Abstract
GPR126 has been identified to be associated with AIS (Adolescent Idiopathic Scoliosis) in different populations, but data on the northern Chinese population are unavailable. Additionally, it is important to know the exact clinical phenotypes associated with specific genetic polymorphisms. Fourteen SNP (single nucleotide polymorphism) loci in GPR126 were genotyped in 480 northern Chinese Han AIS patients and 841 controls. These patients were classified into three types based on the PUMC classification system. Luciferase assays were used to investigate their regulation of GPR126 transcription activity. Combined and stratified genotype–phenotype association analyses were conducted. The alleles rs225694, rs7774095 and rs2294773 were significantly associated with AIS (P = 0.021, 0.048 and 0.023, respectively). rs225694 and rs7774095 potentially have regulatory functions for the GRP126 gene. Correlation analysis revealed that allele A of rs225694 was a risk allele only for PUMC type II AIS (P = 0.036) and allele G of rs2294773 was a risk allele only for PUMC type I AIS (P = 0.018). In summary, rs225694, rs7774095 and rs2294773 are significantly associated with disease in northern Chinese Han AIS patients. The SNPs rs225694 and rs2294773 are associated with different AIS PUMC classifications.
Keywords: adolescent idiopathic scoliosis, GPR126/ADGRG6, single nucleotide polymorphism, PUMC classification
Background
AIS is the most common spinal deformity, affecting approximately 1–3% of children throughout the world 1, 2. There are many classification systems, including the King classification 3, Lenke classification 4 and the Peking Union Medical College (PUMC) classification systems 5. According to the PUMC classification, AIS can be classified into single (I), double (II), or triple (III) curve types. The PUMC classification is a three‐dimensional classification system that is relatively simple to use and has corresponding recommended surgical approaches 5. Despite decades of research on AIS, its exact aetiological and pathological causes remain unclear.
Previous studies have suggested that genetic polymorphisms play a pivotal role in the pathogenesis of AIS 6, 7. Although a recent genomewide association study (GWAS) and other studies have identified several susceptible gene variants, such as GPR126 8, LBX1 9, PAX1 10, BCN2 11 and PTK7 12, few of these variants have been replicated in different populations, and even fewer have been analysed with regard to different AIS subtypes.
GPR126 (encoding G protein‐coupled receptor 126), also known as ADGRG6, located at 6q24.1, is 144,348 bp in length and consists of 25 exons. According to Monk et al. 13, gpr126 is essential for mouse survival due to Schwann cell myelination. Courtney et al. also found 14 that a gpr126 deletion in cartilage could lead to idiopathic scoliosis and pectus excavatum in mice. It has been shown that GPR126 gene SNPs are associated with AIS. Kou et al. 8 identified rs6570507 as a susceptibility locus with AIS in 1,819 Japanese cases and 25,939 controls and found that the developing spine of a gpr126 knockdown zebrafish model had delayed ossification. Subsequently, Xu et al. 15 found this locus and two other loci (rs7774095 and rs7755109) in 352 southern Chinese cases and 149 controls. Recently, Qin et al. 16 found another functional locus (rs9403380) that regulates GPR126 expression in the paraspinal muscles of southern Chinese AIS patients.
As far as we know, there has not been an association analysis of the GPR126 gene with AIS in the northern Chinese Han population. It will be of great significance to identify the specific subgroups of AIS that are associated with GPR126 genetic polymorphisms. We suggested that GPR126 risk SNPs are potentially associated with the AIS PUMC classification system. Therefore, we enrolled a cohort from the northern Chinese Han population and corresponding controls. SNPs in GRP126 were genotyped with subsequent phenotypic association analysis based on their PUMC classification.
Materials and methods
Participants
A total of 480 patients diagnosed with AIS and 841 healthy in‐house controls from a northern Chinese Han population at Peking Union Medical College Hospital were enrolled between July 2011 and August 2016. The inclusion and exclusion criteria were as follows:
Inclusion criteria
Diagnosed as AIS with a Cobb angle > 20 degrees.
Age of onset between 10 and 18 years.
Origin was from the northern Chinese Han population as identified by a Native Place questionnaire.
Having complete imaging data, including X‐ray, three‐dimensional imaging of the spine CT or spinal MRI.
Exclusion criteria
Scoliosis secondary to a known aetiology, including congenital scoliosis, neuromuscular scoliosis, scoliosis secondary to skeletal dysplasia or connective tissue abnormalities.
