Abstract
Large genomic deletions are potential candidate for loss-of-function, which could be lethal as homozygote. Analysing whole genome data of 175 cattle, we report 8,480 large deletions (199 bp–773 KB) with an overall false discovery rate of 8.8%; 82% of which are novel compared with deletions in the dbVar database. Breakpoint sequence analyses revealed that majority (24 of 29 tested) of the deletions contain microhomology/homology at breakpoint, and therefore, most likely generated by microhomology-mediated end joining. We observed higher differentiation among breeds for deletions in some genic-regions, such as ABCA12, TTC1, VWA3B, TSHR, DST/BPAG1, and CD1D. The genes overlapping deletions are on average evolutionarily less conserved compared with known mouse lethal genes (P-value = 2.3 × 10−6). We report 167 natural gene knockouts in cattle that are apparently nonessential as live homozygote individuals are observed. These genes are functionally enriched for immunoglobulin domains, olfactory receptors, and MHC classes (FDR = 2.06 × 10−22, 2.06 × 10−22, 7.01 × 10−6, respectively). We also demonstrate that deletions are enriched for health and fertility related quantitative trait loci (2-and 1.5-fold enrichment, Fisher’s P-value = 8.91 × 10−10 and 7.4 × 10−11, respectively). Finally, we identified and confirmed the breakpoint of a ∼525 KB deletion on Chr23:12,291,761-12,817,087 (overlapping BTBD9, GLO1 and DNAH8), causing stillbirth in Nordic Red Cattle.
Keywords: dairy cattle, structural variants, whole genome sequence, population genetics, loss-of-function
1. Introduction
Embryonic lethality has become a challenge to cattle breeders, especially for dairy cattle where a limited number of bulls were extensively used in breeding for fast genetic progress in economic traits like milk and protein yield.1 An estimated yearly loss of ∼$10.74 million is attributed to known recessive lethals in four dairy cattle breeds from USA only, where Holstein accounts for ∼70% of the total losses, followed by Jersey, Brown Swiss, and Ayrshire.2 Hence, understanding the genomic architecture of cattle populations is important, now more than ever, for optimizing genetic gain while constraining negative impact of deleterious mutations responsible for genetic defects and inbreeding depression.
Unlike single nucleotide polymorphism (SNP) and small insertion or deletion (indel), structural variants (SVs), i.e. DNA alterations larger than 50 base pairs (bp) that include insertions, deletions, duplications, inversions, and translocations,3 are the least explored polymorphisms in cattle. SVs contribute substantially to phenotypic variations and have a wide-spectrum of impact ranging from beneficial to lethal in both humans3,4 and animals.5 The phenotypic impact of SVs in cattle is well evident from numerous studies. For example, Xu et al.6 showed that a combination of SNPs with SVs could explain additional genetic variance underlying milk production traits, while Charlier et al.,7 Schutz et al.,8 and Kadri et al.,9 showed the lethal effect of large deletions in dairy cattle. Furthermore, a ∼525 KB deletion on chromosome 23 is reported to be associated with stillbirth in Nordic Red Cattle.10
Earlier SV studies on cattle were mostly SNP-array based, such as array-comparative genomic hybridization,11 50 K BovineSNP50 BeadChip12 or 777 K BovineHD BeadChip (BovineHD chip)13 based. But, using these approaches a substantial portion of the genome could not be explored and breakpoint resolution is still an issue.14 However, whole-genome sequence (WGS) based techniques could improve resolution as well as power to capture SVs in a wide size and frequency spectrum.14 For example, majority of the novel SVs in humans15,16 and mouse17 were detected using WGS approaches. Besides, breakpoint sequences could also be assembled with high accuracy from sequencing reads,18 which are necessary for elucidating the mechanisms underlying SV formation.19
In the advent of next-generation sequencing (NGS) techniques, hundreds of cattle (bull or cow) genomes were re-sequenced in collaborative initiative such as 1000 Bull Genomes Project (1KBGP)20 (and other independent projects, e.g.21,22) to build a comprehensive database of sequence variants, mainly SNPs and indels. This NGS data provides a unique opportunity to study SVs in cattle. However, few studies23,24 utilized these (and/or other) NGS resources so far for studying SVs in cattle.
Therefore, in this study we scanned the WGSs of 175 cattle from three dairy breeds, namely Holstein, Jersey, and Nordic Red Cattle, to discover large deletions segregating in the population, and analyse their population-genetic properties. In particular, we focused on understating the population diversity, stratification, and plausible functional effects. We also explored the probable mechanisms of SV formation for a set of breakpoint-resolved deletions.
2. Materials and methods
2.1. Animal samples and ethics
This study was performed on WGS of 175 dairy cattle from three breeds, e.g. 67 Holstein, 27 Jersey, and 81 Nordic Red Cattle. The sample included 7 Holstein cows and 168 bulls from these three breeds−144 animals from Run 5 of 1KBGP20 and 31 animals from Nordic sequence data.21 Genome sequences were generated using Illumina paired-end sequencing to an average coverage of 10-fold.
Here, we did not include any experimentation on animals and only dealt with analysis-ready WGS data; hence, no ethical approval was required.
2.2. Sequence alignment to reference genome and SNPs/indels calling
Raw sequencing reads were filtered and ‘FASTQ’ files were aligned to bovine reference genome assembly ‘UMD3.1’ using ‘BWA’ software25 to produce BAM files for subsequent variant calling. In 1KBGP, SNPs and indels were called using ‘SAMtools 0.1.18 mpileup’ software,26 while ‘GATK v1.6’ software27 was used for Nordic WGS data (detailed method in20 and,21 respectively). For all the analysis, bovine genome assembly ‘UMD3.1’ was used as the reference genome.
2.3. Discovery and genotyping of deletions
SVs can be detected from NGS data based on sequence signatures such as (discordant) read-pair (RP), split-read (SP), and read-depth (RD), as well as de novo assembly of reads.14 However, approaches based on only one sequence signature could be constrained by high false discovery rate (FDR),28 hence we employed a population scale SV detection method called ‘Genome STRucture in Populations (Genome STRiP)’28—which leverages technical (e.g. RP and RD signals) and population-level sequence features (e.g. coherence around shared alleles, and heterogeneity of evidentiary sequences in different genomes) for accurate discovery of deletions, and determines genotype (allelic state) of each locus from RD using a Gaussian mixture model.
2.3.1. Genome STRiP
For deletion discovery and genotyping ‘Genome STRiP’ software version 2.00.167828 was used. Following the documentation, we built a custom reference metadata bundle for cattle samples that includes alignability mask, copy-number mask (CN2 mask), ploidy map, gender map. Alignability mask represents sites on the reference genome that are uniquely alignable by sequence read of a certain length (readLength). Our WGS data was a mixture of different ‘Illumina’ paired-end reads ranging from 90 to 101 bp (Q1 = 90, median = 100, and Q3 = 100), hence genome alignability mask was prepared with readLength value of 90 using ‘ComputeGenomeMask’ utility from Genome STRiP. Copy-number mask (CN2 mask), i.e. regions on the reference genome unlikely to be copy-number variable in most individuals, was produced for the bovine assembly UMD3.1 excluding sex chromosome X, unplaced contigs, and repeat sequences (retrieved from RepeatMasker track of UCSC Table Browser,29 accessed on 4 July 2016).
