Abstract
Astrocytes are the most abundant cell type in the brain, where they perform a wide array of functions, yet the nature of their cellular heterogeneity and how it oversees these diverse roles remains shrouded in mystery. Using an intersectional fluorescence-activated cell sorting–based strategy, we identified five distinct astrocyte subpopulations present across three brain regions that show extensive molecular diversity. Application of this molecular insight toward function revealed that these populations differentially support synaptogenesis between neurons. We identified correlative populations in mouse and human glioma and found that the emergence of specific subpopulations during tumor progression corresponded with the onset of seizures and tumor invasion. In sum, we have identified subpopulations of astrocytes in the adult brain and their correlates in glioma that are endowed with diverse cellular, molecular and functional properties. These populations selectively contribute to synaptogenesis and tumor pathophysiology, providing a blueprint for understanding diverse astrocyte contributions to neurological disease.
Cellular heterogeneity is a defining feature of all organ systems, in which homeostatic function relies on diverse cell populations operating in concert to ensure proper physiological activities. The mammalian brain is the most complex organ in the body and comprised of an extraordinary array of diverse cell populations, namely neurons, astrocytes and oligodendrocytes. Proper brain function relies on the interplay between these principal cell types, with oligodendrocytes and astrocytes subserving a wide array of key neuronal functions1,2. Understanding neuronal diversity has been a focal point of developmental neuroscience, with numerous studies identifying possibly hundreds of molecularly and functionally distinct subtypes of neurons across the mammalian brain3–5. That neurons demonstrate extensive cellular heterogeneity raises the question of whether the other cell types in the brain also demonstrate cellular and functional diversity.
Among the principal cell types in the brain, astrocytes are the most abundant and have vital roles in all facets of physiology, ranging from neurotransmission and synaptogenesis to metabolic support and blood–brain barrier formation6,7. This immense functional diversity suggests the existence of heterogeneous astrocyte populations throughout the brain. In fact, Cajal initially described extensive morphological heterogeneity of astrocyte populations in the brain over 100 years ago8. Since then, our understanding of the molecular and cellular heterogeneity of astrocytes has remained stagnant, with astrocytes being grouped into two broad, morphological categories, fibrous and protoplasmic9,10. Despite the broad reach of astrocytes across numerous brain functions, the question of whether their diverse functions are executed by distinct subpopulations of astrocytes in the adult brain remains very poorly defined. Moreover, astrocytes are intimately involved in a wide range of neurological disorders, and whether changes in specific subpopulations selectively contribute to specific pathologies remains undefined.
Cellular diversity is often viewed through the lens of developmental patterning, a key organizing principle responsible for the generation of diverse cell populations across most tissues11,12. The embryonic spinal cord is the archetype for cellular diversity and patterning in the central nervous system (CNS), containing at least 12 molecularly distinct subtypes of neurons and three subtypes of white matter, which are fibrous astrocytes that are segmentally organized along the dorsal–ventral axis11,13,14. Despite the power of developmental patterning for the generation of cellular diversity, it does not provide a comprehensive census of all cell populations. Moreover, applying these patterning principles toward understanding cellular diversity requires an intimate knowledge of region-specific patterning mechanisms and the associated mouse tools to gain access to these populations. Currently, these mechanisms remain very poorly defined across diverse brain regions, which is a major barrier for understanding the nature of astrocyte diversity in the normal and diseased brain, further highlighting the need for new tactics to address this fundamental question.
To begin dissecting the cellular heterogeneity of astrocytes in the adult brain, we developed an intersectional, fluorescence-activated cell sorting (FACS)-based approach that combines the specificity of the astrocyte reporter mouse line (Aldh1l1–GFP; bacteria artificial chromosome (BAC) transgenic expression of GFP under control of the Aldh1l1 promoter) with the diversity afforded by cell surface markers. We identified five subpopulations of Aldh1l1–GFP-expressing astrocytes present across five brain regions that demonstrate both molecular and functional diversity. Application of these tactics toward malignant glioma revealed the presence of analogous astrocyte populations in both mouse and human malignant glioma that directly contribute to tumor pathophysiology. Our study reveals new molecular insight into the nature of astrocyte diversity in the brain and provides a blueprint for how these principles can be applied toward delineating diverse astroglial contributions to neurological disease.
RESULTS
Prospective identification of diverse astrocyte populations
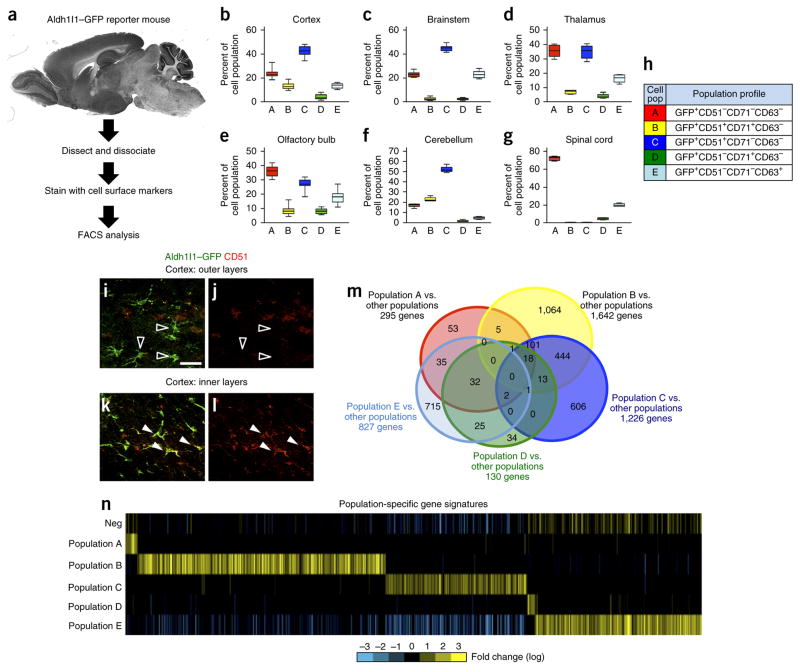
Strategies for dissecting the cellular diversity of astrocytes in the brain are limited in part by the lack of available mouse tools to access these populations. Recently aldehyde dehydrogenase 1 family member L1 (Aldh1l1) has emerged as a broad marker of astrocytes across the adult brain15. By using this as an entry point for dissecting the cellular heterogeneity of astrocytes in the brain, we combined the specificity of the Aldh1l1–GFP reporter mouse with the diversity of cell surface markers to create an intersectional, FACS-based approach (Fig. 1a). We then screened a panel of 81 cell surface antigens for their ability to fractionate the bulk Aldh1l1–GFP+ astrocyte population within the olfactory bulb (OB) and cortex from 12- to 16-week-old mice. Selection criteria included antibodies that marked between 10% and 50% of the total Aldh1l1–GFP+ population, identified using FACS, in these brain regions over three independent screening experiments. By using these screening parameters we identified antibodies specific for CD51, CD63 and CD71 as those that consistently subdivided the Aldh1l1–GFP bulk population across these brain regions (see Supplementary Table 1 for complete antibody list, Supplementary Fig. 1 for FACS plots and Supplementary Table 1 for statistics). We performed FACS analyses using different combinations of these antibodies, considering both antibody-positive and antibody-negative populations, and identified five distinct subpopulations of Aldh1l1–GFP-expressing astrocytes present in the OB, cortex, brainstem, thalamus and cerebellum (hereafter referred to as populations A, B, C, D and E, respectively) (Fig. 1b–h, Supplementary Fig. 2 and Supplementary Table 1). Immunostaining revealed that CD51 colocalizes with Aldh1l1–GFP+ astrocytes in the inner cortical layers but not in the outer cortical layers (Fig. 1a,i–l), suggesting that the CD51+ populations B and C preferentially localize to the inner layers of the cortex, whereas the CD51− populations A, D and E occupy the outer cortical layers. Taken together, our studies suggested the existence of five prospective subpopulations among Aldh1l1–GFP-expressing astrocytes that are present within these five distinct brain regions (Fig. 1b–l).
Figure 1.
Prospective isolation of Aldh1l1–GFP astrocyte subpopulations. (a) Schematic of the intersectional approach used to identify astrocyte subpopulations in the adult brain. (b–g) FACS analysis for each subpopulation that was identified across the cortex (b), brainstem (c), thalamus (d), OB (e), cerebellum (f) and spinal cord (g) through the combinatorial use of anti-CD51, anti-CD63 and anti-CD71. Data are mean ± s.e.m., represented as box and whisker plots (25th–75th percentiles (boxes), 10th–90th percentile (whiskers) and median (horizontal lines)) and derived from eight independent FACS experiments; for each experiment, brain regions from n = 4 mice were pooled and analyzed. Please see Supplementary Figure 3 for associated statistical analysis. (h) Code relating population nomenclature (A–E) to cell surface marker combination. (i–l) Representative images for Aldh1l1–GFP expression (i,k) and immunostaining for CD51 expression (j,l) in the outer (n = 3) (i,j) and inner (n = 3) (k,l) cortical layers in the brains of Aldh1l1–GFP mice. Unfilled arrowheads demonstrate non-overlapping GFP and CD51 expression, and filled arrowheads represent cells with overlapping expression. Please see Supplementary Figure 4 for additional CD71 immunostaining in the cortex. Scale bar, 20 μm. (m) Venn diagram depicting the absolute number of genes whose expression is enriched in each prospective population and their relative overlap with the genes in other populations using the ‘Y ~population + region’ linear model (FDR < 10%). Due to dimensional constraints, only 21 of the possible 31 intersections are depicted (complete quantification of all 31intersection points in Supplementary Table 3). (n) Heat map depicting the expression of gene sets identified for each subpopulation using the ‘Y ~population + region’ linear model (FDR < 10%). “Neg” represents the Aldh1l1–GFP-negative population. Bright yellow and blue represent threefold differences from the median across populations.
