Abstract
Background
Soybean, a major legume crop native to East Asia, presents a wealth of resources for utilization. The basic leucine zipper (bZIP) transcription factors play important roles in various biological processes including developmental regulation and responses to environmental stress stimuli. Currently, little information is available regarding the bZIP family in the legume crop soybean.
Results
Using a genome-wide domain analysis, we identified 160 GmbZIP genes in soybean genome, named from GmbZIP1 to GmbZIP160. These 160GmbZIP genes, distributed unevenly across 20 chromosomes, were grouped into 12 subfamilies based on phylogenetic analysis. Gene structure and conserved motif analyses showed that GmbZIP within the same subfamily shared similar intron-exon organizations and motif composition. Syntenic and phylogenetic analyses identified 40 Arabidopsis bZIP genes and 83 soybean bZIP genes as orthologs. By investigating the expression profiling of GmbZIP in different tissues and under drought and flooding stresses, we showed that a majority of GmbZIP (83.44%) exhibited transcript abundance in all examined tissues and 75.6% displayed transcript changes after drought and flooding treatment, suggesting that GmbZIP may play a broad role in soybean development and response to water stress.
Conclusions
One hundred sixty GmbZIP genes were identified in soybean genome. Our results provide insights for the evolutionary history of bZIP family in soybean and shed light on future studies on the function of bZIP genes in response to water stress in soybean.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4511-6) contains supplementary material, which is available to authorized users.
Keywords: Soybean, bZIP genes, Phylogenetic analysis, Expression, Synteny analysis, Water stress
Background
Plants are exposed to extreme environment during their life cycle. Growth and development would be delayed once plants are exposed to terrible environmental conditions, such as drought, cold, high salt, and heat. To survive these stress conditions, plants regulate the expression of stress related genes. Transcription factors (TFs) play key roles in regulating the expression of functional proteins when plants encounter extreme environment conditions. Among TF families, the basic leucine zipper (bZIP) transcription factor family is a kind of conserved family that is prevalent in plants, animals, and microorganism. They possess a conserved 40–80 amino acid bZIP domain that has two structural features: a basic DNA binding region and a leucine zipper dimerization motif [1, 2]. The basic region is conserved, containing an invariant motif N-× 7-R/K-× 9 with about 18 amino acid residues [3–5]. The leucine zipper has a less conserved dimerization motif composing a heptad repeat of leucine residues, or other bulky hydrophobic amino acids that are involved in recognition and dimerization [4–6].
Considerable evidence has shown that bZIP TFs take part in different biological processes. OsABF1 (Oryza sativa ABA responsive element binding factor 1) can directly regulate and suppress Ehd1 expression and confer a later flowering phenotype in rice [7]. OsABF2, as a transcriptional regulator, can modulate the expression of abiotic stress-responsive genes through an ABA-dependent pathway [8]. OsbZIP23 transcription factor acts as a central regulator in ABA signaling and biosynthesis, and overexpressed OsbZIP23 shows significantly improved drought and salt tolerance, and BiFC assay in rice protoplast indicated that OsbZIP23 could interact with a SnRK2 protein kinase (SAPK2) [9]. ZmFEA4, an ortholog of Arabidopsis PERIANTHIA bZIP transcription factor in maize, promotes differentiation in the meristem periphery by regulating auxin-based responses [10]. The Arabidopsis transcription factor AtbZIP53 regulates seed maturation by influencing heterodimerization and protein complex formation [11]. AtbZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery [12]. The bZIP TFs also play key roles in regulation of plants response to various biotic and abiotic stresses, such as glucose-ABA signaling [13], sugar signaling during metabolism [14], ABA signaling for osmotic stress responses during vegetative growth [15], seed germination and flowering time [16], response to zinc deficiency [17], lipid stress responses [18], floral patterning [19], and auxin- mediated histone acetylation [20].
Due to their crucial role as regulator of responding to biotic and abiotic stresses, bZIP transcription factor families have been identified in various plant species. Based on their DNA binding specificity and amino acid sequences in basic and hinge regions, 89bZIPs were identified in rice [21]; 64 were found in cucumber based on predicted structural features [22], 92 in sorghum through genome-wide identification and characterization [23], 136 in Brassica rapa according to the presence of the UARR and LCRs [24]. Soybean, a major crop, plays an important role in food and industrial production. However, the genome-wide characterization of bZIP transcription factors in soybean has not yet been carried out. Here, we identified 160 bZIP genes from soybean genome and conducted a comprehensive genome-wide analysis of bZIP transcription factors in soybean in comparison with Arabidopsis. To infer the potential functions of these genes, the expression profile of bZIP genes in different soybean tissues and their response to drought and flooding were analyzed.
Methods
Identification of bZIP gene family in soybean and Arabidopsis
The whole bZIP protein sequences from soybean and Arabidopsis were obtained from Phytozome12 (https://phytozome.jgi.doe.gov/pz/portal.html), downloading the HMM profiles of the bZIP domain (PF00170, PF07716, PF03131) (http://pfam.sanger.ac.uk/) as queries to search predicted bZIP proteins in the soybean dataset using HMMER software 3.0 (HMMER: http://hmmer.wustl.edu/) with a threshold of e-value < e− 5 [25]. Subsequently, BLAST searches were performed to identify the predicted bZIPs in soybean database with all the Arabidopsis bZIPs as queries. Finally, candidate genes were examined to confirm the protein sequences derived from the selected soybean bZIPs, using the domain analysis programs of Pfam (Protein family: http://pfam.sanger.ac.uk/) and SMART (Simple Modular Architecture Research Tool: http://smart.embl-heidelberg.de/).
Phylogenetic analysis
Multiple sequence alignments of soybean and Arabidopsis bZIP were performed using muscle (http://www.ebi.ac.uk/Tools/msa/muscle/). Phylogenetic and molecular evolutionary analyses were generated using MEGA 6.0 software (http://www.megasoftware.net) and the FastTree 2.1.7 software (http://www.microbesonline.org/fasttree/).
Protein properties and sequence analyses
The molecular weight and isoelectric points of predicted GmbZIP proteins were detected using the ExPASy proteomics server (http://expasy.org/) [26]. The MEME program (http://meme.nbcr. net/meme/cgi-bin/meme.cgi) was employed to identify the conserved motifs in full-length soybean bZIP proteins with the following parameters: maximum number of motifs was 20 and the optimum width of motifs was set between 10 and 50 [27]. The GmbZIP gene structure analysis was displayed with GSDS (http://gsds.cbi.pku.edu.cn/).
Synteny analysis and chromosome localization
We used BLASTP to search for potential anchors (E < 1e-5, top 3 matches) within the soybean genome and between the soybean and Arabidopsis genomes and those containing soybean and Arabidopsis bZIP genes were identified and analyzed. All data used to analyze the expansion patterns of the GmbZIP family were shown in Additional file 1: Table S3 and Additional file 2: Table S4. Diagrams were generated using the Circos program (version 0.69) (http://circos.ca/). The synonymous (Ks) and non-synonymous (Ka) nucleotide substitutions between orthologous gene pairs were calculated based on the comparative synteny map between the soybean and Arabidopsis genomes, using Clustal W [28], PAL2NAL [29] and yn00 program of the bio-pipeline (https://github.com/ tanghaibao/ bio- pipeli ne).
Expression analysis of GmbZIP genes in different tissues
Transcriptome analysis was performed to identify expression patterns in representative tissues, including roots, root hairs, stems, nodules, leaves, shoot apical meristem, flowers, pods, and seeds. Transcriptome data of soybean tissues in different stages was obtained from Phytozome12 (https://phytozome.jgi.doe.gov/pz/portal.html) (Additional file 3: Table S5). Finally, heatmap of GmbZIP expression profile was produced by the pheatmap packages in R.