2.3.2. Pre-processing, deletion discovery and genotyping
We ran the preprocessing Queue script (dry run) to emit all the commands, prepared bash scripts to run in Portable Batch System job scheduler, and executed these commands proving 175 BAM files (one for each sample) as input.
Large deletions (100 bp ≤ size ≤ 1 MB) were discovered and filtered using SVDiscovery Queue script. Discovered sites were filtered (default filters) if (i) the site contained too high or too low read pileup, (ii) RPs spacing was inconsistent with a single segregating deletion, (iii) RD and RP evidences were inconsistent across samples, (iv) RD differences were not significant, and (v) RP evidence was thinly distributed across samples (Genome STRiP Tutorial—GATK Workshop 2013, http://software.broadinstitute.org/software/genomestrip/workshop-presentations, accessed on 26 August 2016).
All passed sites were genotyped by SVGenotyper with default parameters. Genotyped deletion calls were then filtered based on following criteria, e.g. (i) sites with excess number of heterozygote calls (inbreeding coefficient ≤ −0.15), (ii) non-variant site based on genotype likelihood (parameter: non-variance score ≥ 13.0), (iii) sites with too low or too high RD (parameter: 0.5 ≥ GSM1 ≥ 2.0), (iv) sites with less than 30% uniquely alignable bases, (v) potential duplicate of another site (parameter: duplicateOverlapThreshold 0.5 and duplicateScoreThreshold ≥ 0.0), (vi) start/end position of a deletion call within 150 bp of assembly gap, (vii) all samples homozygous for reference allele (95% CI), and (viii) sites with ≥ 10% missing genotype.
2.4. Validation of deletions
2.4.1. Validation using 777K BovineHD BeadChip intensity data
We validated deletion calls using 777K BovineHD BeadChip (Illumina, San Diego, CA, USA) intensity data on 26 Holstein samples that were both WGS and 777K chip typed. We calculated FDR for the deletion call-set using IntensityRankSum (IRS) test implemented in Genome STRiP. Intensity file was prepared from raw chip intensity data following the guideline for IRS test. Overall FDR for the call-set was calculated as two times the fraction of sites with IRS P-value ≥ 0.5 (i.e. sites with IRS P-value ≥ 0.5 to the sites with valid P-value). Details of IRS test could be found in.15,28
2.4.2. Validation by targeted assembly of breakpoint
Targeted iterative graph routing assembler (TIGRA-0.4.3) software18 was used, with default parameters, for assembling deletion breakpoint sequences from a set of randomly selected deletions along with three previously known deletions segregating in the study populations. TIGRA extracted all reads mapped to 500 bp upstream and 50 bp downstream of start coordinate, and 50 bp upstream and 500 bp downstream of end coordinate of a given deletion; and reads were then assembled iteratively using de Bruijn graph assembler with multiple k-mers (e.g. 15 bp followed by 25 bp). We aligned the assembled contigs to UMD3.1 using Cow BLAT Search30 from UCSC Genome Browser (https://genome.ucsc.edu/cgi-bin/hgBlat) to visualize and infer breakpoints from the alignments.
2.4.3. Validation by PCR and amplicon sequencing
We validated a previously reported ∼525 KB deletion segregating in Nordic Red Cattle10 using PCR and amplicon sequencing. Genomic DNA was extracted as described previously by Miller et al.31 from semen sample of two bulls carrying the deletion and two non-carriers. The PCR reaction was done with the DyNAzyme II DNA Polymerase (Thermo Fisher, MA, US) in a 30 µl volume of 1× PCR buffer, 0.2 mM dNTPs, 10 pmol primer mix (forward primer: 5′- AAGCCACCACAATGAGAAGC -3′ and reverse primer: 5′- TTTGGGGTAGGAGAAGTAGGG -3′) and 50 ng of genomic DNA. The cycling conditions were the following: (i) an initial denaturation at 95 °C for 3 min, (ii) 35 cycles of 30 s denaturation (94 °C), 30 s hybridization (65.2 °C), 30 s elongation (72 °C), and a final 3 min elongation (72 °C). PCR products were separated on a 2% agarose gel, purified and directly sequenced using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, CA, US). Electrophoresis of sequencing reactions was performed on ‘3500xL Genetic Analyzers’ (Applied Biosystems, CA, US), and sequences were visualized with Sequencher 5.4.6 (Gene Codes Corporation, MI, USA). A 977 bp control amplification, with a primer pair within the deletion (forward primer: 5′- CCCAATGCAAAATCACAAAA -3′ and reverse primer: 5′- CCAGAAAAGCTACACTTGAACTGA -3′), was performed using the same reaction conditions as above except hybridization was performed at 59.8 °C.
2.5. Analysis of population genetic properties
The population genetic properties of deletions, among the three breeds, were studied in terms of population diversity, population structure, and population differentiation. Population diversity was calculated using ‘VariantsPerSampleAnnotator’ from ‘Genome STRiP’ software, which provides distribution of variants across samples and populations. We performed principal component analysis (PCA) using PLINK (v1.90p) software32 to distinguish three cattle breeds (details in Supplementary Material). We calculated VST33—a population stratification measure of SVs (highly correlated with Wright’s fixation index, FST34), for each deletion locus using variant allele frequency (VAF) and genotypes from pairwise comparison of one breed with the rest, such as Holstein vs Jersey + Nordic Red Cattle, and vice versa.
2.6. Functional annotation and enrichment analysis
Functional annotation of deletions were performed using ‘Variant Effect Predictor (VEP-87)’ software,35 and enrichment of protein domains (InterPro36 and Pfam37) and pathways (KEGG38) were analysed using ‘STRING-v10 database’.39
Selective constraints on genes were measured from the ratio of non-synonymous (dN) to synonymous (dS) substitution rate, i.e. dN/dS ratio, between cow-mouse 1-to-1 orthologues downloaded from Ensembl database40 (release 87, last accessed on 21 February 2017) using BioMart.41 Here we analysed whether dN/dS of genes overlapping deletions are higher (i.e. less constrained) than that of mouse lethal genes (from Dickinson et al.42) using Wilcoxson test. Reported causal genes for cattle were also retrieved from OMIA database (http://omia.angis.org.au/, last accessed on 10 May 2016) for dN/dS comparison.
We retrieved cattle quantitative trait loci (QTL) from QTLdb database43 (release 31; accessed on 6 January 2017); autosomal QTL from Holstein, Jersey, Nordic Red Cattle and Ayrshire, associated to any of the six trait classes, e.g. ‘Reproduction’, ‘Milk’, ‘Production’, ‘Exterior’, ‘Meat and Carcass’, and ‘Health’, were considered for QTL enrichment analysis. We calculated fold enrichment for a trait, such as for Health related QTL: (No. of Health QTL on Deletions ∕ Total QTL on deletions) / (Total Health QTL ∕ Total QTL in the dataset), and statistical significance using ‘Fisher’s exact test’ (two sided).
2.7. Data manipulation, visualization, and statistical analysis
All statistical analyses and plots were generated in RStudio software44 running R software version 3.3.2,45 unless mentioned otherwise. BEDTools (v2.26.0) software46 is used for identifying the overlap between deletion calls and other genomic features, such as, ‘UMD3.1’ assembly gaps (from ‘UCSC Table Browser’), CNVs from dbVar database47, three known deletions from7,9,10, QTL from QTLdb. VCFtools (v0.1.15) software and PLINK (v1.90p) software were used for analysing the VCF file.