Heterogeneity of Aldh1l1–GFP+ astrocyte subpopulations
A key attribute of our intersectional, FACS-based approach is that we can isolate each of the prospective Aldh1l1–GFP+ astrocyte subpopulations from these diverse brain regions and directly interrogate their underlying molecular heterogeneity. To delineate the molecular profiles of these five prospective Aldh1l1–GFP+ subpopulations across different brain regions, we performed whole-transcriptome, RNA sequencing (RNA-seq) expression analysis on FACS-isolated populations from the OB, cortex and brainstem. To confirm that these populations demonstrated molecular features of astrocytes, we cross-compared our data sets with established astrocyte expression profiles from other studies16,17 and found that the prospective Aldh1l1–GFP+ astrocyte subpopulations were strongly enriched for pan-astrocyte gene signatures observed in mice and humans (Supplementary Fig. 3 and Supplementary Table 2).
To determine whether these 15 prospective subpopulations (five subpopulations from each of three sites) contain unique molecular features, we implemented multiple bioinformatics approaches. First, we performed unsupervised clustering analysis, which revealed two broad group molecular groups, one containing populations A–C and the other containing populations D and E (Supplementary Fig. 3). Furthermore, this analysis revealed that each of the populations, regardless of region, generally clustered together, suggesting that each population contains a conserved molecular signature across these distinct brain regions. To independently assess whether these prospective astrocyte subpopulations are endowed with unique gene-enrichment signatures, we applied two linear analytical models to our RNA-seq data sets. The first model (‘Y ~neg + region’; Online Methods) compared the prospective subpopulations with the Aldh1l1–GFP-negative reference population. Application of statistical cut-offs of false discovery rate (FDR) <10% and <5% revealed widespread gene expression differences between these subpopulations, with populations B and C demonstrating the most robust differences in terms of the absolute number of genes (Supplementary Fig. 4 and Supplementary Tables 3 and 4). The second model (‘Y ~population + region’; Online Methods) compared the prospective populations with each other, in an ordered pairwise series of comparisons between the populations. Next we applied similar statistical cut-offs (FDR < 10% and FDR < 5%) and also identified widespread differences in gene expression, with each population demonstrating unique molecular signatures (Fig. 1m,n and Supplementary Tables 5 and 6). Similar to that seen with the first model, populations B and C both demonstrated the largest number of unique gene signatures (Fig. 1m,n and Supplementary Fig. 4). This analysis of our transcriptome data revealed that each of these five prospective subpopulations is endowed with unique gene-enrichment profiles that are conserved across these brain regions; moreover, these population-specific enrichment sets contain unique gene ontologies (Supplementary Tables 5 and 7). Finally, we used immunostaining to confirm the expression of highly expressed candidate genes from the gene sets identified in populations B and C and validated expression of the transcription factors Zbtb20 and Erg1 and the cell signaling factors Enrb and TROY in subsets of Aldh1l1–GFP+ astrocytes from the cortex and OB (Supplementary Fig. 4). Taken together, our bioinformatics analysis of the transcriptome indicates that these prospective Aldh1l1–GFP+ astrocyte subpopulations show multiple layers of molecular diversity, suggesting that they are also endowed with diverse functional properties.
Astrocyte populations demonstrate diverse developmental functions
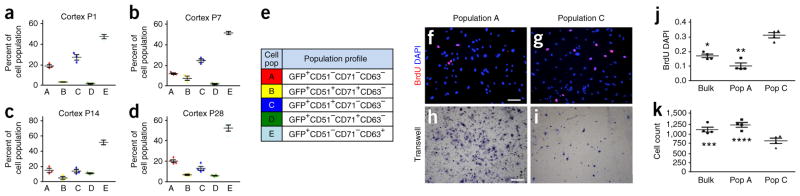
Having identified these diverse astrocyte subpopulations in the adult brain, we next sought to determine their developmental origins and properties. We used the developing cortex as a model, in which the spatial and temporal patterns of astrocyte generation have been characterized and examined the emergence of the subpopulations during postnatal development. Astrocyte production in the cortex commences at approximately postnatal day 1(P1) and continues until ~P28. Therefore, we screened the cortex of Aldh1l1–GFP mice for the presence of these subpopulations during this interval and found that populations A, C and E were the predominant subpopulations present early in development and that populations B and D emerged later during cortical development (Fig. 2a–e, Supplementary Fig. 2 and Supplementary Table 1). After we identified these populations in the developing cortex, we next determined whether they exhibited diverse functional properties. During the P1–P28 interval, astrocytes migrate out of the ventricular zone (VZ) and proliferate locally to populate the gray matter of the cortex; therefore, we assessed the migratory and proliferative properties of these astrocyte subpopulations in the developing cortex. Because of the technical constraints of our combinatorial, FACS-based approach, coupled with the relative abundance of populations A and C, we FACS-isolated populations A and C, as well as bulk Aldh1l1–GFP+ astrocytes, from Aldh1l1–GFP mice at P14 and performed acute culture, 5-bromo-2′-deoxyuridine (BrdU) labeling to assess proliferation and Transwell assays to determine migratory potential. These studies revealed that the cells in population C were more proliferative than those of population A or than bulk Aldh1l1–GFP+ astrocytes, whereas the cells of population A demonstrated more migratory potential than the cells of population C or bulk Aldh1l1–GFP+ astrocytes (Fig. 2f–k). These functional studies, combined with our population dynamics time course, indicated that these astrocyte populations were present in the developing cortex and had diverse developmental functions.
Figure 2.
Astrocyte populations demonstrate diverse developmental functions. (a–d) Plots showing the mean and standard error of FACS analysis for each subpopulation in the cortex across postnatal time points P1 (a), P7 (b), P14 (c) and P28 (d); Data are mean ± s.e.m. For each time point, data were derived from 4 independent FACS experiments; for each experiment, the cortices from 4 mice were pooled and analyzed (associated statistical analysis in Supplementary Fig. 3). (e) Code between population nomenclature (A–E) and cell surface marker combination. (f–k) Representative images for BrdU staining (f,g) or Transwell assays (h,i) from the acute culture of FACS-isolated populations A (f,h) and C (g,i) and quantification for BrdU staining (j) or migration through the matrix in the Transwell assays (k). DAPI was used to stain nuclei. Data are mean ± s.e.m. Quantification was from four independent cell cultures derived from four independent FACS isolation procedures; each experiment was performed in triplicate. *P = 5.22 × 10−4, **P = 2.29 × 10−5, ***P = 0.02784 and ****P = 0.00391; F2,11 = 41.95 (j) and F2,11 = 10.62 (k); by one-way analysis of variance (ANOVA) and followed by Tukey’s test for between-group comparisons. Scale bars, 100 μm (f,g) and 200 μm (h,i).
Astrocyte subpopulations differentially support synaptogenesis
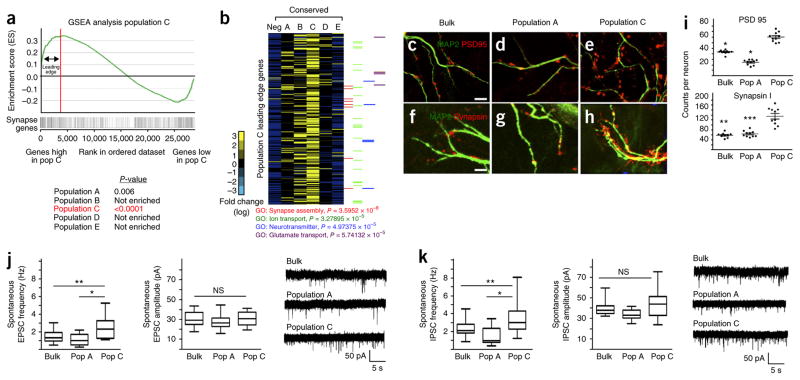
Our developmental studies suggested that the astrocyte subpopulations are endowed with diverse functions. Another key role for astrocytes is synapse formation and function. Therefore, we compared the expression profiles of each Aldh1l1–GFP+ subpopulation to a comprehensive list of genes that encode factors associated with synapse formation and function (hereafter referred to as synaptic genes) using gene set enrichment analysis (GSEA). This analysis revealed that the cells in population C are strongly enriched for genes associated with synapse formation and function (Fig. 3a and Supplementary Fig. 4). Further analysis of the ‘leading edge’ of the GSEA revealed that the expression levels of numerous synaptic genes were significantly higher in population C than in the other Aldh1l1–GFP+ astrocyte subpopulations, and we identified several genes associated with glutamate transport, ion transport and links to synaptic assembly (Fig. 3b and Supplementary Table 7). Taken together, the GSEA and gene ontology analysis suggest that population C astrocytes selectively promote synapse-associated functions onto neurons, which we directly tested by co-culturing cortical neurons with FACS-isolated population A and C cells, as well as bulk Aldh1l1–GFP+ astrocytes, from the cortex of Aldh1l1–GFP mice. Expression analysis of the presynaptic marker synapsin and the postsynaptic marker PSD95 revealed that synapse formation was significantly enhanced in neurons that were cocultured with population C cells, as compared to those cocultured with population A cells or with bulk Aldh1l1–GFP+ cells (Fig. 3c–i). Whole-cell patch clamp recordings of spontaneous excitatory and inhibitory postsynaptic currents (sEPSCs and sIPSCs, respectively) on these neurons revealed significant changes in the frequency but not amplitude of the postsynaptic events, consistent with an increase in number of functional synapses (Fig. 3j,k). Combined with our transcriptomic analysis, these data indicate that we have identified molecularly and functionally distinct astrocyte subpopulations in the brain and that a subset of these populations differentially support synapse formation between neurons.
Figure 3.
