Abstract
Drosophila melanogaster Bag-of-marbles (Bam) promotes germline stem cell (GSC) differentiation by repressing the expression of mRNAs encoding stem cell maintenance factors. Bam interacts with Benign gonial cell neoplasm (Bgcn) and the CCR4 deadenylase, a catalytic subunit of the CCR4–NOT complex. Bam has been proposed to bind CCR4 and displace it from the CCR4–NOT complex. Here, we investigated the interaction of Bam with the CCR4–NOT complex by using purified recombinant proteins. Unexpectedly, we found that Bam does not interact with CCR4 directly but instead binds to the CAF40 subunit of the complex in a manner mediated by a conserved N-terminal CAF40-binding motif (CBM). The crystal structure of the Bam CBM bound to CAF40 reveals that the CBM peptide adopts an α-helical conformation after binding to the concave surface of the crescent-shaped CAF40 protein. We further show that Bam-mediated mRNA decay and translational repression depend entirely on Bam's interaction with CAF40. Thus, Bam regulates the expression of its mRNA targets by recruiting the CCR4–NOT complex through interaction with CAF40.
Keywords: deadenylation, mRNA decay, translational repression, germ cell differentiation
INTRODUCTION
The CCR4–NOT deadenylase complex is a major downstream effector complex in post-transcriptional mRNA regulation in eukaryotes (Wahle and Winkler 2013). Beyond its role in global mRNA degradation, the complex regulates the expression of a large number of specific mRNAs, to which it is recruited via interactions with RNA-associated proteins. These proteins include the GW182 family, which is involved in miRNA-mediated gene silencing in animals (Chen et al. 2014a; Mathys et al. 2014); tristetraprolin (TTP), a protein required for the degradation of mRNAs containing AU-rich elements (Fabian et al. 2013); the germline determinant Nanos (Suzuki et al. 2012; Bhandari et al. 2014; Raisch et al. 2016); and human (Hs) and Drosophila melanogaster (Dm) Roquin proteins (Leppek et al. 2013; Sgromo et al. 2017).
In metazoans, the CCR4–NOT complex comprises a core of seven proteins, which bind to independently folding α-helical domains in the central NOT1 scaffold subunit, forming four subcomplexes or modules: the catalytic module, the CAF40 module, the NOT module, and the NOT10-NOT11 module (Wahle and Winkler 2013). The catalytic module comprises two deadenylases, namely CAF1 or its paralog POP2 (also known as CNOT7 and CNOT8, respectively, in humans), and CCR4a or its paralog CCR4b (also known as CNOT6 and CNOT6L, respectively, in humans). CAF1 (or POP2) binds to a central domain of NOT1 that is structurally related to the middle portion of eIF4G (termed the NOT1 MIF4G domain) (Basquin et al. 2012; Petit et al. 2012). CAF1 or POP2 also bind to a leucine-rich repeat domain (LRR) in CCR4a/b, thus bridging the interaction of CCR4 paralogs with NOT1, and consequently with the assembled CCR4–NOT complex (Draper et al. 1994, 1995; Dupressoir et al. 2001; Basquin et al. 2012; Petit et al. 2012; Bawankar et al. 2013). The NOT1 MIF4G domain also serves as a binding platform for the DEAD-box protein DDX6 (also known as RCK), which functions as a translational repressor and decapping activator (Chen et al. 2014a; Mathys et al. 2014). The CAF40 module consists of the highly conserved CAF40 subunit (also known as NOT9) bound to the NOT1 CAF40/NOT9-binding domain (CN9BD), which is located C-terminal to the MIF4G domain (Chen et al. 2014a; Mathys et al. 2014). The NOT module consists of the NOT2–NOT3 heterodimer bound to the C-terminal NOT1 superfamily homology domain SHD (Bhaskar et al. 2013; Boland et al. 2013), whereas the NOT10 and NOT11 subunits bind to the N-terminal end of NOT1 (Bawankar et al. 2013; Mauxion et al. 2013).
Bag-of-marbles (Bam) is a key differentiation factor that determines the fate of germline stem cells (GSCs) (Cooley et al. 1988; McKearin and Spradling 1990; Carreira and Buszczak 2014). Loss of Bam results in uncontrolled stem cell proliferation, thus giving rise to germ cell tumors that characterize the mutant phenotype (McKearin and Ohlstein 1995). In contrast, ectopic Bam expression causes stem cell loss (Ohlstein and McKearin 1997). Bam is conserved in Drosophila and other dipteran species and contains several predicted α-helices (Fig. 1A), thus suggesting that it is mainly a folded protein. However, Bam does not display detectable similarity to other known proteins or domains.
FIGURE 1.
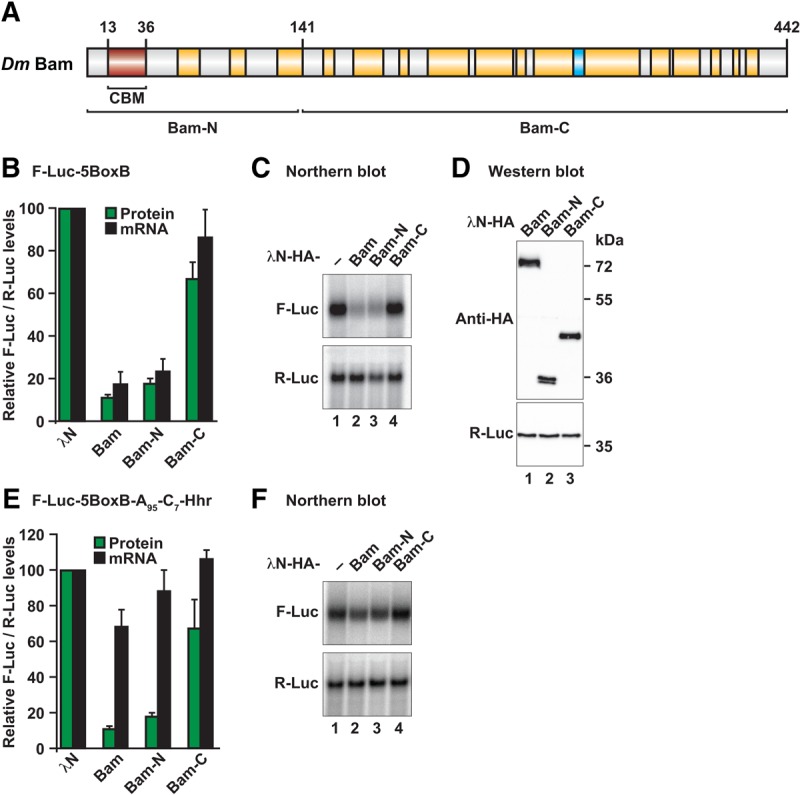
Bam induces degradation of bound mRNAs through its N-terminal region. (A) Bam consists of several predicted α-helices (shown in yellow) and a β-strand (shown in cyan). The position of the CAF40-binding motif (CBM, in red) as well as the boundaries of the Bam-N and Bam-C fragments are indicated. (B) Tethering assay using the F-Luc-5BoxB reporter and λN-HA-tagged Bam (full-length or the indicated fragments) in Dm S2 cells. A plasmid expressing R-Luc mRNA served as a transfection control. For each experiment, F-Luc activity and mRNA levels were normalized to those of the R-Luc transfection control and set to 100% in cells expressing the λN-HA peptide. (C) Northern blot of representative RNA samples shown in B. (D) Western blot analysis showing the equivalent expression of the λN-HA-tagged proteins used in the tethering assays shown in B and C. Protein size markers (kDa) are shown on the right of the panel. Full-length Bam and Bam-N display an aberrant mobility in SDS–PAGE, thus resulting in a higher apparent molecular weight. (E,F) Tethering assay using the F-Luc-5BoxB-A95-C7-HhR reporter and λN-HA-tagged Bam (full-length or the indicated fragments) in Dm S2 cells. The samples were analyzed as described in B and C. In B and E, bars represent mean values and error bars represent standard deviations from at least three independent experiments.
Bam controls GSC differentiation by post-transcriptionally repressing the expression of nanos and E-cadherin mRNAs (Li et al. 2009). Bam function requires the assembly of a protein complex, which includes Benign gonial cell neoplasm (Bgcn), a putative DEXH RNA helicase protein, and additional proteins such as Tumorous testis (Tut) (Chen et al. 2014b), Sex-lethal (Sxl) (Chau et al. 2012) and Mei-P26 (Neumüller et al. 2008; Li et al. 2013). All of these proteins have been implicated in germ cell differentiation in Dm, but their individual contributions to mRNA binding and repression, as well as their interaction modes are not well understood. Bam has also been shown to interact with the translation initiation factor eIF4A and to antagonize its role in translation initiation (Shen et al. 2009).
Although the mechanism through which Bam-containing complexes repress the expression of specific mRNA targets has not been fully elucidated, it apparently involves interaction with the CCR4 deadenylase subunit of the CCR4–NOT complex (Fu et al. 2015). Bam has been proposed to compete with CAF1/POP2 for direct binding to the CCR4 LRR domain, thereby displacing CCR4 from the CCR4–NOT complex. In this model, CCR4 participates in Bam-mediated repression as an isolated deadenylase and not as an integral component of the CCR4–NOT complex. The model was proposed on the basis of the observation that mutations in the CCR4 LRR domain disrupt binding to both Bam and CAF1/POP2 (Fu et al. 2015). However, the mutated residues are located in the hydrophobic core of the LRR domain (Basquin et al. 2012) and most probably destabilize the domain fold. Therefore, it remains unclear whether free CCR4 or the assembled CCR4–NOT complex is required for Bam-mediated repression.