3. Results and discussion
3.1. Discovery and genotyping of deletions
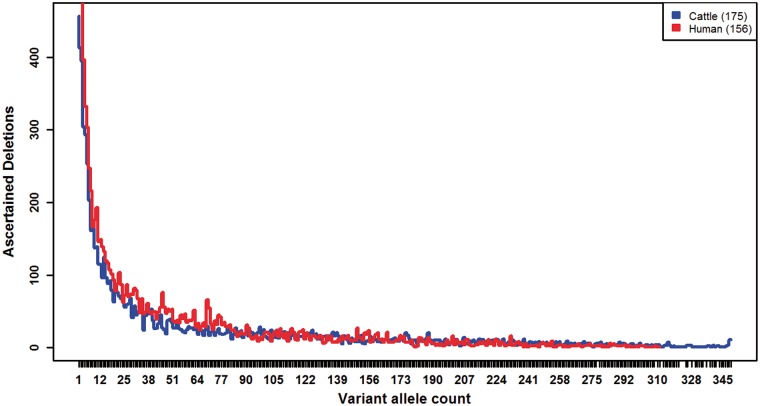
Deletion discovery and genotyping were carried out using Genome STRiP. After filtering, we report 8,480 large deletions with genotypes in 67 Holsten, 27 Jersey, and 81 Nordic Red Cattle. The deletion size ranged from 199 bp to 773 KB with a mean of 4.5 KB (median = 1 KB), which is approximately 10 times smaller compared with 184 deletion-CNVs (mean = 44.5 KB, median = 7.7 KB) reported in a recent 777K BovineHD BeadChip (BovineHD chip) based study,13 reflecting the resolution of our sequence-based calls. Only 18% of the deletion calls have overlap (≥1 bp) with previously reported bovine deletions (or CNV-loss) in the dbVar database (accessed on 27 January 2017), while remaining 82% are novel. However, ∼72% of our deletion regions remained unique when compared with all CNVs (gain or loss) and copy number variable regions in the database. Interestingly, majority (∼80%) of these overlapping regions are from an earlier WGS-based study, where genome sequences of 27 Holstein, 17 Montbéliarde, and 18 Normande bulls were analysed.23 Nonetheless, we were able to broaden the accessible deletion size-range, more importantly towards smaller one unascertainable by usual SNP-array based approaches. We also report high quality genotypes for all the 8,480 deletions. Apparently, there are more low frequency variants than that of high frequency one, and the frequency distribution is very similar to humans16 (Fig. 1).
Figure 1.
Number of ascertained deletions relative to variant allele count. Here, VAF is expressed in terms of variant allele count. Deletions down to an allele count of 1 (VAF = 0.0026 and 0.0032, in cattle and humans, respectively) are also represented here. Human deletion calls by Mills et al.16 were downloaded from 1K Genomes Project FTP server (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/pilot_data/paper_data_sets/companion_papers/mapping_structural_variation).
Previous NGS-based studies on cattle were mostly limited to SV discovery, while copy-number states (genotypes) were inferred using BovineHD chip,24 or custom SNPs array.23 However, in this study we estimated the copy-number at each deletion locus (per sample) from RD within the region using a constrained Gaussian mixture model with three classes, e.g. copy-number zero (i.e. homozygous deletion), one (i.e. heterozygous deletion) and two (i.e. homozygous reference). It is known from human studies that majority of the (bi-allelic) common SVs segregate on specific SNP haplotypes,48,49 which could be imputed with high accuracy.28,50 Thus, this approach has the potential, albeit with large reference, for accurate haplotype phasing and imputation of SVs to large cohorts of low-density chip-typed animals with no additional cost.
3.2. Validation of deletions
We validated the results using three approaches: (i) using BovineHD chip intensity data, (ii) breakpoint assembly and alignment, and (iii) PCR + sequencing of amplicons.
3.2.1. BovineHD chip intensity
We used 777K BovineHD chip intensity data of 26 Holstein animals, both chip-typed and sequenced, to validate the deletion calls using Genome STRiP’ IRS test. We had partial power to investigate all deletions due to the sparsity of the array-probes (one probe per ∼3.5 KB), and were underpowered to accurately verify small deletions (e.g. overlapping one-probe). Furthermore, we could only test a deletion for which at least one of the 26 samples had non-reference allele. Therefore, an estimate of FDR for the deletion call-set is provided here from the overall P-value distribution. In this approach, we were able to interrogate ∼8.3% of the total call, majority of which contain a single array-probe within the region (Table 1 and Supplementary Table S1). We found that deletions overlapping only one array-probe had higher FDR (11.3%) compared with two or more probes. And finally, we showed that our deletion call-set had an overall FDR of 8.8%, which is within our chosen threshold of FDR ≤ 10%.
Table 1.
FDR estimates of Genome STRiP’ deletion calls using 777K BovineHD BeadChip intensity data
| Array-probe Overlap | Aa | Bb | FDRc |
|---|---|---|---|
| One-probe | 497 | 28 | 11.3% |
| >1 array-probe | 206 | 3 | 2.9% |
| >2 array-probe | 113 | 1 | 1.8% |
| Overall | 703 | 31 | 8.8% |
aA, No. of sites with P-value.
bB, No. of sites with P-value ≥ 0.5
cFDR estimates were based on ‘Wilcoxson rank sum test’ using BovineHD chip intensity data of 26 Holstein animals. FDR was calculated as (B/A × 2 × 100).
3.2.2. Targeted breakpoint assembly
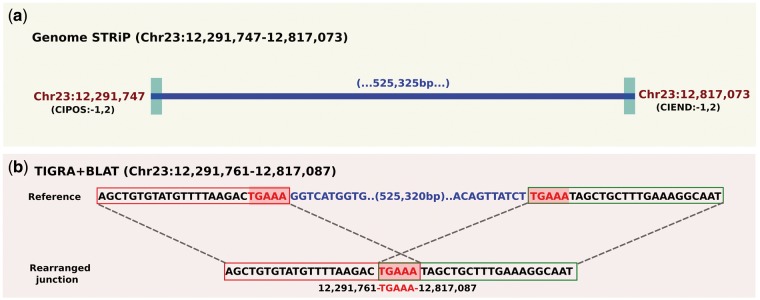
We next validated three randomly chosen set of ten-deletions each by assembling breakpoint sequences using TIGRA18: 10 deletions ≤ 500 bp, 10 deletions > 500 bp but ≤ 1 KB, and 10 deletions with VAF ≤ 0.10. Out of the thirty, we successfully resolved breakpoints of 26 deletions (∼87% success rate) using a combination of TIGRA and BLAT search30 (Supplementary Table S2). Additionally, we assembled breakpoints of three previously reported deletions that were also segregating in our study populations, such as a ∼662 KB deletion on chromosome 12 encompassing RNASEH2B, GUCY1B2, and FAM124A, a ∼3 KB deletion on chromosome 21 encompassing FANCI, and a ∼525 KB deletion on chromosome 23 encompassing BTBD9, GLO1, and DNAH8. Breakpoints of the former two deletions were previously reported in,7,9 which exactly matched with our predicted breakpoint sequences (Supplementary Figs S1a–c and S2a–c). Although for the later, we resolved breakpoint sequences in this study (Fig. 2a and b). Overall, the success rate of our deletion-breakpoint assembly was better than the reported success rate of TIGRA on similar sized read-length.18 And Genome STRiP’s breakpoint predictions were on average within 20 bp of the validated breakpoint, which is within the tool’s reported estimate of 1–20 bp.28
Figure 2.
