Abstract
The recent breakthroughs in NMR probe technologies resulted in the development of MAS NMR probes with rotation frequencies exceeding 100 kHz. Herein, we explore dramatic increases in sensitivity and resolution observed at MAS frequencies of 110–111 kHz in a novel 0.7 mm HCND probe that enable structural analysis of fully protonated biological systems. Proton-detected 2D and 3D correlation spectroscopy under such conditions requires only 0.1 – 0.5 mg of sample and a fraction of time compared to conventional 13C-detected experiments. We discuss the performance of several proton- and heteronuclear- (13C-,15N-) based correlation experiments in terms of sensitivity and resolution, using a model microcrystalline fMLF tripeptide. We demonstrate the applications of ultrafast MAS to a large, fully protonated protein assembly of the 231-residue HIV-1 CA capsid protein. Resonance assignments of protons and heteronuclei, as well as 1H-15N dipolar and 1HN CSA tensors are readily obtained from the high sensitivity and resolution proton-detected 3D experiments. The approach demonstrated here is expected to enable the determination of atomic-resolution structures of large protein assemblies, inaccessible by current methodologies.
Keywords: MAS NMR, protein assemblies, proton-detected MAS NMR correlations, HIV-1 capsid protein
Graphical abstract
1. Introduction
Since the advent of magic angle spinning as a primary method for averaging anisotropic spin interactions in solid-state NMR spectroscopy [1; 2; 3], there has been a sustained effort in the field to push the upper limit of accessible MAS frequencies. A major motivation for the development of faster MAS spinners and achieve rotation frequencies that exceed the magnitude of 1H-1H homonuclear dipolar interaction is to enable high-resolution 1H-based spectroscopy, for maximum detection sensitivity. Early seminal work of Samoson resulted in the development of MAS probes capable of spinning at 40 kHz [4; 5], and several groups demonstrated that 1H-detected experiments are feasible above 30 kHz, even in fully protonated samples [6; 7; 8]. Extensive deuteration yields further resolution and sensitivity benefits [7; 9; 10; 11]. These reports spurred further developments by Samoson and by instrument manufacturers, and reports of 1H-detected spectroscopy of large biomolecules at MAS frequencies of 60–62 kHz, in samples which are deuterated and fully back-protonated at exchangeable sites [7; 12; 13; 14; 15], amino acid-specifically protonated [16], or even fully protonated [8]. Using MAS probes spinning at frequencies of up to 111 kHz [8; 17] showed early promise to revolutionize the field of biological solid-state NMR. Nishiyama, Ishii, and others observed that at these rotation frequencies proton linewidths narrow considerably (to 100–150 Hz in fully protonated samples) with the added benefit of greatly enhanced spectral sensitivity [18]. At MAS frequencies of 100 kHz and above, 1H-1H correlation experiments are accessible in organic solids [19]. Under these conditions, deuteration still remains beneficial [20; 21], with 1H line widths down to 20 Hz in extensively deuterated samples, yielding spectra similar to those observed for small proteins in solution [22]. In fully protonated samples, unprecedented high sensitivity and resolution was observed in 1H-detected experiments at 110–111 kHz, permitting multidimensional spectroscopy of protein assemblies, in a fraction of time (hours vs. days/weeks) and with a fraction of amounts (0.1 – 0.3 mg) normally required for conventional experiments conducted at MAS frequencies of 10–60 kHz [23; 24]. These results set the stage for further exploration of the ‘ultrafast’ MAS conditions for applications to structural biology.
In this study, we examined the performance and benefits of 2D and 3D correlation spectroscopy at MAS frequencies of 110–111 kHz in applications to fully protonated biological solids: the microcrystalline U-13C,15N-labeled fMLF tripeptide, a frequently used MAS NMR standard, and tubular assemblies of the 231-residue U-13C,15N-labeled HIV-1 capsid (CA) protein. 1H linewidths in both systems were found to be of the order of 100 – 170 Hz. For microcrystalline fMLF, 1H-13C, and 13C-13C, and 1H-1H single- and multiple-quantum correlation experiments afford excellent sensitivity and resolution, and permitted resonance assignments for aliphatic, amide, aromatic and formyl protons. In the tubular assemblies of U-13C,15N-labeled HIV-1 CA, the resolution of the 2D and 3D spectra permitted ready resonance assignments of heteronuclei (13C and 15N) and protons are readily accomplished; to our knowledge this is the only example of 1H assignments in a fully protonated protein of this size from MAS NMR experiments. Most gratifyingly, 1H-15N dipolar and 1H chemical shift anisotropy (CSA) tensors, which are indispensable probes of structure and dynamics, are readily measured using RN-symmetry recoupling [25; 26; 27; 28; 29] implemented in proton-detected 3D experiments. Our overall results demonstrate the promise of proton-based spectroscopy at MAS frequencies of 110–111 kHz for structural and dynamics analysis of large biological assemblies.
2. Materials and Methods
2.1. Materials and Sample Preparation
U-13C,15N-fMLF tripeptide was purchased from CortecNet and used without further recrystallization. For MAS NMR experiments conducted at 19.96 T, U-13C,15N-fMLF was used; for experiments at 11.74 T, U-13C,15N-fMLF was mixed with the natural abundance fMLF in a 1:5 ratio. 0.5 mg of material were packed into 0.7 mm rotors and used for MAS NMR experiments.
HIV-1 CA protein was expressed and purified as reported previously [30; 31]. In brief, uniformly 13C, 15N-enriched CA was expressed in modified M9 media, containing 15NH4Cl and U-13C6-glucose as sole nitrogen and carbon sources, respectively. Expression was induced with 0.8 mM IPTG at 18–23 °C for 16 h. Cells were harvested by centrifugation, resuspended in 25 mM sodium phosphate buffer (pH 7.0) and opened by sonication. The lysate was centrifuged at 26,892 × g at 4 °C for 1 h, the pH of the supernatant was adjusted to 5.8 with acetic acid, and the conductivity reduced by dilution to below 2.5 ms/cm. After a second centrifugation step at 26,892 × g at 4 °C for 1 h, the final supernatant was loaded onto a cation exchange column and protein was eluted with a 0–1 M NaCl gradient in a buffer containing 25 mM sodium phosphate (pH 5.8), 1 mM DTT, and 0.02% NaN3. Concentrated protein fractions were further purified over a size-exclusion column equilibrated 25 mM sodium phosphate buffer (pH 7.0), 1 mM DTT, and 0.02% NaN3.