In the present study, we investigated the role of the CCR4–NOT complex in Bam-mediated mRNA regulation. We found that Bam promotes translational repression and degradation of bound mRNAs and that these activities depend on the N-terminal region of Bam, which does not contain the previously identified Bgcn-binding region and putative CCR4-binding site (Supplemental Fig. S1; Pan et al. 2014; Fu et al. 2015). We further show that this N-terminal region contains a CAF40-binding motif (CBM) that interacts directly with CAF40. A crystal structure of the Bam CBM peptide bound to CAF40 reveals a binding mode similar to that observed for the Dm Roquin CBM (Sgromo et al. 2017). However, in contrast to Dm Roquin, which recruits the CCR4–NOT complex through multiple redundant binding sites, Bam relies entirely on the interaction with CAF40. Disruption of the Bam–CAF40 interaction also disrupts the interaction with CCR4 and the additional subunits of the CCR4–NOT complex and abolishes Bam activity. Thus, Bam recruits the assembled CCR4–NOT deadenylase complex through a direct interaction with CAF40 and this interaction is essential for Bam to repress bound mRNAs.
RESULTS
The Bam N-terminal region mediates translational repression and degradation of mRNA targets
Bam promotes stem cell differentiation by repressing the expression of specific mRNA targets through a mechanism that involves interaction with the CCR4 deadenylase (Fu et al. 2015). To obtain detailed mechanistic insights into this repressive mechanism and more precisely define the Bam sequences responsible for its repressive activity, we used a λN-based tethering assay in Drosophila melanogaster S2 cells (Behm-Ansmant et al. 2006). On the basis of sequence alignments, we designed Bam N- and C-terminal fragments for tethering assays (Fig. 1A; Supplemental Fig. S1; Supplemental Table S1). Full-length Bam and the Bam-N and Bam-C fragments were expressed with λN-HA tags that bind to a coexpressed firefly luciferase mRNA reporter containing five λN-binding sites (BoxB hairpins) in the 3′ UTR (F-Luc-5BoxB mRNA). An mRNA encoding Renilla luciferase (R-Luc) served as a transfection control.
λN-HA-tagged Bam decreased the F-Luc expression level to 10% relative to the λN-HA fusion protein, which was used as a negative control (Fig. 1B). The decrease in F-Luc activity was predominantly explained by a corresponding decrease in the mRNA abundance (Fig. 1B,C) and a shortening of the mRNA half-life (Supplemental Fig. S2A,B), thus indicating that Bam induces mRNA degradation in S2 cells. Furthermore, the Bam-N fragment retained the activity of the full-length protein, whereas the activity of the Bam-C fragment was strongly impaired (Fig. 1B,C). All proteins were expressed at comparable levels (Fig. 1D), and none of the proteins affected the expression of an F-Luc reporter lacking the BoxB hairpins (Supplemental Fig. S2C,D), thus confirming that Bam must bind to the mRNA to induce degradation.
To determine whether Bam might repress translation independently of mRNA degradation, we used an mRNA reporter containing a 3′ end generated by a self-cleaving hammerhead ribozyme (HhR) that consequently lacks a poly(A) tail (F-Luc-5BoxB-A95C7-HhR) (Zekri et al. 2013). Additionally, the reporter contains an internal, DNA-encoded, oligo(A) stretch of 95 nucleotides and a 3′ oligo(C) stretch of seven nucleotides upstream of the ribozyme cleavage site. This reporter is resistant to deadenylation and subsequent degradation and is efficiently translated in S2 cells (Zekri et al. 2013). Full-length Bam and the Bam-N fragment repressed the expression of this reporter in S2 cells (Fig. 1E,F). This repression occurred mainly at the translational level, because mRNA levels were not decreased to a similar extent as with the polyadenylated reporter. Together, our results indicated that Bam promotes the degradation of polyadenylated mRNAs and also represses translation independently of mRNA degradation when deadenylation is blocked. Furthermore, the Bam activity resides primarily in the Bam-N fragment, which does not contain the putative CCR4-binding region (Supplemental Fig. S1; Fu et al. 2015).
Bam directs bound mRNAs to the 5′-to-3′ decay pathway
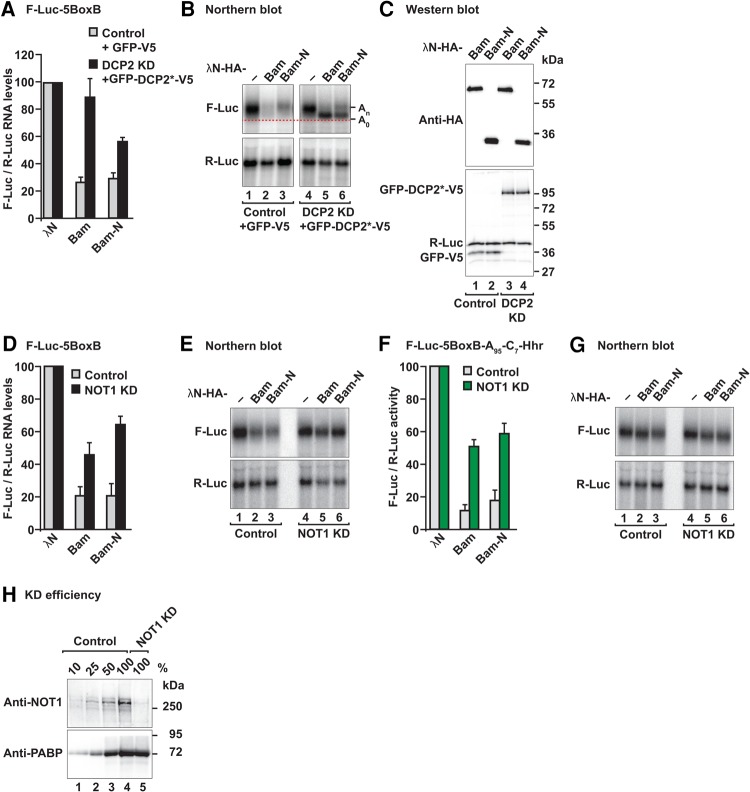
We next investigated whether Bam elicits mRNA degradation via the 5′-to-3′ decay pathway. In this pathway, deadenylation is followed by decapping and 5′-to-3′ exonucleolytic degradation of the mRNA body. We therefore performed tethering assays in S2 cells depleted of the decapping enzyme DCP2 and overexpressing a catalytically inactive DCP2 mutant (DCP2 E361Q), which inhibits decapping in a dominant negative manner (Chang et al. 2014). In these cells, degradation of the F-Luc-5BoxB mRNA by tethered Bam or the Bam-N fragment was impaired (Fig. 2A). The F-Luc-5BoxB mRNA accumulated as a fast-migrating form corresponding to a deadenylated decay intermediate (A0; Fig. 2B, lanes 5 and 6). Despite the restoration of mRNA levels, F-Luc activity was not restored (Supplemental Fig. S2E), most likely because deadenylated transcripts are translated less efficiently. The expression of the tethered proteins was not affected by coexpression of the DCP2 mutant (Fig. 2C). Together, these results indicated that Bam directs mRNA targets to the 5′-to-3′ decay pathway.
FIGURE 2.
Bam degrades mRNAs through the 5′-to-3′ mRNA decay pathway. (A) Tethering assay using the F-Luc-5BoxB reporter was performed in control S2 cells or cells depleted of the decapping enzyme DCP2 (DCP2 KD). The transfection mixture included plasmids expressing either GFP-V5 or a GFP-tagged catalytically inactive DCP2 mutant (DCP2*; E361Q). The F-Luc-5BoxB mRNA levels were normalized to those of the R-Luc transfection control and set to 100% in control and knockdown cells expressing the λN-HA peptide. The gray bars represent the normalized F-Luc-5BoxB mRNA values in control cells expressing GFP-V5. The black bars represent the normalized F-Luc-5BoxB mRNA values in DCP2-depleted cells expressing GFP-DCP2*-V5. (B) Northern blot of representative RNA samples shown in A. The positions of the polyadenylated (An) and deadenylated (A0, dashed red line) mRNA reporter are indicated on the right of the panel. (C) Western blot analysis showing equivalent expression of λN-HA-tagged proteins in the experiments shown in A and B. (KD) Knockdown. (D,E) Tethering assay using the F-Luc-5BoxB reporter in control S2 cells or in NOT1-depleted cells. Samples were analyzed as described in Figure 1B–D. (F,G) Tethering assay using the F-Luc-5BoxB-A95-C7-HhR reporter in control cells and in NOT1-depleted cells. Samples were analyzed as described in Figure 1B–D. In A, D, and F, bars represent mean values and error bars represent standard deviations from at least three independent experiments. (H) Western blot analysis showing the efficiency of NOT1 depletion in the experiments shown in D–G. Dilutions of control cell lysates were loaded in lanes 1–4. PABP served as a loading control. Protein size markers (kDa) are shown on the right in each panel.
Bam recruits the CCR4–NOT complex to induce mRNA degradation
Our results indicated that Bam promotes deadenylation-dependent decapping. To determine whether Bam-mediated deadenylation requires the assembled CCR4–NOT complex or, alternatively, whether only the CCR4 subunit acts in isolation, as suggested previously (Fu et al. 2015), we disrupted CCR4–NOT complex assembly by depleting NOT1, the scaffold subunit of the complex (Wahle and Winkler 2013). NOT1 depletion partially suppressed degradation of F-Luc-5BoxB mRNA mediated by Bam and Bam-N (Fig. 2D,E; Supplemental Fig. S2F), thus suggesting that the assembled CCR4–NOT complex is required for Bam's repressive activity. Furthermore, NOT1 depletion also suppressed Bam-mediated translational repression of the reporter that was resistant to deadenylation and decay (F-Luc-5BoxB-A95C7-HhR; Fig. 2F,G; Supplemental Fig. S2G). Western blot analysis indicated that NOT1 levels were indeed decreased to <25% of the control levels in the knockdown cells (Supplemental Fig. S2H).