A ∼525-KB deletion on chromosome 23 discovered using Genome STRiP (a), and resolved breakpoint sequences from TIGRA and BLAT search (b). Shaded bases are a 5-bp microhomology at breakpoint junction. (This figure was drawn and modified using Inkscape version 0.91.)
3.2.3. PCR and amplicon sequencing
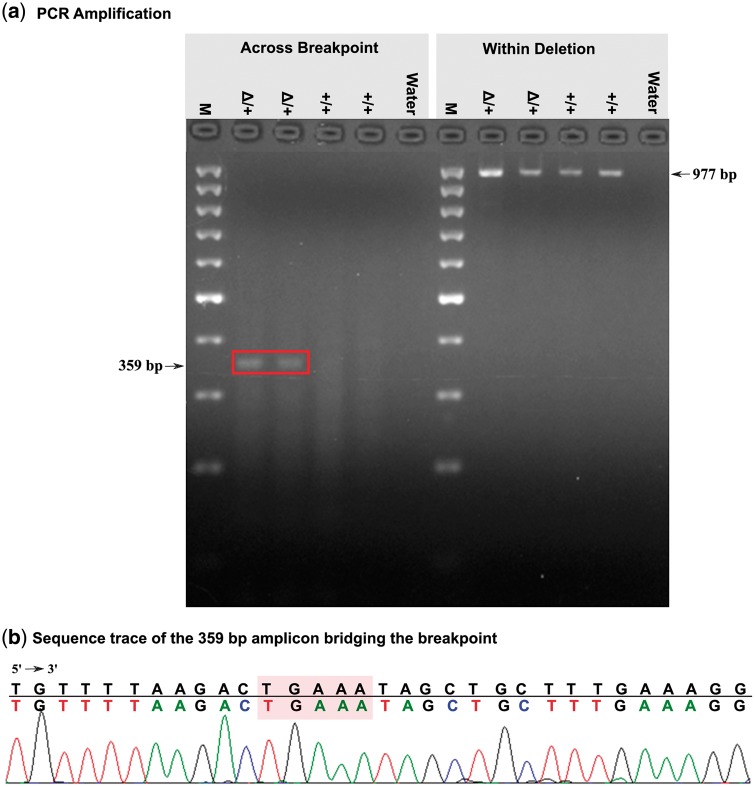
We then experimentally validated breakpoints for ‘Chr23:12,291,761-12,817,087’ deletion, previously reported to be associated with stillbirth in Nordic Red Cattle.10 Four animals were used for PCR validation: two carriers and two non-carriers. The breakpoint spanning PCR products of 359 bp were only observed in the carrier animals, while no amplicon was seen for non-carriers (Fig. 3a). The 359 bp amplicon was then sequenced, and exact breakpoint sequences were observed (Fig. 3b), thus confirming the breakpoint for this deletion.
Figure 3.
Experimental validation of the ∼525 KB deletion on chromosome 23. (a) PCR amplification across (left) and within the deletion (right) for two carrier (Δ/+) and two homozygous wild-type (+/+) animals. Water, negative control; M, molecular weight marker (GeneRuler 100 bp DNA ladder, Fermentas). (b) Sequence trace of the 359 bp amplicon bridging the breakpoint. Shaded bases are a 5-bp microhomology at breakpoint junction. (This figure was drawn and modified using Inkscape version 0.91.)
3.3. Population genetic properties of deletions
3.3.1. Population diversity
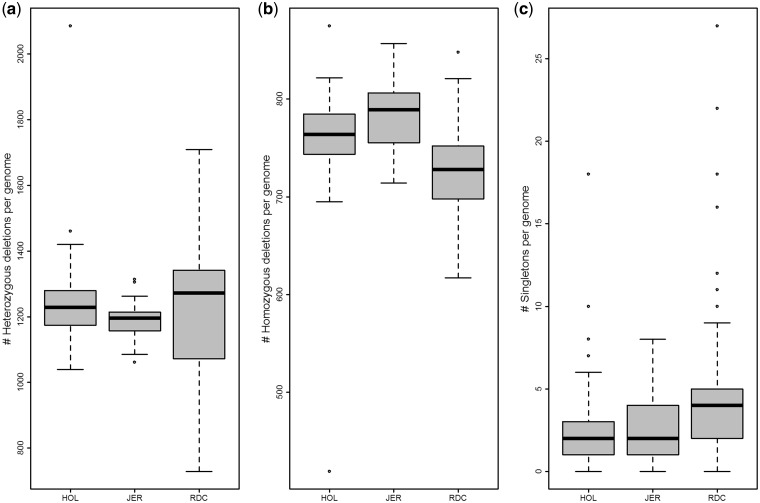
We explored population diversity among the three dairy cattle breeds from per-individual deletion-heterozygosity and homozygosity. We found that individuals from Nordic Red Cattle exhibits 3.5 and 6.4% higher deletion-heterozygosity than in Holstein and Jersey, respectively. Median numbers of heterozygote-deletion were 1,272, 1,229 and 1,196, in Nordic Red Cattle, Holstein, and Jersey, respectively (Fig. 4a). In contrast to heterozygosity, Jersey animals showed highest levels of deletion-homozygosity, followed by Holstein (Fig. 4b). Similar estimates of genetic diversity were also reported for these breeds in a SNP heterozygosity and runs-of-homozygosity analysis—where Jersey exhibited lowest (genome-wide) average nucleotide diversity (and higher number/size of runs-of-homozygosity) followed by Holstein and Nordic Red Cattle.51 These differences could be understood from the current effective population size (Ne) of these breeds, e.g. Ne of Jersey, Holstein, and Nordic Red Cattle are 73, 99, and 106, respectively52; this entailed higher diversity in Nordic Red Cattle, and Holstein, than in Jersey. From singletons estimate it is also evident that Nordic Red Cattle has more rare deletions compared with Holstein and Jersey (Fig. 4c); partly could be due to incorrect ascertainment of rare ones.
Figure 4.
Population diversity. (a) Heterozygous deletions per genome. (b) Homozygous deletions per genome. (c) Singletons per genome. Only high-confidence genotype calls are included. The y-axis in (a–c) represents the number of heterozygous, homozygous, and singleton deletions per genome, respectively. HOL, Holstein; JER, Jersey; RDC, Nordic Red Cattle.