Tubular assemblies of CA were prepared by incubation of 26 mg/ml protein solution in 25 mM phosphate buffer (pH 5.5), containing 2.4 M NaCl, at 37 °C for one hour and 4 °C overnight [31]. The morphology was characterized by transmission electron microscopy (TEM). TEM analysis was performed with a Zeiss Libra 120 transmission electron microscope operating at 120 kV. Assemblies were stained with uranyl acetate (0.5–1% w/v), deposited onto 400 mesh, formval/carbon-coated glow discharged copper grids, and dried for 45 min in the air. NMR sample was prepared by centrifugation of the tubular assemblies for 15 min at 10,000 × g. The pellet was transferred to the 0.7 mm MAS rotor; the amount of hydrated material used in the experiments was estimated to be 0.2–0.3 mg. This estimate is based on the nominal volume that the 0.7 mm probe can hold and extrapolated from our weight-based measurements when packing 1.3 and 1.9 mm probes.
2.2. Solid-State NMR Spectroscopy
MAS NMR experiments were conducted on a 19.96 T Bruker Avance III standard bore spectrometer. A 0.7 mm Bruker HDCN MAS probe was employed. The MAS frequency was set to 110 or 111 kHz and controlled to within ±5 Hz using a Bruker MAS III controller. Additionally, J-based HSQC and TQSQ spectra of fMLF were acquired on a 11.74 T Bruker Avance NEO wide bore spectrometer. A 0.7 mm Bruker HCN MAS probe was employed. The actual sample temperature was calibrated for the different MAS frequencies by recording the temperature dependence of the 79Br T1 relaxation time in KBr [32]; the nitrogen gas flow for bearing and drive settings were kept at the same values for the samples under investigation. The typical 90° pulse lengths were 1.00 μs (1H), 1.6–2.00 μs (13C, depending on the experiment), and 2.73 μs (15N). The 1H-13C and 1H-15N CP employed a linear amplitude ramp of 90–110% on 1H, and the center of the ramp matched to Hartmann-Hahn conditions at the first spinning sideband, with contact times of 1.1–1.5 and 1.5–1.8 ms, respectively. CP was performed as double quantum CP with a 13C spinlock rf-field between 2/3 and ¾*ωr and ¼ to 1/3*ωr or as zero quantum CP employing the +1 condition (1.667 – or 1.75*ωr) for 1H.
All experiments on fMLF were conducted at the temperature of 26 °C. For 2D 13C-13C RFDR spectrum, the RFDR mixing time was 1.45 ms, the radiofrequency (rf) power for the RFDR pulses was 125 kHz; the spectrum was acquired with one scan. 13C-1H ((H)CH) HETCOR spectra were acquired with 4 scans, and the contact time of the last CP step was either 200 μs or 1 ms. WALTZ-16 decoupling (rf field strength 10 kHz) was applied on the 13C and 15N channels during the acquisition. The 13C-1H J-based HSQC spectra were collected with 16 scans and 256 t1 points; the recycle delay was 2 s. The 13C-1H triple quantum – single quantum (TQ-SQ) and double quantum – single quantum (DQ-SQ) correlation experiments employed the SPC53 sequence [33] for excitation and reconversion of the double quantum coherence; the triple quantum coherence was selected by cycling the phases of the pulses by 60°. The 1H rf power was 370 kHz. The DQ and TQ efficiencies depend on the spin system. For the fMLF sample these were found to be of the order of 20%. For the 2D 1H-1H RFDR spectrum, the RFDR mixing time was 0.27 ms, and the rf power for the RFDR recoupling pulse was 166.5 kHz using fpRFDR conditions. The spectrum was acquired with 16 scans for the DQSQ experiment and 24 scans for the TQSQ experiment, and 256 t1 points; the recycle delay was 3 s.
For CA assemblies, the sample temperature was 26 °C. The recycle delay in all experiments was 2 s. The 2D (H)CH and (H)NH [9] HETCOR spectra were acquired with 32/64 scans and 440/128 t1 points, respectively. The 3D (H)CANH experiment was conducted as reported in [8]. 36 and 64 points were collected in t2 (15N) and t1 (13C) dimensions respectively, and 64 transients were added; the total experiment time was 3.4 days.
1H-15N dipolar tensors were measured using the PARS pulse sequence [34] incorporated into the (H)NH HETCOR experiment. 72 and 16 points were collected in t2 (15N) and t1 (PARS) dimensions respectively, and 56 transients were added; the total experiment time was 1.6 days.
1H CSA tensors were measured using an R1887-symmetry recoupling sequence incorporated into the (H)NH HETCOR experiment. 72 and 24 points were collected in t2 (15N) and t1 (RN) dimensions respectively, and 48 transients were added; the total experiment time was 2.1 days.
fMLF data sets were processed in TopSpin with no apodization (13C-13C RFDR and (H)CH HETCOR), or Lorentzian-to-Gaussian apodization (HSQC, TQ-SQ, and DQ-SQ). Spectra of tubular assemblies of HIV-1 CA were processed in NMRPipe with linear prediction in the indirect dimension(s) to twice the number of data points, followed by sinebell apodization (30-degree for 2D data sets, 60-degree for the 3D (H)CANH spectrum).
2.3. Simulations of 1H-15N Dipolar and 1H CSA Lineshapes
Numerical simulations of 1H-15N dipolar and 1H CSA lineshapes were performed with the SIMPSON software package [35], version 1.1.2. 320 REPULSION angles {α,β} and 13 γ angles were used to calculate a powder average for all simulations. All experimental and processing parameters (i.e., Larmor frequency, MAS frequency, RF field strength, number of t1 points, finite pulse lengths, zero-filling, line broadening, etc.) were taken into account in the simulation for the fitting of 1H-15N dipolar parameters and 1H CSA. The lineshapes were extracted automatically using a series of home-written C++ programs and shell scripts, and were inspected manually to ensure the correctness of the assignments.
