Abstract
Claudins, the integral tight junction (TJ) proteins that regulate paracellular permeability and cell polarity, are frequently dysregulated in cancer; however, their role in neoplastic progression is unclear. Here, we demonstrated that knockout of Cldn18, a claudin family member highly expressed in lung alveolar epithelium, leads to lung enlargement, parenchymal expansion, increased abundance and proliferation of known distal lung progenitors, the alveolar epithelial type II (AT2) cells, activation of Yes-associated protein (YAP), increased organ size, and tumorigenesis in mice. Inhibition of YAP decreased proliferation and colony-forming efficiency (CFE) of Cldn18–/– AT2 cells and prevented increased lung size, while CLDN18 overexpression decreased YAP nuclear localization, cell proliferation, CFE, and YAP transcriptional activity. CLDN18 and YAP interacted and colocalized at cell-cell contacts, while loss of CLDN18 decreased YAP interaction with Hippo kinases p-LATS1/2. Additionally, Cldn18–/– mice had increased propensity to develop lung adenocarcinomas (LuAd) with age, and human LuAd showed stage-dependent reduction of CLDN18.1. These results establish CLDN18 as a regulator of YAP activity that serves to restrict organ size, progenitor cell proliferation, and tumorigenesis, and suggest a mechanism whereby TJ disruption may promote progenitor proliferation to enhance repair following injury.
Keywords: Pulmonology, Stem cells
Keywords: Adult stem cells, Lung cancer, Tight junctions
Introduction
Apical junctional complexes (AJCs) in polarized epithelia are composed of adherens junctions (AJs) and tight junctions (TJs) (1, 2) that mediate intercellular adhesion and paracellular permeability to ions and solutes, respectively. TJs, the most apical component of AJCs, are composed of integral membrane proteins (3, 4) linked to the actin cytoskeleton via accessory proteins (5). Claudins are a family of integral membrane proteins essential for TJ formation and integrity (3, 6–8). Unique permeability properties of various epithelia are determined by cell-specific patterns of claudin expression. Targeted deletions in mice as well as human mutations have revealed novel tissue-specific biological functions of claudins in several organs including kidney (9, 10), skin (11), ear (12), and peripheral nervous system (13). Interestingly, mice with knockout of Cldn15 demonstrated an unexpected phenotype of upper small intestine enlargement (megaintestine) and increased crypt cell proliferation without evidence of inflammation (14). Both up- and downregulation of claudin expression have been observed in a number of cancers, although how these changes contribute to carcinogenesis and/or neoplastic progression remains controversial (15–18). These studies suggest that, in addition to traditional roles in regulating epithelial barrier function and polarity, claudins also regulate cell functions such as proliferation that might contribute to tumorigenesis.
CLDN18 is one of the most highly expressed claudin family members in lung alveolar epithelium (19, 20). It is expressed at low levels in airway epithelium and is not expressed in lung endothelium (21). Cldn18 has 2 promoters, each with a unique exon 1 spliced to common exons 2 through 5. Alternative promoter usage leads to production of lung- and stomach-specific isoforms (22). Deletion of the stomach-specific Cldn18.2 isoform in mice leads to loss of TJ strands and increased paracellular H+ leakage in the stomach, resulting in atrophic gastritis and metaplasia but without evidence of tumor formation (23). Recently generated Cldn18–/– mice with deletion of both isoforms showed increased lung permeability to ions and solutes, consistent with known roles of claudins in regulation of barrier function (24, 25).
The conserved Hippo signaling pathway is a key regulator of organ size, stem/progenitor cell function, and tumorigenesis that exerts opposing effects on cell proliferation and apoptosis by controlling cellular localization of the downstream transcriptional coactivator Yes-associated protein (YAP) (26, 27). Cellular localization of YAP is determined by activity of the core Hippo kinases mammalian sterile 20–like 1/2 (MST1/2) and large tumor suppressor homolog 1/2 (LATS1/2), which phosphorylate YAP on serine residues leading to its cytoplasmic retention by 14-3-3 proteins and proteasomal degradation (26, 28). Dephosphorylated YAP translocates to the nucleus where it functions as a transcriptional coactivator of target genes (29) primarily via interactions with transcription enhancer factors 1–4 (TEF/TEAD 1–4) (30). Uncontrolled YAP activation leads to tissue overgrowth (31, 32), while increased YAP activity has been demonstrated in several cancers including lung adenocarcinoma (LuAd) (33, 34). YAP localization and activity are also regulated through interactions with membrane-associated proteins important for maintenance of cell polarity (e.g., Crb3 [ref. 35] and angiomotins [AMOTs] [refs. 36, 37]) and cell-cell contact (e.g., E-cadherin and α-catenin [refs. 38–40]) in both a Hippo-dependent and -independent manner.