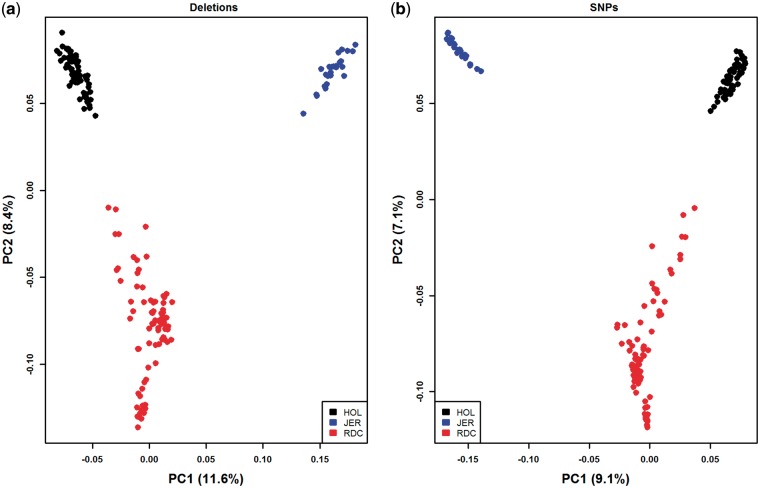
3.3.2. Principal component analysis
We performed PCA using the deletion genotypes of the samples. Around 6 K deletions with VAF between 0.02 and 0.90 were used in the analysis. For comparison, we also performed PCA on ∼168 K bi-allelic SNPs randomly selected from 29 autosomes of the same individuals. The first two principal components (PCs) from both deletion and SNP-based PCA clearly distinguished the three breeds, and jointly explained 20 and 16.2% of the variance, respectively (Fig. 5a and b). In addition, PC3 and PC4 recapitulated substructures within Nordic Red cattle (Supplementary Fig. S3), and first five PCs cumulatively explained 33.6% (with the deletions) and 28.4% (with the SNPs) of the variance (Supplementary Fig. S4). Our deletion results agree with the known population structure of the three breeds. Similar population structure (and substructure within Nordic Red Cattle) has been reported using genome-wide SNPs.53 Nordic Red Cattle from Denmark showed closer relationship with the Holstein in our WGS samples (Supplementary Fig. S5). This is consistent with the known history of Holstein interrogation in Danish Red cattle, as previously reported based on imputed WGS SNP analysis on a larger sample.53 It is also known from admixture analysis that genomes of Nordic Red Cattle are a mosaic of multiple ancestral populations, i.e. more ancestral components in Nordic Red Cattle than in Holstein and Jersey52,54; our deletion-based PCA largely corroborate that.
Figure 5.
PCA depicting three dairy cattle breeds. The analysis is based on (a) ∼6 K deletions (0.02 < VAF < 0.9), and (b) ∼168 K bi-allelic SNPs randomly selected from 29 bovine autosomes. First two PCs from deletions and SNPs are plotted here; jointly explained 20 and 16.2% of the variance, respectively. HOL, Holstein; JER, Jersey; RDC, Nordic Red Cattle.
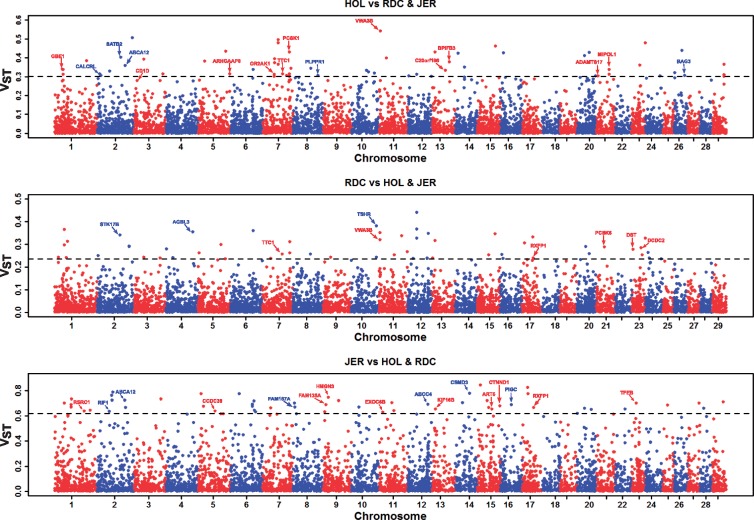
3.3.3. VST analysis
We analysed population stratification in terms of VST,33 a measure highly correlated with Wright’s fixation index (FST),34 to identify population differentiation. We calculated VST for each deletion pairwise amongst the breeds (e.g. Holstein vs Jersey + Nordic Red Cattle) from VAFs. We identified 158 highly stratified deletions (pairwise VST ≥ mean + 4 s.d.) among the breeds (Fig. 6). Around 27% of these deletions overlap genic elements, i.e. exons, introns, or (upstream/downstream) untranslated regions (UTRs), and remaining 73% are intergenic variants (Supplementary Tables S3–S5). There were eleven sites shared between Holstein and Nordic Red Cattle, two sites between Holstein and Jersey, and one site between Nordic Red Cattle and Jersey. Among these sites were gene variants, such as ABCA12 (Chr2:103,682,772-103,684,297 in Holstein & Jersey) associated with growth and development,55,56TTC1 (Chr7:73,725,513-73,725,918 in Holstein & Nordic Red Cattle) with cold tolerance,57VWA3B (Chr11:3,521,329-3,522,551 in Holstein & Nordic Red Cattle) with milk glycosylated kappa-casein percentage,58 and were intergenic variants, such as Chr15:41,393,393-41,393,780 (in Jersey and Nordic Red Cattle) and Chr20:26,812,159-26,812,834 (in Holstein and Jersey) overlap QTL associated with calving traits,59,60 Chr20:45,816,245-45,820,519 (in Holstein & Nordic Red Cattle) with meat and carcass trait,61 and Chr23:49,778,653-49,782,567 (in Holstein and Nordic Red Cattle) with body weight.62 We also identified a highly differentiated fertility associated gene DST/BPAG163,64 (Chr23:3,486,232-3,486,603) in Nordic Red Cattle. One differentially selected deletion (VST = 0.28) of chromosome 3 (Chr3:12,141,822-12,170,916) overlapping ENSBTAG00000047776 and ENSBTAG00000024960 genes (human orthologue CD1D), drawn our attention (though it marginally failed our selection threshold); Holsteins exhibited VAF of 24.63% (have both homozygous and heterozygous deletion), while it is mostly homozygous for the reference allele in Nordic Red cattle (VAF = 0.62%) and Jersey (VAF = 0.0%). The CD1D gene has known function in host immune response and parasite resistance,65,66 and also reported differentially expressed post intra-mammary infection.67 Majority of these stratified deletion regions are novel compared with previous CNV studies in cattle, and therefore are interesting targets to investigate large deletions undergoing genetic drift or artificial selection.
Figure 6.
Population stratification based on VST (a measure of differentiation for SV, highly correlated with Wright’s fixation index, FST). Horizontal dash line indicates highly stratified deletion regions (VST ≥ Mean + 4 s.d.). Highly stratified genic-deletions, e.g. overlapping exons, introns, or UTRs, are highlighted with HGNC gene symbol.
3.4. Functional impact of deletions
We annotated all the deletions using Variant Effect Predictor (Ensembl 87). Around 71% (6,019 SVs) variants were intergenic and remaining 29% (2,461 SVs) overlapped genic elements, such as exons, introns, and UTR. On average, high frequency gene disrupting deletions were somewhat depleted compared with intergenic variants (VAFintergenic > VAFgenic, P-value = 0.04; one-sided Wilcoxon test). Furthermore, we observed many common genic deletions. These genes are relatively less conserved, and majority has multiple paralogs (discussed later). However, deletions on known essential genes were only observed as heterozygote with relatively low VAF (<3%), and generally were private to a specific breed. For example, FANCI deletions (cause brachyspina7) were only observed in Holstein, and RNASEH2B deletions (cause embryonic lethality9) in Nordic Red Cattle.