3. Results and Discussion
3.1. 13C and 1H-Detected Correlation Spectroscopy at 110 kHz: fMLF Tripeptide
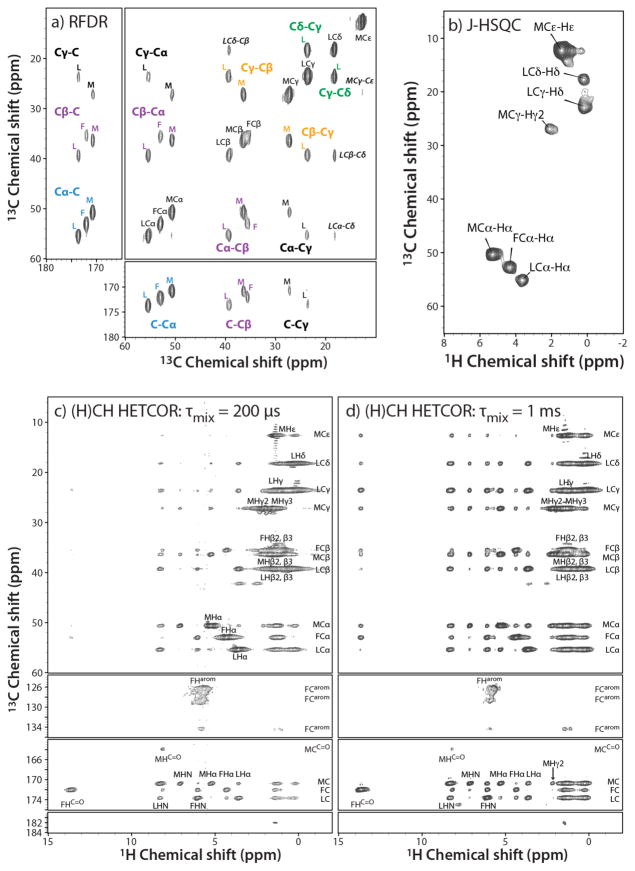
13C-13C RFDR, 13C-1H J-HSQC, and 13C-1H (H)CH HETCOR spectra of U-13C,15N-fMLF are shown in Figure 1, exhibiting 13C and 1H line widths are of the order of 100–150 Hz, consistent with sample’s polycrystallinity. The sensitivity in the dipolar-based RFDR and (H)CH HETCOR experiments acquired at 19.96 T is remarkably high, which permitted the spectra to be recorded with 1 and 4 transients, respectively, thus in a short time and with a fraction of amount normally required for obtaining the equivalent data sets with larger-diameter MAS rotors (1.3 – 3.2 mm). The signal-to-noise ratios (SNRs) for the first FID is 15 for the RFDR spectrum, and 56 (or 88) - for the (H)CH HETCOR spectra, acquired with a back-CP contact time of 200 μs (or 1 ms). The SNR for the first FID of the J-HSQC data set acquired at 11.74 T with 16 scans was 14. The remarkably high resolution in the proton dimension, readily permitted completion of 1H resonance assignments using J-HSQC and (H)CH HETCOR data sets, as illustrated in Figure 1b–d.
Figure 1.
2D 13C- and 1H-detected correlation spectra of f-MLF: a) 13C-13C RFDR spectrum, acquired with 1 scan and 1024 t1 points (total experiment time 0.7 hours); b) 13C-1H HSQC spectrum, acquired with 16 scans and 256 t1 points (total experiment time 2.27 hours); and c) (H)CH HETCOR spectrum, acquired with 4 scans and 1024 t1 points (total experiment time 2.84 hours). RFDR and (H)CH HETCOR spectra were acquired at 19.96 T (850 MHz), J-HSQC spectrum – at 11.74 T (500 MHz), using a 0.7 mm probe, with the MAS frequency of 111 kHz.
Table 1 summarized 13C and 1H chemical shifts of fMLF, and the 13C shifts agree with the previously reported values [36].
Table 1.
13C and 1H Isotropic NMR Chemical Shifts for the fMLF Tripeptide.
| HC=O | C′ | Cα | Cβ | Cγ | Cδ | Cε | Carom | |
|---|---|---|---|---|---|---|---|---|
| M | 163.9 | 170.7 | 49.0 | 36.4 | 27.2 | -- | 12.6 | -- |
| L | -- | 173.6 | 55.4 | 39.3 | 23.6 | 18.2 | -- | -- |
| F | -- | 172.0 | 53.0 | 35.6 | see Carom | see Carom | 126.3, 128.6, 134.7 |
| HC=O | HN | Hα | Hβ2 | Hβ3 | Hγ2 | Hγ3 | Hδ | Hε | Harom | O=C-OH | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| M | 8.2 | 7.1 | 5.3 | 1.4 | 1.2 | 2.1 | 1.8 | -- | 1.4 | -- | -- |
| L | -- | 8.3 | 3.6 | 1.5 | 0.5 | 1.2 | -- | 0.2 | -- | -- | -- |
| F | -- | 6.1 | 4.3 | 1.4 | 1.1 | -- | -- | see Harom | see Harom | 5.6, 5.8 | 13.6 |
The (H)CH HETCOR spectra contained both one-bond and long-range correlations, which could be distinguished from the data sets acquired with the contact times of 200 μs and 1 ms (Figure 1c–d). Long-range correlations between the backbone Cα and C′ carbon atoms and HN and Hα protons within the same amino acids and between neighboring residues were particularly informative for corroborating the resonance assignments.
