrospective cohorts, and trends of AR and AR-signaling (higher in the luminal subtypes), CD49f signature (higher in the basal subtype), and NKX3.1 and KRT19 (both higher in the luminal subtypes) gene expression are conserved (Figure 3). A total of 737 samples (33.3%) are classified as luminal A, 723 (32.6%) as luminal B, and 755 (34.1%) as basal (eTable 4 in the Supplement). We also confirmed MYC and KRAS expression patterns, which are both increased in the luminal subtypes (eFigure 3 in the Supplement). Finally, while clinical outcomes are not available for patients in the Decipher GRID cohort, the luminal B subtype demonstrates the highest Gleason scores, as well as rates of SVI, ECE, and LNI, consistent with clinical outcomes and clinicopathologic data in our retrospective cohorts (eTable 4 in the Supplement). This independent prospective validation increases our confidence that these associations are accurate and applicable in a large contemporary cohort of patients.
Figure 3. Prospective Validation in GRID.
A, PAM50 clusters demonstrate the same pattern of expression in a prospective validation cohort of 2215 prostate cancer samples run on a commercial clinical platform, with 3 subtypes: luminal A (LumA), luminal B (LumB), and basal. B, As in the retrospective cohorts, AR is increased in luminal samples (P < .001). C, The basal lineage CD49f signature is increased in the basal subtype (P < .001). D, NKX3-1 (P < .001) and KRT18 (P < .001) are increased in the luminal subtypes. Bar graphs show the mean (SE) of median-centered gene expression, and P values are determined via analyses of variance.
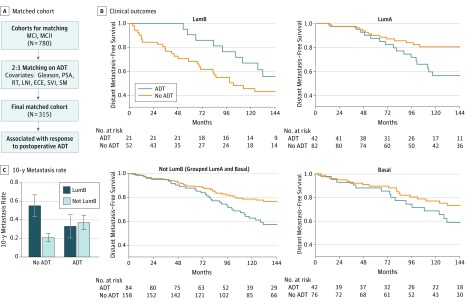
The association between androgen signaling and luminal-like prostate cancer is of particular interest given the importance of ADT in treating prostate cancer. We investigated whether these subtypes could predict response to hormonal therapy in an exploratory subgroup analysis by first designing a postprostatectomy subcohort of patients (n = 315) who were either treated with ADT (n = 105) or not treated with ADT (n = 210) matched by clinicopathologic factors (Gleason score, PSA, LNI, ECE, SVI, and positive surgical margin status), and postoperative radiotherapy (Figure 4A and eTables 5 and 6 and the eAppendix in the Supplement). The matched cohort had a median follow-up of 13 years. In this analysis, we pooled the luminal A and basal subtypes to compare with the luminal B subtype because the luminal A and basal subtypes have similar outcomes for ADT and no ADT in the matched cohorts. In the luminal B subtype, which has the poorest prognosis of the 3 subtypes and contains patients with increased expression of AR-signaling genes, patients treated with ADT had improved DMFS compared with those who did not receive ADT (10-year metastasis rates: ADT, 33% vs no ADT, 55%; Figure 4B and C). However, in the patients with non–luminal B subtypes, patients treated with ADT had poorer DMFS compared with untreated patients (10-year metastasis rates: ADT, 37% vs no ADT, 21%; Figure 4B and C), with a similar trend in patients with the luminal A or basal subtypes. Separating patients receiving adjuvant or salvage therapy in the matched cohort results in similar trends, although the P values are insignificant owing to the reduced numbers (eFigure 5 in the Supplement). Finally, we used interaction analysis in a Cox proportional hazards model of these matched patients to demonstrate a statistically significant interaction term between ADT and the luminal B subtype. Prognostic signatures, such as Decipher and the microarray version of the Cell Cycle and Progression signature, did not predict response to postoperative ADT (eFigure 6 in the Supplement), suggesting that it is not simply more aggressive disease that responds better to postoperative ADT. The luminal B subtype represents a subgroup of prostate cancers with poor prognosis combined with biological differences in AR-signaling that result in improved response to postoperative ADT.
Figure 4. Predicting Response to Androgen Deprivation Therapy (ADT).