Origin was not from the northern Chinese Han population.
Incomplete imaging data.
Having a chronic disease that influenced skeletal development.
All patients diagnosed as AIS were classified into three types based on the PUMC classification system 5 by at least two experienced orthopaedic surgeons. Written informed consent was obtained from all participants or their parents. The Ethics Committee of Peking Union Medical College Hospital, Chinese Academy of Medical Sciences approved this study.
SNP selection and genotyping
Fourteen candidate SNPs around GPR126 were selected by the following criteria from a literature review and the NCBI SNP database (www.ncbi.nlm.nih.gov/SNP; Table S1): (i) SNP with a minor allele frequency (MAF) above 5%; (ii) Tag SNPs were preferred; (iii) reported SNPs associated with idiopathic scoliosis; and (iv) potential functional SNPs predicted by HaploReg (v4.1) 17.
Genomic DNA was extracted from the peripheral blood of each participant using DNeasy Blood & Tissue Kits (QIAGEN, Eastwin Scientific, Inc. Beijing, China). SNP genotyping was performed using the MassArray system from Sequenom. All of the details were the same as in our previous studies 18, 19, 20.
Luciferase assay
We used luciferase assays to test the influence of significantly associated SNPs on transcription. We cloned oligonucleotides around each SNP (Table S2) and cloned them into a pGL3 promoter luciferase reporter vector (Promega, Madison, WI, USA). Human embryonic kidney 293a cells (HEK293a) and HeLa cells were transfected with the reporter vector by lip3000 (Invitrogen, Carlsbad, CA, USA). After a 48‐hr incubation, cells were harvested and luciferase activity was measured using a Dual‐Luciferase Reporter Assay System (Promega, Madison, WI, USA).
Data analysis
Hardy–Weinberg equilibrium (HWE) tests and primary analyses, including allelic, genotypic, haplotypic analyses and linear regression controlling for sex were conducted using PLINK software (v1.07, 2009 Shaun Purcell) and the Haploview program (version 4.2, Broad Institute of MIT and Harvard, Cambridge, MA, USA). The baseline characteristics of AIS participants presenting different PUMC types were compared through SPSS software (16.0 version, SPSS Inc., Chicago, IL, USA). Intergroup differences were assessed with the chi‐square test for categorical variables and one‐way anova for continuous normally distributed variables.
Results
Baseline characteristics of participants
Seventy‐eight males and 402 females were enrolled in the case group, and 483 males and 358 females were enrolled in the control group (Table 1). All patients included in our study were classified into three different PUMC AIS types based on their radiology data (Fig. S1). There were no significant differences in the main Cobb angle, age or gender between each of the PUMC groups (Table 1).
Table 1.
Characteristics of three PUMC types of AIS patients
| Variables | Casesa | Control | |||
|---|---|---|---|---|---|
| PUMC type I | PUMC type II | PUMC type III | In total | ||
| Number | 146 | 273 | 61 | 480 | 841 |
|
Gender M:F |
27:119 | 40:233 | 11:50 | 78:402 | 483:358 |
| Mean age | 13.57 ± 1.93 | 13.37 ± 1.96 | 13.59 ± 1.93 | 13.46 ± 1.94 | NA |
| Main Cobb angle | 49.22 ± 16.95 | 50.55 ± 14.16 | 53.23 ± 14.53 | 50.49 ± 15.12 | NA |
P value of Gender/Mean age/main Cobb angel was greater than 0.05 within groups; NA, not available.
Genotyping and Hardy–Weinberg equilibrium test
Among all participants, 14 SNPs in or around GPR126 were genotyped. rs6929442 was excluded from subsequent analyses because of the low call rate (Table S1). The other 13 SNPs were successfully genotyped with a minimum call rate of 98%. None of the 13 SNPs deviated from HWE and, thus, were subjected to subsequent analyses 21.
Association analysis
Of the 13 SNPs, rs225694, rs7774095 and rs2294773 presented significant allelic differences between the case and control groups. This result was demonstrated with LocusZoom, which showed that the alleles of rs225694, rs2294773 and rs7774095 had the highest association signal with AIS (http://csg.sph.umich.edu/locuszoom/) (Fig. 1). Controlling for sex, allele A of rs225694, allele A of rs774095 and allele G of rs2294773 had risk effects based on the different test models (P = 0.021, 0.048 and 0.023, respectively; Table 2).