Astrocyte populations differentially support synaptogenesis. (a) GSEA plot, comparing genes whose expression is enriched in population (Pop) C to an established set of genes associated with synapse formation and function. P values (by GSEA method, with 1,000 gene-set permutations) evaluate enrichment of synapse gene expression within each respective population. Red line and associated arrows depict the ‘leading edge’, which can be interpreted as the core set of genes that accounts for the enrichment signal. (b) Heat map analysis for expression of population C leading-edge genes (270 genes represented) across all astrocyte populations. Note that ‘neg’ represents the Aldh1l1–GFP-negative control population; the experiment was done once, and its bioinformatics were compared between populations and synaptic genes. P values were derived using the Fisher exact test. (c–i) Representative images for expression of synaptic markers PSD95 (c–e) and synapsin (f–h), as well as their quantification (i) after co-culture of bulk astrocytes (c,f,i), population A cells (d,g,i) or population C cells (e,h,i), with neurons (marked by MAP2). Data are mean ± s.e.m. Quantification is from three independent cell cultures (neurons from three different mice and astrocytes derived from three independent FACS experiment). A total of ten neurons were each quantified for both PSD95 and synapsin expression. *P < 10−7, **P = 2.72 × 10−8 and ***P = 2.3 × 10−8; F2,29 =142 (PSD95) and F2,29 = 37.79 (synapsin). P-values derived using one-way ANOVA. Scale bars, 5 μm. Scale bars in c also apply to d and e; scale bars in f also apply to g and h. (j,k) Whole-cell recordings from neurons; frequency (left) and amplitude (middle) of sEPSC (bulk, n = 20 neurons; population A, n = 23 neurons; population C, n = 18 neurons) (j) and sIPSC (bulk, n = 21 neurons; population A, n = 23 neurons; population C, n = 21 neurons) (k), and the associated traces (right), are shown. The recording was done from three independent cell cultures (neurons from three mice and astrocytes derived from three independent FACS experiments). Data are mean ± s.e.m.; data are represented as box and whisker plots (25th–75th percentiles (boxes), 10th–90th percentiles (whiskers) and medians (horizontal lines)). *P < 0.01 (P = 0.0051 (j) or P = 0.0056 (k), *P = 1.8 × 10−5, **P < 1.26 × 10−6 and NS, not significant; F2,60=12.57 (j) or F2,63=16.03 (k); by one-way ANOVA followed by Tukey’s test for between-group comparisons.
Molecular and cellular correlates of astrocyte populations in malignant glioma
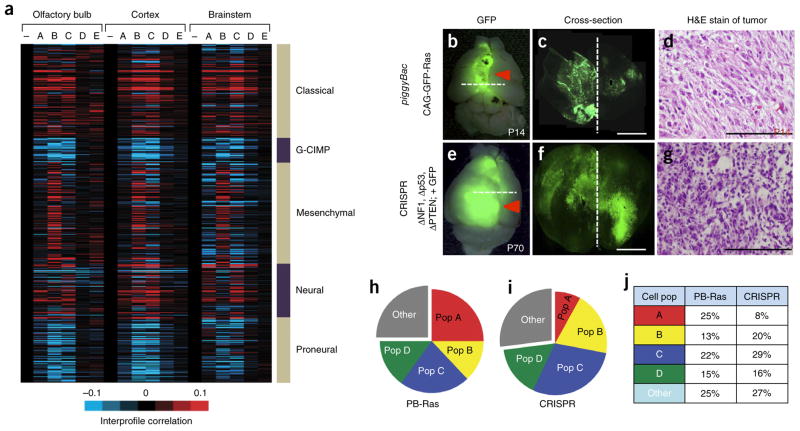
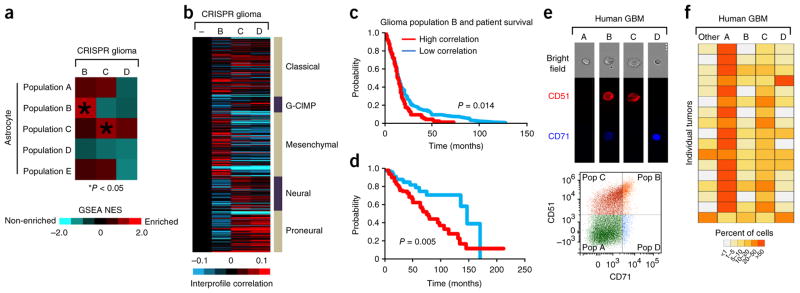
Astrocytes also have central roles in brain malignancies, raising the question of how astrocyte heterogeneity might drive or participate in these disease-associated phenotypes. To address this question, we focused on malignant glioma because these tumors demonstrate extensive cellular heterogeneity. Comparison of the global expression profiles of each astrocyte subpopulation with the profiles of different subtypes of human glioblastoma (GBM)18,19 revealed that the expression profiles of populations A–D are highly correlated with the classical subtypes, those of populations A–C are correlated with the neural subtype and that the mesenchymal subtype is exclusively aligned with expression profile of population B (Fig. 4a). Conversely, none of the astrocyte populations demonstrated a strong correlation with the glioma–CpG island methylator phenotype (GCIMP) or proneural subtypes. These molecular profile correlations between our mouse astrocyte subpopulations and human GBM subtypes suggest that multiple, analogous astroglial subpopulations may exist within human GBM.
Figure 4.
Identification of analogous populations in malignant glioma. (a) Heat map showing intersample Pearson’s correlations (red, positive; bright red or blue denotes r-value > 0.1 or < −0.1, respectively, across ~7,000 genes) between mRNA profiles of astrocyte populations (columns; genes within each region first centered on corresponding control group) and mRNA profiles of human glioblastoma (rows; from the Cancer Genome Atlas (TCGA), n = 528 cases). Note that minus sign represents the Aldh1l1–GFP-negative control population. (b–d) Representative images P14 from the piggyBac-Ras model of malignant glioma (n = 6). (e–g) Representative images at P70 from the CRISPR model of malignant glioma (n = 6). Images in b,e are from whole brain; dashed lines represent cortical midline. The dashed lines in b and e are the planes of section in c and f, respectively. Scale bars, 2,500 μm (c,f) and 250 μm (d,g). H&E, hematoxylin and eosin. (h–j) Pie charts showing the representative subdivision of bulk, GFP+ tumor cells through the combinatorial use of the CD51 and CD71 cell surface markers. Data are representative of eight independent FACS experiments. Gray fraction represents CD133+ GSCs and CD31+ endothelial cells; cell population and marker coding is the same as described in Figure 1c. PB-Ras, piggyback-Ras. Filled red arrows denote tumor.
Because we initially identified these astrocyte subpopulations in the mouse brain, we sought to determine whether their malignancy correlates were present in mouse models of malignant glioma. Recently, we developed two such models. One makes use of in utero electroporation (IUE) of the cortex of mouse fetuses at embryonic day 16.5 (E16.5) to target glial lineages with the Ras oncogene and produce infiltrative malignant gliomas by age P14 (Fig. 4b–d). The second utilizes E16.5 IUE in combination with CRISPR-mediated gene editing, in which we use IUE to introduce guide RNA (gRNA)-expressing vectors to delete the genes encoding neurofibromin 1 (NfI), phosphatase and tensin homolog (Pten) and p53 (Trp53) and to express a reporter construct encoding the endonuclease Cas9 and GFP (herein referred to as the ‘CRISPR’ model), resulting in the highly reproducible generation of malignant glioma at P70 (Fig. 4e–g and Supplementary Fig. 5)20–22. By using GFP expression to distinguish tumors from normal brain tissue, along with FACS-based selection against the glioma stem cell (GSC) and endothelial cells, we screened the tumors in our mouse models for the presence of these prospective astroglial populations in the non-GSC and endothelial fractions of these tumors (Supplementary Fig. 5). As indicated in Figure 4h–j, both the Ras and CRISPR models contain FACS profiles consistent with populations A–D in the non-GSC fraction, suggesting that analogous populations are present in mouse models of malignant glioma.
Prospective subpopulations are present in human glioma
To ascertain whether the astrocyte subpopulations present in our mouse model of glioma are analogous to astrocyte subpopulations identified in the native mouse brain, we performed RNA-seq analysis on populations B, C and D that were isolated from the CRISPR model of glioma and found a strong molecular correlation between glioma populations B and C and their astrocyte counterparts. (Fig. 5a, Supplementary Fig. 6 and Supplementary Tables 8 and 9). Population D in glioma did not demonstrate a molecular correlation with its corresponding astrocyte population, indicating differential conservation among these astrocyte populations in glioma. Next we determined whether these mouse glioma populations show a molecular correlation with those in human GBM and found strong association between each population and the classical subtype (Fig. 5b). Similar to its astrocyte counterpart, population B cells from glioma also demonstrate a specific correlation with the mesenchymal subtype (Figs. 4a and 5b). Consistent with the mesenchymal subtype being the most aggressive form of malignant glioma, we also found that human glioma samples (both malignant and lower grade) that were globally most similar to glioma subpopulation B cells, in terms of expression patterns, were associated with worse prognosis and decreased survival compared to those samples that did not strongly correlate with glioma subpopulation B (Fig. 5c,d and Supplementary Fig. 6).
Figure 5.
Glioma subpopulations are present in human glioma. (a) GSEA results comparing each indicated glioma-population-specific gene set (Supplementary Table 10) with global gene rankings according to indicated astrocyte population comparisons. NES, GSEA normalized enrichment score scale is shown in color bar. Red denotes enrichment of glioma gene set within the astrocyte population profile. (b) Heat map showing intersample Pearson’s correlations (red, positive; bright red or blue denotes r-value > 0.1 or < −0.1, respectively, across ~7,000 genes) between mRNA profiles of mouse glioma populations (columns; genes first centered on control group) and mRNA profiles of human glioblastoma (rows; from TCGA, n = 528 cases). Minus sign represents the Aldh1l1–GFP-negative control population. (c,d) Differences in overall survival for patients with glioblastoma (n = 523 cases in total) (c) and for patients with lower-grade glioma (n = 515 cases) (d) that is associated with global correlation with population B. Red, brain cancer cases in top one-third of correlations (n = 175 (c); n = 172 (d)); blue, cases in bottom one-third of correlations. Log-rank p-value comparing the top one-third cases with the bottom one-third cases is 0.014 (c) and 0.005 (d). In c, chi square = 6.09515907; in d, chi square = 7.75721882233268. (e) Representative FACS analysis of primary human malignant glioma and the associated plot (n = 5). (f) Heat map analysis showing the constitution of the subpopulations (A–D and ‘other’) in primary human glioma across 17 samples. The ‘other’ population is comprised of CD133+ and CD31+ subpopulations.