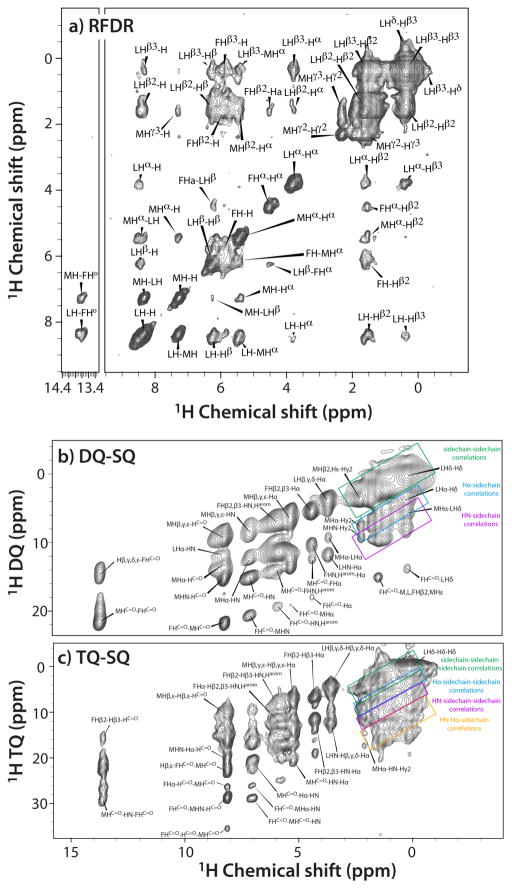
We also examined the performance of 1H-1H dipolar-based experiments, RFDR, as well as of spectra containing double- and triple-quantum filtered to single-quantum (DQ-SQ and TQ-SQ) correlations. The multiple-quantum filtered experiments are beneficial for resolution enhancement and spectral editing [37]. Exemplary 2D spectra are shown in Figure 2a–c. The resolution of the RFDR spectra is high, except for the region containing aliphatic sidechain protons, rendering assignments of the 1H resonances straightforward. The DQ-SQ and TQ-SQ spectra are somewhat more crowded, yet many of the cross peaks were assigned based on chemical shifts available from (H)CH HETCOR, J-HSQC, and RFDR data sets. It is important to note that all prior reports on 1H-1H correlation experiments on unlabeled material at ultrafast MAS focused on small molecules with many fewer protons (crystalline amino acids). The high resolution observed for fMLF was very encouraging and prompted us to investigate larger systems. Taken together, our results suggest that the approach and the suite of experiments discussed here, as well as their higher-dimensional versions, that help to alleviate spectral congestion, such as 3D SQ-DQ(TQ)-SQ correlation experiments [38], will be particularly valuable for molecules that cannot be isotopically labeled, e.g., natural products, pharmaceuticals, and proton-containing solid materials.
Figure 2.
2D 1H-1H correlation spectra of f-MLF: a) RFDR; b) double quantum – single quantum, and c) triple quantum – single quantum spectra. All spectra were recorded at 11.74 T (500 MHz) using a 0.7 mm probe, and a MAS frequency of 111 kHz.
3.2. 1H-Based Correlation Spectroscopy of HIV-1 CA Capsid Protein Assemblies
Mature HIV-1 virions contain conical capsids assembled from ~1,500 copes of a 231-residue CA capsid protein [39; 40]. Capsid cores enclose the RNA genome and several proteins, essential for virus replication. Capsid cores are pleomorphic, exhibiting varied curvature and appearance. They are made up from a hexagonal lattice comprising ca. 216 CA hexamers, which is closed into the ovoid shape by incorporating 12 pentamers [41]. In vitro, CA can form tubular assemblies, whose morphology closely mimics the hexagonal lattice of the CA cores [31; 41]. The structure of CA tubes has been characterized by various methods [41], illuminating details of the capsid architecture and its relationship to viral function. Our team has extensively studied structure and dynamics of these assemblies by heteronuclear- based MAS NMR spectroscopy at frequencies of 10–20 kHz [31; 42; 43; 44]. The samples employed possessed high conformational homogeneity and yielded excellent-resolution spectra, but, alas, at these spinning speeds 1H chemical shifts have not been accessible.
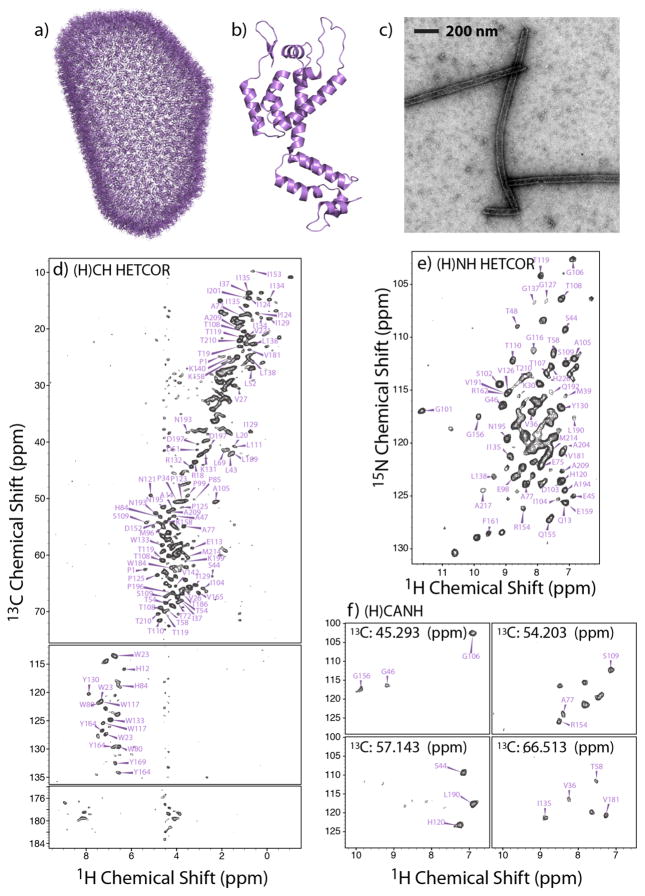
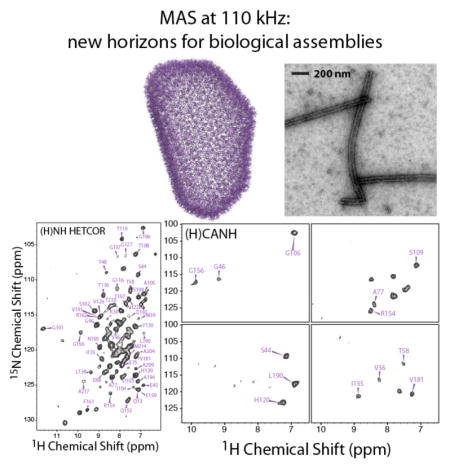
With the 0.7 mm ultrafast MAS HDCN probe, we have explored 2D and 3D 1H-detected heteronuclear correlation experiments for fully protonated tubular assemblies of the U-13C,15N-CA protein. 1H-detected spectra acquired at the MAS frequency of 111 kHz are illustrated in Figure 3: 2D (H)CH HETCOR, 2D (H)NH HETCOR, and representative planes from a 3D (H)CANH experiment are shown, demonstrating exceptional sensitivity and resolution. The SNR of the first FID in the (H)CH and (H)NH HETCOR data sets is 30 (32 scans) and 40 (64 scans); in the (H)CANH spectrum the SNR is 6 (64 scans). The SNR of the (H)CH and (H)NH HETCOR 2D with respect to the most and least intense peaks is 88 and 7, and 32 and 6; in the (H)CANH spectrum the SNR of the four planes shown in Figure 3f is 10, 11, 11, and 10. The 1H line widths range from 120 to 170 Hz. Given this excellent resolution, even in the 2D spectra many individual cross peaks are non-overlapped (Figure 3). Overall, 285 resolved peaks are present in the 2D (H)CH (aliphatic and aromatic regions) and 85 in the (H)NH (amide region) spectra, respectively. In the (H)CH data set, given the well-resolved peaks in the aromatic region, assignments of sidechain carbon and protons for four Trp (out of 5 total), three Tyr (out of 5 total), and one His (out of 5 total) were obtained, on the basis of 13C chemical shifts assigned in our previous heteronuclear-based studies [30; 31; 42].