Figure 1.
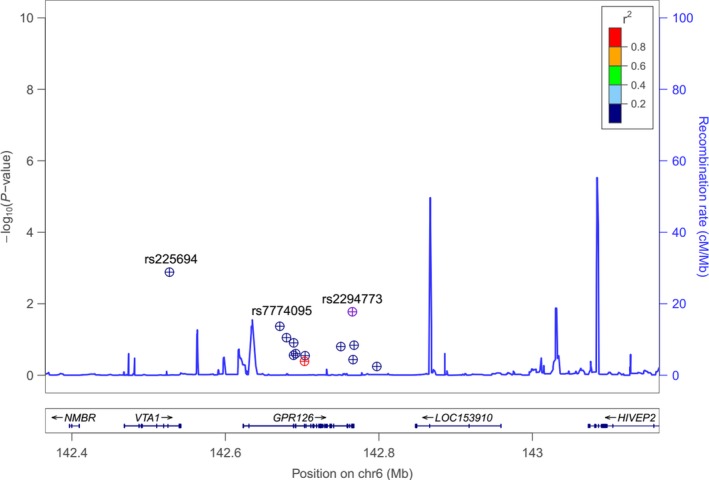
Regional Association Plot for the GPR126 candidate loci on 6q24. The ‐log10 (P value) of the candidate SNP loci association with adolescent idiopathic scoliosis was plotted with LocusZoom software. The SNPs with the highest association signal were rs225694, rs2294773 and rs7774095.
Table 2.
Association results of tested SNPs with AIS whilst controlling for sex
| SNP | Allele (1/2) | Test | OR | P * |
|---|---|---|---|---|
| rs225694 | A/G | ADD | 0.8749 | 0.8378 |
| DOMDEV | 2.409 | 0.211 | ||
| GENO 2DF | NA | 0.0208 | ||
| rs7774095 | A/C | ADD | 1.224 | 0.0481 |
| DOMDEV | 1.02 | 0.885 | ||
| GENO 2DF | NA | 0.08038 | ||
| rs6570507 | A/G | ADD | 1.187 | 0.08712 |
| DOMDEV | 1.01 | 0.9426 | ||
| GENO 2DF | NA | 0.169 | ||
| rs35699755 | T/C | ADD | 1.563 | 0.4273 |
| DOMDEV | 0.7156 | 0.5784 | ||
| GENO 2DF | NA | 0.6481 | ||
| rs17280293 | G/A | ADD | NA | NA |
| DOMDEV | NA | NA | ||
| GENO 2DF | NA | NA | ||
| rs11155242 | C/A | ADD | 0.9789 | 0.9444 |
| DOMDEV | 0.8723 | 0.6919 | ||
| GENO 2DF | NA | 0.6573 | ||
| rs2143390 | T/C | ADD | 1.241 | 0.1668 |
| DOMDEV | 0.888 | 0.5333 | ||
| GENO 2DF | NA | 0.3362 | ||
| rs4896582 | G/A | ADD | 0.8017 | 0.1006 |
| DOMDEV | 1.175 | 0.3417 | ||
| GENO 2DF | NA | 0.2575 | ||
| rs7755109 | G/A | ADD | 1.121 | 0.2292 |
| DOMDEV | 1.092 | 0.5039 | ||
| GENO 2DF | NA | 0.2732 | ||
| rs2294773 | G/C | ADD | 1.191 | 0.2659 |
| DOMDEV | 1.193 | 0.348 | ||
| GENO 2DF | NA | 0.0233 | ||
| rs2294775 | G/C | ADD | 0.8286 | 0.5173 |
| DOMDEV | 1.109 | 0.7514 | ||
| GENO 2DF | NA | 0.7197 | ||
| rs3748069 | G/A | ADD | 1.134 | 0.1949 |
| DOMDEV | 1.087 | 0.5332 | ||
| GENO 2DF | NA | 0.224 | ||
| rs7763064 | A/G | ADD | 1.126 | 0.2386 |
| DOMDEV | 0.7834 | 0.08444 | ||
| GENO 2DF | NA | 0.1994 |
ADD, additive effects of allele dosage test. DOMDEV tests a variable coded 0,1,0 for the three genotypes A1A1, A1A2, A2A2. GENO 2DF tests the coefficients for ADD and DOMDEV together. OR, odds ratio. NA, not available. *Chi‐square test, P‐values for each line reflect the effect of the entity under the TEST column. The bold values mean “statistically significantly different”.