The molecular correlation between the mouse glioma subpopulations and human GBM subtypes suggests that these populations are also present in primary human GBM tumors, which we tested using freshly resected human malignant glioma samples and a FACS-based approach to screen for their presence. Similar to our mouse studies, we selected against the GSC population and found cell surface profiles within the bulk tumors that were consistent with the presence of populations A–D in these primary tumors (Fig. 5e,f and Supplementary Fig. 6). Taken together, we applied comparative bioinformatics to infer the existence of analogous astrocyte subpopulations in malignant glioma and used mouse models and human tissue to validate the presence of subsets of these populations in tumors.
Emergence of astrocyte subpopulations in glioma correlates with the onset of seizures
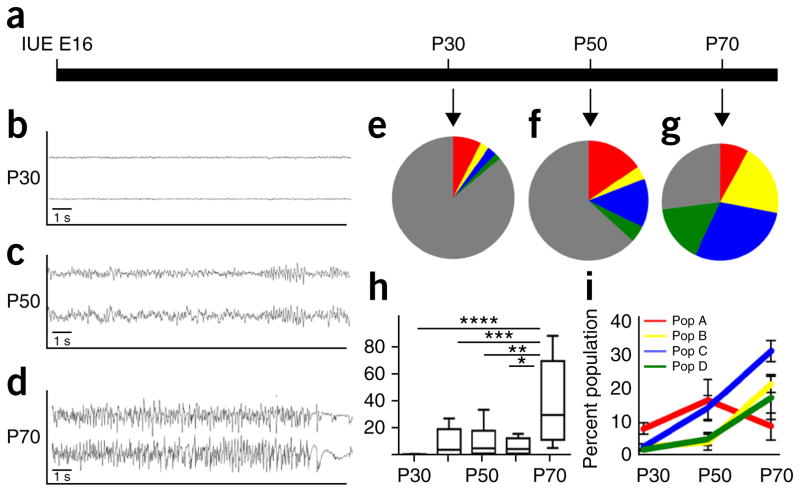
Tumors have a vast array of functions that support malignant growth, and our identification of diverse glioma subpopulations raises the question of whether these populations perform distinct functions23. Because astrocyte population C is enriched for genes whose products are associated with synaptic function and promote synaptogenesis (Fig. 3), we investigated whether its glioma analog is similarly correlated with changes in the neural microcircuitry in the peritumoral margin. Seizures are a pathophysiological hallmark of glioma and are due in part to an imbalance of synaptic activity24,25; therefore, we determined whether there was a link between seizure onset in our mouse model and the emergence of population C. First, we determined the evolution of seizure onset by performing video electroencephalography (EEG) recordings from our mouse CRISPR model and found a progressive pattern of increased aberrant network hyperexcitability beginning at P40 that uniformly culminated with spontaneous cortical seizures at P70 (Fig. 6a–d,h). After EEG monitoring, we harvested tumors from these mice and found that populations B–D were nominally present in tumors from mice at P30 and that their numbers gradually increased over the course of tumor progression (Fig. 6e–g,i). The emergence of these populations was tightly correlated with increased EEG activity (Fig. 6h,i), suggesting that changes in tumor composition contribute to the progressive increases in network hyperexcitabilty, which culminates in glioma-induced seizures. Alternatively, it is also possible that changes in network hyperexcitability cause the differentiation or production of these subpopulations in glioma.
Figure 6.
Emergence of glioma subpopulations correlates with seizures. (a) Schematic for time course of tumor progression in the CRISPR model. (b–d) Representative EEG recordings from mice containing tumors at P30 (b), P50 (c) and P70 (d); note the progressive cortical hyperexcitability during tumor progression (n = 6 for each timepoint). y axes denote millivolts. (e–g) Pie charts showing the representative subdivision of the GFP+ tumor cells into populations A–D (and other) during the P30–P70 progression interval (n = 6 for each timepoint). (h) Quantification of the interictal spike activity at each time point during tumor progression (n = 6 animals for each time point). *P = 0.0049, **P = 0.0143, ***P = 0.0041 and ****P = 0.0014; by one-way ANOVA and followed by Tukey’s test for between-group comparisons. (i) Quantification of the glioma subpopulation constitution (n = 5 tumors for each time point. Throughout, data are mean ± s.e.m.
Glioma subpopulations execute distinct tumorigenic functions
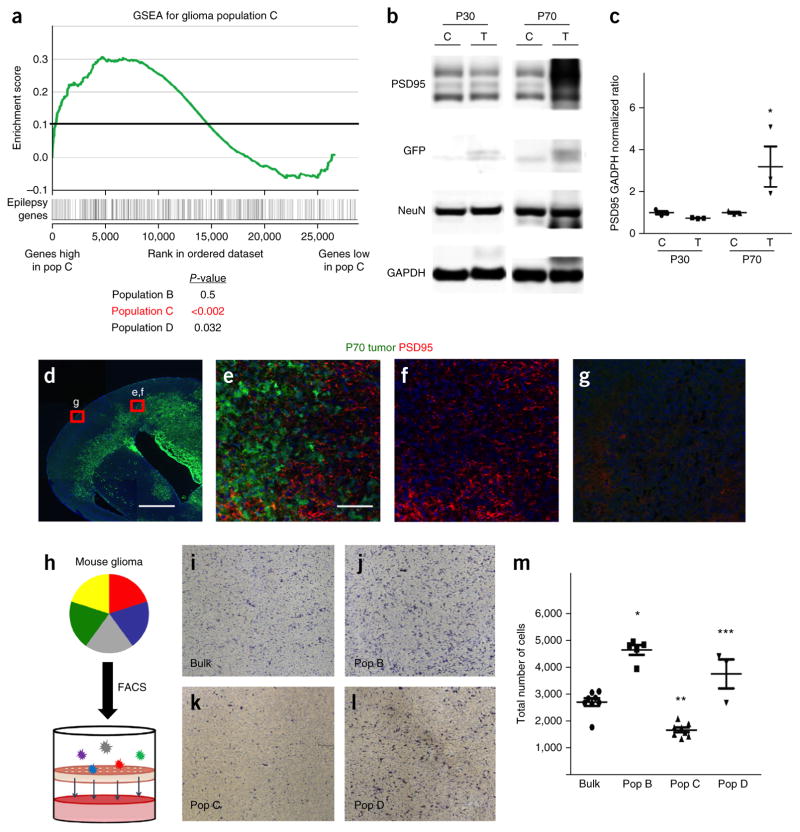
Our observation that the emergence of populations B, C and D over the course of tumor progression corresponds with increased EEG activity raises the question of whether their molecular profiles are similarly linked to seizure- or epilepsy-associated gene networks. We performed GSEA, comparing the molecular signatures of each population with the expression of a cohort of established epilepsy-associated genes26, and found that glioma populations C and D are enriched for the expression of these genes, whereas glioma population B was not (Fig. 7a and Supplementary Table 9). Moreover, glioma population C was also enriched for expression of many synaptic genes found in its astrocyte analog (Fig. 4b, Supplementary Fig. 6 and Supplementary Table 9), suggesting that the abnormal cortical EEG activity in brain tumors from mice at P70 is the result of increased synaptogenesis in the neighboring neuronal milieu. Therefore, we performed immunoblots on cortical hemispheres from tumors from mice at P30 and P70 and found an increase in the amount of PSD95 in the P70 tumor samples (Fig. 7b,c). Immunostaining of tumors from mice at P70 revealed significant increases in the extent of PSD95 staining in regions adjacent to the tumor (Fig. 7d–g). These data suggest that an aberrant growth of synapses in the brains of mice at P70 contributes to increased EEG excitability and, in conjunction with our transcriptomic and epileptogenesis time course studies, support a model in which increases in synapse formation and the onset of seizures correspond to the emergence of populations C and D in P70 tumors.
Figure 7.
Glioma subpopulations show functional diversity. (a) GSEA plot comparing the molecular signature of glioma population C (versus negative control) to an established cohort of genes associated with epilepsy. P-values (by GSEA method, with 1,000 gene-set permutations) evaluate enrichment of epilepsy-associated genes within each respective population. (b,c) Representative immunoblot for PSD95 expression in tumor (T) and control (C) cortex from mice at P30 and P70 (b), and quantification from three independent immunoblots from three independent tumors for each time point (c). *P = 0.054 by independent t-test to analyze the difference between control and tumor group. NeuN is included to normalize for neuronal numbers and GAPDH is the loading control. (d–g) Representative low-magnification (d) and high-magnification (e–g) images of PSD95 staining (red) in a brain tumor (green; GFP+) at P70 (from n = 3 for each). Red boxes in d indicate tumor-adjacent and tumor-distal areas that are magnified in e–g. Scale bars, 1,000 μm (d) and 100 μm (e–g). (h) Schematic showing the combined FACS and Transwell invasion assay. (i–m) Representative images showing cells that have migrated through the Transwell onto the chamber for bulk glioma astrocytes (i) or for FACS-isolated glioma populations B (j), C (k), D (l) and their quantification derived from three independent experiments using three independently derived tumors (m). Images in i–l are from a 96-well plate ChemoTx Chemotaxis System (106-8, NeuroProbe). *P = 1.49 × 10−6, **P = 9.23 × 10−4 and ***P = 0.0128; F2,11 = 10.62; by one-way ANOVA followed by Tukey’s test for between-group comparisons. Throughout, data are mean ± s.e.m. For the purpose of presentation, the blot images were cropped from the original images. Full-length blots can be found in Supplemental Figure 6.