Figure 3.
a) All-atom model of the mature HIV-1 capsid core, determined by an integrated cryo-EM, cryo-ET, solution NMR, and MD approach (PDBID: 3J3Y [41]). b) 3D structure of an HIV-1 CA monomer (PDBID: 3NTE [45]). c) Transmission electron microscopy (TEM) image of the NMR sample of tubular assemblies of U-13C,15N CA (NL4-3 variant). d–f) 2D (H)CH HETCOR (d) and (H)NH HETCOR (e) spectra, and representative 13C planes of a 3D (H)CANH 1H-detected spectrum (f) of tubular assemblies of U-13C,15N CA, recorded at 19.96 T (850 MHz) and a MAS frequency of 110 kHz.
In the 3D (H)CANH spectrum, 131 resolved peaks were identified. A number of peaks were not resolved. Several stretches of residues were missing in the spectrum. These are the N- and C- terminal residues (1-20 and 217-231) the CypA binding loop (85-95), and the NTD-CTD flexible linker region (residues 142-149). These residues exhibit motions spanning timescales slower than nanoseconds and as slow as milliseconds [42; 44; 46], and the absence of the corresponding resonances in the spectra is not surprising. We note that these peaks are present in the 13C-detected experiments conducted at MAS frequencies of 10–20 kHz, because these were performed at temperatures of 4–15 °C, while the lowest temperature attainable in the current study with the 0.7 mm probe was 26 °C.
In another study, we have acquired additional 3D 1H-detected ultrafast MAS experiments, (H)CONH, (H)N(CO)CAHA, (H)NCAHA, (H)(CO)CA(CO)NH, (H)CCH, and (H)CHH. On the basis of these experiments and our prior 13C and 15N chemical shift assignments [31; 43], we have assigned the majority of 1H, 13C, and 15N chemical shifts; this work will be reported elsewhere.
3.3. 1H-Based Measurements of 1H-15N Dipolar and 1H CSA Tensors in HIV-1 CA Capsid Protein Assemblies
Encouraged by the high resolution and sensitivity in the 2D and 3D correlation experiments of CA assemblies at 111 kHz, we explored whether 1H-15N dipolar and 1H CSA tensors could be extracted. HIV-1 capsids are highly dynamic entities [41; 42; 44; 46; 47], with motions occurring over many orders of magnitudes (nano to milliseconds and slower). Functionally, dynamic processes play important roles in capsid’s assembly, uncoating, and interactions with host factors [44]. We have recently interrogated nano- to microsecond timescale dynamics in HIV-1 CA assemblies using 1H-15N dipolar tensors [42]. RN-symmetry based experiments and their improved versions developed by our group for accurate measurements of heteronuclear dipolar and 1H CSA tensors at MAS frequencies of 40 kHz and below have been applied to organic and biological systems, including proteins and protein assemblies [27; 28; 29; 34; 42; 48]. In these reports, we have presented in-depth analysis of the performance of the RN-symmetry based experiments and validated their applications in protein assemblies, including the HIV-1 CA assemblies, using various means, such as empirical correlations between 1H CSA and hydrogen bond length using electrostatic models [29], quantum chemical (DFT) calculations of CSA tensors [49], as well as a combined MD/DFT approach [44]. Others have demonstrated that 1H CSA tensors can be recorded in small molecules at the MAS frequency of 65 kHz [50], albeit no validation was reported on the measured CSAs from quantum chemical calculations. Indeed, 1H CSA tensors, are another sensitive probe of dynamics, which also report on hydrogen bonding interactions [29; 49]. To extract dynamics information from CSA tensors, quantum mechanical Density Functional Theory (DFT) calculations as well as combined MD/DFT calculations can be employed, as we have demonstrated recently [44].
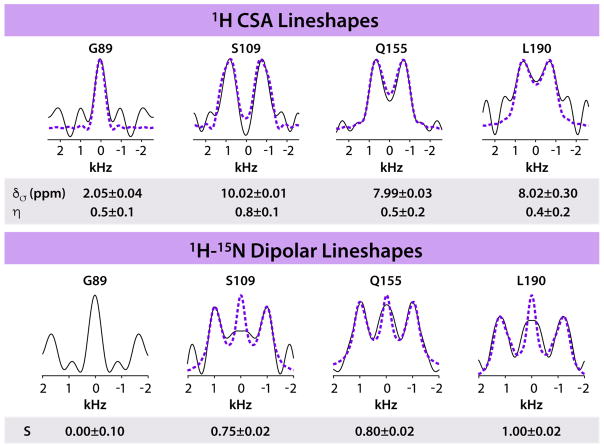
Representative 1H-15N dipolar and 1H CSA lineshapes are shown for four residues in the CA assemblies in Figure 4: G89 (in the Cyclophilin A (CypA) binding loop), S109 (in a short loop connecting helices 5 and 6), Q155 (at the end of loop connecting helices 310 and 8), and L190 (in helix 10). The lineshapes were recorded in 3D 1H-detected experiments, where a dipolar/CSA dimension was combined with two isotropic 15N-1H chemical shift dimensions. The sensitivity in these experiments is remarkably high: it took only 1.6 and 2.1 days to record dipolar and CSA tensors, respectively. In comparison, it typically takes 4–7 days to collect the corresponding data sets by heteronuclear-based spectroscopy with 1.9 – 3.2 mm probes.
Figure 4.
Representative 1H CSA (top) and 1H-15N dipolar (bottom) lineshapes for G89, S109, Q155, and L190 in tubular assemblies of HIV-1 CA protein, extracted from RN-symmetry based 3D experiments. The spectra were recorded at 19.96 T (850 MHz) using a 0.7 mm probe, and a MAS frequency of 110 kHz. Features outside of ±1.5 kHz range are spectral noise.