3.4.1. Selective constraints on genes overlapping deletions
The relative abundance of high-frequency genic and intergenic variants indicate that majority of these intersected genes are non-essential, and thus did not affect the viability or fecundity of the carriers. To test this hypothesis, we analysed the selective constraints between deleted genes (overlap of any genic element) and known mouse lethal genes (from Dickinson et al.42) in terms of dN/dS ratio of cow-mouse 1-to-1 orthologues (Fig. 7). Here, high dN/dS values indicate low selective constraints on genes, and low value indicates high constraints. We found that genes in deletions have significantly higher dN/dS ratios than lethal genes (P-value 2.3 × 10−6; one-sided Wilcoxon test), and thus are evolutionarily less conserved. This is consistent with the rate of evolution seen in essential and non-essential genes—where mutations in essential genes were under strong purifying selection and thus evolved slowly (low dN/dS ratio), while non-essential genes were under relaxed selection, and hence, evolved faster (high dN/dS ratio).68 Nonetheless, robustness of these processes is also evident in the evolution of human essential genes. Interestingly, ∼77% human essential genes could even be traced back to pre-metazoans.69
Figure 7.
Difference between dN/dS ratios of mouse-lethal and deletion-overlapped genes in cattle. Cow genes for which one-to-one mouse orthologues available were considered for a one-sided Wilcoxon rank-sum test. Mouse lethal genes are from Dickinson et al.42
3.4.2. Nonessential genes in cattle
In total, we found 5,000 deletions for which at least one individual was homozygous. In the set, we analysed homozygous deletions in genes to find natural gene knockouts. We found 167 deleted genes (transcript-ablation or complete deletion) corresponding to 115 independent deletions that are apparently nonessential based on the occurrences of live homozygote individuals. This is ∼45% more than the previous report.23 Nonetheless, we found ∼44% fewer genes compared with in humans (240 nonessential genes),15 which could be due to the differences in sample size (175 vs 2,504 individuals) and study populations (3 vs 26 populations in human).
Among these genes, ∼83% (139 genes) are protein-coding, 12% pseudogenes, and the rest are different types of small RNAs (Supplementary Table S6). Most of these genes belong to multigenic families and are not highly conserved (median cow-mouse dN/dS of 0.17 vs OMIA genes dN/dS of 0.11; Supplementary Fig. S6), as expected for homozygous deletion.15 Moreover, this set of genes are functionally enriched in immunoglobulin domains, olfactory receptors, and MHC classes (FDR = 2.06 × 10−22, 2.06 × 10−22, 7.01 × 10−6, respectively), along with other related domains (Supplementary Tables S7–S9). Similar functional enrichment of nonessential genes was also seen in humans.15 Olfactory receptor related genes are well known for extensive gains and losses in mammalian evolution.70 And population specific copy-number variations of olfactory receptor genes were also reported in human (deletions)71 and cattle (gains).72 Nevertheless, this is the first report, to our knowledge, of homozygous deletion of olfactory receptor genes in cattle.
3.4.3. QTL enrichment
We next explored the enrichment (or depletion) of QTL on deleted regions (at least 1 bp overlap with deletion). We retrieved ∼24 K autosomal QTL from QTLdb reported to be associated with any of the six trait classes, e.g. ‘Health’, ‘Reproduction’, ‘Milk’, ‘Exterior’, ‘Production’, and ‘Meat and Carcass’. The association of deletions with diseases, fitness or fertility related traits is well evident.3 Hence, we suspected enrichment of fitness and fertility related traits for our deletions. As expected, health (2-fold) and reproduction (1.5-fold) related QTL were significantly enriched, while other trait classes were highly depleted (Table 2). Higher enrichment of health related QTL could be driven by immune-system genes, which were also highly enriched in our dataset (discussed earlier).
Table 2.
Enrichment of QTL on deletions
| Trait classesa | Fold enrichment | P value (Fisher’s test) |
|---|---|---|
| Health | 2 | 8.91 × 10−10 |
| Reproduction | 1.5 | 7.4 × 10−11 |
| Milk | 0.8 | 2.45 × 10−7 |
| Exterior | 0.5 | 1.85 × 10−4 |
| Production | 0.5 | 0.002 |
| Meat and Carcass | 0.5 | 0.058 |
aTrait classes are from cattleQTLdb.42 QTL from autosomes of Holsteins, Jersey, Nordic Red Cattle, and Ayrshires were considered for Fisher’s exact test (two-sided).
3.5. Deletion formation mechanisms
Finally we explored the probable mechanisms of deletion formation. There are two key mechanisms of SVs formation (for detail see review19,73); for example, recurrent SVs often result from ‘non-allelic homologous recombination’ between large low-copy repeats (LCRs), and thus, contain extensive sequence homology provided by LCRs, such as segmental duplicates, at the flanking regions.19 In contrast, non-recurrent SVs often form either by ‘microhomology-mediated end joining’ or ‘non-homologus end joining’, which requires limited to no sequence homology, and thus could be characterized by microhomologies or simple blunt ends at the breakpoint junction.73
Breakpoint information is crucial for understanding the mechanism, and therefore, we analysed 29 breakpoint resolved deletions from our validation set. We found that 24 of 29 deletions contain microhomology ranging from 2 to 31 bp at the breakpoint, and two of which also contain insertions (Supplementary Table S2). In addition, four deletions exhibited non-reference insertion at breakpoint junctions, and one deletion with no apparent homology. However, the number of breakpoint sequences analysed here were not a robust representation of our deletion call-set (<0.5% deletions), though selected randomly (for validation), we were able to demonstrate that majority of deletions contain microhomoloy at breakpoint, followed by few insertions, and rarely with no homology. Our results largely agree with the trend reported for large deletions in humans, e.g. 70.8% deletions exhibited microhomology/homology and 16.1% insertions at the breakpoint.16
3.6. Limitations
This study only focused on identifying deletions in cattle because of their potential relevance to loss-of-function and embryonic lethality. However, we had limited success to identify small deletions, such as <200 bp due to reduced sensitivity of the SV caller. It is also not a comprehensive list of deletions for these samples, since we could have missed many true deletions due to sensitivity, coverage, or stringent filtering (among other reasons). Furthermore, the short read length (∼100 bp) in our WGS dataset also made it difficult to resolve breakpoints from regions of long repeats.
3.7. Conclusions
Loss-of-function variants are responsible for a substantial yearly-economic loss in dairy industry, where a limited number of elite sires are in extensive use for rapid genetic gains. Mapping of such variants is essential for effective breeding planning and genomic selection. Here we showed an NGS-based analytical framework suitable for population-scale mapping of large deletions in cattle, leveraging the available WGSs. Here we described population-genetic, functional, and evolutionary properties of discovered deletions. We identified and confirmed a ∼525 KB deletion on chromosome 23, causing stillbirth in Nordic Red Cattle. We demonstrated that Nordic Red Cattle had higher population diversity than Holstein and Jersey, and deletion-genotype could recapitulate genetic structure of these breeds. Natural gene knockouts are enriched for immune-related and olfactory receptor genes. We also showed that deletions are significantly enriched for health and fertility related QTL, while depleted for production related QTL. Our population genetic and functional analysis showed promise for inclusion of SVs in genomic studies in dairy cattle. This deletion catalog will facilitate discovery, genotyping, and imputation of deletions in large cohorts of animals, and subsequent studies for gene mapping and genomic prediction of breeding values.