Our time course studies indicated that the onset of seizures was associated with tumor progression. Another key feature of glioma progression is invasion, in which populations of tumor cells migrate into the surrounding brain parenchyma27,28. Given that glioma populations B, C and D emerge over the course of tumor progression and also correlate with invasion, we next determined whether these populations differentially supported migration. To test this, we FACS-isolated populations B, C and D from our glioma model and performed Transwell assays; we found that populations B and D showed greater migration than population C or the bulk tumor cells (Fig. 7h–m). These findings indicate that the glioma subpopulations demonstrate differential activities toward cell migration and, combined with our seizure data, suggest that these populations execute different functions associated with tumor progression.
DISCUSSION
The nature of astrocyte diversity in the adult brain has remained enigmatic, due, in part, to the lack of molecular entry points. By using our newly developed tactics, we identified a set of astrocyte subpopulations across diverse adult brain regions that demonstrate cellular, molecular and functional diversity. To our knowledge, these studies represent the first comprehensive characterization of diverse astrocyte subpopulations in the adult mouse brain, and the correlation with human astrocyte gene signatures suggest that these diverse populations may also be present in the human brain (Supplementary Fig. 3).
The approach we used to identify these astrocyte subpopulations opens the door for additional exploration into their molecular and functional diversity through further subfractionation analyses using cell surface markers and/or single cell RNA sequencing approaches. Moreover, we can also leverage our molecular profiling data and the associated cell surface markers toward the development of specific genetic reagents so that we can better access and define these subpopulations during development and into adulthood. These tools have been useful in defining astrocyte populations in the embryonic spinal cord and can now be used to interrogate both local and regional astrocyte heterogeneity in the brain29,30. One limitation of our approach is that it does not easily allow the precise anatomical arrangement of these cells to be localized in the adult brain, reinforcing the need to develop more selective mouse reporter lines based on these molecular profiles. Notably, we will also point out that altering the original selection criteria (Online Methods) would result in the possible identification of additional, or fewer, subpopulations. Further exploration of our original antibody screen (Supplementary Table 1) may provide insight into additional layers of astrocyte diversity in the brain.
We can also use these expression profiles, in conjunction with our newly developed methods, as entry points to systematically examine the diverse functions of these populations using a litany of established assays to address one of the key questions in neuroscience: do all astrocyte perform identical glial functions, or is the biology defined by distinct subpopulations? Using synapse formation as a prototype, we found a subset of astrocytes that are endowed with an enhanced capacity to support synapse formation, a finding that suggests important functional diversity amongst astrocyte subpopulations in the brain. This concept of functional diversity was further reinforced with our developmental studies demonstrating differential migratory and proliferative properties. Moving forward, it will be critical to further decode these cellular and functional relationships across the diverse array of physiological, metabolic and developmental roles filled by astrocytes.
Our identification of astrocyte populations with differential molecular and functional properties for synaptogenesis raises the question of how such populations are regulated. In the developing cortex, >50% of astrocytes are produced between P1 and P28, in regions outside the germinal centers31,32. This robust, postnatal production of astrocytes is coincident with a major wave of synaptogenesis in the neocortex33,34. One possibility is that these properties are influenced by local neuronal activity, in which astrocyte function is sculpted by the surrounding neuronal milieu, allowing neuronal excitability to drive the differentiation of adjacent astrocytes into those that sustain higher levels of synaptic transmission. Indeed, recent studies have shown that sonic hedgehog (Shh) signaling in neurons influences astrocyte gene expression and associated functions in the cerebellum35. Such neuron–astrocyte interactions may explain the population dynamics during development, where it is possible that extrinsic signaling mechanisms from neurons and associated circuits are influencing astrocyte subpopulation identity or function36.
Linking diverse astrocyte subpopulations with specific diseases represents a potentially new way of thinking about the cellular basis of neurological disease. As proof of concept for this approach, we chose to focus on malignant glioma, as these tumors contain diverse cell populations that execute a wide range of functions and are notoriously heterogeneous at the cellular level37,38, and found that malignant glioma contains cellular populations that correlate with astrocyte subpopulations in the brain. Notably, our study used an Nf1, Trp53 and Pten prototype model of glioma, and thus these parallels between astrocyte populations and glioma populations may only be directly applicable to human tumors with this mutation spectrum. However, the fact that we identified analogous populations across a cohort of primary human tumors suggests that these populations may occur in diverse genetic backgrounds. Moving forward, it will be important to generate different genetic mouse models of glioma and cross-correlate the cellular constituency in human samples with the same spectrum of prototypical driver mutations.
This discovery lends critical insight into the cellular diversity of malignant glioma, as we have identified four populations of cells that comprise the non-GSC fraction of the bulk tumor. Although much attention has been given to the GSC, it only comprises <20% of the bulk tumor39,40, whereas the remaining ~80% of the tumor is comprised of pathological glia, whose identities and functions toward tumorigenesis remain poorly defined. Recent single-cell RNA-seq studies in malignant glioma have shown that tumors show extensive molecular heterogeneity, and it has been postulated that each tumor contains multiple, prospective populations with molecular profiles consistent with each of the GBM subtypes41. Consistent with this, we found that each of these populations contained molecular profiles that also correlated with these human GBM subtypes (Fig. 4), further supporting the notion that each tumor contains numerous cell populations with differential GBM-subtype signatures.
The identification of discrete glioma subpopulations, coupled with the tractability of our mouse model systems, allowed us to query the dynamics and functional effect of these populations during tumor progression. Time-course analysis indicated that these populations emerged over the course of tumor progression and that this emergence was correlated with two key features of glioma progression, tumor invasion and the onset of seizures. Causes of glioma-induced seizures have traditionally been viewed as a function of the intracortical invasion by the primary tumor, along with the potential for inflammatory-signal-induced synaptogenesis and increased glutamate release by glioma cells at the margins42,43. Notably, the critical underlying astrocytic correlates that drive this pathophysiological response within the heterogeneous bulk glioma tumor remain undefined. At the cellular level, our results add novel evidence that increasing hyperexcitabilty and, ultimately, seizure onset is correlated with the emergence of neosynaptogenesis that is engendered by a specific and expanding population of pathological glia within the tumor. At the molecular level, we found that a subset of these emerging populations (populations C and D) had gene expression signatures that were enriched for epilepsy-associated genes, suggesting a glial, as well as neuronal, basis for tumor-driven epileptogenesis. These findings suggest a model in which, over the course of tumor progression, cell populations emerge that support increased and aberrant synaptic biology, which contributes to seizure onset.
The brain represents a unique microenvironment, and the mechanism by which the glioma interfaces with resident neuronal populations to propagate malignancy remains poorly defined. Our data suggest that over the course of tumor progression, specific glioma subpopulations contribute to an aberrant neuronal microenvironment to promote synaptic activity. Of note, recent studies have shown that increased neuronal activity promotes glioma tumor proliferation44. From these two observations, we propose a model for the complex interplay between the evolving tumor, the surrounding neurons and glioma growth. In this model, the evolving glioma alters the neuronal milieu to provoke synaptic activity, which in turn, feeds back to glioma cells and stimulates proliferation. The net result of this putative feedback cycle would be glioma-induced seizures and increased tumor proliferation, two hallmarks of advanced and progressive glioma.
Drawing on the molecular parallels between the astrocyte subpopulations and their malignant analogs, we found that population C in adult brain astrocytes and in glioma is enriched for the expression of genes linked to synapse formation and epilepsy. These conserved properties across these systems parallels what has been observed for cancer stem cells (CSCs), in which the malignant cell population contains key features of its normal, nonmalignant counterpart. For stem cells and their associated CSCs, these conserved properties include multipotency and self-renewal. For population C, this property could involve modulating neuronal activity through synapse formation. The logic underlying this conservative principle is that synapse formation, in particular, is a highly specialized process that may require the tumor to reuse existing programs and/or cell populations to engender tumor propagation in the unique brain microenvironment. This observation suggests the existence of other functional parallels between these astrocyte subpopulations and their malignant analogs that contribute to tumorigenesis and/or alterations to the brain microenvironment that engender malignancy.
ONLINE METHODS
Mice, FACS analysis and cell culture
Both male and female BAC Aldh1l1–eGFP mice and wild-type mice (CD-1) were used. The care and handling of animals in all experiments were in compliance with NIH guidelines and were approved by the institutional animal care committee at Baylor College of Medicine.
To identify subpopulations of astrocytes, we prepared 12- to 16-week-old Aldh1l1–eGFP mice and dissected the OB, cortex, hypothalamus, cerebellum and brainstem from the mice. The dissociated cells from these different brain regions from the Aldh1l1–eGFP mice were pelleted and incubated with antibodies to cell surface molecules, diluted in FACS buffer (Leibowitz Media, supplemented with 1 mg/ml BSA, 10 mM HEPES pH 7.05, pen/strep, and 25 μg/ml DNaseI), for 90 min at room temperature. The following antibodies were used at the manufacturer’s recommended dilutions: phycoerythrin (PE)-conjugated rat anti-mouse-CD51 (RMV7; BD Biosciences), allophycocyanin (APC)-conjugated rat anti-mouse-CD63 (NVG-2; eBioscience) and rat anti-mouse-CD71 (C2; BD Biosciences). For cell sorting, a BD FACSAria III instrument (100-μm nozzle and 20-p.s.i. setting) was used with FACSDIVA software, and astrocytes were sorted into a 15-ml tube containing sorting medium (Dulbecco’s modified Eagle’s medium (DMEM)+F12 supplemented with 20% FBS, penicillin–streptomycin and nonessential amino acids).
For in vitro studies, FACS-isolated astrocyte subpopulations were cultured on poly-d-lysine-coated flasks with DMEM+F12, with 10% FBS and penicillin–streptomycin, before they were used for cortical neuron co-culture experiments. Dissociated cortical neurons from wild-type P0–P1 pups was prepared using published procedures45.