Stem/progenitor cell populations in adult lung have been identified in region-specific niches along a proximal-distal axis, functioning as facultative progenitors that become activated for respiratory epithelial repair following injury (41–43). AT2 cells have been identified as progenitors of distal lung epithelium (44, 45) that are relatively quiescent under physiological conditions, becoming activated following injury. Molecular mechanisms that maintain homeostasis or activate endogenous lung stem/progenitor cells to promote adult lung repair are not yet well defined. In particular, a role for sites of cell-cell contact in modulating signals to regulate lung progenitor cell function has not been previously explored. We report in this study that, in addition to previously reported increases in lung epithelial permeability to ions and solutes, Cldn18–/– mice show enlargement of lung, stomach, and kidney, sites of CLDN18 expression. Lung parenchyma in Cldn18–/– mice is expanded with increased abundance and proliferation of AT2 cells together with activation of YAP signaling. Inhibition of YAP and overexpression of CLDN18 inhibit AT2 cell proliferation and progenitor capacity. Interestingly, aged Cldn18–/– mice showed increased propensity to develop LuAd, with stage-specific downregulation of CLDN18 in human LuAd. These results identify CLDN18 as a potentially novel regulator of YAP activity that acts to restrict progenitor cell proliferation and suggest a tumor suppressor role for CLDN18. Furthermore, since TJs become established during normal tissue morphogenesis and are frequently disrupted following injury, they suggest a mechanism whereby extracellular signals are transduced from sites of cell-cell contact to control organ size during development and regulate stem/progenitor cell proliferation and expansion following injury.
Results
Loss of Cldn18 causes hypercellularity and increased organ size.
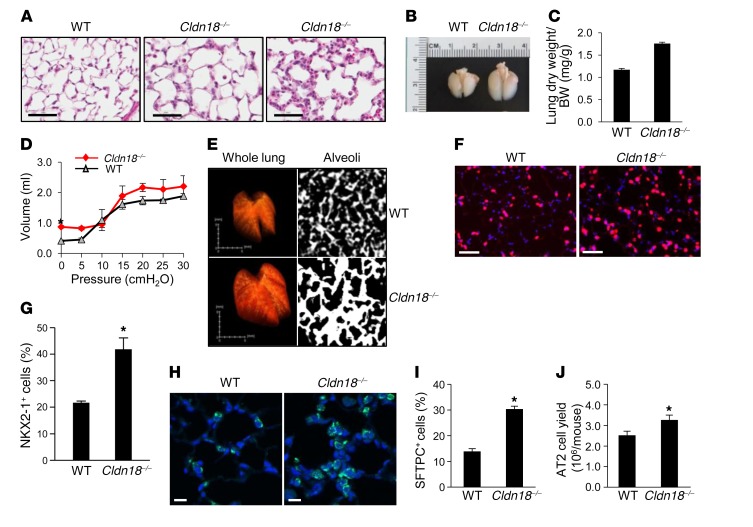
We previously generated mice with total knockout of Cldn18 (24). In addition to alterations in epithelial permeability and ion transport, we observed increased lung cellularity from embryonic day 18 (E18) onward (Figure 1A and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI90429DS1) with areas of more marked alveolar wall hypercellularity (Figure 1A), as well as airspace enlargement (Figure 1A and Supplemental Figure 1, A and B) as previously reported (25). Lungs of Cldn18–/– mice are visibly enlarged (Figure 1B) with unchanged body size and weight (Supplemental Figure 2, A and B). Further characterization revealed increased lung dry weight relative to body weight (Figure 1C) and increased lung volume (Figure 1D) with unchanged compliance. Micro-computerized tomographic (micro-CT) imaging shows lung enlargement with significantly expanded parenchyma (Figure 1E and Supplemental Table 1). Examination of other CLDN18-expressing organs (Supplemental Figure 3A; see complete unedited blots in the supplemental material) revealed visible enlargement of stomach, duodenum, and kidney (Supplemental Figure 3, B and C). The size of non–CLDN18-expressing organs (Supplemental Figure 3D) was similar between WT and Cldn18–/– mice at 2 months of age, with progressive enlargement of spleen, heart, and liver with increased age in Cldn18–/– mice (data not shown), perhaps due to circulatory changes accompanying increased lung size. Consistent with the visible increase in stomach size, gastric mucosal thickness was increased in Cldn18–/– compared with WT mice (Supplemental Figure 4, A and B).