Plant material and treatments
The soybean seeds were obtained from center for genomics and biotechnology in Fujian Agriculture and Forestry University (FAFU), and were grown in a greenhouse in the pots containing nutritional soil. Two-week-old soybean seedlings were transferred to the drought and flooding conditions for a week, and drought stress was imposed by withholding water for 7 days, and flooding stress was imposed by placing the pots into a bigger pot with a trashcan liner filled up to a water level of 4 cm above the soil surface for 7 days. After 7 days of treatment, all the leaves were sampled for experiment [30].
Quantitative real-time PCR
Total RNA was extracted from the soybean leaves under the stress-treatment and unstressed using an RNA plant extraction kit (OMEGA, China) according to the manufacture’s instruction, and approximately 1 μg of purified total RNA was reverse transcribed to cDNA in a 20 μL reaction volume using AMV reverse transcriptase (Takara) according to the supplier’s instructions. To analyze the relative transcript levels of selected genes, real-time PCR was performed with specific primers according to the Bio-Rad Real-time PCR system (Foster city, CA, USA) and the SYBR Premix Ex II system (TakaRa Perfect Real Time). The qRT-PCR program was: 95 °C for 30 s;40 cycles of 95 °C for 5 s and 60 °C for 34 s; 95 °C for 15 s. The primers used for RT-PCR are listed in Additional file 4: Table S9.In each case, three technical replicates were performed for each of at least three independent biological replicates [31]. Quantities of standard RNA were prepared by diluting cDNA (1, 10− 1, 10− 2, 10− 3, 10− 4, and 10− 5) and only Ct values less than 40 were used to calculate correlation coefficients (R2 values) and amplification efficiencies (E) from the slope generated in Microsoft Excel 2013, based on the eq. E = [10−(1/slope) − 1] × 100%. All PCR assays showed efficiency values between 95% and 110% (Additional file 4: Table S9) [32].
Results
Identification of the soybean bZIP gene family
To identify GmbZIP genes, BLAST and Hidden Markov Model searches were used to search the soybean genome database with bZIP sequences from Arabidopsis as query. A total of 160GmbZIP genes were identified from soybean genome, named GmbZIP1 to GmbZIP160. The 160GmbZIP genes were unevenly distributed among 20 soybean chromosomes, with chromosome 11 containing the most (8.75%), followed by chromosome 12 and 13 with about 7.5%, chromosomes 7 and 9 less than 2% (Fig. 1). The full-length of 160 GmbZIP proteins varied from 95 (GmbZIP52) to 853 (GmbZIP44) amino acid residues with CDS ranging from 288 to 2562 bp, relative molecular mass distributing from 10.93 (GmbZIP95) to 157.28 (GmbZIP47) kDa, following with isoelectric points ranged from 4.76 to 10.63 (Additional file 5: Table S1; Additional file 6: Table S2).
Fig. 1.
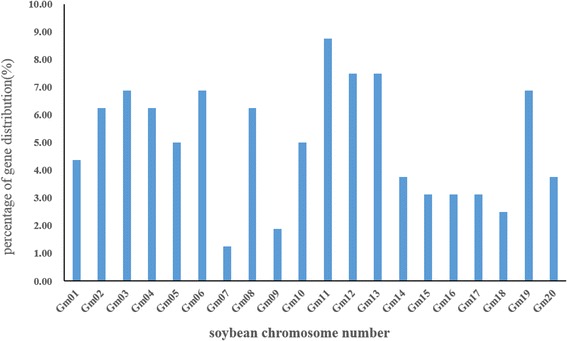
The distribution of bZIP genes on soybean chromosomes, shown as percentages
Expansion patterns of the soybean bZIP family
Segmental and tandem duplications provide two important mechanisms for the expansion of gene families and evidence of both could be found in an analysis of the GmbZIP gene family [33]. As displayed in Fig. 2, some GmbZIP gene pairs were distributed close to each other on the chromosomes, whose gene structure and coding sequence show high similarity (Fig. 3); for example, GmbZIP47 and GmbZIP49 on chromosome 6,GmbZIP114and GmbZIP115 on chromosome13.The segmental duplication blocks among genome were compared to the chromosomal location of all the GmbZIP genes. In total, more than 190 pairs of GmbZIP genes, such as GmbZIP1/GmbZIP72, GmbZIP24/GmbZIP75, GmbZIP153/GmbZIP159, as well as others (Additional file 1: Table S3), were located at two different chromosomes. In general, these results showed that the expansion of GmbZIP genes might result from the tandem and segmental duplications among these genes.
Fig. 2.
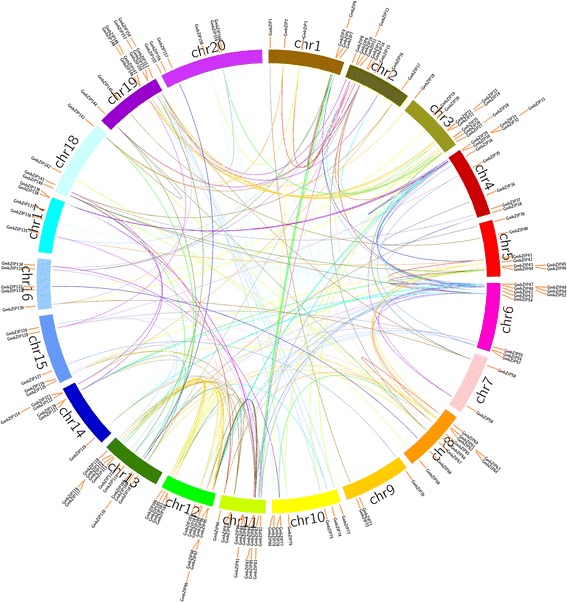
Distribution and synteny analysis of soybean bZIP genes. In the picture, the 20 soybean chromosomes are represented in different color partial circles and Chromosome numbers are indicated at the top of each bar. GmbZIP genes in different chromosomes are indicated by vertical red lines. Colored bars instruct bZIP syntenic regions on the soybean bZIP gene family
Fig. 3.
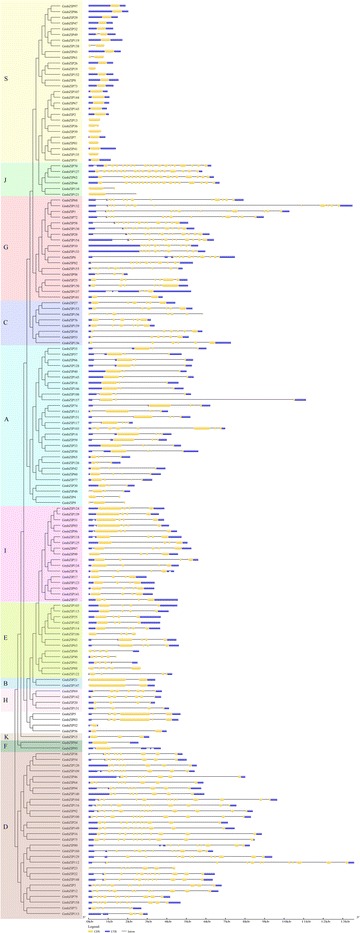
The exon-intron structure of GmbZIP genes based on the evolutionary relationship. The evolutionary tree was carried out with FastTree. Exon-intron analyses of GmbZIP genes were created with GSDS. Lengths of introns and exons of GmbZIP genes were exhibited proportionally
Gene structures and phylogenetic relationships of GmbZIP genes
To analyze the evolutionary history of the GmbZIP gene family, an unrooted phylogenetic tree was generated by a multiple sequence alignment of predicted GmbZIP proteins (Fig. 3). The result showed that bZIP proteins from the same group tended to cluster together, which is consistent with the two-species analysis (Fig. 4). To further understand the evolutionary relationships among GmbZIP genes, we then analyzed intron-exon organizations. As shown in Fig. 3, 25 bZIP genes have no intron, all of which belong to group S, accounting for 15.6% of the total number of bZIP genes. Among the intron containing genes, the number of introns in open reading frames varied from 1 to 18 and its intron number had a considerable variation among the different GmbZIP groups. For example, subfamilies A, B, C, E, F, H, K and I had 1–6 introns, whereas subfamilies D, G and J contained 7–18 introns, except for GmbZIP71, 113, 123, 101, 110 and 121. We thus propose that both exon loss and gain occurred during the evolution of the GmbZIP gene family.