Because Bam activity depends on the integrity of the CCR4–NOT complex and it resides in the N-terminal fragment, which does not contain the putative CCR4-binding region (Fu et al. 2015), we re-examined Bam interactions with subunits of the CCR4–NOT complex. We expressed Bam with an HA tag in S2 cells and tested for interactions with GFP-tagged subunits of the CCR4–NOT complex in coimmunoprecipitation assays. Bam interacted with NOT1, NOT2, NOT3, CCR4, and CAF40 (Supplemental Fig. S3A–E), thus suggesting that it interacts with the assembled CCR4–NOT complex. All of these interactions were observed in the presence of RNaseA. Together, our results indicated that the CCR4–NOT complex is an important downstream effector of Bam-mediated mRNA regulation.
Bam interacts with the CAF40 subunit of the CCR4–NOT complex
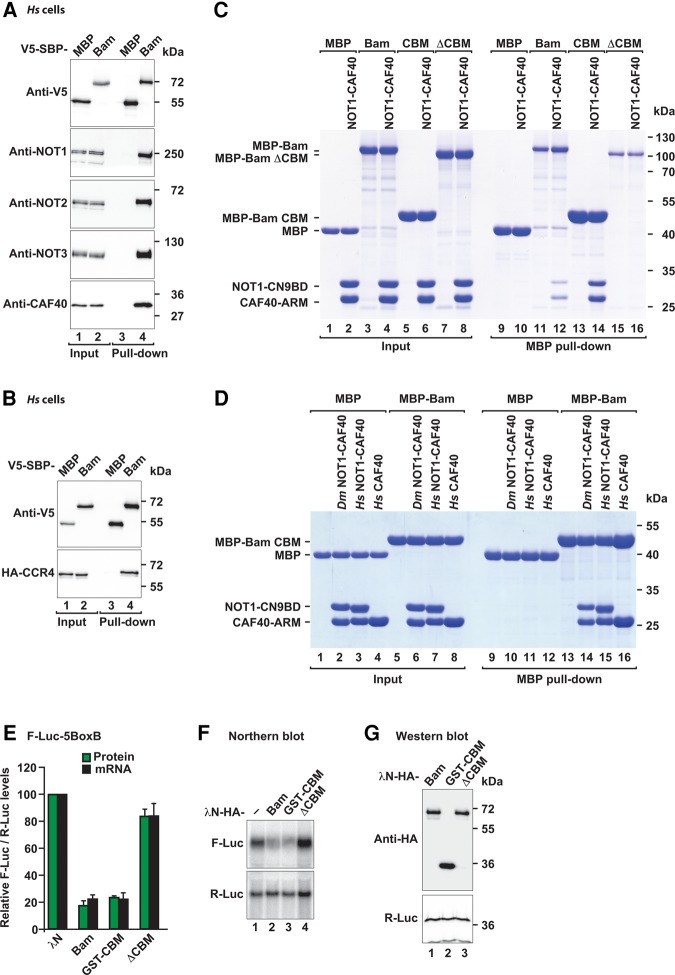
To discriminate between direct and indirect interactions between Bam and the subunits of the CCR4–NOT complex, we performed pull-down assays in vitro, using purified recombinant proteins expressed in Escherichia coli. Because some Dm NOT1 domains are not expressed in a soluble form in bacteria, we first tested whether Bam could also interact with the human CCR4–NOT complex. To this end, we expressed Bam with a V5-SBP tag in human HEK293T cells and tested for interactions with endogenous subunits of the CCR4–NOT complex in pull-down assays. Bam pulled down all of the tested subunits of the endogenous CCR4–NOT complex (NOT1, NOT2, NOT3 and CAF40; Fig. 3A, lane 4) as well as HA-tagged CCR4 (Fig. 3B, lane 4) in the presence of RNaseA, thereby indicating that the Bam-binding surface on the CCR4–NOT complex is conserved.
FIGURE 3.
Bam binds directly to CAF40 by using an N-terminal CAF40-binding motif (CBM). (A) SBP pull-down assay in HEK293T cells expressing V5-SBP-tagged full-length Bam. V5-SBP-tagged MBP served as negative control. Input (1.5% for the V5-SBP tagged proteins and 1% for endogenous CCR4–NOT subunits) and bound fractions (10% for the V5-SBP tagged proteins and 30% for the CCR4–NOT subunits) were analyzed by western blotting using the indicated antibodies. (B) SBP pull-down assay in HEK293T cells expressing V5-SBP-tagged full-length Bam and HA-CCR4. V5-SBP-tagged MBP served as negative control. Samples were analyzed as described in A. (C) MBP pull-down assay testing the interaction of MBP-tagged full-length Bam, the CBM or Bam ΔCBM with the Dm CN9BD–CAF40 complex. MBP served as a negative control. The inputs (10%) and bound fractions (50%) were analyzed by SDS–PAGE and subsequent Coomassie staining. (D) MBP pull-down assay showing the interaction of MBP-tagged Bam CBM with the Dm and Hs CN9BD–CAF40 complex and Hs CAF40. Samples were analyzed as in C. (E) Tethering assay using the F-Luc-5BoxB reporter and λN-HA-tagged Bam (full-length or the indicated fragments) in S2 cells. The samples were analyzed as described in Figure 1B–D. The mean values ±SD from three independent experiments are shown. (F) Northern blot of representative RNA samples shown in E. (G) Western blot showing the equivalent expression of λN-HA-tagged proteins used in E and F. Protein size markers (kDa) are shown on the right in each panel.
This result allowed us to test for interactions with individual purified human CCR4–NOT subcomplexes in vitro, including the NOT1-10-11 module, the catalytic module comprising the NOT1 MIF4G domain bound to CAF1 and CCR4a, the NOT1 CN9BD domain bound to CAF40, a C-terminal connector domain of unknown function (CD), the NOT module comprising the NOT1 SHD and the C-terminal regions of NOT2 and NOT3, and an N-terminal coiled coil domain of NOT3 (Supplemental Fig. S3F). MBP-tagged Bam interacted only with the CN9BD–CAF40 module but not with any other subcomplex (Supplemental Fig. S3G, lane 25). The CN9BD–CAF40 module is highly conserved between Hs and Dm (CN9BD and CAF40 display 50% and 75% sequence identity, respectively). Accordingly, Bam also interacted with the Dm CN9BD–CAF40 module in pull-down assays (Fig. 3C, lane 12).
The CAF40-binding motif (CBM) is required for Bam repressive activity
To more precisely define the region of Bam that interacts with the CAF40 module, we performed a series of pull-down assays using various Bam fragments, which led to the identification of a CAF40-binding motif (CBM, residues D13–E36) within the Bam N-terminal fragment. The Bam CBM was sufficient for binding to the Dm and human CN9BD–CAF40 modules in pull-down assays (Fig. 3C, lane 14 and Fig. 3D, lanes 14 and 15). Furthermore, the CBM interacted directly with the isolated Hs CAF40 subunit in the absence of the NOT1 CN9BD (Fig. 3D, lane 16). Importantly, deletion of the CBM in the context of full-length Bam abolished the interaction with the Dm CN9BD–CAF40 module in vitro (Fig. 3C, lane 16), thereby indicating that the CBM is the principal CAF40-binding site in Bam.
To determine the contribution of the CBM to Bam's repressive activity, we performed tethering assays in S2 cells. Remarkably, the CBM alone (fragment D13–E36 fused to GST) was sufficient to induce the repression and degradation of the F-Luc-5BoxB mRNA to a similar extent as full-length Bam (Fig. 3E,F). Furthermore, deletion of the CBM was sufficient to abolish the repressive activity of Bam in tethering assays (Fig. 3E,F). All proteins were expressed at comparable levels (Fig. 3G) and did not affect the expression of an F-Luc reporter lacking the BoxB hairpins (Supplemental Fig. S4A,B). We therefore concluded that the CBM is essential for Bam's repressive activity.
Crystal structure of the Bam CBM bound to CAF40
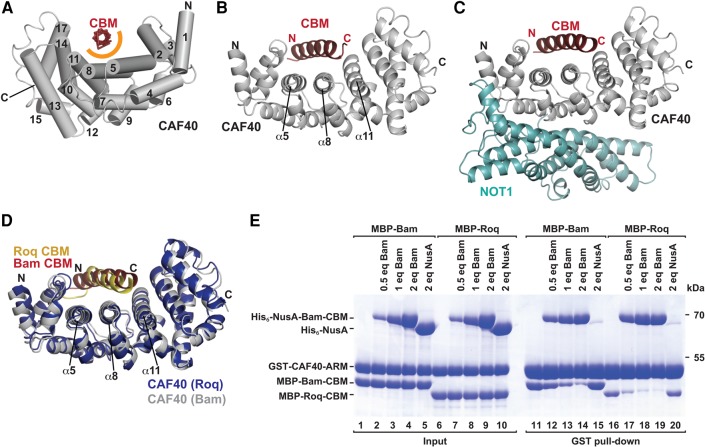
To elucidate the molecular principles underlying the interaction of Bam with the CAF40 module, we sought to determine the crystal structure of the CBM peptide (residues D13–E36) bound to the Dm and Hs CAF40 armadillo (ARM) domain (Dm CAF40 E25–G291 and Hs CAF40 R19–E285) as well as to the CAF40 modules containing the NOT1 CN9BD (residues Dm NOT1 Y1468–T1719 and Hs NOT1 V1351–L1588). However, only the complexes containing the human proteins yielded well-diffracting crystals. We solved the structures of the Bam CBM peptide bound to CAF40 and to the CN9BD–CAF40 complex and refined them to 3.0 Å and 2.7 Å resolution, respectively (Table 1; Fig. 4A–C).
TABLE 1.