As demonstrated by us previously [31; 44], G89 is remarkably dynamic on timescales of nano- to microseconds, exhibiting essentially isotropic 1H-15N and 1H-13C dipolar lineshapes. In contrast, S109, Q155, and L190 are rigid. The dipolar coupling constants obtained from the fits of the experimental lineshapes (Figure 4) are in excellent agreement with the values reported by us previously [31], including the isotropic lineshape observed for G89. This validates the PARS experiment under ultrafast MAS conditions. Remarkably, the 1H CSA tensor for G89 is also averaged by motion, while the 1H CSA tensors for S109, Q155, and L190 suggest a rigid backbone. Theoretical validation of these experimental 1H CSA values requires a hybrid quantum mechanics/molecular mechanics/molecular dynamics approach, as we have demonstrated recently [44], and this work will be reported elsewhere.
4. Conclusions
At MAS frequencies of 110–111 kHz dramatic sensitivity and resolution enhancements permit structure and dynamics characterization of fully protonated biological solids, requiring only nanomoles of material and a few hours of measuring times. The 2D 1H- and 13C-detected correlation experiments established on fMLF will be particularly useful for analysis of small organic and biological molecules at natural abundance, such as natural products. The 3D experiments for resonance assignments and measurements of dipolar and CSA tensors demonstrated for HIV-1 CA protein assemblies can be extended to complexes with host factors and, more broadly, for analysis of a wide range of complex systems. Thus, the approach presented here opens new avenues for tackling questions in structural biology of large assemblies.
Highlights.
Large sensitivity and resolution enhancements observed in NMR experiments at MAS of 110–111 kHz enable structural analysis of fully protonated biological systems
Resonance assignments of protons, heteronuclei and extraction of site-specific 1H-15N dipolar and 1H CSA tensors for HIV-1 capsid CA protein assemblies using proton-detected 2D and 3D correlation experiments at 111 kHz
Acknowledgments
This work was supported by the National Institutes of Health (NIH Grant P50 GM082251), acknowledge the support of the National Science Foundation (NSF Grant CHE0959496) for the acquisition of the 850 MHz NMR spectrometer, of the National Institutes of Health (NIH Grant P30GM110758) for the support of core instrumentation infrastructure at the University of Delaware.
References
- 1.Andrew ER, Bradbury A, Eades RG. Nuclear Magnetic Resonance Spectra from a Crystal Rotated at High Speed. Nature. 1958;182:1659–1659. [Google Scholar]
- 2.Andrew ER, Bradbury A, Eades RG. Removal of Dipolar Broadening of Nuclear Magnetic Resonance Spectra of Solids by Specimen Rotation. Nature. 1959;183:1802–1803. [Google Scholar]
- 3.Lowe IJ. Free Induction Decays of Rotating Solids. Phys Rev Lett. 1959;2:285–287. [Google Scholar]
- 4.Samoson A, Tuherm T, Gan Z. High-Field High-Speed MAS Resolution Enhancement in 1H NMR Spectroscopy of Solids. Solid State Nucl Magn Reson. 2001;20:130–136. doi: 10.1006/snmr.2001.0037. [DOI] [PubMed] [Google Scholar]
- 5.Ernst M, Samoson A, Meier BH. Low-power XiX decoupling in MAS NMR experiments. J Magn Reson. 2003;163:332–339. doi: 10.1016/s1090-7807(03)00155-1. [DOI] [PubMed] [Google Scholar]
- 6.Zhou DH, Rienstra CM. High-performance solvent suppression for proton detected solid-state NMR. J Magn Reson. 2008;192:167–172. doi: 10.1016/j.jmr.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbet-Massin E, Pell AJ, Retel JS, Andreas LB, Jaudzems K, Franks WT, Nieuwkoop AJ, Hiller M, Higman V, Guerry P, Bertarello A, Knight MJ, Felletti M, Le Marchand T, Kotelovica S, Akopjana I, Tars K, Stoppini M, Bellotti V, Bolognesi M, Ricagno S, Chou JJ, Griffin RG, Oschkinat H, Lesage A, Emsley L, Herrmann T, Pintacuda G. Rapid Proton-Detected NMR Assignment for Proteins with Fast Magic Angle Spinning. J Am Chem Soc. 2014;136:12489–12497. doi: 10.1021/ja507382j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchetti A, Jehle S, Felletti M, Knight MJ, Wang Y, Xu ZQ, Park AY, Otting G, Lesage A, Emsley L, Dixon NE, Pintacuda G. Backbone Assignment of Fully Protonated Solid Proteins by 1H Detection and Ultrafast Magic-Angle-Spinning NMR Spectroscopy. Angew Chem-Int Edit. 2012;51:10756–10759. doi: 10.1002/anie.201203124. [DOI] [PubMed] [Google Scholar]
- 9.Paulson EK, Morcombe CR, Gaponenko V, Dancheck B, Byrd RA, Zilm KW. Sensitive High Resolution Inverse Detection NMR Spectroscopy of Proteins in the Solid State. J Am Chem Soc. 2003;125:15831–15836. doi: 10.1021/ja037315+. [DOI] [PubMed] [Google Scholar]
- 10.Akbey U, Lange S, Franks WT, Linser R, Rehbein K, Diehl A, van Rossum BJ, Reif B, Oschkinat H. Optimum levels of exchangeable protons in perdeuterated proteins for proton detection in MAS solid-state NMR spectroscopy. J Biomol NMR. 2010;46:67–73. doi: 10.1007/s10858-009-9369-0. [DOI] [PubMed] [Google Scholar]
- 11.Chevelkov V, Rehbein K, Diehl A, Reif B. Ultrahigh resolution in proton solid-state NMR spectroscopy at high levels of deuteration. Angew Chem-Int Edit. 2006;45:3878–3881. doi: 10.1002/anie.200600328. [DOI] [PubMed] [Google Scholar]