Fig. 4.
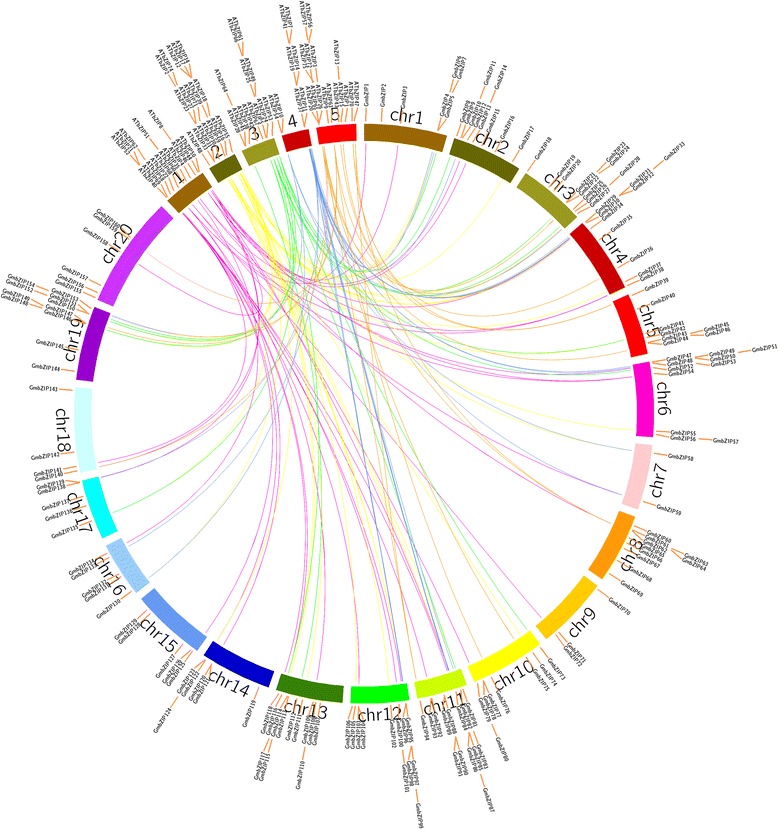
Synteny analysis of bZIP genes between soybean and Arabidopsis. In the picture, the 20 soybean chromosomes are represented in different color partial circles and Chromosome numbers are indicated at the top of each bar. Soybean and Arabidopsis bZIP genes are instructed by vertical red lines. Colored lines connecting two chromosomal regions indicate syntenic regions between soybean and Arabidopsis chromosomes
To obtain insight into the divergence and function of GmbZIP proteins, a total of 20 conserved motifs in the GmbZIPs were captured by the MEME software (Additional file 7: Figure S1) [34]. Most of the GmbZIP proteins within the same subfamily displayed similar motif compositions (Fig. 5), although there was high variance among the different subgroups. For instance, all the subfamilies contain motif1, which was annotated as basic-leucine zipper (bZIP) domain; subgroup A possesses motifs 1, 4, 7, 10 and 12, but different subgroup A members have diversity motifs; subfamily B shares motifs 1, 4 and 17; subfamily C harbors motifs 1, 4 and 9; subfamily D shows motifs 1, 2, 3, 5, 7, 8, and 11; subfamily E has motifs 1, 4 and 6; motifs 1, 4, 13, 15 and 16 appear in the subfamily G; motifs1, 4, 6, and 14 present in the subfamily I; motifs 4, 17, 18, 19 and 20 appear in the subfamily J; motifs 1, 4 and 9 appear in the subfamily S; all the members of subfamilies F and H have motif1 and 4; subfamily K only owns motif1.
Fig. 5.
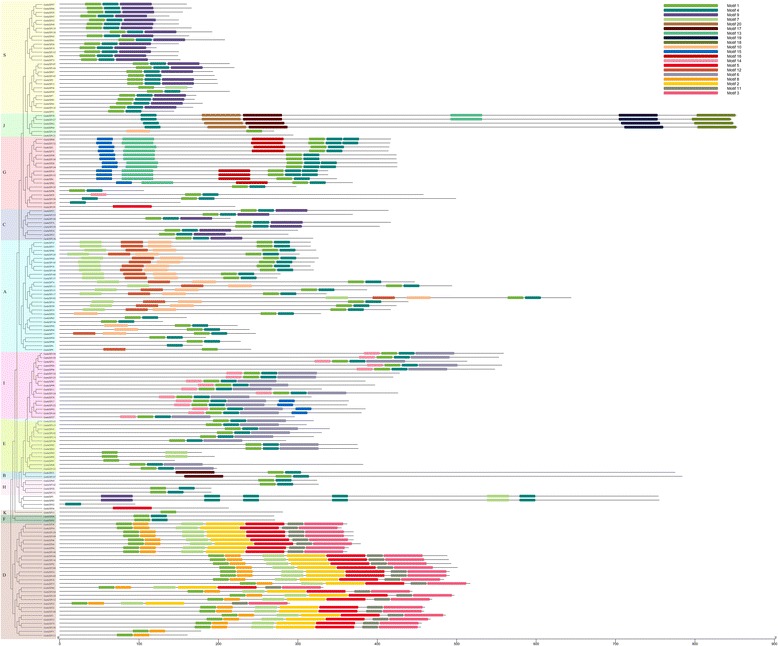
Conserved motifs of soybean bZIP proteins according to the evolutionary relationship. The conserved motifs in the GmbZIP proteins were identified with MEME software. Grey lines denote the non-conserved sequences, and each motif is indicated by a colored box numbered at the top. The length of motifs in each protein was presented proportionally
Evolutionary and phylogenetic relationship of soybean and Arabidopsis bZIP TFs
A comparison of the genomes of different organisms can be an effective mean to deduce the evolutionary history, origin, and function of uncharacterized genes [35]. Since Arabidopsis is one of the best-studied model plant species and the functions of several Arabidopsis bZIP genes have been well characterized, we generated a comparative bZIP synteny map of the soybean and Arabidopsis genomes. Large-scale syntenies identified 40 Arabidopsis genes and 83 soybean genes were orthologous (Fig.4 and Additional file 2: Table S4). Among them, three pairs of syntenic orthologous genes (one to one) were identified: GmbZIP33-AtbZIP15, GmbZIP134-AtbZIP51and GmbZIP112-AtbZIP45. Therefore, we proposed that these genes are derived from the same ancestor of Arabidopsis and soybean. We also identified that one soybean gene corresponds to multiple Arabidopsis genes, such as: GmbZIP117-AtbZIP39/AtbZIP67 and GmbZIP135- AtbZIP5/AtbZIP6. There are also syntenic orthologous gene pairs with one Arabidopsis gene corresponding to multiple soybean genes, such as: AtbZIP17-GmbZIP147/GmbZIP21, AtbZIP30-GmbZIP31/GmbZIP96, AtbZIP18-GmbZIP137/GmbZIP123/GmbZIP17. There are also gene pairs with two Arabidopsis genes correspond to the same four soybean genes: AtbZIP5/AtbZIP6-GmbZIP135/GmbZIP7/ GmbZIP81/GmbZIP4 and AtbZIP54/AtbZIP55- GmbZIP130/GmbZIP154/GmbZIP28/GmbZIP58. Numbers of synteny events suggested that many bZIP genes arose before the divergence of the Arabidopsis and soybean lineages. For further evolutionary studies, Ka and Ks values can be used to predict the selective pressure on duplicated genes, such that Ka/Ks < 1 indicates negative selection, Ka/Ks = 1 indicates neutral selection, and Ka/Ks ratio > 1 indicates positive selection [36]. To explore the divergence of orthologous gene pairs between soybean and Arabidopsis, the Ka, Ks and Ka/Ks of the orthologous gene pairs were calculated based on the comparative synteny map. Most of the soybean genes had a Ka/Ks ratio <0.25, with the highest in the ATbZIP54-GmbZIP130pair (Ka/Ks ratio = 0.35).