Haplotypic analysis
We constructed haplotypes based on genotype data from thirteen SNPs using Haploview software (version 4.2). The pairwise linkage disequilibrium r‐square values between SNPs and the LD plots are presented in Figure S2. According to the Four Gamete Rule, we identified two haplotype blocks: rs7774095 and rs6570507 were in one block and rs35699755, rs17280293, rs11155242, rs2143390, rs4896582, rs7755109, rs2294773, rs2294775 and rs3748069 were in another block. There was no significant association between these two haplotype blocks and AIS when controlling for sex (Table 3).
Table 3.
Conditional haplotype‐based association testing in GPR126 gene with AIS whilst controlling for sex
| Haplotype | Frequency | OR | P Value | |
|---|---|---|---|---|
| Block 1 | AA | 0.337 | (‐ref‐) | 0.0668 |
| CA | 0.0159 | 0.6605 | ||
| CG | 0.646 | 0.8171 | ||
| Block 2 | TAACAGCCG | 0.0397 | (‐ref‐) | 0.0134 |
| CAATAGGCG | 0.203 | 0.85 | ||
| CAACAACCA | 0.365 | 0.7271 | ||
| CAACGACCA | 0.218 | 0.6555 | ||
| CGCCAGCGG | 0.0194 | 0.5834 | ||
| CACCAGCGG | 0.0522 | 0.6356 | ||
| CAACAGCCA | 0.0221 | 0.5847 | ||
| CAACAGGCG | 0.0143 | 2.666 | ||
| CAACGGCGG | 0.0101 | 0.8706 |
Block 1, rs7774095‐rs6570507, three common haplotypes (MHF, minimum haplotype frequency ≥0.01) from four possible. Block 2, rs35699755‐rs17280293‐rs11155242‐rs2143390‐rs4896582‐rs7755109‐rs2294773‐rs2294775‐rs3748069, nine common haplotypes (MHF ≥ 0.01) from 37 possible (‐ref‐), haplotype been selected to be the baseline, reference category. Unadjusted P‐value < Bonferroni‐corrected P‐value was considered statistically significant.
Luciferase assay
Among the three SNPs with positive results in association analyses, rs225694 and rs7774095 had potential enhancer activity. The luciferase activity of the risk allele and no‐risk allele was compared. The construct containing risk allele A of rs225694 had a 1.97‐fold higher level of luciferase activity than non‐risk allele C (P = 0.007). The construct containing risk allele A of rs7774095 had a 0.86‐fold lower level of luciferase activity than the non‐risk allele C (P = 0.015; shown in Fig. 2). Experiments using the HeLa cell line showed similar results (Fig. S3).
Figure 2.

Relative Luciferase Activity for different allele of rs225694 and rs7774095. (A) Transcriptional enhancer activities of rs225694 constructs. The construct containing the risk allele (R) had 1.97‐fold higher level relative luciferase activity than the non‐risk allele. *P value < 0.05. Error bars indicate stand error. The assay was repeated three times. (B) Transcriptional enhancer activities of rs7774095 constructs. The construct containing the risk allele (R) had 0.86‐fold lower level of relative luciferase activity than the non‐risk allele. *P value < 0.05. Error bars indicate stand error. The assay was repeated three times.
PUMC subgroup analysis
We conducted PUMC subgroup analysis in the three SNPs with positive association results. The analysis indicated that the genotypic association of rs225694 had positive results for the PUMC type II subgroups (P = 0.036). rs2294773 presented a positive result for the PUMC type I group (P = 0.018), and the other subgroups showed no significant difference (Table 4).
Table 4.