Supplementary Material
Acknowledgements
Md Mesbah-Uddin benefited from a joint grant from the European Commission within the framework of the Erasmus-Mundus joint doctorate ‘EGS-ABG’.
Funding
This research was supported by the Center for Genomic Selection in Animals and Plants (GenSAP) funded by Innovation Fund Denmark (grant 0603-00519B).
Data availability
All relevant results are within the paper and its Supplementary data files. VCF file with deletion calls could be found at https://github.com/MMesbahU/Deletions_in_cattle. WGSs of 44 samples (out of 175) are available from the NCBI Sequence Read Archive (project accession numbers SRP039339 and SRP065105). Among the 175 samples, 144 are from Run 6 of 1KBGP. Rest of data are available only upon agreement with the commercial breeding organization and should be requested directly from the senior author (G.S.: goutam.sahana@mbg.au.dk) or the Center Director (M.S.L.: mogens.lund@mbg.au.dk).
Conflict of interest: None declared.
Supplementary data
Supplementary data are available at DNARES online.
References
- 1. Charlier C., Li W., Harland C., et al. 2016, NGS-based reverse genetic screen for common embryonic lethal mutations compromising fertility in livestock. Genome Res., 26, 1333–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cole J.B., Null D.J., VanRaden P.M.. 2016, Phenotypic and genetic effects of recessive haplotypes on yield, longevity, and fertility. J Dairy Sci., 99, 7274–88. [DOI] [PubMed] [Google Scholar]
- 3. Weischenfeldt J., Symmons O., Spitz F., Korbel J. O.. 2013, Phenotypic impact of genomic structural variation: insights from and for human disease. Nat. Rev. Genet., 14, 125–138. [DOI] [PubMed] [Google Scholar]
- 4. Zarrei M., MacDonald J.R., Merico D., Scherer S.W.. 2015, A copy number variation map of the human genome. Nat Rev Genet, 16, 172–183. [DOI] [PubMed] [Google Scholar]
- 5. Bickhart D.M., Liu G.E.. 2014, The challenges and importance of structural variation detection in livestock. Front. Genet., 5, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xu L., Cole J.B., Bickhart D.M., et al. 2014, Genome wide CNV analysis reveals additional variants associated with milk production traits in Holsteins. BMC Genomics, 15, 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Charlier C., Agerholm J. S., Coppieters W., et al. 2012, A deletion in the bovine FANCI gene compromises fertility by causing fetal death and brachyspina. PLoS One, 7, e43085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schutz E., Wehrhahn C., Wanjek M., et al. 2016, The holstein friesian lethal haplotype 5 (HH5) results from a complete deletion of TBF1M and cholesterol deficiency (CDH) from an ERV-(LTR) insertion into the coding region of APOB. PLoS One, 11, e0154602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kadri N.K., Sahana G., Charlier C., et al. 2014, A 660-Kb deletion with antagonistic effects on fertility and milk production segregates at high frequency in Nordic Red cattle: additional evidence for the common occurrence of balancing selection in livestock. PLoS Genet., 10, e1004049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sahana G., Iso-Touru T., Wu X., et al. 2016, A 0.5-Mbp deletion on bovine chromosome 23 is a strong candidate for stillbirth in Nordic Red cattle. Genet. Sel. Evol., 48, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu G. E., Hou Y., Zhu B., et al. 2010, Analysis of copy number variations among diverse cattle breeds. Genome Res, 20, 693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hou Y., Liu G. E., Bickhart D. M., et al. 2011, Genomic characteristics of cattle copy number variations. BMC Genomics, 12, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu L., Hou Y., Bickhart D. M., et al. 2016, Population-genetic properties of differentiated copy number variations in cattle. Sci. Rep., 6, 23161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alkan C., Coe B. P., Eichler E. E.. 2011, Genome structural variation discovery and genotyping. Nat. Rev. Genet., 12, 363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sudmant P. H., Rausch T., Gardner E. J., et al. 2015, An integrated map of structural variation in 2,504 human genomes. Nature, 526, 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mills R. E., Walter K., Stewart C., et al. 2011, Mapping copy number variation by population-scale genome sequencing. Nature, 470, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yalcin B., Wong K., Agam A., et al. 2011, Sequence-based characterization of structural variation in the mouse genome. Nature, 477, 326–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen K., Chen L., Fan X., Wallis J., Ding L., Weinstock G.. 2014, TIGRA: a targeted iterative graph routing assembler for breakpoint assembly. Genome Res., 24, 310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carvalho C. M., Lupski J. R.. 2016, Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet, 17, 224–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daetwyler H. D., Capitan A., Pausch H., et al. 2014, Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet., 46, 858–865. [DOI] [PubMed] [Google Scholar]
- 21. Brondum R. F., Guldbrandtsen B., Sahana G., Lund M. S., Su G.. 2014, Strategies for imputation to whole genome sequence using a single or multi-breed reference population in cattle. BMC Genomics, 15, 728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jansen S., Aigner B., Pausch H., et al. 2013, Assessment of the genomic variation in a cattle population by re-sequencing of key animals at low to medium coverage. BMC Genomics, 14, 446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boussaha M., Esquerre D., Barbieri J., et al. 2015, Genome-wide study of structural variants in bovine holstein, montbeliarde and normande dairy breeds. PLoS One, 10, e0135931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen L., Chamberlain A. J., Reich C. M., Daetwyler H. D., Hayes B. J.. 2017, Detection and validation of structural variations in bovine whole-genome sequence data. Genet. Select. Evol., 49, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li H., Durbin R.. 2009, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H., Handsaker B., Wysoker A., et al. 2009, The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McKenna A., Hanna M., Banks E., et al. 2010, The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res., 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Handsaker R. E., Korn J. M., Nemesh J., McCarroll S. A.. 2011, Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat. Genet., 43, 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karolchik D., Hinrichs A. S., Furey T. S., et al. 2004, The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kent W. J. 2002, BLAT–the BLAST-like alignment tool. Genome Res., 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miller S. A., Dykes D. D., Polesky H. F.. 1988, A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res., 16, 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Purcell S., Neale B., Todd-Brown K., et al. 2007, PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Redon R., Ishikawa S., Fitch K. R., et al. 2006, Global variation in copy number in the human genome. Nature, 444, 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wright S. 1931, Evolution in Mendelian Populations. Genetics, 16, 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McLaren W., Gil L., Hunt S. E., et al. 2016, The Ensembl Variant Effect Predictor. Genome Biol., 17, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Finn R. D., Attwood T. K., Babbitt P. C., et al. 2017, InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res., 45, D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Finn R. D., Coggill P., Eberhardt R. Y., et al. 2016, The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res., 44, D279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K.. 2017, KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res., 45, D353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Szklarczyk D., Franceschini A., Wyder S., et al. 2015, STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res., 43, D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yates A., Akanni W., Amode M. R., et al. 2016, Ensembl 2016. Nucleic Acids Res., 44, D710–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kinsella R. J., Kahari A., Haider S., et al. 2011, Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford), 2011, bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dickinson M. E., Flenniken A. M., Ji X., et al. 2016, High-throughput discovery of novel developmental phenotypes. Nature, 537, 508–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hu Z. L., Park C. A., Reecy J. M.. 2016, Developmental progress and current status of the Animal QTLdb. Nucleic Acids Res., 44, D827–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. RStudio Team 2016, RStudio: integrated development environment for R. RStudio, Inc: Boston, MA. [Google Scholar]