- 12.Knight MJ, Webber AL, Pell AJ, Guerry P, Barbet-Massin E, Bertini I, Felli IC, Gonnelli L, Pierattelli R, Emsley L, Lesage A, Herrmann T, Pintacuda G. Fast Resonance Assignment and Fold Determination of Human Superoxide Dismutase by High-Resolution Proton-Detected Solid-State MAS NMR Spectroscopy. Angewandte Chemie International Edition. 2011;50:11697–11701. doi: 10.1002/anie.201106340. [DOI] [PubMed] [Google Scholar]
- 13.Knight MJ, Pell AJ, Bertini I, Felli IC, Gonnelli L, Pierattelli R, Herrmann T, Emsley L, Pintacuda G. Structure and backbone dynamics of a microcrystalline metalloprotein by solid-state NMR. Proc Natl Acad Sci U S A. 2012;109:11095–100. doi: 10.1073/pnas.1204515109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dannatt HRW, Felletti M, Jehle S, Wang Y, Emsley L, Dixon NE, Lesage A, Pintacuda G. Weak and Transient Protein Interactions Determined by Solid-State NMR. Angewandte Chemie International Edition. 2016;55:6638–6641. doi: 10.1002/anie.201511609. [DOI] [PubMed] [Google Scholar]
- 15.Saurel O, Iordanov I, Nars G, Demange P, Le Marchand T, Andreas LB, Pintacuda G, Milon A. Local and Global Dynamics in Klebsiella pneumoniae Outer Membrane Protein a in Lipid Bilayers Probed at Atomic Resolution. J Am Chem Soc. 2017;139:1590–1597. doi: 10.1021/jacs.6b11565. [DOI] [PubMed] [Google Scholar]
- 16.Sinnige T, Daniëls M, Baldus M, Weingarth M. Proton Clouds to Measure Long-Range Contacts between Nonexchangeable Side Chain Protons in Solid-State NMR. J Am Chem Soc. 2014;136:4452–4455. doi: 10.1021/ja412870m. [DOI] [PubMed] [Google Scholar]
- 17.Barbet-Massin E, Felletti M, Schneider R, Jehle S, Communie G, Martinez N, Jensen RM, Ruigrok WHR, Emsley L, Lesage A, Blackledge M, Pintacuda G. Insights into the Structure and Dynamics of Measles Virus Nucleocapsids by (1)H-detected Solid-state NMR. Biophysical Journal. 2014;107:941–6. doi: 10.1016/j.bpj.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wickramasinghe A, Wang SL, Matsuda I, Nishiyama Y, Nemoto T, Endo Y, Ishii Y. Evolution of CPMAS under fast magic-angle-spinning at 100 kHz and beyond. Solid State Nucl Magn Reson. 2015;72:9–16. doi: 10.1016/j.ssnmr.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang RC, Pandey MK, Nishiyama Y, Ramamoorthy A. A Novel High-Resolution and Sensitivity-Enhanced Three-Dimensional Solid-State NMR Experiment Under Ultrafast Magic Angle Spinning Conditions. Sci Rep. 2015;5:9. doi: 10.1038/srep11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agarwal V, Tuherm T, Reinhold A, Past J, Samoson A, Ernst M, Meier BH. Amplitude-modulated low-power decoupling sequences for fast magic-angle spinning NMR. Chem Phys Lett. 2013;583:1–7. [Google Scholar]
- 21.Agarwal V, Penzel S, Szekely K, Cadalbert R, Testori E, Oss A, Past J, Samoson A, Ernst M, Bockmann A, Meier BH. De Novo 3D Structure Determination from Sub-milligram Protein Samples by Solid-State 100 kHz MAS NMR Spectroscopy. Angew Chem-Int Edit. 2014;53:12253–12256. doi: 10.1002/anie.201405730. [DOI] [PubMed] [Google Scholar]
- 22.Penzel S, Smith AA, Agarwal V, Hunkeler A, Org ML, Samoson A, Böckmann A, Ernst M, Meier BH. Protein resonance assignment at MAS frequencies approaching 100 kHz: a quantitative comparison of J-coupling and dipolar-coupling-based transfer methods. J Biomol NMR. 2015;63:165–186. doi: 10.1007/s10858-015-9975-y. [DOI] [PubMed] [Google Scholar]
- 23.Andreas LB, Jaudzems K, Stanek J, Lalli D, Bertarello A, Le Marchand T, Paepe DCD, Kotelovica S, Akopjana I, Knott B, Wegner S, Engelke F, Lesage A, Emsley L, Tars K, Herrmann T, Pintacuda G. Structure of fully protonated proteins by proton-detected magic-angle spinning NMR. Proc Natl Acad Sci U S A. 2016;113:9187–9192. doi: 10.1073/pnas.1602248113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanek J, Andreas LB, Jaudzems K, Cala D, Lalli D, Bertarello A, Schubeis T, Akopjana I, Kotelovica S, Tars K, Pica A, Leone S, Picone D, Xu Z-Q, Dixon NE, Martinez D, Berbon M, El Mammeri N, Noubhani A, Saupe S, Habenstein B, Loquet A, Pintacuda G. NMR Spectroscopic Assignment of Backbone and Side-Chain Protons in Fully Protonated Proteins: Microcrystals, Sedimented Assemblies, and Amyloid Fibrils. Angewandte Chemie International Edition. 2016;55:15504–15509. doi: 10.1002/anie.201607084. [DOI] [PubMed] [Google Scholar]
- 25.Zhao X, Eden M, Levitt MH. Recoupling of heteronuclear dipolar interactions in solid-state NMR using symmetry-based pulse sequences. Chem Phys Lett. 2001;342:353–361. [Google Scholar]
- 26.Levitt MH. Symmetry in the design of NMR multiple-pulse sequences. J Chem Phys. 2008;128:25. doi: 10.1063/1.2831927. [DOI] [PubMed] [Google Scholar]
- 27.Hou GJ, Byeon IJL, Ahn J, Gronenborn AM, Polenova T. H-1-C-13/H-1-N-15 Heteronuclear Dipolar Recoupling by R-Symmetry Sequences Under Fast Magic Angle Spinning for Dynamics Analysis of Biological and Organic Solids. J Am Chem Soc. 2011;133:18646–18655. doi: 10.1021/ja203771a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou GJ, Byeon IJL, Ahn J, Gronenborn AM, Polenova T. Recoupling of chemical shift anisotropy by R-symmetry sequences in magic angle spinning NMR spectroscopy. J Chem Phys. 2012;137:10. doi: 10.1063/1.4754149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou GJ, Paramasivam S, Yan S, Polenova T, Vega AJ. Multidimensional Magic Angle Spinning NMR Spectroscopy for Site-Resolved Measurement of Proton Chemical Shift Anisotropy in Biological Solids. J Am Chem Soc. 2013;135:1358–1368. doi: 10.1021/ja3084972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han Y, Ahn J, Concel J, Byeon IJL, Gronenborn AM, Yang J, Polenova T. Solid-State NMR Studies of HIV-1 Capsid Protein Assemblies. J Am Chem Soc. 2010;132:1976–1987. doi: 10.1021/ja908687k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han Y, Hou GJ, Suiter CL, Ahn J, Byeon IJL, Lipton AS, Burton S, Hung I, Gor’kov PL, Gan ZH, Brey W, Rice D, Gronenborn AM, Polenova T. Magic Angle Spinning NMR Reveals Sequence-Dependent Structural Plasticity, Dynamics, and the Spacer Peptide 1 Conformation in HIV-1 Capsid Protein Assemblies. J Am Chem Soc. 2013;135:17793–17803. doi: 10.1021/ja406907h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beckett P, Denning MS, Carravetta M, Kalda A, Heinmaa I. Field dependence of the relaxation of Br-79 in KBr and its use as a temperature calibrant. J Magn Reson. 