A, A matched cohort was obtained from the Mayo Clinic I (MCI) and II (MCII) cohorts, which matched ADT-untreated and ADT-treated patients in a 2:1 ratio based on Gleason score, prostate-specific antigen (PSA), lymph node invasion (LNI), extracapsular extension (ECE), seminal vesicle invasion (SVI), positive surgical margin status (SM), and radiotherapy (RT), resulting in 315 total patients. B, Kaplan-Meier curves for the patients with luminal B (LumB) and non–luminal B subtypes, which group the patients with luminal A (LumA) and basal subtypes. Patients with luminal B subtype who received ADT have lower rates of metastasis than patients who did not receive ADT. However, in the patients with non–luminal B subtype, this trend is reversed. C, Comparison of the 10-year metastasis rates for treated and untreated patients in the patients with luminal B and non–luminal B subtypes (with the interaction term Wald P = .006).
Discussion
We demonstrated in 3782 prostate cancer samples that patients with prostate cancer can be classified into luminal- and basal-like subtypes by the PAM50 algorithm. The luminal A, luminal B, and basal subtypes had consistent gene expression patterns among 6 retrospective cohorts and 1 prospectively collected cohort and were correlated with clinical outcomes. Although PAM50 subtyping has been applied to other tumor types, such as lung and bladder cancer, to our knowledge this is the first reported use in prostate cancer and suggests that luminal and basal features are a unifying biological concept across multiple tumor types. The PAM50 gene expression patterns in prostate cancer demonstrate concordance with breast cancer. This finding is perhaps not surprising given the similarities between prostate and breast cancer. In both tumor types, gonadal steroid hormones (testosterone in prostate and estrogen and progesterone in breast) play a large role in tumor growth and progression. In addition, both tumors respond to antihormonal therapy. Moreover, circulating androgens and estrogens are present in both men and women, and the role of androgens in breast cancer and estrogens in prostate cancer is an area of active research and may indicate further commonalities between the 2 tumor types.
Despite these similarities, we identified that, in prostate cancer, patients with luminal B disease have the poorest prognosis in contrast to the basal subtype in breast cancer. While luminal subtypes are driven by the estrogen and progesterone receptors in breast cancer, luminal subtypes in prostate cancer have increased androgen receptor expression and signaling activity. Furthermore, in breast cancer, luminal subtypes are associated with response to hormonal therapy, which is unsurprising given the high correlation of luminal subtypes and estrogen receptor status. Our findings again parallel this in prostate cancer, as we demonstrated that patients with luminal B tumors benefit more from ADT than do those with non–luminal B tumors.
We did not find the same benefit from ADT for patients with luminal A tumors, perhaps because these patients already have a better prognosis; thus, aggressive treatment may make little difference in the eventual outcome. We also showed that patients with luminal B tumors have an elevated PAM50 proliferation score compared with those with luminal A tumors. Proliferation genes, such as MKI67, have been associated with poorer prognosis in both prostate and breast cancer. The high proliferation score may be in part why patients with luminal B tumors tend to have poorer clinical outcomes compared with those with luminal A tumors despite their biological similarities. Furthermore, one effect of ADT is to reduce proliferation in androgen-sensitive prostate cancers. Thus, luminal B tumors may represent a high-proliferation, androgen-driven subset, which may explain why they derive the most benefit from ADT. Notably, the luminal B subtype has a proliferation score similar to the basal subtype, indicating that proliferation genes alone are not the defining drivers of poor prognosis and the association with response to ADT.
Androgen deprivation therapy forms the backbone of treatment of metastatic prostate cancer and has been shown to add to the effect of radiotherapy. However, in a systematic review, adjuvant ADT in the postoperative setting has not been shown to improve overall survival, suggesting that perhaps only a subset of men have androgen-responsive tumors that are also aggressive enough to require additional postoperative therapy. In our study, we demonstrated that not only does the luminal B subtype appear to identify such a subset from retrospectively collected patients, but it also can identify these men among contemporary patients undergoing radical prostatectomy on a commercially available genome-wide expression analysis that was ordered as part of their routine clinical care. Prior work in this area has focused on identifying clinical and genomic markers for response to ADT and has identified clinical factors, such as Gleason score, PSA, and metastatic disease status, as well as certain genetic polymorphisms that are associated with outcomes in men treated with ADT. However, Gleason score, PSA, and metastatic disease status are prognostic factors in all men, even those who do not receive ADT. Because these studies did not include a similar group of men who were not treated with ADT, it is difficult to ascertain whether these variables are predictive for response to ADT or if they are simply prognostic markers in all prostate cancers regardless of ADT treatment.
Limitations
This study has some limitations. Owing to the retrospective nature of the cohorts, treatment selection was inevitably affected by baseline risk. Although we attempted to adjust for baseline risk with strict matching criteria, in the non–luminal B arm of the matched cohort, patients treated with ADT had poorer DMFS, indicating that the matching was not adjusted perfectly for all treatment selection confounders. Also, we grouped adjuvant and salvage ADT together because stratification by these variables separately in the matching process would have reduced the number of patients and statistical power even further. Therefore, these results should be considered hypothesis generating and should be independently validated, ideally in a randomized clinical trial. We are currently in the process of obtaining samples from one such trial, RTOG 96-01, which will allow us to definitively test this hypothesis.