In order to further study the evolution of bZIP TFs, an unrooted neighbor-joining (NJ) tree was constructed using the predicted full-length GmbZIP protein sequences and the AtbZIP protein sequences obtained in previous study [37] (Figs. 4 and 6). The grouping in soybean was found to comply with the classification in Arabidopsis, and the Arabidopsis group nomenclature (A, B, C, D, E, F, G, H, I, and S) proposed by Jakoby and his colleagues [6] was adopted, along with the two extra groups (J and K) classified by Correa et al. [37]. The result showed that all the GmbZIP proteins were grouped into 12 subfamilies, except for GmbZIP5, GmbZIP83, GmbZIP52 and GmbZIP56. Subfamily A, D and S have nearly 30 GmbZIPs, 28, 29 and 28 respectively; whereas subfamily B, F, H, J and K own less than 6 GmbZIPs, suggesting that the bZIP family differentiated in soybean with diverse functions. Evolutionary analysis also identified that some orthologous bZIPs differentiated between soybean and Arabidopsis, indicating that some ancestral bZIP existed prior to the divergence of soybean and Arabidopsis.
Fig. 6.
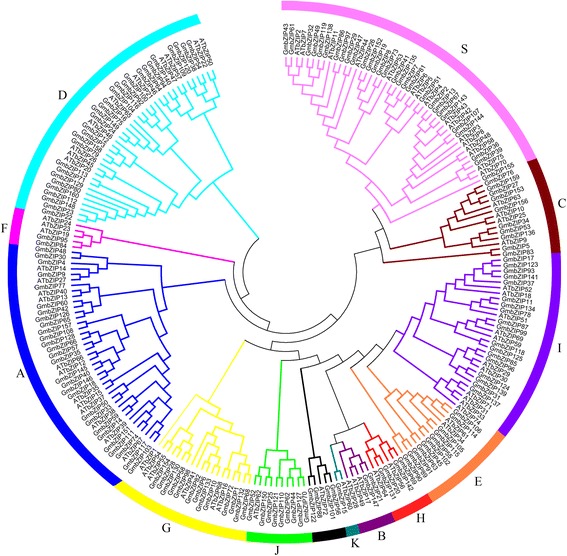
Phylogenetic analysis of soybean and Arabidopsis bZIP proteins
Expression profiles of GmbZIP genes in different tissues of soybean
To investigate the organ expression pattern of bZIP genes in soybean development, we performed transcriptome analysis of soybean tissues at different stages. The result showed that a total of 157 GmbZIP genes expressed in different tissues, while the rest of 3GmbZIP genes (GmbZIP19, GmbZIP110, GmbZIP156) were not detected in the RNA-seq libraries (Fig. 7, Additional file 3: Table S5). 131 (83.44%) genes exhibited transcript abundance in all tissues, in which 12 genes had high expression levels (value>10). Furthermore, 44 genes show high expression levels in flowers; 28 genes show high expression levels in leaves; 29 genes show high expression levels in pods; 29 genes show high expression levels in seeds; 46 genes show high expression levels in roots; 38 genes show high expression levels in root hair; 38 genes show high expression levels in shoot apical meristem; 37 genes show high expression levels in stem, 49 genes show high expression levels in nodules. Among these genes, some express only in reproductive tissues while some others express only in vegetative tissues. Interestingly, GmbZIP74 and GmbZIP111 show high expression level only in seed and barely no expression in vegetative tissues; GmbZIP6 shows high expression level only in flower; and GmbZIP109, GmbZIP41 and GmbZIP135 show high expression level in different vegetative tissues while no expression in reproductive tissues.
Fig. 7.
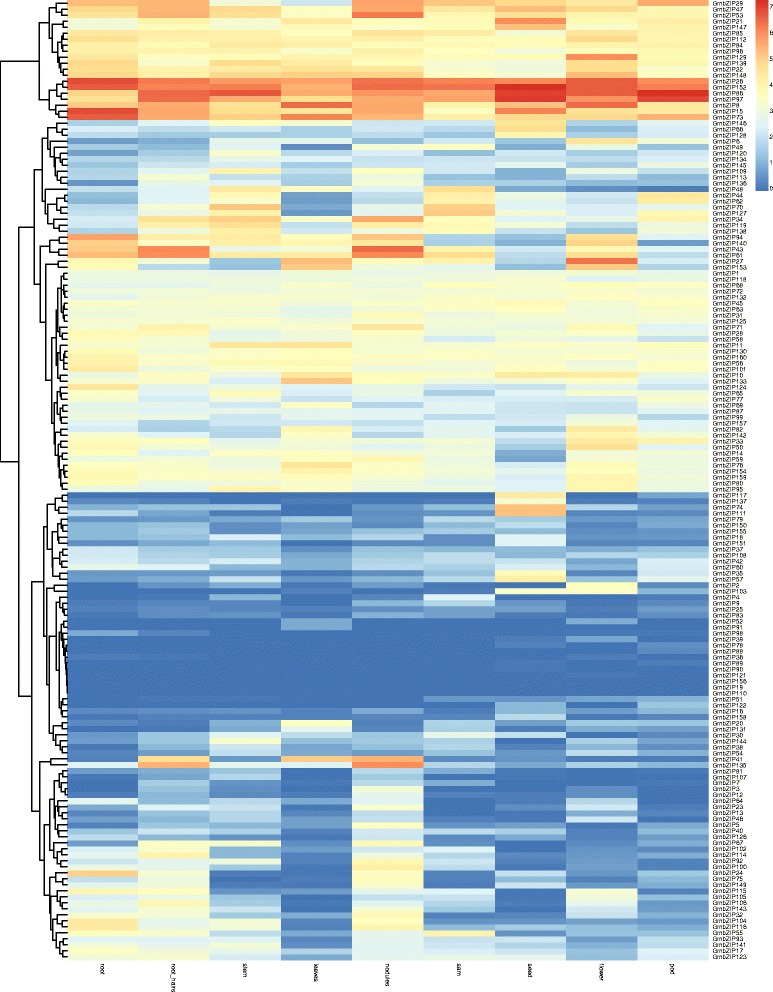
Expression profiles of 160 GmbZIP genes in 9 different tissues of soybean. Different colors in map represent gene transcript abundance values as shown in bar at top of figure
Expression profiles of bZIP genes in response to abiotic stress
To date, many bZIP genes in response to abiotic stress have been identified; some bZIP genes were induced by drought, such as in sheep grass (Leymuschinensis), apple and rice [7, 38, 39], while some bZIP genes were induced by flooding in soybean and Taxodium [40, 41]. To investigate the roles of bZIP genes in soybean in response to abiotic stress, an expression analysis of these genes against drought and flood were displayed with the RNA-seq data obtained from Chen et al. [30] (Additional file 8: Table S6 and Additional file 9: Table S7). According to the transcriptome data, 124 out of 160 GmbZIP genes were involved in drought stress response. 21GmbZIP genes were downregulated significantly (|log2(fold change)|>1), while 31GmbZIP genes were upregulated significantly in drought stress. Genes downregulated after drought treatment mainly distribute in the subfamilies A, D and S, while upregulated in subfamilies A, E, G, I and S. 122 out of 160 GmbZIP genes were involved in flooding stress response. 19 GmbZIP genes showed downregulated significantly; 10 GmbZIP genes behaved upregulated significantly in flooding (Fig. 8). Genes downregulated after flooding treatment mainly distribute in the subfamilies A, D, E, H and S, while upregulated in subfamilies C, G and S (Fig. 8). Expression of many bZIP genes is induced by both drought and flooding treatment, such as, GmbZIP42,58,153,27,61,73 and 8 are upregulated significantly in both abiotic treatment, and GmbZIP49, 131, 30, 120, 20, 119, 14 and 48 are downregulated significantly. GmbZIP102 is specifically upregulated after drought treatment, while downregulated after flooding treatment. To verify the accuracy of these data, quantitative real-time PCR analysis was performed to detect the expression of randomly selected bZIP genes following abiotic stress. These selected genes contain four parts: up- and down-regulated after drought and flooding treatments (Additional file 10: Figure S2 and Additional file 11: Figure S3). The results are consistent with the gene profiling data.