Peking Union Medical College (PUMC) subgroup analysis of three associated SNPs whilst controlling for sex
| SNP | Allele (1/2) | Test model | ALL | PUMC type I | PUMC type II | PUMC type III | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR | P | OR | P | OR | P | OR | P | |||
| rs225694 | A/G | ADD | 0.8749 | 0.8378 | 6.10E‐05 | 0.9991 | 4.52E‐05 | 0.999 | 2.456 | 0.168 |
| DOMDEV | 2.409 | 0.211 | 3.50E+04 | 0.999 | 4.99E+04 | 0.9989 | 0.8687 | 0.8636 | ||
| GENO 2DF | NA | 0.0208 | NA | 0.1246 | NA | 0.03641 | NA | 0.1399 | ||
| rs7774095 | A/C | ADD | 1.224 | 0.0481 | 1.323 | 0.05793 | 1.213 | 0.1166 | 1.038 | 0.8686 |
| DOMDEV | 1.02 | 0.885 | 1.014 | 0.9441 | 1.06 | 0.7272 | 0.8856 | 0.6925 | ||
| GENO 2DF | NA | 0.08038 | NA | 0.1199 | NA | 0.1598 | NA | 0.9247 | ||
| rs2294773 | G/C | ADD | 1.191 | 0.2659 | 1.332 | 0.1859 | 1.019 | 0.9233 | 1.272 | 0.3996 |
| DOMDEV | 1.193 | 0.348 | 1.271 | 0.3587 | 1.379 | 0.1696 | 0.7072 | 0.359 | ||
| GENO 2DF | NA | 0.0233 | NA | 0.01841 | NA | 0.09374 | NA | 0.6177 | ||
ADD, additive effects of allele dosage test. DOMDEV tests a variable coded 0,1,0 for the three genotypes A1A1, A1A2, A2A2. GENO 2DF tests the coefficients for ADD and DOMDEV together. OR, odds ratio. NA, not available. *Chi‐square test, P‐values for each line reflect the effect of the entity under the TEST column. The bold values mean “statistically significantly different”.
Discussion
In this study, we investigated the association of fourteen SNP loci in the GPR126 gene with AIS in a northern Chinese Han population. We found a significant association signal of rs7774095 with AIS. Moreover, we identified two novel SNP loci (rs225694 and rs2294773) that not only contributed to predisposition but were also associated with different types of AIS.
rs225694 was first identified to be associated with height 22. It is located in an intronic area of VTA1, but is in the same topologically associated domain (TAD) as GPR126 (Fig. 3). TAD is the compartment of the 3D chromatin space in which genomes of many bilaterian animals are organized. It is universally recognized that TAD plays an important role in gene regulation 23 and has potential gene‐enhancer interactions 24. Thus, rs225694 is supposed to have enhancer activity for the gene GPR126. Association analysis showed that this allele was most significantly associated with AIS, controlling for sex. Luciferase assays confirmed its enhancer function, including up‐regulation of transcription activity. Considering the PUMC classification, rs225694 had a powerful association only with type II AIS. We therefore suggested that the risk allele of rs225694 up‐regulated transcription of GPR126 and increased the risk of PUMC type II AIS.
Figure 3.
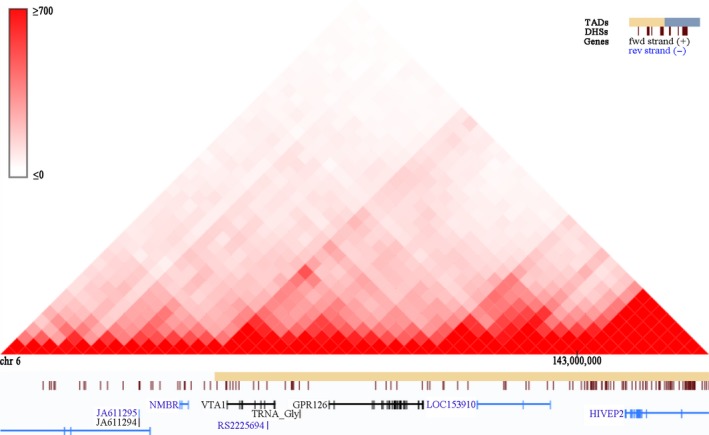
Topologically associated domain (TAD) of GPR126 and VTA1. Topologically associated domains were plotted by Hi‐C website. Genes GPR126 and VTA1 were in the same topologically associated domain.
rs7774095, an intronic SNP of GPR126, was first reported to be associated with AIS in a Japanese population in 2013 8. Other studies in southern Chinese populations replicated the association of this locus 15, 16. However, there was no clear functional evidence provided for this conclusion. We also observed this association and showed that it down‐regulates transcription activity. It has been shown that deletion of gpr126 in cartilage leads to an idiopathic scoliosis deformity in a mouse model 14. However, detailed clinical phenotype analyses found no significant association with any PUMC AIS subgroup, which may be due to the limited sample size. Additional studies with larger sample sizes are needed to identify the relationship between rs7774095 and PUMC AIS subgroups.