Despite the fact that the PAM50 algorithm was derived in breast cancer, its classification of basal and luminal subtypes in prostate cancer samples is correlated with known prostate luminal and basal markers. We have established that the basal-like subtype is associated with the basal lineage CD49f signature and that the luminal-like subtype is associated with the luminal markers NKX3.1, KRT18, and AR. A recent study by Zhang et al described a 100-gene set representing the 50 most overexpressed genes in basal and luminal benign prostate cells. However, when sorting our samples into luminal and basal subtypes based on these 100 genes, we did not find an association with clinical outcomes in our data set (eFigure 7 in the Supplement). These differences with our findings may be due to the biological differences in benign tissue used in the study by Zhang et al vs the malignant prostate cancer samples in our study. These 100 genes were also nominated based on samples from only 3 patients, which may not adequately capture the heterogeneity across prostate cancer. In contrast, the PAM50 clustering was derived specifically in malignant tissue and has been widely validated in breast cancer, bladder cancer, and now in several thousand prostate tumors. Nonetheless, the work by Zhang et al further illustrates the interest in the field of exploring the biological and clinical significance of luminal and basal prostate cancer.
Although the luminal A and basal subtypes are similar to each other in clinical outcomes, they are divergent with respect to basal and luminal lineage markers and androgen receptor signaling, as well as the oncogenic drivers MYC and KRAS, in which luminal A is much more similar to luminal B. The luminal A and basal subtypes also differ in proliferation scores. Our findings suggest that luminal A and basal subtypes are biologically distinct. Although these differences do not always translate into differences in prognosis (eg, ERG [OMIM: 165080] positive vs negative), they are nevertheless important in understanding disease biology and potentially for therapeutic selection.
Conclusions
We have illustrated the clinical and genetic differences between luminal and basal subtypes in prostate cancer across nearly 4000 samples from retrospective and prospective cohorts on a commercial, high-throughput clinical platform. We believe this work not only represents a significant step forward in our understanding of prostate cancer heterogeneity but also is a potential classifier that may identify patients who benefit from postoperative ADT on a clinical-grade platform and provide guidance in personalizing patient care.
eAppendix. Methods
eFigure 1. PAM50 in Retrospective Prostate Cohorts
eFigure 2. PAM50 in Breast vs Prostate Cancer
eFigure 3. Luminal Subtypes Are Associated With MYC and KRAS
eFigure 4. PAM50 Proliferation Score Across Subtypes
eFigure 5. Survival Outcomes for the Matched Cohort, Separating Patients Receiving Adjuvant and Salvage ADT
eFigure 6. Decipher and mCCP Are Not Predictive for Response to Postoperative ADT in the Matched Cohort
eFigure 7. A Previously Published 100-Gene Set Is Not Associated With Clinical Outcomes in Prostate Cancer
eTable 1. Demographics for Pooled Retrospective Cohort (n = 1567)
eTable 2. Univariable and Multivariable Analysis in Pooled Retrospective Cohort (n = 1567)
eTable 3. Univariable and Multivariable Analysis in Pooled Retrospective Cohort (n = 1567) to Examine Independence of Subtypes from D’Amico Risk Classification
eTable 4. Demographics for GRID (n = 2215)
eTable 5. Demographics for Matched Cohort (n = 315)
eTable 6. Number of Patients Receiving ADT and RT in the Matched Cohort (n = 315)
References
- 1.Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON. Identification of a cell of origin for human prostate cancer. Science. 2010;329(5991):568-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi N, Zhang B, Zhang L, Ittmann M, Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell. 2012;21(2):253-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Kruithof-de Julio M, Economides KD, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461(7263):495-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith BA, Sokolov A, Uzunangelov V, et al. A basal stem cell signature identifies aggressive prostate cancer phenotypes. Proc Natl Acad Sci U S A. 2015;112(47):E6544-E6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang ZA, Toivanen R, Bergren SK, Chambon P, Shen MM. Luminal cells are favored as the cell of origin for prostate cancer. Cell Rep. 2014;8(5):1339-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoyanova T, Cooper AR, Drake JM, et al. Prostate cancer originating in basal cells progresses to adenocarcinoma propagated by luminal-like cells. Proc Natl Acad Sci U S A. 2013;110(50):20111-20116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi W, Porten S, Kim S, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25(2):152-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747-752. [DOI] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallden B, Storhoff J, Nielsen T, et al. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med Genomics. 2015;8:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chia SK, Bramwell VH, Tu D, et al. A 50-gene intrinsic subtype classifier for prognosis and prediction of benefit from adjuvant tamoxifen. Clin Cancer Res. 2012;18(16):4465-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damrauer JS, Hoadley KA, Chism DD, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci U S A. 2014;111(8):3110-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffey DS. Similarities of prostate and breast cancer: evolution, diet, and estrogens. Urology. 2001;57(4)(suppl 1):31-38. [DOI] [PubMed] [Google Scholar]
- 14.López-Otín C, Diamandis EP. Breast and prostate cancer: an analysis of common epidemiological, genetic, and biochemical features. Endocr Rev. 1998;19(4):365-396. [DOI] [PubMed] [Google Scholar]
- 15.Risbridger GP, Davis ID, Birrell SN, Tilley WD. Breast and prostate cancer: more similar than different. Nat Rev Cancer. 2010;10(3):205-212. [DOI] [PubMed] [Google Scholar]
- 16.Erho N, Crisan A, Vergara IA, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8(6):e66855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karnes RJ, Bergstralh EJ, Davicioni E, et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol. 2013;190(6):2047-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakagawa T, Kollmeyer TM, Morlan BW, et al. A tissue biomarker panel predicting systemic progression after PSA recurrence post–definitive prostate cancer therapy. PLoS One. 2008;3(5):e2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein EA, Yousefi K, Haddad Z, et al. A genomic classifier improves prediction of metastatic disease within 5 years after surgery in node-negative high-risk prostate cancer patients managed by radical prostatectomy without adjuvant therapy. Eur Urol. 2015;67(4):778-786. [DOI] [PubMed] [Google Scholar]
- 20.Prensner JR, Zhao S, Erho N, et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 2014;15(13):1469-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross AE, Johnson MH, Yousefi K, et al. Tissue-based genomics augments post-prostatectomy risk stratification in a natural history cohort of intermediate- and high-risk men. Eur Urol. 2016;69(1):157-165. [DOI] [PubMed] [Google Scholar]
- 22.Den RB, Feng FY, Showalter TN, et al. Genomic prostate cancer classifier predicts biochemical failure and metastases in patients after postoperative radiation therapy. Int J Radiat Oncol Biol Phys. 2014;89(5):1038-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freedland SJ, Choeurng V, Howard L, et al. Utilization of a genomic classifier for prediction of metastasis following salvage radiation therapy after radical prostatectomy. Eur Urol. 2016;70(4):588-596. [DOI] [PubMed] [Google Scholar]
- 24.Den RB, Santiago-Jimenez M, Alter J, et al. Decipher correlation patterns post prostatectomy: initial experience from 2 342 prospective patients. Prostate Cancer Prostatic Dis. 2016;19(4):374-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alshalalfa M, Verhaegh GW, Gibb EA, et al. Low PCA3 expression is a marker of poor differentiation in localized prostate tumors: exploratory analysis from 12,076 patients [published online February 7, 2017]. Oncotarget. 2017. doi: 10.18632/oncotarget.15133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benzon B, Zhao SG, Haffner MC, et al. Correlation of B7-H3 with androgen receptor, immune pathways and poor outcome in prostate cancer: an expression-based analysis. Prostate Cancer Prostatic Dis. 2017;20(1):28-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piccolo SR, Sun Y, Campbell JD, Lenburg ME, Bild AH, Johnson WE. A single-sample microarray normalization method to facilitate personalized-medicine workflows. Genomics. 2012;100(6):337-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ullén A, Lennartsson L, Harmenberg U, et al. Prostate cancer cell lines lack amplification: overexpression of HER2. Acta Oncol. 2005;44(5):490-495. [DOI] [PubMed] [Google Scholar]
- 30.D’Amico AV, Whittington R, Malkowicz SB, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280(11):969-974. [DOI] [PubMed] [Google Scholar]