Fig. 8.
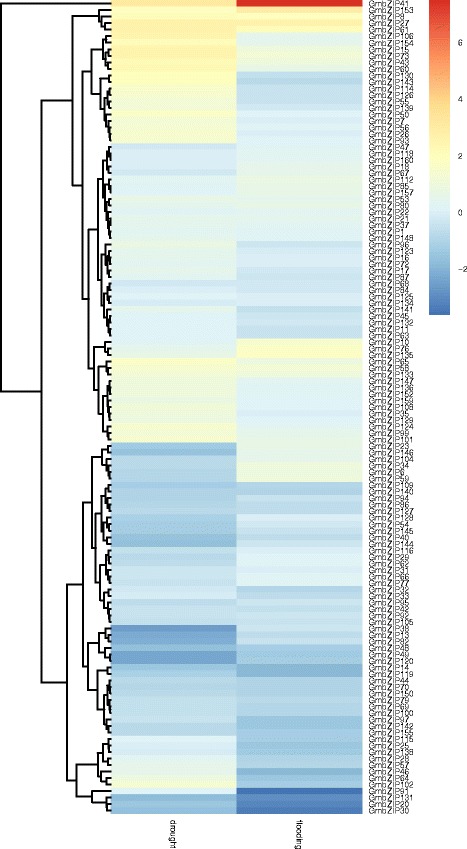
Heat map showing GmbZIP expression patterns in soybean under drought and flooding stress. Different colors in map represent gene transcript abundance values after dividing wild type as shown in bar at top of figure
Identification of cis-elements in GmbZIP promoters
To explore the mechanisms of GmbZIP response to the abiotic stresses, we searched for 16 stress-related cis-elements, such as W-box [42], HSE [43], GARE [44] in the GmbZIP promoters. The result showed that more than two cis-elements were located in the promoters of all GmbZIP genes with the least two cis-elements in the promoters of GmbZIP11, 51, 69, 85 and 141 and the most ten in the promoter of GmbZIP121. About 60% (93 out of 160) GmbZIP promoters contain both HSE and MBS (Additional file 12: Table S8).
Discussion
Soybean, a major legume crop native to east Asia, presents a wealth of resources for utilization, including oils, proteins, mineral nutrients, and natural products such as isoflavonoids that impact human health and nutrition [45]. Compared to other important crops, research progress on soybean has developed slowly, particularly with the aspect of the responses to biotic and abiotic stresses. The bZIP family, one of the most important transcription factor family, has been reported to participate in diverse biological processes, such as plants development, organ differentiation, as well as response to environmental stresses. Although the bZIP family has been identified in many plant species, the genome-wide identification of GmbZIPs has not yet been reported.
In this study, 160 bZIP family genes were identified in the soybean genome. Previous studies have identified 75 bZIPs in Arabidopsis [6], 77 in cassava [3], 85 in rice [46], 64 in cucumber [22], 125 in maize [5]. These data suggested that bZIP in soybean had expanded compared to that in Arabidopsis, cassava, rice, cucumber, and maize. Evolutionary analysis indicated soybean bZIP family could be divided into 12 subgroups based on cluster analysis and a comparison with Arabidopsis bZIP genes. The phylogenetic analysis was also supported by gene structure and conserved motif analyses. Gene structure analysis showed that GmbZIPs possess intron with number varying from 0 to 12, and each subfamily displayed similar intron-exon organizations (Fig. 3). We determined that about 15% of the genes had no introns, which is similar to the status in apple, rice and sorghum [21, 23]. We found that the two genes in a duplicated gene pair tended to be clustered into one group, when phylogenetic analysis was performed, such that segmental pairs GmbZIP17-GmbZIP123 (3introns), GmbZIP21-GmbZIP147 (1introns) and GmbZIP119-GmbZIP138 (no intron) were clustered to group I, B and S, respectively. This was also true for the tandem duplicated pairs. Some duplicated genes had the same number of exons and nearly identical lengths, indicating that segmental or tandem duplication expanded the bZIP family. The members of subfamilies D and G harbor more than 8 introns, while other subfamilies present fewer introns. According to a recent study in rice, the rate of intron loss is faster than the rate of intron gain after segmental duplication [46]. Thus, it can be deduced that the subfamilies G and D might contain the original genes. Exon/intron gain/loss is one of mechanism for diversification of multiple gene families [47], and was also observed in this study. For example, GmbZIP92 contained 12 exons, while the paralogous gene, GmbZIP103, had 11 (Fig. 3), indicating a loss of a exons during evolution, a similar pattern is reported for the apple bZIP family [39]. These gain/losses may be the results of chromosomal rearrangements and fusions, and can potentially lead to the generation of functionally distinct paralogs [48]. Conserved motif analysis indicated that all the GmbZIPs shared the typical bZIP domain, and each subgroup harbored similar motifs. We found motif 17 and 20 share the same domain, function in phosphatidylcholine protein transfer, so does the motif 18 and 19, which function in signal transduction, and they are very conserved, only contained in the subfamily J, so the subfamily J may be also conserved and functional (Additional file 7: Figure S1). Gene structure and conserved motif analyses showed that the same subfamily harbored similar intron-exon organizations and conserved motifs, indicating the relationship of GmbZIPs in the same group was closer during the gene evolution process [49].
Comparative genomics relies on the structuring of genomes into syntenic blocks that display conserved features across the genomes [50]. The synteny analysis denotes functional and evolutionary connections between soybean and Arabidopsis syntenic genes [51]. Many Arabidopsis bZIP genes response to biotic and abiotic stresses. For example, AtbZIP1 can mediate sugar signaling, and when carbon nutrients are limited, gain or loss of function of AtbZIP1 causes changes in the rates of early seedling establishment [52]. AtbZIP34 is required for Arabidopsis pollen wall patterning and can regulate lipid metabolism and cellular transportin developing pollen [53]. In this study, both syntenic and phylogenetic analyses were performed to assess the relationship to the model plant, Arabidopsis, and to infer the functions of orthologous genes. A total of 40 Arabidopsis bZIP genes and 83 soybean bZIP genes were identified as orthologs. Among these genes, three single orthologous gene pairs between soybean and Arabidopsis were identified, indicating that these genes might present in the genome of the last common ancestor of the two species. The remaining gene combinations showed a more severe complex relationship, with many pairs of single Arabidopsis genes corresponding to multiple soybean genes and single soybean genes corresponding to multiple Arabidopsis genes. Since some of the bZIP genes present in these two species that could not be mapped into any syntenic blocks, this may be explained by the fact that after the speciation of soybean and Arabidopsis, their genomes have experienced multiple rounds of important chromosomal rearrangement and fusions, followed by selective gene loss, which can obscure the identification of chromosomal syntenies [54, 55]. In general, we deduce that some of the soybean genes seemed to share a common ancestor with their Arabidopsis bZIP counterparts, and since all the Ka, Ks, and Ka/Ks of the syntenic orthologous gene-pairs had Ka/Ks ratios < 1, they might undergo a strong Darwinian purifying positive selection.