Figure 1. Increased cellularity, alveolar epithelial type II (AT2) cell abundance, and parenchymal expansion in Cldn18–/– mice.
(A) H&E staining shows increased cellularity in Cldn18–/– lungs (age 1 month) with variability among mice (middle and right panels). Scale bar: 50 μm. n ≥ 7 each genotype. (B) Lungs of Cldn18–/– mice (age 3 weeks) are enlarged. Representative of 3 mice for each genotype. (C) Lung dry weight/body weight (BW) ratios of Cldn18–/– mice are increased. n ≥ 33 mice of each genotype, age 5–9 months. Bar graphs represent means ± SEM. Unpaired 2-tailed t test. *P < 0.05. (D) Pressure-volume curves show increased lung volume in Cldn18–/– mice with unchanged compliance. Each data point represents the mean ± SEM of 3 mice of each genotype, age 7–8 months. Two-way ANOVA with Bonferroni’s correction. *P < 0.05 for Cldn18–/– versus WT lungs at zero pressure. (E) Left panel (whole lung): Texture-based volume rendering shows lung enlargement in Cldn18–/– mice. Right panel (alveoli): High-resolution CT shows increased parenchymal thickness in Cldn18–/– mice. Representative of 3 mice for each genotype. (F and G) Representative immunofluorescence and quantification show increased numbers of NKX2-1+ cells (pink) in Cldn18–/– versus WT mice. Nuclei labeled with DAPI (blue). n = 3 age-matched mice of each genotype, age 2–9 months. Unpaired 2-tailed t test. *P < 0.05. Scale bars: 50 μm. Representative immunofluorescence (H) and quantification (I) show increased numbers of SFTPC+ cells (green) in Cldn18–/– versus WT mice. Nuclei labeled with DAPI (blue). n = 3 age-matched mice of each genotype. Unpaired 2-tailed t test. *P < 0.05. Scale bars: 50 μm. (J) AT2 cell yield is increased in Cldn18–/– mice. n = 10 mice per group with 8 independent cell isolations. Unpaired 2-tailed t test. *P < 0.05. Bar graphs represent the mean ± SEM for C, G, I, and J.
Loss of Cldn18 leads to expansion of distal lung epithelial progenitors.
Organ enlargement is known to result from dysregulated progenitor cell homeostasis (26, 46). AT2 cells, identified by expression of NK2 homeobox 1 (NKX2-1) (Figure 1, F and G) and surfactant protein C (SFTPC) (Figure 1, H and I) in the alveolar compartment, are significantly increased (41.6% ± 3.9% vs. 21.9% ± 0.4% and 30.3% ± 1.1% vs. 13.9% ± 1.1% of total cells, respectively) in lungs of Cldn18–/– mice versus WT mice. In addition, numbers of AT2 cells isolated from Cldn18–/– mice are significantly greater than from WT lungs (3.3 ± 0.2 × 106 vs. 2.5 ± 0.2 × 106 cells, respectively, Figure 1J), with unchanged cell size (Supplemental Figure 5). Furthermore, SFTPC protein is significantly increased in Cldn18–/–lungs (Supplemental Figure 6, A and B) with a decrease in alveolar epithelial type I (AT1) cell markers compared with WT lung (Supplemental Figure 6, C and D), suggesting that increased AT2 cell number underlies parenchymal expansion in Cldn18–/– mice. We also generated mice with tamoxifen-inducible AT2 cell–specific deletion of Cldn18 by crossing Sftpc+/creERT2, Cldn18fl/fl, and ROSATm/Tm mice, resulting in mice with the genotype Sftpc+/creERT2;Cldn18fl/fl;ROSA+/Tm in which AT2 cells are labeled with a Tomato (Tm) fluorescent reporter. Administration of tamoxifen (Tmx) resulted in an approximately 70% reduction in Cldn18 expression in isolated AT2 cells (Supplemental Figure 7A). Following Tmx treatment, Tm+ AT2 cells increased approximately 1.6-fold in Sftpc+/creERT2;Cldn18fl/fl;ROSA+/Tm (14.3% ± 0.89%) compared with control Sftpc+/creERT2;ROSA+/Tm (9.2% ± 1.2 %) mice (Supplemental Figure 7B). AT2 cell expansion in mice with both ubiquitous and AT2 cell–specific knockout of Cldn18 suggests a potentially novel role for this TJ protein in regulating lung epithelial stem/progenitor cell homeostasis and indicates that effects are epithelial cell specific.