2012;223:61–63. doi: 10.1016/j.jmr.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Hohwy M, Rienstra CM, Griffin RG. Band-selective homonuclear dipolar recoupling in rotating solids. J Chem Phys. 2002;117:4973–4987. [Google Scholar]
- 34.Hou GJ, Lu XY, Vega AJ, Polenova T. Accurate measurement of heteronuclear dipolar couplings by phase-alternating R-symmetry (PARS) sequences in magic angle spinning NMR spectroscopy. J Chem Phys. 2014;141:11. doi: 10.1063/1.4894226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bak M, Rasmussen JT, Nielsen NC. SIMPSON: A general simulation program for solid-state NMR spectroscopy. J Magn Reson. 2000;147:296–330. doi: 10.1006/jmre.2000.2179. [DOI] [PubMed] [Google Scholar]
- 36.Rienstra CM, Hohwy M, Hong M, Griffin RG. 2D and 3D N-15-C-13-C-13 NMR chemical shift correlation spectroscopy of solids: Assignment of MAS spectra of peptides. J Am Chem Soc. 2000;122:10979–10990. [Google Scholar]
- 37.Schnell I, Spiess HW. High-resolution H-1 NMR spectroscopy in the solid state: Very fast sample rotation and multiple-quantum coherences. J Magn Reson. 2001;151:153–227. doi: 10.1006/jmre.2001.2336. [DOI] [PubMed] [Google Scholar]
- 38.Zhang RC, Mroue KH, Ramamoorthy A. Proton-Based Ultrafast Magic Angle Spinning Solid-State NMR Spectroscopy. Accounts Chem Res. 2017;50:1105–1113. doi: 10.1021/acs.accounts.7b00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Briggs JAG, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 2004;11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 40.Sundquist WI, Krausslich HG. HIV-1 Assembly, Budding, and Maturation. Csh Perspect Med. 2012;2 doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao GP, Perilla JR, Yufenyuy EL, Meng X, Chen B, Ning JY, Ahn J, Gronenborn AM, Schulten K, Aiken C, Zhang PJ. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature. 2013;497:643–646. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu MM, Hou GJ, Zhang HL, Suiter CL, Ahn J, Byeon IJL, Perilla JR, Langmead CJ, Hung I, Gor’kov PL, Gan ZH, Brey W, Aiken C, Zhang PJ, Schulten K, Gronenborn AM, Polenova T. Dynamic allostery governs cyclophilin A-HIV capsid interplay. Proc Natl Acad Sci U S A. 2015;112:14617–14622. doi: 10.1073/pnas.1516920112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perilla JR, Zhao GP, Lu MM, Ning JY, Hou GJ, Byeon IJL, Gronenborn AM, Polenova T, Zhang PJ. CryoEM Structure Refinement by Integrating NMR Chemical Shifts with Molecular Dynamics Simulations. J Phys Chem B. 2017;121:3853–3863. doi: 10.1021/acs.jpcb.6b13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang HL, Hou GJ, Lu MM, Ahn J, Byeon IJL, Langmead CJ, Perilla JR, Hung I, Gor’kov PL, Gan ZH, Brey WW, Case DA, Schulten K, Gronenborn AM, Polenova T. HIV-1 Capsid Function Is Regulated by Dynamics: Quantitative. Atomic-Resolution Insights by Integrating Magic-Angle-Spinning NMR, QM/MM, and MD. J Am Chem Soc. 2016;138:14066–14075. doi: 10.1021/jacs.6b08744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du SC, Betts L, Yang RF, Shi HB, Concel J, Ahn J, Aiken C, Zhang PJ, Yeh JI. Structure of the HIV-1 Full-Length Capsid Protein in a Conformationally Trapped Unassembled State Induced by Small-Molecule Binding. J Mol Biol. 2011;406:371–386. doi: 10.1016/j.jmb.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Byeon IJL, Hou GJ, Han Y, Suiter CL, Ahn J, Jung J, Byeon CH, Gronenborn AM, Polenova T. Motions on the Millisecond Time Scale and Multiple Conformations of HIV-1 Capsid Protein: Implications for Structural Polymorphism of CA Assemblies. J Am Chem Soc. 2012;134:6455–6466. doi: 10.1021/ja300937v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bayro MJ, Chen B, Yau WM, Tycko R. Site-Specific Structural Variations Accompanying Tubular Assembly of the HIV-1 Capsid Protein. J Mol Biol. 2014;426:1109–1127. doi: 10.1016/j.jmb.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu XY, Zhang HL, Lu MM, Vega AJ, Hou GJ, Polenova T. Improving dipolar recoupling for site-specific structural and dynamics studies in biosolids NMR: windowed RN-symmetry sequences. Phys Chem Chem Phys. 2016;18:4035–4044. doi: 10.1039/c5cp07818k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hou G, Gupta R, Polenova T, Vega AJ. A Magic-Angle Spinning NMR Method for the Site-Specific Measurement of Proton Chemical-Shift Anisotropy in Biological and Organic Solids. Israel journal of chemistry. 2014;54:171–183. doi: 10.1002/ijch.201300099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miah HK, Bennett DA, Iuga D, Titman JJ. Measuring proton shift tensors with ultrafast MAS NMR. J Magn Reson. 2013;235:1–5. doi: 10.1016/j.jmr.2013.07.005. [DOI] [PubMed] [Google Scholar]