In plants, basic region/leucine zipper motif (bZIP) transcription factors regulate processes including pathogen defense, light and stress signaling, seed maturation and flower development. The function of some bZIP genes in soybean has been reported, for instance, overexpression of GmbZIP110 can improve the tolerance of salt [56]. GmbZIP44, GmbZIP62 andGmbZIP110 are the negative regulators of ABA signaling [57]. Based on the expression data (Additional file 8: Table S6), all investigated soybean organs expressed at least one bZIP gene, indicating that they may play a broad role in soybean development. Most of the expressed genes were detected to some degree in roots, stems, leaves, as well as in flowers and seeds, which is consistent with previous studies in Arabidopsis [6]. In this study, no less than 75.6% bZIP genes displayed transcriptional changes after abiotic stress treatment, including drought and flooding (Fig. 8 and Additional file 3: Table S5 and Additional file 8: Table S6). Gene expression analysis has showed that more genes changed after drought treatment than flooding, so the bZIP family may play more dominantly role in regulating drought stress. Collectively, these findings indicated that bZIP genes might contribute to the robust resistance to water stress and provide a foundation for further research on the bZIP-mediated water stress response in soybean. Some GmbZIP genes were not detected in the database, and the similar circumstance was observed in apple and grape [39, 58], we therefore hypothesize that these genes may have been silenced or their expression is too weak to be detected. In our study, most of GmbZIP genes were responsive to drought and flood, and consistent with expression, their promoters contained at least one out of three drought-relative cis-elements (ABRE, MBS and W-box).
Conclusions
In general, we performed the first genome-wide identification of the soybean bZIP TF family and conducted a detailed investigation of their structure and organization. We also characterized the expression of some GmbZIP genes following various abiotic stresses. These results may prove useful in developing strategies for the future improvement of stress tolerance in soybean.
Additional files
TableS3. Synteny blocks of bZIP genes within soybean genome. (XLSX 19 kb)
Table S4. Synteny blocks of bZIP genes between soybean and Arabidopsis genomes. (XLSX 19 kb)
Table S5. The expression profiles of the soybean bZIP genes in different tissues. (XLSX 5886 kb)
Table S9. Primers used in qRT-PCR analysis. (XLSX 10 kb)
Table S1. Characteristics of bZIPs in soybean. (XLSX 17 kb)
Table S2. The accession numbers of bZIPs in soybean and Arabidopsis. (XLSX 5633 kb)
Figure S1. 20 conserved motifs and functions of some motifs. (DOCX 720 kb)
Table S6. The expression profiles (log2-based values) of the soybean bZIP genes after drought treatment. In this experiment, two-week-old soybean seedlings were transferred to the drought conditions for a week, and drought stress was imposed by withholding water for 7 days. After 7 days of treatment, the leaves were collected for experiment [30]. (XLSX 18 kb)
Table S7. The expression profiles (log2-based values) of the soybean bZIP genes after flooding treatment. In this experiment, two-week-old soybean seedlings were transferred to the flooding conditions for a week, and flooding stress was imposed by placing the pots into a bigger pot with a trashcan liner filled up to a water level of 4 cm above the soil surface for 7 days. After 7 days of treatment, the leaves were collected for experiment [30]. (XLSX 16 kb)
Figure S2. Validation of drought treatment RNA-seq data. (A) RNA-seq data of downregulated genes after drought treatment. (B) Validation of down regulated genes among drought sequencing data by real-time quantitative PCR. (C) RNA-seq data of upregulated genes after drought treatment. (D) Validation of up regulated genes among drought sequencing data by real-time quantitative PCR. FPKM represents Fragments Per Kilobase per Million fragments according to previously described [59]. (TIFF 203 kb)
Figure S3. Validation of flooding treatment RNA-seq data. (A) RNA-seq data of downregulated genes after flooding treatment. (B) Validation of down regulated genes among flooding sequencing data by real-time quantitative PCR. (C) RNA-seq data of upregulated genes after flooding treatment. (D) Validation of up regulated genes among flooding sequencing data by real-time quantitative PCR. (TIFF 209 kb)
Table S8. Cis-elements in GmbZIP promoters. (XLSX 20 kb)
Acknowledgements
We would like to thank the reviewers for their helpful comments on the original manuscript.
Funding
This work was supported by NSFC (U1605212; 31522009; 31470284 to Y.Q.; 31600249 to L.Z.).
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its Additional files].
Author contributions
MZ performed phylogenetic and expression analysis. YL annotated the genes on chromosomes and conducted the evolution analysis. HS and MG calculated all the data, MC, QH, MY, DC and LZ performed qRT-PCR analysis. HC and YQ designed the research. MZ and YQ wrote the manuscript. All authors have read and approved the manuscript.
Abbreviations
- ABA
Abscisic acid
- At
Arabidopsis thaliana
- bZIP
Basic Leucine Zipper
- Gm
Glycine max
- GSDS
Gene Structure Display Server
- HMM
Hidden Markov Models
- MEGA
Molecular Evolutionary Genetics Analysis
- MEME
Multiple Em for Motif Elicitation
- NJ
Neighbor-joining
- Os
Oryza sativa
- qRT-PCR
Quantitative real-time PCR
- RNA-seq
RNA sequencing
- SMART
Simple Modular Architecture Research Tool
- TF
Transcription factor
- Zm
Zea mays
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4511-6) contains supplementary material, which is available to authorized users.
Man Zhang and Yanhui Liu are co-first authors and contributed equally to this work.
Contributor Information
Man Zhang, Email: zhangman3043@163.com.
Yanhui Liu, Email: yanhuiliu520@gmail.com.
Hang Shi, Email: 1271652262@qq.com.
Mingliang Guo, Email: 1640715085@qq.com.
Mengnan Chai, Email: 695274901@qq.com.
Qing He, Email: 382017115@qq.com.
Maokai Yan, Email: 562640038@qq.com.
Du Cao, Email: 275993304@qq.com.
Lihua Zhao, Email: 308207694@qq.com.
Hanyang Cai, Phone: +86-591-83595382, Email: caihanyang123@163.com.
Yuan Qin, Phone: +86-591-83595382, Email: yuanqin@fafu.edu.cn.