Loss of Cldn18 increases lung AT2 cell proliferation in vivo.
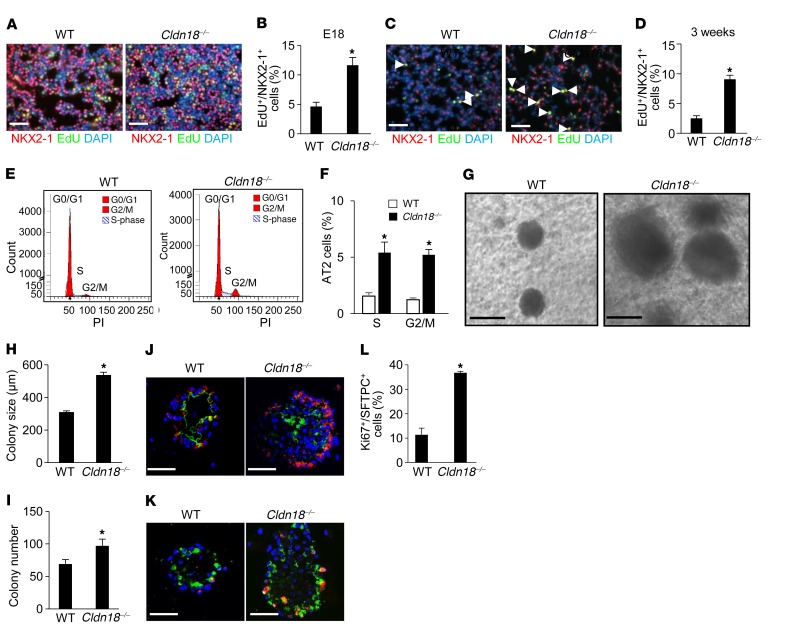
Representative images (Figure 2, A and C) and quantification (Figure 2, B and D) show increased numbers of EdU+/NKX2-1+ cells at E18 (11.6% ± 1.3% vs. 4.6% ± 0.7%) and 3 weeks postnatally (9.0% ± 0.8% vs. 2.5% ± 0.5%) in Cldn18–/– versus WT lungs. Flow cytometry of AT2 cells isolated from Cldn18–/– and WT mice show an approximately 5-fold increase in the percentage of AT2 cells in S and G2/M phase (5.4% ± 0.9% vs. 1.6% ± 0.3% and 5.2% ± 0.5% vs. 1.2% ± 0.3%, respectively) (Figure 2, E and F). TUNEL assay shows low levels of apoptosis in both Cldn18–/– and WT mice (0.2% vs. 0.1%, respectively) (Supplemental Figure 8), indicating that increased cell number is the result of increased proliferation rather than decreased apoptosis. Flow cytometry of Tm+ AT2 cells isolated from Sftpc+/creERT2;Cldn18fl/fl;ROSA+/Tm mice shows an approximately 5-fold increase in the percentage of AT2 cells in S (6.4% ± 1.4% vs. 1.4% ± 0.2%) and G2/M (6.3% ± 1.7% vs. 1.7% ± 1.1%) phase, compared with those from Sftpc+/creERT2;ROSA+/Tm mice (Supplemental Figure 9), indicating that both ubiquitous and AT2 cell–specific loss of Cldn18 activates signaling that promotes AT2 cell proliferation.
Figure 2. Increased alveolar epithelial type II (AT2) cell proliferation in Cldn18–/– lungs in vivo and colony-forming efficiency and proliferation in vitro.