Processing of brain tissue from Aldh1l1–GFP mice
Each of the dissected brain regions was first coarsely chopped into smaller pieces by surgical scissors followed by enzymatic dissociation with papain (with DNase I) at 37 °C on a thermomixer for 20 min (P1000 pipette tip was used to triturate the tissue approximately five times for 15 min, and a P200 pipette tip was used to further triturate the tissue at the end of dissociation procedure). DMEM+F12 with 10% FBS was added to the dissociated cells to neutralize the enzyme, and the cell suspension was spun down. The pellet was washed with PBS twice and resuspended in FACS buffer (Leibowitz medium, supplemented with 1 mg/ml BSA, 10 mM HEPES pH 7.05, penicillin–streptomycin and 25 μg/ml DNaseI) with or without antibodies to cell surface molecules.
FACS screening criteria
One of our criteria was that the antibody should mark between 10% and 50% of the Aldh1l1–GFP+ population. We reasoned that markers showing greater than 50% staining were likely to be pan-astrocytic, whereas those with less than 10% staining were not present at high enough concentrations to reveal rare population when used in combination with other cell surface markers. One marker, CD147, showed greater than 50% staining of the Aldh1l1–GFP+ cells in the OB.
Another criterion was that the markers must also demonstrate similar subfractionation properties across multiple brain regions, in multiple adult mice. Consistent subfractionation profiles in multiple brain regions and in multiple mice was a critical aspect of the screen, with each antibody being subjected to FACS screening on three independent sets of brain tissue, on three separate occasions. For the primary screen, we focused on the OB and cortex from 12- to 14-week-old Ald1l1–GFP mice and tested each antibody in three separate and independent experiments.
Only three antibodies met these criteria—anti-CD51, anti-CD63 and anti-CD71. Anti-CD51 marked 30–40% of Aldh1l1–GFP+ cells in the OB or cortex. Anti-CD71 marked 10–20% of Aldh1l1–GFP+ cells in the OB or cortex. Anti-CD63 marked 15–20% of Aldh1l1–GFP+ cells in the OB or cortex. Other antibodies that resulted in subfractionation but did not meet our criteria included: anti-CD147, anti-CD29 anti-CD316. These markers did not give consistent subfractionation across all brain regions and thus were not selected for further study.
Cell analysis and cell sorting for Aldh1l1–GFP mice
The dissociated cells from the different regions of the brain from the Aldh1l1–GFP mice were pelleted and incubated with antibodies to cell surface molecules, diluted in FACS buffer, for 90 min at room temperature. The following antibodies were used at the manufacturer’s recommended dilutions: PE-conjugated rat anti-mouse-CD51 (RMV7; BD Biosciences), APC-conjugated rat anti-mouse-CD63 (NVG-2; eBioscience) and rat anti-mouse-CD71 (C2; BD Biosciences).
After the incubation, the cells were pelleted again to remove the antibody solutions and washed with PBS twice. The cells were resuspended with FACS buffer and kept on ice before analysis or cell sorting. The chilled cell samples were pipetted through cell strainers right before use to prevent clotting. For cell analysis, a BD LSR II or BD Canto II instrument was used with BD FACSDIVA software. For cell sorting, a BD FACSAria III instrument (100-μm nozzle and 20-p.s.i. setting) was used with FACSDIVA software, and astrocytes were sorted into a 15-ml tube containing sorting medium (DMEM+F12 supplemented with 20% FBS, penicillin–streptomycin and nonessential amino acids). The sorted astrocytes were then pelleted, re-suspended with fresh sorting medium and seeded into poly-d-lysine-precoated flasks.
In the astrocyte and neuron co-culture experiments, ~8 × 104 astrocytes were seeded on a poly-d-lysine-coated 12-mm coverslip in a 24-well culture dish. Approximately 24 h later, ~6 × 104 cortical neurons were seeded on the top of the astrocytes and maintained with serum-free Neurobasal medium supplemented with B27, gentamicin and Glutamax (Gibco).
Electrophysiology
Whole-cell recordings were performed on neurons that were co-cultured with different subpopulations of astrocytes (bulk, population A and population C) in parallel at DIV 7 (days in vitro) and DIV 14. Neurons were recorded in a chamber with continuous flow of external solution at a fixed flow rate controlled by a flow valve. For each recording, data were filtered with a 60-Hz notch filter and analyzed with template-based event detection algorithms in Clampfit 10.6 (Molecular Devices).
Standard artificial cerebral spinal fluid (ACSF) (140 mM of NaCl, 2.4 mM of KCl, 10 mM of HEPES, 10 mM of glucose, 4 mM of MgCl2 and 2 mM CaCl2; pH was adjusted to 7.3 with osmolarity ~300 mOsm) was used as the external solution. The internal solution contained 136 mM of KCl, 17.8 mM of HEPES, 1 mM of EGTA, 0.6 mM of MgCl2, 4 mM of ATP, 0.3 mM of GTP, 12 mM of creatine phosphate and 50 U/ml phosphocreatine kinase. The recording pipettes used in the experiment had a resistance of 3–6 MΩ. Antagonists (3 mM kynurenic acid and 20 μM bicuculline) were applied sequentially with 3 min of washing in between. All the chemicals used were purchased from Sigma-Aldrich,
To determine the glutamatergic or GABAergic identities of the spontaneous activities recorded, the spontaneous synaptic potential was first recorded for 100–150 s for baseline activities, as well as activities under the treatment of 3 mM kynurenic acid and 20 μM bicuculline (100–150 s for each treatment with 3 min of no treatment in between application of the two antagonists). An event with a rise time ~0.5 ms and decay time <20 ms were considered to be a glutamatergic event, whereas an event with a rise time ~0.5 ms and decay time >20 ms were considered to be a GABAergic response.
RNA sequencing
For each population that was subjected to RNA-seq analysis, between 1 × 105 and 2 × 105 FACS-isolated cells were used from pooled samples; each population was sequenced in duplicate. We identified five populations across three brain regions and generated duplicate RNA-seq expression profiles for each of these conditions. For each of the three brain regions, we also generated expression profiles from Aldhl1l–GFP-negative populations (i.e., non-astrocytes) in duplicate.
Each RNA-seq profile was generated from RNA substrates derived from pooled mouse tissue. For one profile we dissected a given brain region (for example, brainstem) from six adult Aldh1l1–GFP mice of the same age, pooled these brainstems and dissociated the tissue. After the tissue was dissociated, we used FACS analysis to isolate a given population (for example, population C) from these dissociated cells. After we isolated the cells via FACS (Aldh1l1–GFP + antibodies to cell surface markers), we then extracted RNA and used this RNA as the substrate for the generation of a single RNA-seq profile. The replicate for the same population from the brainstem was generated in the same manner, in parallel. Extracted RNA was then sent to the Genomic and RNA Profiling Core (GARP) at the Baylor College of Medicine. A ThruPlex DNA-seq library was subsequently prepared, and RNA-seq was performed on an Illumina HS2500.
RNA sequencing at the Genomic and RNA Profiling Core facility at the Baylor College of Medicine
NuGEN Ovation RNA-seq system v.2 protocol
Purified double-stranded cDNA was generated from approximately 300 pg of total RNA and amplified using both oligo-d(T) and random primers. Samples were quantified using the NanoDrop ND-2000 spectrophotometer and the Qubit 2.0 DNA quantitation assay. Five hundred nanograms of each sample’s double-stranded cDNA was sheared using the Covaris S2 focused-ultrasonicator with a 400-bp target size. The sheared samples were quantified using the NanoDrop ND-2000 spectrophotometer and the Qubit 2.0 DNA quantitation assay. The fragment sizes were viewed on the Agilent Bioanalyzer to verify proper shearing.
Rubicon Genomics ThruPlex DNA-seq library preparation protocol
A double-stranded DNA library was generated from 50 ng of sheared, double-stranded cDNA to prepare the fragments for hybridization onto a flow cell. This was achieved by first creating blunt-ended fragments and then ligating stem–loop adapters with blocked 5′ ends to the 5′ end of the double-stranded cDNA, leaving a nick at the 3′ end. Finally, library synthesis extended the 3′ end of the double-stranded cDNA, and Illumina-compatible indexes were incorporated with five amplification cycles. The fragments were purified using the AMPure XP Bead system. The resulting libraries are quantified using the NanoDrop ND-1000 spectrophotometer, and fragment size was assessed with the Agilent Bioanalyzer. qPCR-based quantification was performed on the libraries to determine the concentration of adaptor-ligated fragments using the Applied Biosystems ViiA7 Real-Time PCR System and a KAPA Library Quant Kit.
Cluster generation by bridge amplification
Using the concentration determined using the ViiA7 qPCR machine (as described above), 13 pM of equimolarly pooled library was loaded onto a flow cell and amplified by bridge amplification using the Illumina HS2500 sequencing instrument. A PhiX Control v3 adaptor–ligated library was spiked in at 2.5% to ensure balanced diversity and to monitor clustering and sequencing performance. A paired-end 100-cycle run was used to sequence the library on the flow cell using a HiSeq Sequencing System in Rapid Mode with v2 chemistry.
Bioinformatics analysis
Illumina bcl2fastq 1.8.3 software was used for base-calling and FASTQ file generation. Sequenced reads in FASTQ files were mapped to the mm10 whole genome using Tophat (Galaxy tool version 0.9) with parameters -r 50. University of Santa Cruz (UCSC) transcripts were supplied in a GTF format file. Fragments per kb of transcript per million mapped reads (FPKM) were calculated using Cufflinks (Galaxy tool version 2.2.1.0) with parameters -r 50. UCSC-known transcripts were supplied in a GTF format file. For downstream analyses, we filtered out any transcripts with nonzero expression in only four samples or less from the data set. Quantile normalization was then carried out, using 100 quantiles. Expression heat maps were generated using JavaTreeView46. For comparing a given population with the control group, the following linear regression model was used: ‘Y ~neg+ region’, where Y is expression of a given gene (using log-transformed values), ‘population’ is a categorical variable or factor where negative control is the reference, and ‘region’ is a categorical variable encoding the three brain regions (with region one arbitrarily used as a reference). For comparing a given population with another population, the following linear regression model was used: ‘Y ~population + region’, where Y is expression of a given gene (excluding the negative control group), ‘population’ is a categorical variable or factor where a comparison population of interest is the reference, and ‘region’ is a categorical variable encoding the three brain regions. False discovery rates (FDRs) were estimated from the set of nominal gene-level P values, using the method from Storey and Tibshirini47.