Association analyses of rs2294773 also showed significant results with AIS. rs2294773 is located in the 3′UTR of GPR126. It is postulated that the 3′UTR is a regulatory region that has potential activity in regulating mRNA localization, translation and stability 25. Different alleles at one locus can allow or eliminate miRNA binding. Song et al. found that mRNA containing the susceptibility allele rs4148941 had an enhanced interaction with miR‐513a‐5p, which may contribute to the aetiology of Lumbar disc degeneration (LDD) 26. Zhu et al. found a functional SNP in the 3′UTR region associated with bone mineral density (BMD) 27. As for rs2294773, in silico predictions by three online tools, including TargetScan (http://www.targetscan.org/) 25, 28, 29, miRanda (http://www.microrna.org/) 30 and mirSNP (http://cmbi.bjmu.edu.cn/mirSNP) 31, showed that there are three microRNAs, hsa‐miR‐1343, hsa‐miR‐198 and hsa‐miR‐30b‐3p that can bind to this mRNA. We also performed eQTL analysis in different tissues from the database and found that rs2294773 was associated with GPR126 gene expression in several tissues, including transformed fibroblasts, sun‐exposed skin and so on (Table S3, Fig. S4). Although there was no eQTL data for cartilage or bone, this locus may influence GRP126 gene expression and potentially contribute to AIS formation by these above miRNAs. Regarding the PUMC classification, rs2294773 also had a powerful association with only type I AIS. Additional replication studies with larger sample sizes are needed to provide further evidence for this association.
In our study, we identified two haplotype blocks in the candidate GPR126 gene with AIS. In the first block, we found that the A‐A haplotype of rs7774095‐rs6570507 was not significantly associated with AIS (P = 0.067), although previous studies had identified an association of rs6570507 with AIS. In contrast to previous studies, the lack of evidence of an association between rs6570507 and AIS may be due to different allele frequencies between geographic populations or the limitation of the sample size studied 8, 15, 32. In the second block, we also did not find significant association.
In conclusion, our study found that three SNPs near or in GPR126 were associated with AIS in the northern Chinese Han population. Risk allele A of rs225694 had higher transcription activity, and risk allele A of rs7774095 had lower transcription activity in vitro. We suggested that both overexpression and underexpression of GPR126 increased AIS susceptibility for different types of AIS and that SNPs at microRNA binding sites potentially contribute to AIS. GPR126 risk SNPs had potential associations with the type of AIS, as defined by the PUMC classification system. Rs225694 was functionally associated with PUMC type II AIS, and rs2294773 was mainly functionally associated with type I AIS, which contribute to our understanding of AIS development. SNPs of GRP126 were phenotypically associated with distinct PUMC types of AIS and might be useful as potential biomarkers for AIS classification and further surgical intervention.
Conflict of interest
The authors had no conflict of interest.
Supporting information
Figure S1 X‐ray image of AIS patients with PUMC classification system. A/B/C: adolescent idiopathic scoliosis with PUMC type I/II/III.
Figure S2 Linkage Disequilibrium (LD) structures of the thirteen candidate SNPs genotyped in GPR126 gene. The numbers inside the diamonds indicate the r‐square value for pairwise analysis. The LD strength between paired SNPs are shown in color of the diamonds according to the confidence interval's model.
Figure S3 Relative Luciferase Activity for different allele of rs225694 and rs7774095 in HeLa cell line. (A) Transcriptional enhancer activities of rs225694 constructs. The construct containing risk‐allele (R) had 1.97‐fold higher level of relative luciferase activity than non‐risk allele. *P value <0.05. Error bar, stand error. The assay was repeated three times. (B) Transcriptional enhancer activities of rs7774095 constructs. The construct containing risk‐allele (R) had 0.86‐fold lower level of relative luciferase activity than non‐risk allele. *P value <0.05. Error bar, stand error. The assay was repeated three times.
Figure S4 EQTL box plot of rs2294773 in different tissues.
Table S1. Fourteen target SNPs of GRP126 from literature review.
Table S2. Oligonucleotide sequence for luciferase assay.
Table S3. EQTL analysis result of rs2294773 in different tissues.
Acknowledgements
We give our great thanks to Dr. Jianguo Zhang, Jianxiong Shen, Shugang Li, Yipeng Wang, Hong Zhao and Yu Zhao for the patient enrolment and clinical evaluation; and to the patients for their active participation.