- 45. R Core Team 2016, R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- 46. Quinlan A. R., Hall I. M.. 2010, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lappalainen I., Lopez J., Skipper L., et al. 2013, DbVar and DGVa: public archives for genomic structural variation. Nucleic Acids Res., 41, D936–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McCarroll S. A., Kuruvilla F. G., Korn J. M., et al. 2008, Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet., 40, 1166–74. [DOI] [PubMed] [Google Scholar]
- 49. Conrad D. F., Pinto D., Redon R., et al. 2010, Origins and functional impact of copy number variation in the human genome. Nature, 464, 704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Handsaker R. E., Van Doren V., Berman J. R., et al. 2015, Large multiallelic copy number variations in humans. Nat. Genet., 47, 296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang Q., Guldbrandtsen B., Bosse M., Lund M. S., Sahana G.. 2015, Runs of homozygosity and distribution of functional variants in the cattle genome. BMC Genomics, 16, 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bovine HapMap C., Gibbs R. A., Taylor J. F., et al. 2009, Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science, 324, 528–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mao X., Sahana G., De Koning D. J., Guldbrandtsen B.. 2016, Genome-wide association studies of growth traits in three dairy cattle breeds using whole-genome sequence data. J. Anim. Sci., 94, 1426–37. [DOI] [PubMed] [Google Scholar]
- 54. Brondum R. F., Rius-Vilarrasa E., Stranden I., et al. 2011, Reliabilities of genomic prediction using combined reference data of the Nordic Red dairy cattle populations. J. Dairy Sci., 94, 4700–4707. [DOI] [PubMed] [Google Scholar]
- 55. Xu L., Bickhart D. M., Cole J. B., et al. 2015, Genomic signatures reveal new evidences for selection of important traits in domestic cattle. Mol. Biol. Evol., 32, 711–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cole J. B., Waurich B., Wensch-Dorendorf M., Bickhart D. M., Swalve H. H.. 2014, A genome-wide association study of calf birth weight in Holstein cattle using single nucleotide polymorphisms and phenotypes predicted from auxiliary traits. J Dairy Sci, 97, 3156–3172. [DOI] [PubMed] [Google Scholar]
- 57. Howard J. T., Kachman S. D., Snelling W. M., et al. 2014, Beef cattle body temperature during climatic stress: a genome-wide association study. Int. J. Biometeorol., 58, 1665–1672. [DOI] [PubMed] [Google Scholar]
- 58. Buitenhuis B., Poulsen N. A., Gebreyesus G., Larsen L. B.. 2016, Estimation of genetic parameters and detection of chromosomal regions affecting the major milk proteins and their post translational modifications in Danish Holstein and Danish Jersey cattle. BMC Genet., 17, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McClure M. C., Morsci N. S., Schnabel R. D., et al. 2010, A genome scan for quantitative trait loci influencing carcass, post-natal growth and reproductive traits in commercial Angus cattle. Anim. Genet., 41, 597–607. [DOI] [PubMed] [Google Scholar]
- 60. Sahana G., Guldbrandtsen B., Lund M. S.. 2011, Genome-wide association study for calving traits in Danish and Swedish Holstein cattle. J. Dairy Sci., 94, 479–486. [DOI] [PubMed] [Google Scholar]
- 61. McClure M. C., Ramey H. R., Rolf M. M., et al. 2012, Genome-wide association analysis for quantitative trait loci influencing Warner-Bratzler shear force in five taurine cattle breeds. Anim. Genet., 43, 662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Snelling W. M., Allan M. F., Keele J. W., et al. 2010, Genome-wide association study of growth in crossbred beef cattle. J. Anim. Sci., 88, 837–848. [DOI] [PubMed] [Google Scholar]
- 63. Cole J. B., Wiggans G. R., Ma L., et al. 2011, Genome-wide association analysis of thirty one production, health, reproduction and body conformation traits in contemporary U.S. Holstein cows. BMC Genomics, 12, 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lobago F., Gustafsson H., Bekana M., Beckers J. F., Kindahl H.. 2006, Clinical features and hormonal profiles of cloprostenol-induced early abortions in heifers monitored by ultrasonography. Acta Vet. Scand., 48, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sandri M., Stefanon B., Loor J. J.. 2015, Transcriptome profiles of whole blood in Italian Holstein and Italian Simmental lactating cows diverging for genetic merit for milk protein. J. Dairy Sci., 98, 6119–6127. [DOI] [PubMed] [Google Scholar]
- 66. Araujo R. N., Padilha T., Zarlenga D., et al. 2009, Use of a candidate gene array to delineate gene expression patterns in cattle selected for resistance or susceptibility to intestinal nematodes. Vet. Parasitol., 162, 106–115. [DOI] [PubMed] [Google Scholar]
- 67. Fang L., Sahana G., Su G., et al. 2017, Integrating sequence-based GWAS and RNA-seq provides novel insights into the genetic basis of mastitis and milk production in dairy cattle. Sci. Rep., 7, 45560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hurst L. D., Smith N. G.. 1999, Do essential genes evolve slowly? Curr. Biol., 9, 747–50. [DOI] [PubMed] [Google Scholar]
- 69. Blomen V. A., Majek P., Jae L. T., et al. 2015, Gene essentiality and synthetic lethality in haploid human cells. Science, 350, 1092–6. [DOI] [PubMed] [Google Scholar]
- 70. Niimura Y., Nei M.. 2007, Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS One, 2, e708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Van Ziffle J., Yang W., Chehab F. F.. 2011, Homozygous deletion of six olfactory receptor genes in a subset of individuals with Beta-thalassemia. PLoS One, 6, e17327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lee K., Nguyen D. T., Choi M., et al. 2013, Analysis of cattle olfactory subgenome: the first detail study on the characteristics of the complete olfactory receptor repertoire of a ruminant. BMC Genomics, 14, 596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hastings P. J., Lupski J. R., Rosenberg S. M., Ira G.. 2009, Mechanisms of change in gene copy number. Nat. Rev. Genet., 10, 551–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant results are within the paper and its Supplementary data files. VCF file with deletion calls could be found at https://github.com/MMesbahU/Deletions_in_cattle. WGSs of 44 samples (out of 175) are available from the NCBI Sequence Read Archive (project accession numbers SRP039339 and SRP065105). Among the 175 samples, 144 are from Run 6 of 1KBGP. Rest of data are available only upon agreement with the commercial breeding organization and should be requested directly from the senior author (G.S.: goutam.sahana@mbg.au.dk) or the Center Director (M.S.L.: mogens.lund@mbg.au.dk).
Conflict of interest: None declared.