Established gene lists for synaptic genes are found at SynSysNet (http://bioinformatics.charite.de/synsys/), and epilepsy-related genes are found at EpilepsyGene (http://61.152.91.49/EpilepsyGene)26. GEO accession numbers for the RNA-seq data sets are GSE79437 (CRISPR glioma) and GSE72826 (astrocytes).
Comparative bioinformatics with astrocytes, glioma and gene set enrichment analyses
For selecting genes specific to a given astrocyte population (A–E) and conserved across the three regions (OB, cortex and brainstem), we selected genes that were differentially expressed (FDR < 10%, linear model incorporating both region and population) within the given population across the three regions versus any one of the other populations. Five linear models were computed, one for each population (used as the reference). For a given population, the full set of genes that differed from that of any of the other populations was used as the basis of the population-specific signature. Please see Supplementary Tables 3–6 for the associated gene lists, FDR and P value analysis.
The TCGA GBM (glioblastoma) data set of gene expression profiles (U133A platform, n = 528 cases) was analyzed in conjunction with the astrocyte and glioma subpopulation expression data sets, using a previously described interprofile correlation analysis18,19. GBM profiles were previously classified according to expression-based subtypes. For each gene in the TCGA GBM data set, expression values were centered on the median and divided by the s.d. across tumor profiles; within each of the astrocyte and glioma subpopulation data sets from our own study, values were centered on the average of the corresponding control group. Using the centered data sets, for each TCGA and brain subpopulation mRNA profile, we computed the global interprofile correlation (by Pearson’s correlation), using all ~7,000 genes that were common to the two data sets. For visual display, global correlation profiles of biological duplicates within each subpopulation group were averaged together. A similar analysis was carried out for the TCGA lower-grade glioma (LGG) RNA-seq expression data set (n = 516 cases). For the TCGA interprofile correlations with glioma population B, both GBM and LGG cases were subdivided according to average correlation (top third, bottom third and middle third of correlations) and evaluated for differences in overall survival (patient survival data being compiled in August 2015).
Gene set enrichment analysis (GSEA) was carried out48 using signal-to-noise as the ranking metric and with the ‘weighted’ scoring scheme. For cases in which multiple transcripts in the brain subpopulation data set referred to the same gene, the most variable transcript across the cases was used to represent the gene. Note that in the GSEA in Figure 3a,b the leading-edge subset in a gene set are those genes that appear in the ranked list at or before the point at which the running sum reached its maximum deviation from zero. The leading-edge subset can be interpreted as the core that accounts for the gene set’s enrichment signal48. The values in the heat map in Figure 3b represent average log-fold changes from Neg, normalized to s.d. from the median across the six groups. These represented the aggregated data used in analyzing the astrocyte subpopulations.
Mouse models of glioma
All mouse gliomas were generated in the CD-1 IGS mouse background. In utero electroporations (IUEs) were performed as previously described49. CRISPR-IUE GBM were generated by electroporating the three pX330 constructs targeting Nf1, Trp53 and Pten. Guide sequences for these CRISPRs were selected from previously published sequences21,22 and validated in our IUE model. CRISPR-IUE GBM were also electroporated with the pGlast-PBase and PBCAG-GFPt2aLuc vectors to help visualize tumors through fluorescence (GFP) and bioluminescence (Luc). CRISPR-IUE tumors were harvested at P30–P90 from both male and female mice. All mouse experiments were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee.
Procurement of glioblastoma surgical specimens
All fresh tumor specimens were obtained from patients who provided written consent at the participating institutions under an Institutional Review Board (IRB)-approved protocol, H15280. After intraoperative frozen-section consultation confirmed the tumor pathology, specimens were removed fresh, moistened with saline and transported on ice for immediate FACS analysis. Please see Supplementary Table 10 for the age and gender of the 17-patient cohort.
In vivo video-electroencepholography (EEG) recordings
Mice bearing tumors were anesthetized using avertin (250 mg per kg body weight) and surgically implanted with a bilateral silver wire electrode (0.005-inch diameter) attached to a microminiature connector. EEG and behavioral activity in freely moving mice were analyzed using simultaneous video-EEG monitoring (Haramonie software version 6.1c, Stellate systems). All EEG signals were filtered using a 60-Hz notch filter, 0.3-Hz high-pass filter and 70-Hz low-pass filter. Control and tumor bearing mice were monitored every 10 d from 30–70 d for a 17-h period. Seizure activity was quantified by visual inspection of the EEG waveform and corresponding video-recorded behavior. The frequency of interictal spike activity was calculated from EEG waveform data of each 17-h recording period using Matlab scripts (MathWorks, Natick, MA)50.
Statistical analyses
One-way ANOVA was used to analyze the proliferation, migration, electrophysiology and EEG experiments to determine the differences between group means, followed by Tukey’s test to compare individual means, and it is reported as asterisks in the associated figure graphs. An independent t-test was used to compare means between the control and the tumor group in the PSD95 protein-quantification assay. All statistical tests were two-sided. No statistical methods were used to pre-determine sample sizes, but our sample sizes were similar to those reported in previous publications45,50. Data distribution was assumed to be normal, but this was not formally tested. Randomization of animal studies was used in the data analysis. Data collection and analysis were not performed in a blinded manner to the conditions of the experiments. No animal or data points were excluded from the analyses. A Supplementary Methods Checklist is available with the supplementary materials.
Immunocytochemistry
Neurons that were co-cultured with different subpopulations of astrocytes seeded on 12-mm coverslips were rinsed with PBS and fixed with 4% paraformaldehyde for 15 min, then rinsed again with PBS three times before there were treated with blocking buffer (10% goat serum, 0.2% Triton X-100 in PBS) for 1 h at room temperature. Next, they were treated with primary antibodies prepared in the blocking solutions overnight at 4 °C. The following antibodies were used: chicken anti-MAP2 (1:1,000; EnCor Biotech. Inc.), rabbit anti-synapsin-I (1:1,000; Millipore) and mouse anti-PSD95 (1:1,000; UC Davis K28/43). The primary antibodies were then removed, and the coverslips were washed with PBS three times on an orbital shaker for 5 min each wash. After three washes, the coverslips were incubated with secondary antibodies to chicken, mouse, or rabbit, that complement the primary antibody, prepared in blocking solution (1:1,000; Alexa Fluor 488 and Alexa Fluor 647, BD Biosciences) for 1 h at room temperature, followed by removal of the secondary antibodies and three washes with PBS. At the end, they were treated with Hoechst prepared in blocking solution (1:10,000; Sigma-Aldrich) for 2 min and washed with PBS three times. The coverslips were mounted with VECTA anti-fading mounting medium (Vector Laboratories). Additional antibodies used: anti-TROY (goat, Santa Cruz Biotechnology, 1:100), anti-ZBTB20 (rabbit, Sigma, 1:1,000), anti-Egr-1 (rabbit, Santa Cruz Biotechnology, 1:150), anti-Ednrb (ETBr) (rabbit, Sigma-Aldrich, 1:150), anti-NDRG2 (rabbit, Cell Signaling,1:1,000), anti-transferrin-receptor (anti-CD71; mouse, Invitrogen, 1:500), anti-integrin-αV (anti-CD51; rabbit, EMD Millipore, 1:500). All the images were taken with Leica TCS SPE confocal microscope and Leica Application Suit X. Please see Supplementary Table 11 for detailed antibody information.
Tumor dissociation and cell-surface-antigen staining
Tumors derived from IUE models were removed from whole mouse brains using a fluorescence dissecting scope. Patient samples were received from patient biopsy on IRB-approved protocols. Tumor samples were then dissected into small pieces of tissue approximately 1–2 mm3 in size and incubated in a 50% papain solution (Worthington Biochemical) diluted with PBS. This mixture was incubated at 37 °C for 10 min and gently triturated with a pipette. This process was repeated until there were no visible pieces of tissue. After the enzymatic–mechanical dissociation, the papain was neutralized, and the cells were resuspended in FACS buffer. Cells were incubated with cell surface antibodies according to the manufacturer’s instructions in a photo-protected tube rotator. The following antibodies were used for flow cytometry and FACS-based isolation at the manufacturer’s recommended dilutions: mouse anti-human-CD31 (WM59; BD Bioscience), mouse anti-human-CD51 (NKI-M9; BioLegend), mouse anti-human-CD63 (H5C6; BD Biosciences), mouse anti-human-CD71 (M-A712; BD Biosciences), mouse anti-human-CD133 (AC133; Miltenyi Biotec), rat anti-mouse-CD31 (MEC 13.3; BD Biosciences), rat anti-mouse-CD51 (RMV-7; BD Biosciences), rat anti-mouse-CD71 (C2; BD Biosciences) and rat anti-mouse-CD133 (13A4; eBioscience). After incubation, cells were pelleted and resuspsended in FACS buffer for flow cytometry analysis or FACS-based isolation. SYTOX Red viability dyes (ThermoFisher) were included during FACS to enrich for healthy, live cells. Cells were sorted on a BD FACSAria III instrument with a 100-μm nozzle and a 20-p.s.i. setting.
Tumor cell invasion assay
To assay for cell migration, cells were FACS-isolated and seeded onto the ChemoTx Chemotaxis System (106-8, NeuroProbe). Specifically, one side of the Transwell filter was coated with fibronectin (Biomedical Technologies Inc) for 30 min followed by two PBS washes. Wells were filled with DMEM + 10% FBS. After the filter layer was carefully placed such that the fibronectin-coated side was face down into the well, 5,000 sorted cells (resuspended in DMEM + 10% FBS) were seeded on top of the membrane on the noncoated side. Chemotaxis plates were placed in 5% CO2 at 37 °C for 12 h. Filter membranes were then fixed, stained and mounted onto slides. Quantifications represent the averages of six individual assays.