Funding source: This research was funded by National Natural Science Foundation of China (81501852, 81472046, 81472045, 81772299, 81772301), Beijing Natural Science Foundation (7172175), Beijing Nova Program (Z161100004916123), Beijing Nova Program Interdisciplinary Collaborative Project (xxjc201717), 2016 Milstein Medical Asian American Partnership Foundation Fellowship Award in Translational Medicine, The Central Level Public Interest Program for Scientific Research Institute (2016ZX310177), PUMC Youth Fund & the Fundamental Research Funds for the Central Universities (3332016006), CAMS Initiative for Innovative Medicine (2016‐I2M‐3‐003, 2016‐I2M‐2‐006), the Distinguished Youth Foundation of Peking Union Medical College Hospital (JQ201506) the 2016 PUMCH Science Fund for Junior Faculty (PUMCH‐2016‐1.1) and the National Key Research and Development Program of China (No. 2016YFC0901501).
Contributor Information
Nan Wu, Email: dr.wunan@pumch.cn.
Guixing Qiu, Email: qiuguixingpumch@126.com.
References
- 1. Weinstein SL. Adolescent idiopathic scoliosis: prevalence and natural history. Instr Course Lect. 1989; 38: 115–28. [PubMed] [Google Scholar]
- 2. Yawn BP, Yawn RA, Hodge D, et al A population‐based study of school scoliosis screening. JAMA. 1999; 282: 1427–32. [DOI] [PubMed] [Google Scholar]
- 3. King HA, Moe JH, Bradford DS, et al The selection of fusion levels in thoracic idiopathic scoliosis. J Bone Joint Surg Am. 1983; 65: 1302–13. [PubMed] [Google Scholar]
- 4. Lenke LG, Betz RR, Harms J, et al Adolescent idiopathic scoliosis: a new classification to determine extent of spinal arthrodesis. J Bone Joint Surg Am. 2001; 83‐A: 1169–81. [PubMed] [Google Scholar]
- 5. Qiu G, Li Q, Wang Y, et al Comparison of reliability between the PUMC and Lenke classification systems for classifying adolescent idiopathic scoliosis. Spine (Phila Pa 1976). 2008; 33: E836–42. [DOI] [PubMed] [Google Scholar]
- 6. Kesling KL, Reinker KA. Scoliosis in twins. A meta‐analysis of the literature and report of six cases. Spine (Phila Pa 1976). 1997; 22: 2009–14. [DOI] [PubMed] [Google Scholar]
- 7. Miller NH. Genetics of familial idiopathic scoliosis. Clin Orthop Relat Res. 2007; 462: 6–10. [DOI] [PubMed] [Google Scholar]
- 8. Kou I, Takahashi Y, Johnson TA, et al Genetic variants in GPR126 are associated with adolescent idiopathic scoliosis. Nat Genet. 2013; 45: 676–9. [DOI] [PubMed] [Google Scholar]
- 9. Liu S, Wu N, Zuo Y, et al Genetic polymorphism of LBX1 is associated with adolescent idiopathic scoliosis in Northern Chinese Han population. Spine (Phila Pa 1976). 2017; 42: 1125–9. [DOI] [PubMed] [Google Scholar]
- 10. Sharma S, Londono D, Eckalbar WL, et al A PAX1 enhancer locus is associated with susceptibility to idiopathic scoliosis in females. Nat Commun. 2015; 6: 6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ogura Y, Kou I, Miura S, et al A functional SNP in BNC2 is associated with adolescent idiopathic scoliosis. Am J Hum Genet. 2015; 97: 337–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grimes DT, Boswell CW, Morante NF, et al Zebrafish models of idiopathic scoliosis link cerebrospinal fluid flow defects to spine curvature. Science. 2016; 352: 1341–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Monk KR, Oshima K, Jors S, et al Gpr126 is essential for peripheral nerve development and myelination in mammals. Development. 2011; 138: 2673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karner CM, Long F, Solnica‐Krezel L, et al Gpr126/Adgrg6 deletion in cartilage models idiopathic scoliosis and pectus excavatum in mice. Hum Mol Genet. 2015; 24: 4365–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu JF, Yang GH, Pan XH, et al Association of GPR126 gene polymorphism with adolescent idiopathic scoliosis in Chinese populations. Genomics. 2015; 105: 101–7. [DOI] [PubMed] [Google Scholar]
- 16. Qin X, Xu L, Xia C, et al Genetic variant of GPR126 gene is functionally associated with adolescent idiopathic scoliosis in Chinese population. Spine (Phila Pa 1976). 2017; 42: E1098–103. [DOI] [PubMed] [Google Scholar]