Data collection and refinement statistics
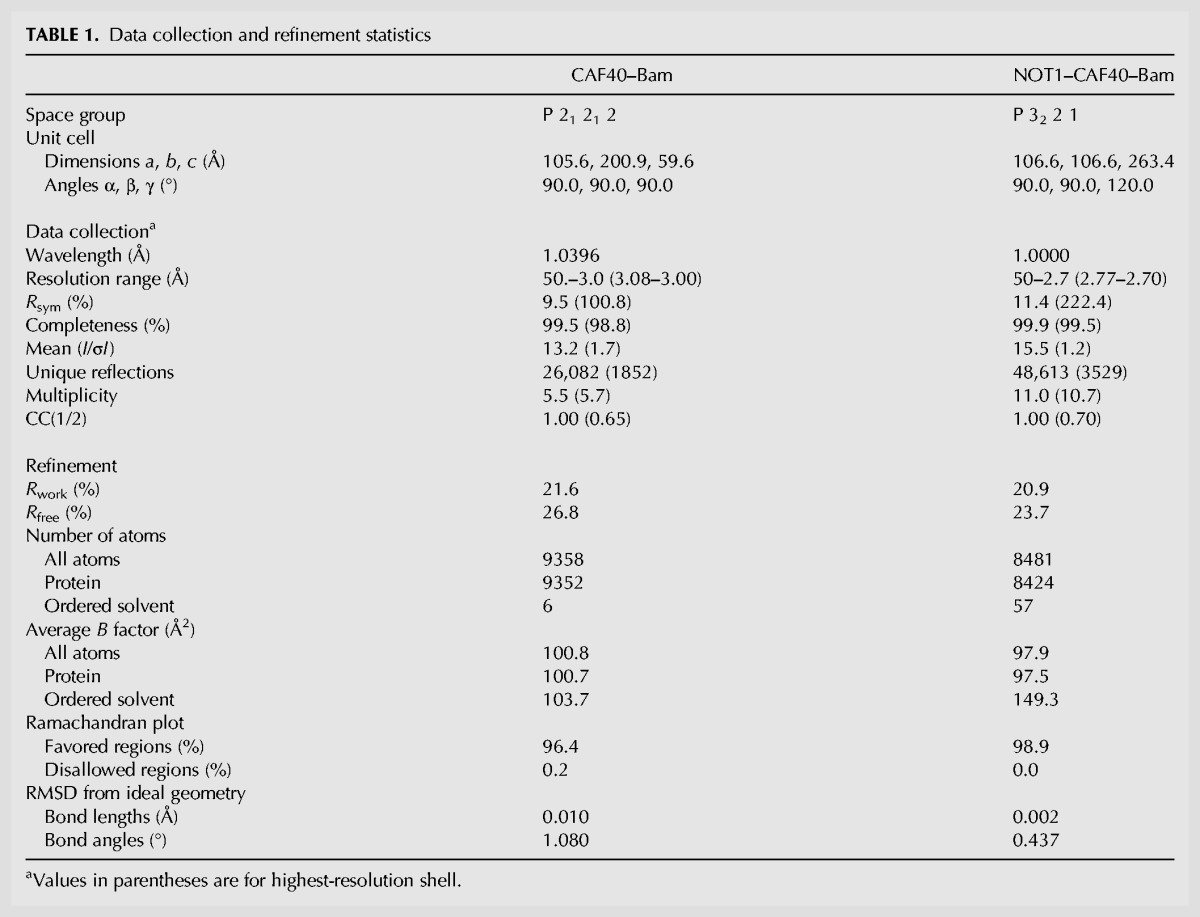
FIGURE 4.
Structure of the Bam CBM bound to CAF40 and to the NOT1 CN9BD–CAF40 module. (A) The Bam CBM peptide (red, backbone shown in ribbon representation) bound to Hs CAF40 (gray). CAF40 helices are depicted as tubes and numbered in black. The orange semicircle marks the predominantly hydrophobic interface between the CBM peptide and CAF40. (B) Cartoon representation of the Bam CBM peptide bound to Hs CAF40. Selected CAF40 secondary structure elements are labeled in black. (C) Structure of the CBM peptide bound to the NOT1 CN9BD–CAF40 complex. (D) Superposition of the CAF40–Bam CBM structure with the structure of CAF40 bound to the Roq CBM (PDB 5LSW; Sgromo et al. 2017). The Roq CBM is shown in yellow and CAF40 from the Roq complex in blue. (E) In vitro competition assay. GST-tagged Hs CAF40 was incubated with equimolar amounts of MBP-tagged Bam or Roq CBMs and increasing amounts of His6-NusA-tagged Bam CBM. His6-NusA was used as a negative control. Proteins bound to GST-CAF40 were pulled down and analyzed by SDS-PAGE and subsequent Coomassie staining. Molar equivalents (eq) are relative to GST-CAF40.
The asymmetric unit of the CAF40-CBM crystal contained four complexes that were highly similar to each other (RMSDs between 0.31 and 0.75 Å; over 237–278 Cα atoms) and that were arranged as two pairs of dimers (Supplemental Fig. S5A,B). The dimer interface corresponds to the one previously observed in the structure of free CAF40 (Supplemental Fig. S5C; Garces et al. 2007). The asymmetric unit of the CN9BD–CAF40-CBM crystals contained two almost identical complexes (RMSD of 0.28 Å over 457 Cα atoms; Supplemental Fig. S5D,E). In all complexes, the interaction of the CBM peptide with the CAF40 concave surface was found to be almost identical (Fig. 4B,C; Supplemental Fig. S5F), and the CBM does not contact the NOT1 CN9BD (Fig. 4A–C), thus confirming that the CBM interacts exclusively with CAF40. Superposition of the CAF40 dimer bound to the CBM with the previously determined ligand-free CAF40 dimer (Supplemental Fig. S5C; RMSD of 0.90 Å over 509 Cα atoms; Garces et al. 2007) or with CAF40 bound to the NOT1 CN9BD (Supplemental Fig. S5G; RMSD of 0.94 Å over 416 Cα atoms; Chen et al. 2014a), indicated that binding of the CBM peptide does not induce any major conformational changes in the CAF40 ARM domain.
The CBM peptide folds into an amphipathic α-helix that is bound centrally across the concave surface of the crescent-shaped CAF40 ARM domain, which consists of 17 α-helices arranged into six armadillo (ARM) repeats (Fig. 4A–C; Supplemental Fig. S6A,B; Garces et al. 2007; Chen et al. 2014a; Mathys et al. 2014). The α-helix binds to a conserved hydrophobic patch close to the previously proposed nucleic acid-binding groove (Garces et al. 2007). In the structure of the Bam CBM bound to the CAF40 module, the NOT1 CN9BD binds to the convex surface of CAF40 and prevents CAF40 dimerization, as previously observed (Chen et al. 2014a; Mathys et al. 2014). Importantly however, the NOT1 CN9BD does not interfere with Bam CBM binding on the concave surface of CAF40, thus indicating that Bam can interact with CAF40 also in the context of the fully assembled CCR4–NOT complex (Fig. 4C).
The Bam CBM competes with the Roquin CBM for binding to CAF40
Remarkably, the Bam CBM occupies the same binding surface as the previously described CBM of Dm Roquin (Roq) (Sgromo et al. 2017) and binds CAF40 in a similar manner (Fig. 4D). The two CBM peptides fold into amphipathic helices that bind via their hydrophobic sides along a groove on the concave face of CAF40. Consequently, the two peptides cannot bind CAF40 simultaneously and compete for binding to CAF40 when tested in vitro in a competition assay. In this assay, GST-tagged CAF40 was incubated with equimolar amounts of MBP-tagged Bam or Roq CBMs and increasing concentrations of His6-NusA-tagged Bam CBM. The peptides bound to CAF40 were pulled down by using glutathione-agarose beads. Increasing concentrations of the His6-NusA-Bam CBM competed with the two MBP-tagged CBMs for CAF40 binding (Fig. 4E). Interestingly, the Roquin CBM was competed out more efficiently than the Bam CBM (Fig. 4E, e.g., lane 12 versus 17), thus suggesting that Bam has a competitive advantage.
To obtain information on the affinities of the CBM peptides for CAF40, we performed isothermal titration calorimetry (ITC) experiments. The Bam CBM bound to the Dm CN9BD–CAF40 complex with a dissociation constant (KD) in the nanomolar range (183 ± 44 nM; Supplemental Fig. S7A). In contrast, the affinity of the Roq CBM was too low to be measured by ITC (i.e., the necessary peptide concentrations for measurement could not be reached), thereby explaining why the Roq CBM competed rather poorly with the Bam CBM.
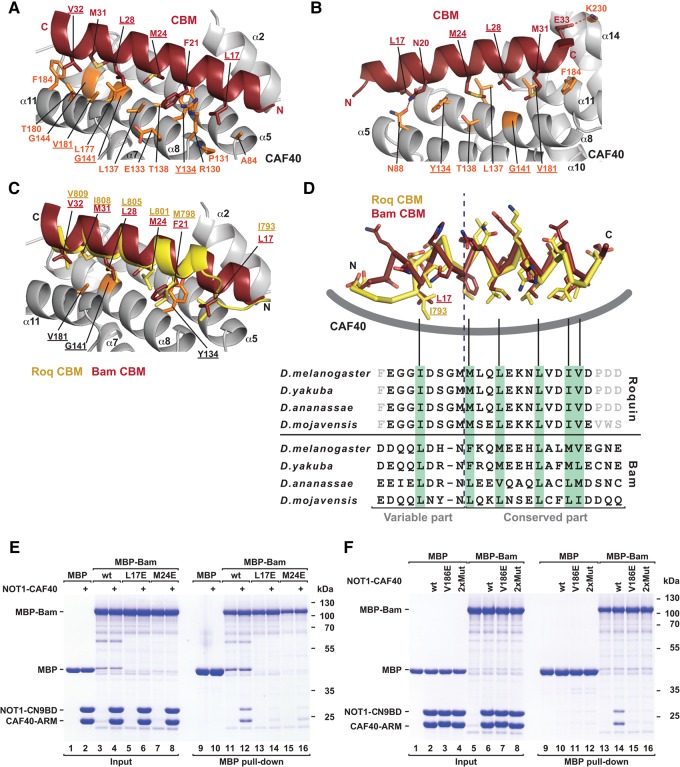
The Bam and Roquin CBMs use similar binding modes
The Bam CBM forms a single amphipathic α-helix extending through residues D13–E33 and the hydrophobic face of this helix binds in a groove formed by helices α5, α8 and α11 on the CAF40 concave surface (Fig. 5A,B). The interaction buries a total surface area of 1638 Å2 and involves the side chains of Bam residues L17, F21, M24, L28, M31 and V32, which engage the hydrophobic CAF40-binding surface consisting of residue A84 in helix α5, residues R130, Y134, L137, T138, G141 and G144 in helix α8, and residues L177, T180, V181 and F184 in helix α11 on the CAF40 side (Fig. 5A,B). In addition, the N and C termini of the CBM helix contact the CAF40 surface through hydrogen bonds between Bam N20 and CAF40 N88, and Bam E33 and CAF40 K230, respectively (Fig. 5B). However, these interactions were not observed in all six complexes, thus indicating some degree of flexibility of the helix ends.