- 31.Subramanian A, Tamayo P, Mootha VK, et al. Gene Set Enrichment Analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuzick J, Berney DM, Fisher G, et al. ; Transatlantic Prostate Group . Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br J Cancer. 2012;106(6):1095-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siegfried JM, Lin Y, Diergaarde B, et al. Expression of PAM50 genes in lung cancer: evidence that interactions between hormone receptors and HER2/HER3 contribute to poor outcome. Neoplasia. 2015;17(11):817-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrelli F, Viale G, Cabiddu M, Barni S. Prognostic value of different cut-off levels of Ki-67 in breast cancer: a systematic review and meta-analysis of 64,196 patients. Breast Cancer Res Treat. 2015;153(3):477-491. [DOI] [PubMed] [Google Scholar]
- 35.Agus DB, Cordon-Cardo C, Fox W, et al. Prostate cancer cell cycle regulators: response to androgen withdrawal and development of androgen independence. J Natl Cancer Inst. 1999;91(21):1869-1876. [DOI] [PubMed] [Google Scholar]
- 36.Loblaw DA, Virgo KS, Nam R, et al. ; American Society of Clinical Oncology . Initial hormonal management of androgen-sensitive metastatic, recurrent, or progressive prostate cancer: 2006 update of an American Society of Clinical Oncology practice guideline. J Clin Oncol. 2007;25(12):1596-1605. [DOI] [PubMed] [Google Scholar]
- 37.Denham JW, Steigler A, Lamb DS, et al. ; Trans-Tasman Radiation Oncology Group . Short-term androgen deprivation and radiotherapy for locally advanced prostate cancer: results from the Trans-Tasman Radiation Oncology Group 96.01 randomised controlled trial. Lancet Oncol. 2005;6(11):841-850. [DOI] [PubMed] [Google Scholar]
- 38.Pilepich MV, Winter K, Lawton CA, et al. Androgen suppression adjuvant to definitive radiotherapy in prostate carcinoma—long-term results of phase III RTOG 85-31. Int J Radiat Oncol Biol Phys. 2005;61(5):1285-1290. [DOI] [PubMed] [Google Scholar]
- 39.Kumar S, Shelley M, Harrison C, Coles B, Wilt TJ, Mason MD. Neo-adjuvant and adjuvant hormone therapy for localised and locally advanced prostate cancer. Cochrane Database Syst Rev. 2006;(4):CD006019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross RW, Oh WK, Xie W, et al. Inherited variation in the androgen pathway is associated with the efficacy of androgen-deprivation therapy in men with prostate cancer. J Clin Oncol. 2008;26(6):842-847. [DOI] [PubMed] [Google Scholar]
- 41.Ross RW, Xie W, Regan MM, et al. Efficacy of androgen deprivation therapy (ADT) in patients with advanced prostate cancer: association between Gleason score, prostate-specific antigen level, and prior ADT exposure with duration of ADT effect. Cancer. 2008;112(6):1247-1253. [DOI] [PubMed] [Google Scholar]
- 42.Ballman KV. Biomarker: predictive or prognostic? J Clin Oncol. 2015;33(33):3968-3971. [DOI] [PubMed] [Google Scholar]
- 43.Shipley WU, Seiferheld W, Lukka HR, et al. ; NRG Oncology RTOG . Radiation with or without antiandrogen therapy in recurrent prostate cancer. N Engl J Med. 2017;376(5):417-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang D, Park D, Zhong Y, et al. Stem cell and neurogenic gene-expression profiles link prostate basal cells to aggressive prostate cancer. Nat Commun. 2016;7:10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomlins SA, Alshalalfa M, Davicioni E, et al. Characterization of 1577 primary prostate cancers reveals novel biological and clinicopathologic insights into molecular subtypes. Eur Urol. 2015;68(4):555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eAppendix. Methods
eFigure 1. PAM50 in Retrospective Prostate Cohorts
eFigure 2. PAM50 in Breast vs Prostate Cancer
eFigure 3. Luminal Subtypes Are Associated With MYC and KRAS
eFigure 4. PAM50 Proliferation Score Across Subtypes
eFigure 5. Survival Outcomes for the Matched Cohort, Separating Patients Receiving Adjuvant and Salvage ADT
eFigure 6. Decipher and mCCP Are Not Predictive for Response to Postoperative ADT in the Matched Cohort
eFigure 7. A Previously Published 100-Gene Set Is Not Associated With Clinical Outcomes in Prostate Cancer
eTable 1. Demographics for Pooled Retrospective Cohort (n = 1567)
eTable 2. Univariable and Multivariable Analysis in Pooled Retrospective Cohort (n = 1567)
eTable 3. Univariable and Multivariable Analysis in Pooled Retrospective Cohort (n = 1567) to Examine Independence of Subtypes from D’Amico Risk Classification
eTable 4. Demographics for GRID (n = 2215)
eTable 5. Demographics for Matched Cohort (n = 315)
eTable 6. Number of Patients Receiving ADT and RT in the Matched Cohort (n = 315)