References
- 1.Talanian RV, McKnight CJ, Kim PS. Sequence-specific DNA binding by a short peptide dimer. Science. 1990;249(4970):769–771. doi: 10.1126/science.2389142. [DOI] [PubMed] [Google Scholar]
- 2.Hurst HC. Transcription factors. 1: bZIP proteins. Protein Profile. 1994;1(2):123–168. [PubMed] [Google Scholar]
- 3.Hu W, Yang H, Yan Y, Wei Y, Tie W, Ding Z, Zuo J, Peng M, Li K. Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci Rep. 2016;6:22783. doi: 10.1038/srep22783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu X, Chu Z. Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium Distachyon. BMC Genomics. 2015;16:227. doi: 10.1186/s12864-015-1457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei K, Chen J, Wang Y, Chen Y, Chen S, Lin Y, Pan S, Zhong X, Xie D: Genome-wide analysis of bZIP-encoding genes in maize. DNA research: an international journal for rapid publication of reports on genes and genomes 2012, 19(6):463–476. [DOI] [PMC free article] [PubMed]
- 6.Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F, Ziprg B. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7(3):106–111. doi: 10.1016/S1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- 7.Zhang C, Liu J, Zhao T, Gomez A, Li C, Yu C, Li H, Lin J, Yang Y, Liu B, et al. A drought-inducible transcription factor delays reproductive timing in Rice. Plant Physiol. 2016;171(1):334–343. doi: 10.1104/pp.16.01691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hossain MA, Cho JI, Han M, Ahn CH, Jeon JS, An G, Park PB. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J Plant Physiol. 2010;167(17):1512–1520. doi: 10.1016/j.jplph.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Zong W, Tang N, Yang J, Peng L, Ma S, Xu Y, Li G, Xiong L. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiol. 2016;171(4):2810–2825. doi: 10.1104/pp.16.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pautler M, Eveland AL, LaRue T, Yang F, Weeks R, Lunde C, Je BI, Meeley R, Komatsu M, Vollbrecht E, et al. FASCIATED EAR4 encodes a bZIP transcription factor that regulates shoot meristem size in maize. Plant Cell. 2015;27(1):104–120. doi: 10.1105/tpc.114.132506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alonso R, Onate-Sanchez L, Weltmeier F, Ehlert A, Diaz I, Dietrich K, Vicente-Carbajosa J, Droge-Laser W. A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell. 2009;21(6):1747–1761. doi: 10.1105/tpc.108.062968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berendzen KW, Weiste C, Wanke D, Kilian J, Harter K, Droge-Laser W. Bioinformatic cis-element analyses performed in Arabidopsis and rice disclose bZIP- and MYB-related binding sites as potential AuxRE-coupling elements in auxin-mediated transcription. BMC Plant Biol. 2012;12:125. doi: 10.1186/1471-2229-12-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matiolli CC, Tomaz JP, Duarte GT, Prado FM, Del Bem LE, Silveira AB, Gauer L, Correa LG, Drumond RD, Viana AJ, et al. The Arabidopsis bZIP gene AtbZIP63 is a sensitive integrator of transient abscisic acid and glucose signals. Plant Physiol. 2011;157(2):692–705. doi: 10.1104/pp.111.181743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dietrich K, Weltmeier F, Ehlert A, Weiste C, Stahl M, Harter K, Droge-Laser W. Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell. 2011;23(1):381–395. doi: 10.1105/tpc.110.075390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida T, Fujita Y, Maruyama K, Mogami J, Todaka D, Shinozaki K, Yamaguchi-Shinozaki K. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015;38(1):35–49. doi: 10.1111/pce.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Li L, Ye T, Lu Y, Chen X, Wu Y. The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J Exp Bot. 2013;64(2):675–684. doi: 10.1093/jxb/ers361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Assuncao AG, Herrero E, Lin YF, Huettel B, Talukdar S, Smaczniak C, Immink RG, van Eldik M, Fiers M, Schat H, et al. Arabidopsis Thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proc Natl Acad Sci U S A. 2010;107(22):10296–10301. doi: 10.1073/pnas.1004788107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stotz HU, Mueller S, Zoeller M, Mueller MJ, Berger S. TGA transcription factors and jasmonate-independent COI1 signalling regulate specific plant responses to reactive oxylipins. J Exp Bot. 2013;64(4):963–975. doi: 10.1093/jxb/ers389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maier AT, Stehling-Sun S, Wollmann H, Demar M, Hong RL, Haubeiss S, Weigel D, Lohmann JU. Dual roles of the bZIP transcription factor PERIANTHIA in the control of floral architecture and homeotic gene expression. Development. 2009;136(10):1613–1620. doi: 10.1242/dev.033647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiste C, Droge-Laser W. The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nat Commun. 2014;5:3883. doi: 10.1038/ncomms4883. [DOI] [PubMed] [Google Scholar]
- 21.Nijhawan A, Jain M, Tyagi AK, Khurana JP. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008;146(2):333–350. doi: 10.1104/pp.107.112821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baloglu MC, Eldem V, Hajyzadeh M, Unver T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS One. 2014;9(4):e96014. doi: 10.1371/journal.pone.0096014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Zhou J, Zhang B, Vanitha J, Ramachandran S, Jiang SY. Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum. J Integr Plant Biol. 2011;53(3):212–231. doi: 10.1111/j.1744-7909.2010.01017.x. [DOI] [PubMed] [Google Scholar]
- 24.Hwang I, Jung HJ, Park JI, Yang TJ, Nou IS. Transcriptome analysis of newly classified bZIP transcription factors of Brassica Rapa in cold stress response. Genomics. 2014;104(3):194–202. doi: 10.1016/j.ygeno.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 25.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7(10):e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31(13):3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown P, Baxter L, Hickman R, Beynon J, Moore JD, Ott S. MEME-LaB: motif analysis in clusters. Bioinformatics. 2013;29(13):1696–1697. doi: 10.1093/bioinformatics/btt248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34(Web Server):W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen W, Yao Q, Patil GB, Agarwal G, Deshmukh RK, Lin L, Wang B, Wang Y, Prince SJ, Song L, et al. Identification and comparative analysis of differential gene expression in soybean leaf tissue under drought and flooding stress revealed by RNA-Seq. Front Plant Sci. 2016;7:1044. doi: 10.3389/fpls.2016.01044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai H, Zhao L, Wang L, Zhang M, Su Z, Cheng Y, Zhao H, Qin Y. ERECTA signaling controls Arabidopsis inflorescence architecture through chromatin-mediated activation of PRE1 expression. The New phytologist. 2017;214(4):1579–1596. doi: 10.1111/nph.14521. [DOI] [PubMed] [Google Scholar]
- 32.Niu X, Qi J, Chen M, Zhang G, Tao A, Fang P, Xu J, Onyedinma SA, Su J. Reference genes selection for transcript normalization in kenaf (Hibiscus Cannabinus L.) under salinity and drought stress. PeerJ. 2015;3:e1347. doi: 10.7717/peerj.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis Thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyons E, Pedersen B, Kane J, Alam M, Ming R, Tang H, Wang X, Bowers J, Paterson A, Lisch D, et al. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 2008;148(4):1772–1781. doi: 10.1104/pp.108.124867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krishnamurthy P, Hong JK, Kim JA, Jeong MJ, Lee YH, Lee SI. Genome-wide analysis of the expansin gene superfamily reveals Brassica Rapa-specific evolutionary dynamics upon whole genome triplication. Molecular genetics and genomics : MGG. 2015;290(2):521–530. doi: 10.1007/s00438-014-0935-0. [DOI] [PubMed] [Google Scholar]