FIGURE 5.
The Bam and Roq CBMs use a similar CAF40-binding mode. (A,B) Close-up views of the CAF40-Bam CBM-binding interface in two orientations. Selected residues of CAF40 and Bam are shown as orange and red sticks, respectively. Hydrogen bonds are indicated by red dashed lines. Residues mutated in this study are underlined. (C) Close-up view of the structural superposition of the CAF40-Bam CBM structure with the structure of the Roq CBM bound to CAF40. The Bam and Roq CBMs are shown in red and yellow, respectively. (D) (Upper panel) Superposition of the Bam and Roq CBM peptides bound to CAF40. The backbones are shown in ribbon representation, and side chains are shown as sticks. CAF40 is indicated as a thick gray line. (Lower panel) Sequence alignment of the Bam and Roq CBMs from the indicated Drosophila species. Hydrophobic residues interacting with CAF40 are highlighted by a light green background. Gray letters indicate residues that were not included in the crystallization setup. (E) MBP pull-down assay testing the interaction of MBP-tagged Bam (wild-type or mutants L17E and M24E) with the Dm NOT1-CN9BD–CAF40 complex. MBP served as a negative control. (F) MBP pull-down assay testing the interaction of MBP-tagged Bam with Dm NOT1-CN9BD–CAF40 complex (containing CAF40 wild-type or the indicated mutants). MBP served as a negative control.
In the Roq CBM, the N-terminal portion (residues E790–M797) is no longer α-helical, owing to the insertion of a glycine (G796), which is conserved among Roq proteins from different Drosophila species (Fig. 5C,D). Instead, the residues form an extended “hook” that is stabilized by internal hydrogen bonds. In contrast, the Bam N-terminal residues (D13–N20) extend the amphipathic α-helix by another two turns. Despite this structural difference, Bam residue L17 engages the same binding pocket as Roq residue I793. Thus, critical contacts are preserved in both peptides despite the fact that Roq is no longer helical (Fig. 5C,D). Overall, the all α-helical conformation of the Bam CBM is likely to be more stable on its own than the more extended conformation of the Roq CBM, which probably does not form in the absence of CAF40. The resulting difference in the binding entropy could contribute to the higher affinity of the Bam CBM for CAF40 and to its competitive advantage over the Roquin CBM. Alternatively, differences in the hydrophobic interface residues may also potentially explain the observed differences in affinity and competition between the two CBMs, e.g., the side chain of residue F21 in the center of the Bam CBM establishes a more extensive network of hydrophobic interactions along the interface than the side chain of residue M798 at the same structural position in the Roq CBM.
The interaction of Bam with CAF40 is required for mRNA repression
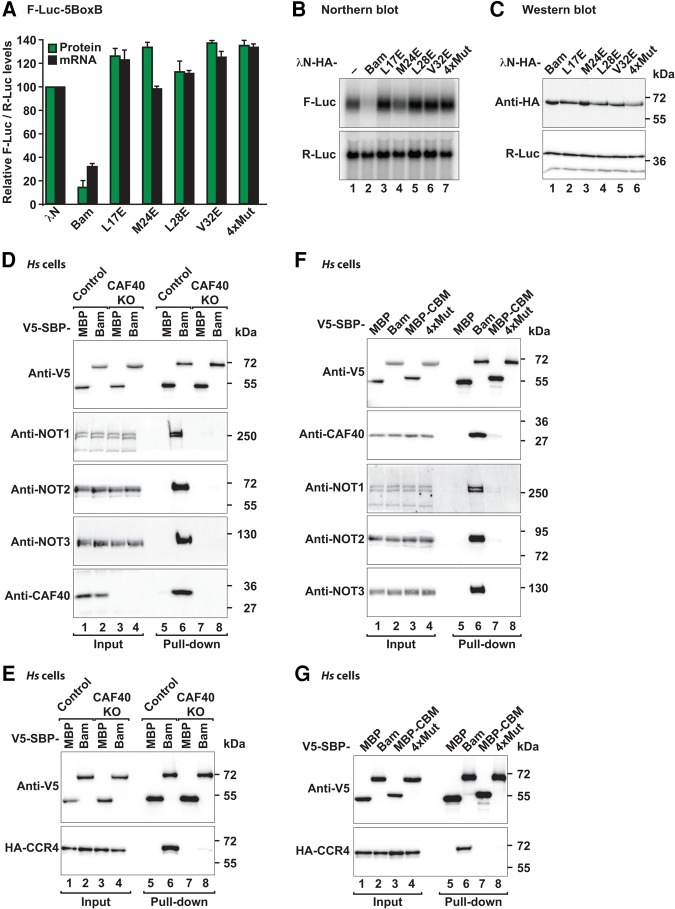
To assess the importance of the interactions observed in the crystal structure, we substituted Bam residues L17 or M24 with glutamic acid. These substitutions abolished the interaction of the MBP-tagged Bam with the Dm CAF40 module in pull-down assays (Fig. 5E; Supplemental Table S1), thus indicating that the CBM is the only CAF40-binding site in Bam. We also analyzed the effects of amino acid substitutions in the CAF40 interface on complex formation. A single V186E substitution or the double Y139D, G146W substitution (2xMut) in Dm CAF40 (corresponding to Hs CAF40 residues V181, Y134 and G141) were sufficient to disrupt the interaction with Bam in vitro (Fig. 5F; Supplemental Table S1). The equivalent substitutions in Hs CAF40 were also sufficient to disrupt binding to the Roq CBM (Sgromo et al. 2017), thus further confirming the similarity in the CBM-binding modes.
Next, we assessed the relevance of the interface in S2 cells. The single amino acid substitution in Dm CAF40 (V186E) was sufficient to abolish binding to full-length Bam in cell lysates (Supplemental Fig. S3E, lane 6). Conversely, substitutions of CBM residues (4xMut, Supplemental Table S1) in the context of full-length Bam abolished binding to Dm CAF40 (Supplemental Fig. S7B).
To assess the functional relevance of the CAF40-Bam interaction in mRNA target repression, we performed tethering assays in S2 and human cells. Single amino acid substitutions in the Bam CBM abolished Bam activity in tethering assays in S2 cells (Fig. 6A,B) a result indicating that the CBM provides a major contribution to Bam's repressive activity. All mutants were expressed at comparable levels (Fig. 6C) and did not affect the expression of a F-Luc mRNA lacking the BoxB hairpins (Supplemental Fig. S7C,D).
FIGURE 6.
The CBM is necessary for Bam-mediated mRNA repression. (A) Tethering assay using the F-Luc-5BoxB reporter and λN-HA-tagged Bam (wild-type or the indicated mutants) in S2 cells. The samples were analyzed as described in Figure 1B. (B) Northern blot of representative RNA samples shown in A. (C) Western blot showing the equivalent expression of λN-HA-tagged proteins used in A and B. (D) SBP pull-down assay in control and CAF40-null HEK293T cells expressing V5-SBP-tagged full-length Bam. V5-SBP-tagged MBP served as a negative control. Input (1.5% for the V5-SBP tagged proteins and 1% for endogenous CCR4–NOT subunits) and bound fractions (10% for the V5-SBP tagged proteins and 30% for the CCR4–NOT subunits) were analyzed by western blotting using the indicated antibodies. (KO) Knockout. (E) SBP pull-down assay in control and CAF40-null HEK293T cells expressing V5-SBP-tagged full-length Bam and HA-CCR4. Samples were analyzed as in D. (F) SBP pull-down assay in HEK293T cells expressing V5-SBP-tagged full-length Bam or the 4xMut. V5-SBP-tagged MBP served as a negative control. Input (1.5% for the V5-SBP tagged proteins and 1% for CCR4–NOT subunits) and the bound fractions (10% for the V5-SBP tagged proteins and 30% for CCR4–NOT subunits) were analyzed by western blotting using the indicated antibodies. (G) SBP pull-down assay in HEK293T cells expressing V5-SBP-tagged full-length Bam or the 4xMut and HA-tagged CCR4.
In human cells, we tethered MS2-HA-tagged full-length Bam (wild-type or the 4xMut) to a β-globin reporter containing six binding sites for the MS2 protein in the 3′ UTR. As observed in Dm cells, wild-type Bam caused degradation of the β-globin-6xMS2bs reporter, whereas the Bam 4xMut was inactive (Supplemental Fig. S7E–G). Furthermore, the CBM alone fused to MBP was as active as full-length Bam (Supplemental Fig. S7E–G). Thus, Bam depends on the integrity of the CBM to repress mRNA expression both in human and S2 cells.