- 17. Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012; 40: D930–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu S, Wu N, Liu J, et al Association between ADAMTS‐4 gene polymorphism and lumbar disc degeneration in Chinese Han population. J Orthop Res. 2016; 34: 860–4. [DOI] [PubMed] [Google Scholar]
- 19. Wu N, Yuan S, Liu J, et al Association of LMX1A genetic polymorphisms with susceptibility to congenital scoliosis in Chinese Han population. Spine (Phila Pa 1976). 2014; 39: 1785–91. [DOI] [PubMed] [Google Scholar]
- 20. Wu N, Chen J, Liu H, et al The involvement of ADAMTS‐5 genetic polymorphisms in predisposition and diffusion tensor imaging alterations of lumbar disc degeneration. J Orthop Res. 2014; 32: 686–94. [DOI] [PubMed] [Google Scholar]
- 21. Stark AE. Hardy‐Weinberg law: asymptotic approach to a generalized form. Science. 1976; 193: 1141–2. [DOI] [PubMed] [Google Scholar]
- 22. Lango AH, Estrada K, Lettre G, et al Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010; 467: 832–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Acemel RD, Maeso I, Gomez‐Skarmeta JL. Topologically associated domains: a successful scaffold for the evolution of gene regulation in animals. Wiley Interdiscip Rev Dev Biol. 2017; 6: e265. [DOI] [PubMed] [Google Scholar]
- 24. Lupianez DG, Kraft K, Heinrich V, et al Disruptions of topological chromatin domains cause pathogenic rewiring of gene‐enhancer interactions. Cell. 2015; 161: 1012–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005; 120: 15–20. [DOI] [PubMed] [Google Scholar]
- 26. Song YQ, Karasugi T, Cheung KM, et al Lumbar disc degeneration is linked to a carbohydrate sulfotransferase 3 variant. J Clin Invest. 2013; 123: 4909–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu DL, Guo Y, Zhang Y, et al A functional SNP regulated by miR‐196a‐3p in the 3'UTR of FGF2 is associated with bone mineral density in the Chinese population. Hum Mutat. 2017; 38: 725–35. [DOI] [PubMed] [Google Scholar]
- 28. Friedman RC, Farh KK, Burge CB, et al Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009; 19: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grimson A, Farh KK, Johnston WK, et al MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007; 27: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. John B, Enright AJ, Aravin A, et al Human MicroRNA targets. PLoS Biol. 2004; 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu C, Zhang F, Li T, et al MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA‐related SNPs in GWAS SNPs and eQTLs. BMC Genom. 2012; 13: 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moonesinghe R, Ioannidis JP, Flanders WD, et al Estimating the contribution of genetic variants to difference in incidence of disease between population groups. Eur J Hum Genet. 2012; 20: 831–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 X‐ray image of AIS patients with PUMC classification system. A/B/C: adolescent idiopathic scoliosis with PUMC type I/II/III.
Figure S2 Linkage Disequilibrium (LD) structures of the thirteen candidate SNPs genotyped in GPR126 gene. The numbers inside the diamonds indicate the r‐square value for pairwise analysis. The LD strength between paired SNPs are shown in color of the diamonds according to the confidence interval's model.
Figure S3 Relative Luciferase Activity for different allele of rs225694 and rs7774095 in HeLa cell line. (A) Transcriptional enhancer activities of rs225694 constructs. The construct containing risk‐allele (R) had 1.97‐fold higher level of relative luciferase activity than non‐risk allele. *P value <0.05. Error bar, stand error. The assay was repeated three times. (B) Transcriptional enhancer activities of rs7774095 constructs. The construct containing risk‐allele (R) had 0.86‐fold lower level of relative luciferase activity than non‐risk allele. *P value <0.05. Error bar, stand error. The assay was repeated three times.
Figure S4 EQTL box plot of rs2294773 in different tissues.
Table S1. Fourteen target SNPs of GRP126 from literature review.
Table S2. Oligonucleotide sequence for luciferase assay.
Table S3. EQTL analysis result of rs2294773 in different tissues.