- 37.Correa LG, Riano-Pachon DM, Schrago CG, dos Santos RV, Mueller-Roeber B, Vincentz M. The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS One. 2008;3(8):e2944. doi: 10.1371/journal.pone.0002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao P, Liu P, Yuan G, Jia J, Li X, Qi D, Chen S, Ma T, Liu G, Cheng L. New insights on drought stress response by global investigation of gene expression changes in Sheepgrass (Leymus Chinensis) Front Plant Sci. 2016;7:954. doi: 10.3389/fpls.2016.00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao J, Guo R, Guo C, Hou H, Wang X, Gao H. Evolutionary and expression analyses of the apple basic Leucine zipper transcription factor family. Front Plant Sci. 2016;7:376. doi: 10.3389/fpls.2016.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nanjo Y, Maruyama K, Yasue H, Yamaguchi-Shinozaki K, Shinozaki K, Komatsu S. Transcriptional responses to flooding stress in roots including hypocotyl of soybean seedlings. Plant Mol Biol. 2011;77(1–2):129–144. doi: 10.1007/s11103-011-9799-4. [DOI] [PubMed] [Google Scholar]
- 41.Qi B, Yang Y, Yin Y, Xu M, Li H. De novo sequencing, assembly, and analysis of the Taxodium 'Zhongshansa' roots and shoots transcriptome in response to short-term waterlogging. BMC Plant Biol. 2014;14:201. doi: 10.1186/s12870-014-0201-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsutsui T, Morita-Yamamuro C, Asada Y, Minami E, Shibuya N, Ikeda A, Yamaguchi J. Salicylic acid and a chitin elicitor both control expression of the CAD1 gene involved in the plant immunity of Arabidopsis. Biosci Biotechnol Biochem. 2006;70(9):2042–2048. doi: 10.1271/bbb.50700. [DOI] [PubMed] [Google Scholar]
- 43.Yoo HY, Chang MS, Rho HM. The activation of the rat copper/zinc superoxide dismutase gene by hydrogen peroxide through the hydrogen peroxide-responsive element and by paraquat and heat shock through the same heat shock element. J Biol Chem. 1999;274(34):23887–23892. doi: 10.1074/jbc.274.34.23887. [DOI] [PubMed] [Google Scholar]
- 44.Gomez-Cadenas A, Zentella R, Sutliff TD, Ho TH. Involvement of multiple cis-elements in the regulation of GA responsive promoters: definition of a new cis-element in the Amy32b gene promoter of barley (Hordeum Vulgare) Physiol Plant. 2001;112(2):211–216. doi: 10.1034/j.1399-3054.2001.1120209.x. [DOI] [PubMed] [Google Scholar]
- 45.Tran LS, Mochida K. Functional genomics of soybean for improvement of productivity in adverse conditions. Functional & integrative genomics. 2010;10(4):447–462. doi: 10.1007/s10142-010-0178-z. [DOI] [PubMed] [Google Scholar]
- 46.Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465(1–2):30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 47.Wang L, Yin X, Cheng C, Wang H, Guo R, Xu X, Zhao J, Zheng Y, Wang X. Evolutionary and expression analysis of a MADS-box gene superfamily involved in ovule development of seeded and seedless grapevines. Molecular genetics and genomics : MGG. 2015;290(3):825–846. doi: 10.1007/s00438-014-0961-y. [DOI] [PubMed] [Google Scholar]
- 48.Xu G, Guo C, Shan H, Kong H. Divergence of duplicate genes in exon-intron structure. Proc Natl Acad Sci U S A. 2012;109(4):1187–1192. doi: 10.1073/pnas.1109047109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu W, Wang L, Tie W, Yan Y, Ding Z, Liu J, Li M, Peng M, Xu B, Jin Z. Genome-wide analyses of the bZIP family reveal their involvement in the development, ripening and abiotic stress response in banana. Sci Rep. 2016;6:30203. doi: 10.1038/srep30203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghiurcuta CG, Moret BM. Evaluating synteny for improved comparative studies. Bioinformatics. 2014;30(12):i9–18. doi: 10.1093/bioinformatics/btu259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang K, Han YT, Zhao FL, Hu Y, Gao YR, Ma YF, Zheng Y, Wang YJ, Wen YQ. Genome-wide identification and expression analysis of the CDPK gene family in grape, Vitis spp. BMC Plant Biol. 2015;15:164. doi: 10.1186/s12870-015-0552-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang SG, Price J, Lin PC, Hong JC, Jang JC. The arabidopsis bZIP1 transcription factor is involved in sugar signaling, protein networking, and DNA binding. Mol Plant. 2010;3(2):361–373. doi: 10.1093/mp/ssp115. [DOI] [PubMed] [Google Scholar]
- 53.Gibalova A, Renak D, Matczuk K, Dupl'akova N, Chab D, Twell D, Honys D. AtbZIP34 is required for Arabidopsis pollen wall patterning and the control of several metabolic pathways in developing pollen. Plant Mol Biol. 2009;70(5):581–601. doi: 10.1007/s11103-009-9493-y. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Mao L, Wang H, Brocker C, Yin X, Vasiliou V, Fei Z, Wang X. Genome-wide identification and analysis of grape aldehyde dehydrogenase (ALDH) gene superfamily. PLoS One. 2012;7(2):e32153. doi: 10.1371/journal.pone.0032153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao M, Zhang H, Guo C, Cheng C, Guo R, Mao L, Fei Z, Wang X. Evolutionary and expression analyses of basic zipper transcription factors in the highly homozygous model grape PN40024 (Vitis Vinifera L.) Plant Mol Biol Report. 2014;32(5):1085–1102. doi: 10.1007/s11105-014-0723-3. [DOI] [Google Scholar]
- 56.Xu Z, Ali Z, Xu L, He X, Huang Y, Yi J, Shao H, Ma H, Zhang D. The nuclear protein GmbZIP110 has transcription activation activity and plays important roles in the response to salinity stress in soybean. Sci Rep. 2016;6:20366. doi: 10.1038/srep20366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao Y, Zou HF, Wei W, Hao YJ, Tian AG, Huang J, Liu YF, Zhang JS, Chen SY. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta. 2008;228(2):225–240. doi: 10.1007/s00425-008-0731-3. [DOI] [PubMed] [Google Scholar]
- 58.Liu J, Chen N, Chen F, Cai B, Dal Santo S, Tornielli GB, Pezzotti M, Cheng ZM. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis Vinifera) BMC Genomics. 2014;15:281. doi: 10.1186/1471-2164-15-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TableS3. Synteny blocks of bZIP genes within soybean genome. (XLSX 19 kb)
Table S4. Synteny blocks of bZIP genes between soybean and Arabidopsis genomes. (XLSX 19 kb)
Table S5. The expression profiles of the soybean bZIP genes in different tissues. (XLSX 5886 kb)
Table S9. Primers used in qRT-PCR analysis. (XLSX 10 kb)
Table S1. Characteristics of bZIPs in soybean. (XLSX 17 kb)
Table S2. The accession numbers of bZIPs in soybean and Arabidopsis. (XLSX 5633 kb)
Figure S1. 20 conserved motifs and functions of some motifs. (DOCX 720 kb)
Table S6. The expression profiles (log2-based values) of the soybean bZIP genes after drought treatment. In this experiment, two-week-old soybean seedlings were transferred to the drought conditions for a week, and drought stress was imposed by withholding water for 7 days. After 7 days of treatment, the leaves were collected for experiment [30]. (XLSX 18 kb)
Table S7. The expression profiles (log2-based values) of the soybean bZIP genes after flooding treatment. In this experiment, two-week-old soybean seedlings were transferred to the flooding conditions for a week, and flooding stress was imposed by placing the pots into a bigger pot with a trashcan liner filled up to a water level of 4 cm above the soil surface for 7 days. After 7 days of treatment, the leaves were collected for experiment [30]. (XLSX 16 kb)
Figure S2. Validation of drought treatment RNA-seq data. (A) RNA-seq data of downregulated genes after drought treatment. (B) Validation of down regulated genes among drought sequencing data by real-time quantitative PCR. (C) RNA-seq data of upregulated genes after drought treatment. (D) Validation of up regulated genes among drought sequencing data by real-time quantitative PCR. FPKM represents Fragments Per Kilobase per Million fragments according to previously described [59]. (TIFF 203 kb)
Figure S3. Validation of flooding treatment RNA-seq data. (A) RNA-seq data of downregulated genes after flooding treatment. (B) Validation of down regulated genes among flooding sequencing data by real-time quantitative PCR. (C) RNA-seq data of upregulated genes after flooding treatment. (D) Validation of up regulated genes among flooding sequencing data by real-time quantitative PCR. (TIFF 209 kb)
Table S8. Cis-elements in GmbZIP promoters. (XLSX 20 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its Additional files].