Bam interaction with CCR4 is indirect and mediated by CAF40
In the pull-down assays using recombinant proteins, we did not observe a direct interaction between Bam and the catalytic module (containing Hs CCR4a, which is 57% identical to Dm CCR4; Supplemental Fig. S3G). Furthermore, Bam did not competitively displace the CAF1-NOT1 subcomplex from CCR4a (Supplemental Fig. S3G, lane 24), as has previously been suggested (Fu et al. 2015). To determine whether Bam interaction with CCR4 was direct or mediated by CAF40, we used CRISPR-Cas9 gene editing to generate a CAF40-null HEK293T human cell line in which the CAF40 levels were decreased below detectable levels (Fig. 6D, lanes 3 and 4 versus 1 and 2 and Supplemental Fig. S7H), whereas the expression of the additional subunits of the CCR4–NOT complex was not affected (Fig. 6D, lane 3 and 4 versus 1 and 2). In this cell line, Bam did not interact with endogenous NOT1, NOT2 and NOT3 (Fig. 6D, lane 8 versus 6) or with HA-tagged CCR4 (Fig. 6E, lane 8 versus 6), thus indicating that the interaction of Bam with the subunits of the CCR4–NOT complex is indeed mediated by CAF40. Similarly, the combined quadruple substitutions in the Bam CBM (4xMut) abrogated the interaction with the endogenous subunits of the CCR4–NOT complex in human cells (Fig. 6F, lane 8 versus 6) as well as the interaction with HA-tagged CCR4 (Fig. 6G, lane 8 versus 6). Similarly, the Bam 4xMut did not interact with CCR4 or NOT2 in S2 cells (Supplemental Fig. S7I,J). Together, our results indicated that the previously reported interaction of Bam with CCR4 (Fu et al. 2015), is most likely indirect and mediated by CAF40 in the context of the fully assembled CCR4–NOT complex.
CAF40 is the only Bam-binding site within the CCR4–NOT complex
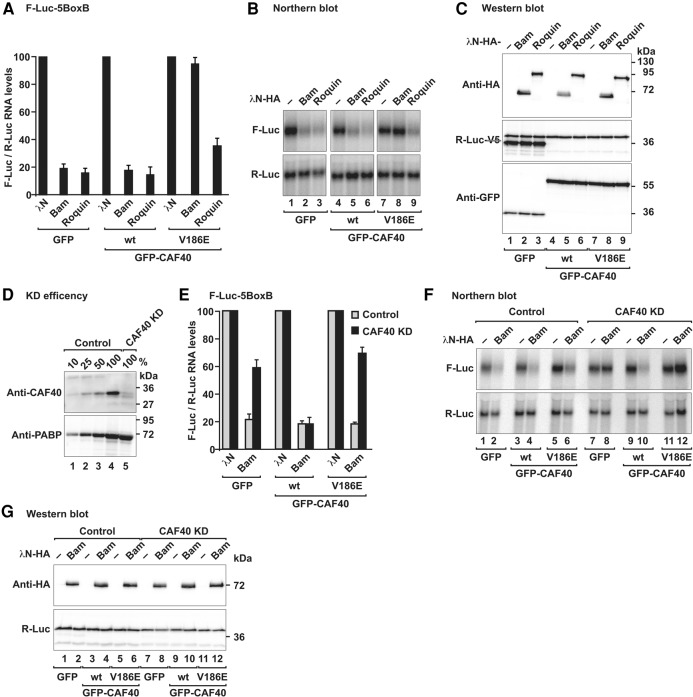
To further validate the relevance of Bam interaction with CAF40 for the recruitment of the CCR4–NOT complex, we performed tethering assays in Dm S2 cells overexpressing CAF40 wild-type or the CAF40 V186E mutant, which does not interact with Bam and was thus expected to suppress Bam activity in a dominant negative manner. Accordingly, Bam activity in tethering assays was suppressed in cells overexpressing the CAF40 V186E mutant but not when CAF40 wild-type was overexpressed (Fig. 7A,B). For a control, we tethered Dm Roq, which in addition to the CBM contains additional binding sites for the CCR4–NOT complex (Sgromo et al. 2017). Consequently, Roq activity was only slightly affected in cells overexpressing the CAF40 mutant (Fig. 7A,B, lane 9). Overexpression of CAF40 did not affect the Bam and Roq expression levels (Fig. 7C). The differential effect of the CAF40 mutant on Bam and Roq activities further confirmed that Bam, in contrast to Roq, depends entirely on its interaction with CAF40 for repression, whereas Roq can recruit the CCR4–NOT complex through additional binding sites.
FIGURE 7.
Bam depends on CCR4–NOT complex recruitment to induce mRNA decay. (A) Tethering assay using the F-Luc-5BoxB reporter and λN-HA-tagged Bam and Roq in S2 cells. The transfection mixtures also contained plasmids for expression of GFP (control) or GFP-CAF40 (wild-type or the V186E mutant) as indicated. The samples were analyzed as described in Figure 1B. (B) Northern blot of representative RNA samples shown in A. (C) Western blot showing the equivalent expression of the λN-HA-tagged proteins in cells expressing GFP or GFP-CAF40 (either wild-type or the V186E mutant) used in A and B. (D) Western blot showing the efficiency of the CAF40 depletion in Dm S2 cells. Dilutions of control cell lysates were loaded in lanes 1–4 to estimate the efficacy of the depletion. PABP served as a loading control. (KD) Knockdown. (E) Complementation assay using the F-Luc-5BoxB reporter and λN-HA-tagged Bam in S2 cells depleted of CAF40 (CAF40 KD) or in control cells (control). Samples were analyzed as described in Figure 1B. (F) Northern blot of representative RNA samples shown in E. (G) Western blot showing the equivalent expression of the λN-HA-tagged Bam constructs used in E and F.
In an independent approach, we tethered Bam in S2 cells depleted of CAF40 in which CAF40 levels were decreased to ∼10% of the control levels (Fig. 7D). CAF40 depletion partially suppressed Bam activity in tethering assays in S2 cells (Fig. 7E,F). The Bam-mediated repression was restored by transient expression of wild-type CAF40 but not by expression of the CAF40 V186E mutant, which does not interact with Bam (Fig. 7E,F), despite comparable expression levels (Fig. 7G). Thus, Bam requires interactions with CAF40 for full repressive activity.
DISCUSSION
In this study, we showed that Bam represses the translation and promotes the degradation of bound mRNAs by directly recruiting the CCR4–NOT deadenylase complex through an interaction with CAF40. This interaction is mediated by a short CAF40-binding motif (CBM) that is necessary and sufficient for Bam's repressive activity. We further elucidated the structural basis of the interaction of the Bam CBM with CAF40 and identified the concave surface of CAF40 as a binding site for amphipathic helices in RNA-associated proteins that recruit the CCR4–NOT complex.
CCR4–NOT complex recruitment is required for Bam repressive activity
The recruitment of the CCR4–NOT complex via the CAF40-CBM interaction is required for Bam to repress the translation of mRNA targets. Unlike other proteins, such as GW182, TTP, Roq and Dm Nanos, that use multiple redundant motifs to recruit the CCR4–NOT complex (Fabian et al. 2013; Chen et al. 2014a; Mathys et al. 2014; Raisch et al. 2016; Sgromo et al. 2017), Bam depends entirely on the interaction between the CBM and CAF40. Indeed, single point mutations in the CBM that abolished the interaction with CAF40 also disrupted the interaction with CCR4 and additional subunits of the CCR4–NOT complex and abrogated Bam's repressive activity. Similar results were obtained in cells depleted of CAF40, thus indicating that the previously reported interaction between Bam and CCR4 is indirect and is mediated by CAF40 in the context of the assembled CCR4–NOT complex. These results also indicated that the CCR4–NOT complex is the main downstream effector complex in Bam-mediated mRNA regulation.
CAF40 serves as a binding platform of the CCR4–NOT complex
Bam adds to the growing number of examples of RNA-associated proteins that directly recruit the CCR4–NOT complex via short linear motifs to down-regulate mRNA targets. To date, the motifs that have been characterized have been found to bind non-overlapping surfaces on the CCR4–NOT complex. For example, vertebrate and Dm Nanos and Saccharomyces cerevisiae (Sc) NOT4 bind to non-overlapping surfaces on the NOT module (Bhandari et al. 2014; Bhaskar et al. 2015; Raisch et al. 2016). The CAF40 subunit also provides interaction sites for RNA-associated proteins within the CCR4–NOT complex. The convex surface of the CAF40 armadillo-repeat domain features two tryptophan-binding sites that are used by proteins of the GW182 protein family, which recruit the CCR4–NOT complex to miRNA targets (Chen et al. 2014a; Mathys et al. 2014). The CAF40 concave surface provides binding sites for the CBM in the Dm Roq protein (Sgromo et al. 2017). Here, we found that this surface also binds to the Bam CBM, thus indicating that Bam and Roq binding to CAF40 is mutually exclusive. However, the Bam and Roq proteins share no apparent sequence similarity and thus their CBMs may have evolved independently to bind to the same surface of CAF40. The functional relevance of this competitive binding remains unclear, because it is not known whether the two proteins are expressed in the same cell type under the same physiological conditions.
The high conservation of CAF40 (75% sequence identity between the Hs and Dm proteins, and 57% identity between Hs and Sc), particularly of the CBM-binding surface, suggests the existence of additional CBM-containing proteins in eukaryotes. Through an in silico search using a consensus pattern derived from the Bam and Dm Roq CBM sequences, we could indeed identify several potential CBMs in other proteins (Supplemental Fig. S8). However, none of the tested sequences interacted with Hs CAF40 in vitro in pull-down assays (data not shown), thus indicating that the tested fragments are not bona fide CBMs and that the rules guiding CAF40 binding are still incompletely understood. From what we know, it is possible and quite likely indeed that if CBMs exist in other proteins, they do not share an evolutionary origin with Bam and Roq and therefore also have no phylogenetic sequence conservation. Indeed, sequence searches conducted with either Bam or Roq did not identify the respective other protein as a CBM-containing protein.
CCR4–NOT complex recruitment is a recurring mechanism for targeted repression of gene expression
With the expanding repertoire of RNA-binding proteins that are known to interact with the CCR4–NOT complex, some underlying principles of recruitment are emerging. First, many RNA-associated proteins use extended peptide motifs embedded in unstructured regions for binding to CCR4–NOT. Interactions of such short linear motifs (SLiMs) are generally characterized by high specificity and at the same time relatively low individual affinity (Tompa 2012; Van Roey et al. 2014). This aspect is important in regulatory complexes such as the CCR4–NOT complex, because the complex must be recruited in a highly specific manner and need to be released again after exerting its specific function. Additionally, these motifs usually show high evolutionary plasticity (Tompa 2012; Van Roey et al. 2014) and are not conserved in orthologous proteins accross species.