Data availability
All data contained within this manuscript will be made available to the public, without any restrictions. RNA-seq data have been deposited into the Gene Expression Omnibus (Accession codes GSE72826 and GSE79437).
Supplementary Material
Acknowledgments
We thank M. Brenner and M. Goodell for assistance with our cell-surface-marker antibody screen. This work was supported by grants from the Sontag Foundation (B.D.), the National Multiple Sclerosis Society (RG-1501-02756; B.D.), the Cancer Prevention Research Institute of Texas (RP510334 and RP160192 awarded to both B.D. and C.J.C.), the American Cancer Society (PF-15-220; K.Y.) and the US National Institutes of Health (NIH) (NS071153 and AG054111 (B.D.), NS089366 (B.D.), NS29709 (J.L.N.) and T32HL902332 (K.Y. and J.C.)). This project was also supported in part by the Genomic and RNA Profiling Core at Baylor College of Medicine with funding from the NIH–NCI grant (P30CA125123) and the expert assistance of L. White, the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the NIH (P30 AI036211, P30 CA125123 and S10 RR024574), the expert assistance of J. Sederstrom and by IDDRC grant number 1U54 HD083092 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
C.-C.J.L., K.Y. and B.D. conceived the project, designed the experiments and wrote the manuscript; C.-C.J.L., K.Y., A.H., T.-W.H., H.K.L., J.C. and W.Z. performed the experiments; M.C.W., C.A.M., N.A., A.J. and B.R.A. provided essential reagents; F.C., Y.Z. and C.J.C. designed and executed the bioinformatics analysis; and J.L.N. and C.J.C. assisted in experimental design.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Barres BA. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 2008;60:430–440. doi: 10.1016/j.neuron.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 2.Ligon KL, Fancy SP, Franklin RJ, Rowitch DH. Olig gene function in CNS development and disease. Glia. 2006;54:1–10. doi: 10.1002/glia.20273. [DOI] [PubMed] [Google Scholar]
- 3.Jiang X, et al. Principles of connectivity among morphologically defined cell types in adult neocortex. Science. 2015;350:aac9462. doi: 10.1126/science.aac9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kepecs A, Fishell G. Interneuron cell types are fit to function. Nature. 2014;505:318–326. doi: 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyoshi G, et al. Prox1 regulates the subtype-specific development of caudal-ganglionic-eminence-derived GABAergic cortical interneurons. J Neurosci. 2015;35:12869–12889. doi: 10.1523/JNEUROSCI.1164-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abbott NJ, Rönnbäck L, Hansson E. Astrocyte–endothelial interactions at the blood–brain barrier. Nat Rev Neurosci. 2006;7:41–53. doi: 10.1038/nrn1824. [DOI] [PubMed] [Google Scholar]
- 7.Chung WS, Allen NJ, Eroglu C. Astrocytes control synapse formation, function, and elimination. Cold Spring Harb Perspect Biol. 2015;7:a020370. doi: 10.1101/cshperspect.a020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramón y Cajal S. Histology of the Nervous System of Man and Vertebrates. Oxford University Press; New York: 1897. [Google Scholar]
- 9.Andriezen WL. The neuroglia elements in the human brain. BMJ. 1893;2:227–230. doi: 10.1136/bmj.2.1700.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kölliker A. Handbuch der Gewebelehre des Menschen. Verlag von Wilhelm Engelmann; 1889. [Google Scholar]
- 11.Bayraktar OA, Fuentealba LC, Alvarez-Buylla A, Rowitch DH. Astrocyte development and heterogeneity. Cold Spring Harb Perspect Biol. 2014;7:a020362. doi: 10.1101/cshperspect.a020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwai Y, et al. Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- 13.Hochstim C, Deneen B, Lukaszewicz A, Zhou Q, Anderson DJ. Identification of positionally distinct astrocyte subtypes whose identities are specified by a homeodomain code. Cell. 2008;133:510–522. doi: 10.1016/j.cell.2008.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai HH, et al. Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science. 2012;337:358–362. doi: 10.1126/science.1222381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anthony TE, Heintz N. The folate metabolic enzyme ALDH1L1 is restricted to the midline of the early CNS, suggesting a role in human neural tube defects. J Comp Neurol. 2007;500:368–383. doi: 10.1002/cne.21179. [DOI] [PubMed] [Google Scholar]
- 16.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lovatt D, et al. The transcriptome and metabolic gene signature of protoplasmic astrocytes in the adult murine cortex. J Neurosci. 2007;27:12255–12266. doi: 10.1523/JNEUROSCI.3404-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bakheit AM, Brennan A, Gan P, Green H, Roberts S. Anarchic hand syndrome following resection of a frontal lobe tumor. Neurocase. 2013;19:36–40. doi: 10.1080/13554794.2011.654213. [DOI] [PubMed] [Google Scholar]
- 19.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de la Iglesia N, et al. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008;22:449–462. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue W, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen F, Rosiene J, Che A, Becker A, LoTurco J. Tracking and transforming neocortical progenitors by CRISPR–Cas9 gene targeting and piggyBac transposase lineage-labeling. Development. 2015;142:3601–3611. doi: 10.1242/dev.118836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 24.Dudek FE. Role of glial cells in seizures and epilepsy: intracellular calcium oscillations sn spatial buffering. Epilepsy Curr. 2002;2:137–139. doi: 10.1046/j.1535-7597.2002.t01-1-00052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ransom BR, Sontheimer H. The neurophysiology of glial cells. J Clin Neurophysiol. 1992;9:224–251. doi: 10.1097/00004691-199204010-00005. [DOI] [PubMed] [Google Scholar]
- 26.Ran X, et al. EpilepsyGene: a genetic resource for genes and mutations related to epilepsy. Nucleic Acids Res. 2015;43:D893–D899. doi: 10.1093/nar/gku943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuddapah VA, Robel S, Watkins S, Sontheimer H. A neurocentric perspective on glioma invasion. Nat Rev Neurosci. 2014;15:455–465. doi: 10.1038/nrn3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paw I, Carpenter RC, Watabe K, Debinski W, Lo HW. Mechanisms regulating glioma invasion. Cancer Lett. 2015;362:1–7. doi: 10.1016/j.canlet.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeh TH, Lee DY, Gianino SM, Gutmann DH. Microarray analyses reveal regional astrocyte heterogeneity with implications for neurofibromatosis type 1 (NF1)-regulated glial proliferation. Glia. 2009;57:1239–1249. doi: 10.1002/glia.20845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey MS, Shipley MT. Astrocyte subtypes in the rat olfactory bulb: morphological heterogeneity and differential laminar distribution. J Comp Neurol. 1993;328:501–526. doi: 10.1002/cne.903280405. [DOI] [PubMed] [Google Scholar]
- 31.Cameron RS, Rakic P. Glial cell lineage in the cerebral cortex: a review and synthesis. Glia. 1991;4:124–137. doi: 10.1002/glia.440040204. [DOI] [PubMed] [Google Scholar]
- 32.Ge WP, Miyawaki A, Gage FH, Jan YN, Jan LY. Local generation of glia is a major astrocyte source in postnatal cortex. Nature. 2012;484:376–380. doi: 10.1038/nature10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tau GZ, Peterson BS. Normal development of brain circuits. Neuropsychopharmacology. 2010;35:147–168. doi: 10.1038/npp.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang X, Nardelli J. Cellular and molecular introduction to brain development. Neurobiol Dis. 2016;92(Pt. A):3–17. doi: 10.1016/j.nbd.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farmer WT, et al. Neurons diversify astrocytes in the adult brain through sonic hedgehog signaling. Science. 2016;351:849–854. doi: 10.1126/science.aab3103. [DOI] [PubMed] [Google Scholar]
- 36.Molofsky AV, et al. Astrocytes and disease: a neurodevelopmental perspective. Genes Dev. 2012;26:891–907. doi: 10.1101/gad.188326.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dallas NA, et al. Endoglin (CD105): a marker of tumor vasculature and potential target for therapy. Clin Cancer Res. 2008;14:1931–1937. doi: 10.1158/1078-0432.CCR-07-4478. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, et al. CD133-negative glioma cells form tumors in nude rats and give rise to CD133-positive cells. Int J Cancer. 2008;122:761–768. doi: 10.1002/ijc.23130. [DOI] [PubMed] [Google Scholar]
- 39.Singh SK, et al. Identification of human brain tumor–initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 40.Stiles CD, Rowitch DH. Glioma stem cells: a midterm exam. Neuron. 2008;58:832–846. doi: 10.1016/j.neuron.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 41.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robel S, Sontheimer H. Glia as drivers of abnormal neuronal activity. Nat Neurosci. 2016;19:28–33. doi: 10.1038/nn.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campbell SL, et al. GABAergic disinhibition and impaired KCC2 cotransporter activity underlie tumor-associated epilepsy. Glia. 2015;63:23–36. doi: 10.1002/glia.22730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venkatesh HS, et al. Neuronal activity promotes glioma growth through neuroligin-3 secretion. Cell. 2015;161:803–816. doi: 10.1016/j.cell.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weston MC, Chen H, Swann JW. Multiple roles for mammalian target of rapamycin signaling in both glutamatergic and GABAergic synaptic transmission. J Neurosci. 2012;32:11441–11452. doi: 10.1523/JNEUROSCI.1283-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 47.Storey JD, Tibshirani R. Statistical significance for genome-wide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glasgow SM, et al. Mutual antagonism between Sox10 and NFIA regulates diversification of glial lineages and glioma subtypes. Nat Neurosci. 2014;17:1322–1329. doi: 10.1038/nn.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barkmeier DT, et al. High inter-reviewer variability of spike detection on intracranial EEG addressed by an automated multichannel algorithm. Clin Neurophysiol. 2012;123:1088–1095. doi: 10.1016/j.clinph.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data contained within this manuscript will be made available to the public, without any restrictions. RNA-seq data have been deposited into the Gene Expression Omnibus (Accession codes GSE72826 and GSE79437).