Another common theme is that RNA binding is often spatially separated from CCR4–NOT complex recruitment. In many cases including Nanos and Roq, RNA binding is mediated by highly conserved RNA-binding domains, whereas CCR4–NOT complex recruitment is mediated by SLiMs in long unstructured regions of up to several hundred amino acids in length. In other cases, RNA binding and CCR4–NOT recruitment are associated with different polypeptides. For example, in the miRNA-induced silencing complexes (miRISCs), RNA binding is achieved by Argonaute proteins (AGOs), whereas CCR4–NOT complex recruitment is mediated by the GW182 proteins that act as adaptor molecules downstream from AGOs (Jonas and Izaurralde 2015). In the case of Bam, it is unknown whether the RNA-binding activity resides in the Bam protein itself or whether additional factors mediate mRNA binding.
Finally, it is interesting to note that some proteins such as Bam and vertebrate Nanos (this study; Bhandari et al. 2014), use a single motif with relatively high affinity to interact with the CCR4–NOT complex, whereas others such as Dm Roq and the GW182 proteins, use avidity effects in a distributive binding mode involving multiple lower-affinity motifs in disordered protein regions for recruitment (Chen et al. 2014a; Mathys et al. 2014; Sgromo et al. 2017). The highly diverse sequence motifs bind to several structured surfaces on the complex. Nevertheless, independently of the mode of interaction, the recruitment of the CCR4–NOT complex by diverse RNA-binding proteins results in a common functional outcome: the repression of the mRNA target through deadenylation-dependent and independent mechanisms and, in cellular contexts in which deadenylation is coupled to decapping, the degradation of the mRNA through the 5′-to-3′ mRNA decay pathway. Thus, the CCR4–NOT complex, through its ability to provide binding sites for diverse sequence motifs, is a major downstream effector complex in post-transcriptional mRNA regulation.
MATERIALS AND METHODS
DNA constructs
The DNA constructs used in this study are described in the Supplemental Material and are listed in Supplemental Table S1. All of the mutants used in this study were generated by site-directed mutagenesis using a QuikChange mutagenesis kit (Stratagene). All constructs and mutations were confirmed by sequencing.
Coimmunoprecipitation and SBP pull-down assays
All coimmunoprecipitation and SBP pull-down assays in S2 and HEK293T cell lysates were performed in the presence of RNaseA as previously described (Sgromo et al. 2017). All western blots were developed using an ECL western blotting detection system (GE Healthcare). The antibodies used in this study are listed in Supplemental Table S2. A detailed description of these assays is included in the Supplemental Material.
Tethering and complementation assays
Knockdown of DCP2, NOT1 and CAF40 in S2 cells using dsRNA was performed as previously described (Behm-Ansmant et al. 2006). For the λN-tethering assays in Dm S2 cells, 2.5 × 106 cells per well were seeded in six-well plates and transfected using Effectene (Qiagen). The transfection mixtures contained the following plasmids: 0.1 µg of Firefly luciferase reporters (F-Luc-5BoxB, F-Luc-V5 or F-Luc-5BoxB-A95C7-HhR), 0.4 µg of the R-Luc transfection control and various amounts of plasmids expressing λN-HA-tagged full-length Bam or Bam fragments (0.05 µg for wild-type or mutant full-length Bam, 0.02 µg for Bam-N, 0.1 µg for Bam-C, 0.1 µg GST-CBM and 0.05 µg of Bam ΔCBM). Cells were harvested 3 days after transfection.
In the experiment shown in Figure 2A, control and DCP2 knockdown cells were additionally transfected with plasmids expressing GFP-V5 (0.08 µg) and GFP-DCP2*-V5 mutant (E361Q; 1 µg), respectively. In the experiment shown in Figure 7A and B, cells were also transfected with plasmids expressing GFP (0.05 µg) or GFP-tagged CAF40 (1.5 µg) either wild-type or mutant. In the complementation assay shown in Figure 7E,F, knockdown cells were also transfected with plasmids expressing GFP (0.002 µg) or GFP-tagged CAF40 (0.005 µg) either wild-type or mutant (V186E). To measure the mRNA half-life, S2 cells were treated with actinomycin D (5 µg/ml final concentration) 3 d after transfection and collected at the indicated time points. RNA samples were analyzed by northern blotting.
A detailed description of the procedure to generate the HEK293T CAF40-null cell line is included in the Supplemental Material. For the Bam tethering assays in human cells, HEK293T cells (0.7 × 106 cells per well) were seeded in six-well plates and transfected using Lipofectamine 2000 (Thermo Fisher Scientific). The transfection mixtures contained 0.5 µg of the β-globin reporter containing six MS2-binding sites (β-globin-6xMS2bs), 0.5 µg of the control plasmid containing the β-globin gene fused to a fragment of the GAPDH gene inserted in the 3′ UTR but lacking MS2-binding sites (control: β-globin-GAP), and various amounts of pT7-MS2-HA plasmids for the expression of MS2-HA-fusion proteins [full-length Bam (1 µg), MBP-Bam CBM (0.2 µg) and Bam 4xMut (0.5 µg)].
Firefly and Renilla luciferase activities were measured 3 d (S2 cells) or 2 d (HEK293T cells) after transfection by using a Dual-Luciferase Reporter Assay System (Promega). The total RNA was isolated using a Trifast Reagent (Peqlab) and analyzed by northern blotting, as previously described (Braun et al. 2011).
Protein expression and purification
All recombinant proteins were expressed in E. coli BL21 (DE3) Star cells (Thermo Fisher Scientific) grown in LB medium overnight at 20°C. The cells were lysed with an EmulsiFlex-C3 homogenizer (AVESTIN) in the indicated lysis buffer supplemented with DNase I (5 µg/mL), lysozyme (1 mg/mL) and complete EDTA-free protease inhibitor cocktail (Roche). Bam constructs were expressed as fusions with an N-terminal, TEV-cleavable MBP tag. The cells were lysed in a buffer containing 50 mM HEPES (pH 7.5), 300 mM NaCl and 2 mM DTT. The proteins were purified from cleared cell lysates by using amylose resin (New England Biolabs), and this was followed by anion chromatography using a HiTrapQ column (GE Healthcare). The Bam constructs were further purified on a Superdex 200 26/600 column (GE Healthcare) in a buffer containing 10 mM HEPES (pH 7.5), 200 mM NaCl and 2 mM DTT.
The purification of Hs CAF40 (ARM domain, residues R19–E285) was as previously described (Sgromo et al. 2017). Briefly, the protein was expressed with an N-terminal His6 tag cleavable by the HRV3C protease. Cells were lysed in a buffer containing 50 mM potassium phosphate (pH 7.5), 500 mM NaCl, 10% (v/v) glycerol, 20 mM imidazole and 2 mM β-mercaptoethanol. The protein was purified from cleared cell lysates with a HiTrap IMAC column (GE Healthcare). The His6 tag was removed by overnight cleavage using HRV3C protease during dialysis in a buffer containing 50 mM Tris–HCl (pH 7.5), 150 mM NaCl and 1 mM DTT. After cleavage of the tag, CAF40 was further purified using a HiTrap Heparin column (GE Healthcare) followed by gel filtration on a Superdex 200 26/600 column (GE Healthcare) in a buffer containing 20 mM Tris–HCl (pH 7.5), 150 mM NaCl and 1 mM DTT.
The purification of the Hs NOT1 CN9BD–CAF40 complex has been previously described (Chen et al. 2014a). The complex was obtained by co-expression of MBP-tagged NOT1-CN9BD (residues V1351–L1588) with His6-tagged CAF40 (R19–E285). The cells were lysed in a buffer containing 50 mM potassium phosphate (pH 7.5), 300 mM NaCl and 2 mM β-mercaptoethanol. The complex was purified from the cleared lysate by using amylose resin, and this was followed by removal of the His6 and MBP tags by cleavage with HRV3C protease overnight at 4°C during dialysis in a buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 10% (v/v) glycerol and 2 mM DTT. The complex was separated from the tags by binding to a HiTrap Heparin column (GE Healthcare), and this was followed by elution with a linear gradient to 1 M NaCl. Finally, size exclusion chromatography was performed using a Superdex 200 26/600 column in a buffer containing 10 mM HEPES (pH 7.5), 150 mM NaCl, 10% (v/v) glycerol and 2 mM DTT.
A detailed description of the purification of the additional modules of the human and Drosophila CCR4–NOT complex can be found in the Supplemental Material.
Crystallization, data collection, and structure determination
A detailed description of the crystallization conditions and the structure determination process are included in the Supplemental Material. Diffraction data sets of the CN9BD–CAF40–Bam CBM complex were recorded on a PILATUS 6M detector at the PXII beamline of the Swiss Light Source at a temperature of 100 K. The best data set of the CAF40–Bam CBM complex was recorded on a PILATUS 6M fast detector (DECTRIS) at the DESY beamline P11. The diffraction data and refinement statistics are summarized in Table 1.
In vitro MBP pull-down assays
Purified MBP (20 µg) or MBP-tagged full-length Bam or fragments (40 µg) were incubated with equimolar amounts of purified CCR4–NOT complex modules or subunits and amylose resin (New England Biolabs) in pull-down buffer containing 50 mM HEPES (pH 7.5), 200 mM NaCl and 2 mM DTT. After 1 h incubation, the beads were washed five times with pull-down buffer and the proteins were eluted with pull-down buffer supplemented with 25 mM D-(+)-Maltose. The eluted proteins were precipitated with trichloroacetic acid and analyzed by SDS-PAGE and subsequent Coomassie staining.
In vitro competition assays
Purified GST-tagged CAF40 (ARM domain, 50 µg) was incubated with equimolar amounts of either MBP-tagged Bam CBM or MBP-Roquin CBM, increasing amounts of His6-NusA-tagged Bam CBM as a competitor, and 50 µL 50% slurry of Protino glutathione agarose 4B (Macherey-Nagel). Purified His6-NusA served as a negative control. The experiment was performed in pull-down buffer. After 1 h of incubation, the beads were pelleted and washed three times with pull-down buffer. The proteins bound to the beads were eluted by boiling the beads in 2× protein sample buffer. The eluted proteins were analyzed by SDS-PAGE and subsequent Coomassie staining.
Isothermal titration calorimetry (ITC) and bioinformatics analysis
The ITC measurements were performed as previously described (Igreja et al. 2014). A detailed description of the ITC conditions and the bioinformatic analysis can be found in the Supplemental Material.
DATA DEPOSITION
The coordinates for the structure of the Bam CBM peptide bound to CAF40 and to the CAF40 module were deposited in the Protein Data Bank (PDB) under ID code 5ONB and 5ONA, respectively.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Heike Budde and Catrin Weiler for excellent technical support. We thank Lara Wohlbold for cloning the Dm Bam construct, Daniel Peter and Stefan Grüner for assistance with the ITC experiments, and Eugene Valkov for comments on the manuscript. We thank the staff at the PX beamlines of the Swiss Light Source, Villigen, and the staff at the P11 beamline of the DESY, Hamburg, for assistance with X-ray data collection.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.064584.117.
Freely available online through the RNA Open Access option.
REFERENCES
- Basquin J, Roudko VV, Rode M, Basquin C, Séraphin B, Conti E. 2012. Architecture of the nuclease module of the yeast Ccr4-not complex: the Not1-Caf1-Ccr4 interaction. Mol Cell 48: 207–218. [DOI] [PubMed] [Google Scholar]
- Bawankar P, Loh B, Wohlbold L, Schmidt S, Izaurralde E. 2013. NOT10 and C2orf29/NOT11 form a conserved module of the CCR4-NOT complex that docks onto the NOT1 N-terminal domain. RNA Biol 10: 228–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. 2006. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20: 1885–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari D, Raisch T, Weichenrieder O, Jonas S, Izaurralde E. 2014. Structural basis for the Nanos-mediated recruitment of the CCR4-NOT complex and translational repression. Genes Dev 28: 888–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar V, Basquin J, Conti E. 2015. Architecture of the ubiquitylation module of the yeast Ccr4-Not complex. Structure 23: 921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar V, Roudko V, Basquin J, Sharma K, Urlaub H, Séraphin B, Conti E. 2013. Structure and RNA-binding properties of the Not1-Not2-Not5 module of the yeast Ccr4-Not complex. Nat Struct Mol Biol 20: 1281–1288. [DOI] [PubMed] [Google Scholar]
- Boland A, Chen Y, Raisch T, Jonas S, Kuzuoğlu-Öztürk D, Wohlbold L, Weichenrieder O, Izaurralde E. 2013. Structure and assembly of the NOT module of the human CCR4-NOT complex. Nat Struct Mol Biol 20: 1289–1297. [DOI] [PubMed] [Google Scholar]
- Braun JE, Huntzinger E, Fauser M, Izaurralde E. 2011. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell 44: 120–133. [DOI] [PubMed] [Google Scholar]
- Carreira-Rosario A, Buszczak M. 2014. A competitive cell fate switch. Dev Cell 31: 261–262. [DOI] [PubMed] [Google Scholar]
- Chang CT, Bercovich N, Loh B, Jonas S, Izaurralde E. 2014. The activation of the decapping enzyme DCP2 by DCP1 occurs on the EDC4 scaffold and involves a conserved loop in DCP1. Nucleic Acids Res 42: 5217–5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau J, Kulnane LS, Salz HK. 2012. Sex-lethal enables germline stem cell differentiation by down-regulating Nanos protein levels during Drosophila oogenesis. Proc Natl Acad Sci 109: 9465–9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Boland A, Kuzuoğlu-Öztürk D, Bawankar P, Loh B, Chang CT, Weichenrieder O, Izaurralde E. 2014a. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell 54: 737–750. [DOI] [PubMed] [Google Scholar]
- Chen D, Wu C, Zhao S, Geng Q, Gao Y, Li X, Zhang Y, Wang Z. 2014b. Three RNA binding proteins form a complex to promote differentiation of germline stem cell lineage in Drosophila. PLoS Genet 10: e1004797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L, Kelley R, Spradling A. 1988. Insertional mutagenesis of the Drosophila genome with single P elements. Science 239: 1121–1128. [DOI] [PubMed] [Google Scholar]
- Draper MP, Liu HY, Nelsbach AH, Mosley SP, Denis CL. 1994. CCR4 is a glucose-regulated transcription factor whose leucine-rich repeat binds several proteins important for placing CCR4 in its proper promoter context. Mol Cell Biol 14: 4522–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper MP, Salvadore C, Denis CL. 1995. Identification of a mouse protein whose homolog in Saccharomyces cerevisiae is a component of the CCR4 transcriptional regulatory complex. Mol Cell Biol 15: 3487–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Morel AP, Barbot W, Loireau MP, Corbo L, Heidmann T. 2001. Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Frank F, Rouya C, Siddiqui N, Lai WS, Karetnikov A, Blackshear PJ, Nagar B, Sonenberg N. 2013. Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat Struct Mol Biol 20: 735–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Geng C, Wang H, Yang Z, Wenig C, Li H, Deng L, Liu L, Liu N, Ni J, et al. 2015. Twin promotes the maintenance and differentiation of germline stem cell lineage through modulation of multiple pathways. Cell Rep 13: 1366–1379. [DOI] [PubMed] [Google Scholar]
- Garces RG, Gillon W, Pai EF. 2007. Atomic model of human Rcd-1 reveals an armadillo-like-repeat protein with in vitro nucleic acid binding properties. Protein Sci 16: 176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igreja C, Peter D, Weiler C, Izaurralde E. 2014. 4E-BPs require non-canonical 4E-binding motifs and a lateral surface of eIF4E to repress translation. Nat Commun 5: 4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S, Izaurralde E. 2015. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 16: 421–433. [DOI] [PubMed] [Google Scholar]
- Leppek K, Schott J, Reitter S, Poetz F, Hammond MC, Stoecklin G. 2013. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell 153: 869–881. [DOI] [PubMed] [Google Scholar]
- Li Y, Minor NT, Park JK, McKearin DM, Maines JZ. 2009. Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc Natl Acad Sci 106: 9304–9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Carreira-Rosario A, Maines JZ, McKearin DM, Buszczak M. 2013. Mei-P26 cooperates with Bam, Bgcn and Sxl to promote early germline development in the Drosophila ovary. PLoS One 8: e58301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys H, Basquin J, Ozgur S, Czarnocki-Cieciura M, Bonneau F, Aartse A, Dziembowski A, Nowotny M, Conti E, Filipowicz W. 2014. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol Cell 54: 751–765. [DOI] [PubMed] [Google Scholar]
- Mauxion F, Prève B, Séraphin B. 2013. C2ORF29/CNOT11 and CNOT10 form a new module of the CCR4-NOT complex. RNA Biol 10: 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKearin D, Ohlstein B. 1995. A role for the Drosophila bag-of-marbles protein in the differentiation of cystoblasts from germline stem cells. Development 121: 2937–2947. [DOI] [PubMed] [Google Scholar]
- McKearin DM, Spradling AC. 1990. bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev 4: 2242–2251. [DOI] [PubMed] [Google Scholar]
- Neumüller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA. 2008. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454: 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, McKearin D. 1997. Ectopic expression of the Drosophila Bam protein eliminates oogenic germline stem cells. Development 124: 3651–3662. [DOI] [PubMed] [Google Scholar]
- Pan L, Wang S, Lu T, Weng C, Song X, Park JK, Sun J, Yang ZH, Yu J, Tang H, et al. 2014. Protein competition switches the function of COP9 from self-renewal to differentiation. Nature 514: 233–236. [DOI] [PubMed] [Google Scholar]
- Petit AP, Wohlbold L, Bawankar P, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O. 2012. The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4-NOT deadenylase complex. Nucleic Acids Res 40: 11058–11072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisch T, Bhandari D, Sabath K, Helms S, Valkov E, Weichenrieder O, Izaurralde E. 2016. Distinct modes of recruitment of the CCR4-NOT complex by Drosophila and vertebrate Nanos. EMBO J 35: 974–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgromo A, Raisch T, Bawankar P, Bhandari D, Chen Y, Kuzuoğlu-Öztürk D, Weichenrieder O, Izaurralde E. 2017. A CAF40-binding motif facilitates recruitment of the CCR4-NOT complex to mRNAs targeted by Drosophila Roquin. Nat Commun 8: 14307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R, Weng C, Yu J, Xie T. 2009. eIF4A controls germline stem cell self-renewal by directly inhibiting BAM function in the Drosophila ovary. Proc Natl Acad Sci 106: 11623–11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Saba R, Miyoshi K, Morita Y, Saga Y. 2012. Interaction between NANOS2 and the CCR4-NOT deadenylation complex is essential for male germ cell development in mouse. PLoS One 7: e33558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. 2012. Intrinsically disordered proteins: a 10-year recap. Trends Biochem Sci 37: 509–516. [DOI] [PubMed] [Google Scholar]
- Van Roey K, Uyar B, Weatheritt RJ, Dinkel H, Seiler M, Budd A, Gibson TJ, Davey NE. 2014. Short linear motifs: ubiquitous and functionally diverse protein interaction modules directing cell regulation. Chem Rev 114: 6733–6778. [DOI] [PubMed] [Google Scholar]
- Wahle E, Winkler GS. 2013. RNA decay machines: deadenylation by the Ccr4-not and Pan2-Pan3 complexes. Biochim Biophys Acta 1829: 561–570. [DOI] [PubMed] [Google Scholar]
- Zekri L, Kuzuoğlu-Öztürk D, Izaurralde E. 2013. GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. EMBO J 32: 1052–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.