Representative immunofluorescence (IF) images (A) and quantification (B) show EdU+NKX2-1+ (yellow) cells are increased in Cldn18–/– mouse lungs at E18. n = 5 mice of each genotype. Unpaired 2-tailed t test. *P < 0.05. Scale bars: 50 μm. Representative IF images (C) and quantification (D) show EdU+NKX2-1+ cells (arrow) are increased in Cldn18–/– mouse lungs 3 weeks postnatally. n = 5 mice of each genotype. Unpaired 2-tailed t test. *P < 0.05. Scale bars: 50 μm. Representative FACS (E) and quantification (F) show a greater percentage of AT2 cells in S and G2/M phase in Cldn18–/– versus WT mice. PI, propidium iodide. n = 3 mice of each genotype (age ~5 months). Two-way ANOVA with Bonferroni’s correction. *P < 0.05 versus WT. Increased size (G and H) and number (I) of colonies generated from Cldn18–/– AT2 cells. n ≥ 3 biological replicates. Unpaired 2-tailed t test. *P < 0.05. Scale bars: 100 μm. (J) SFTPC (red)/AQP5 (green) double staining shows that both WT and Cldn18–/– AT2 cells in 3D culture give rise to cells expressing the AT1 cell marker AQP5. n = 3. Scale bars: 50 μm. Ki67 (red) and SFTPC (green) double staining (K) and quantification (L) demonstrate increased proliferation of Cldn18–/– AT2 cells in organoid culture. DAPI is the nuclear counterstain. n = 3 biological replicates. Unpaired 2-tailed t test. *P < 0.05. Scale bars: 20 μm. Bar graphs represent the mean ± SEM for B, D, F, H, I, and L.
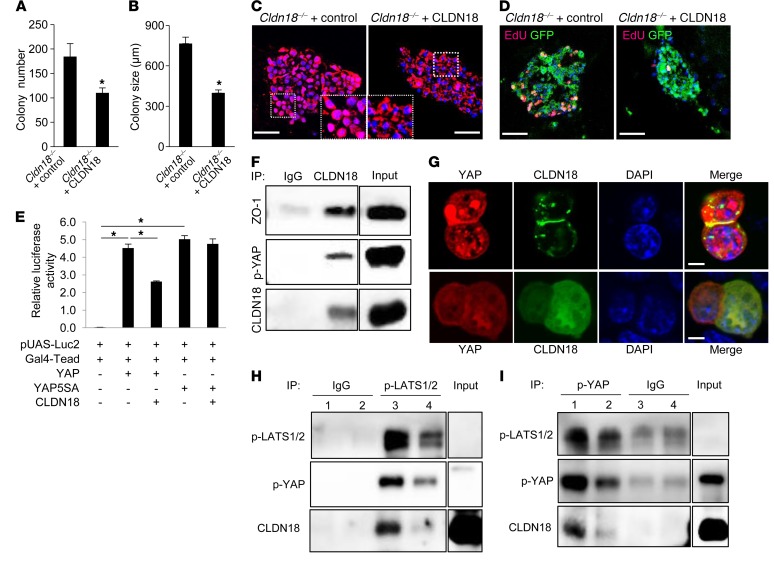
Increased colony formation and proliferation of AT2 cells from Cldn18–/– mice in 3D organoid culture.
We further examined if loss of CLDN18 alters proliferation of lung stem/progenitor cells in vitro using a 3-dimensional (3D) organoid coculture system (44, 47). AT2 cells (CD24–Sca1–; purity 95% ± 5%) isolated from EPCAMhi/CD45–CD34–CD31– populations (Supplemental Figure 10) were cocultured with MLg lung fibroblasts for 14 days. Cldn18–/– AT2 cells showed an increase in colony-forming efficiency (CFE) and sphere size compared with WT cells (Figure 2, G–I). Mixed cultures of Tm+ AT2 cells and unlabeled AT2 cells generated colonies consisting of exclusively labeled or unlabeled cells, indicating clonality (Supplemental Figure 11). Aquaporin-5+ (AQP5+) cells are located in the center of colonies, while SFTPC+ cells are located at the perimeter (Figure 2J), as previously reported (44). Western analysis and linear regression analysis show similar acquisition of AT1 cell markers by Cldn18–/– and WT AT2 cells grown on polycarbonate filters (Supplemental Figure 12), indicating similar transdifferentiation into AT1-like cells. Labeling with anti-Ki67 antibody (Ab) showed increased proliferation of Cldn18–/– compared with WT AT2 cell colonies (36.8% ± 0.3% vs. 11.4% ± 1.1%) (Figure 2, K and L). Additionally, primary AT2 cell colonies were dissociated by enzymatic digestion after approximately 2 weeks in spheroid culture and EPCAM+ cells were replated with fresh MLg cells. Increased CFE of Cldn18–/– cells was maintained following passage, indicating maintenance of stem cell function (Supplemental Figure 13). These data suggest that knockout of Cldn18 increases AT2 cell proliferation in a cell-autonomous manner, while differentiation capacity remains unchanged.
Activation of YAP signaling in whole lung and AT2 cells from Cldn18–/– mice.
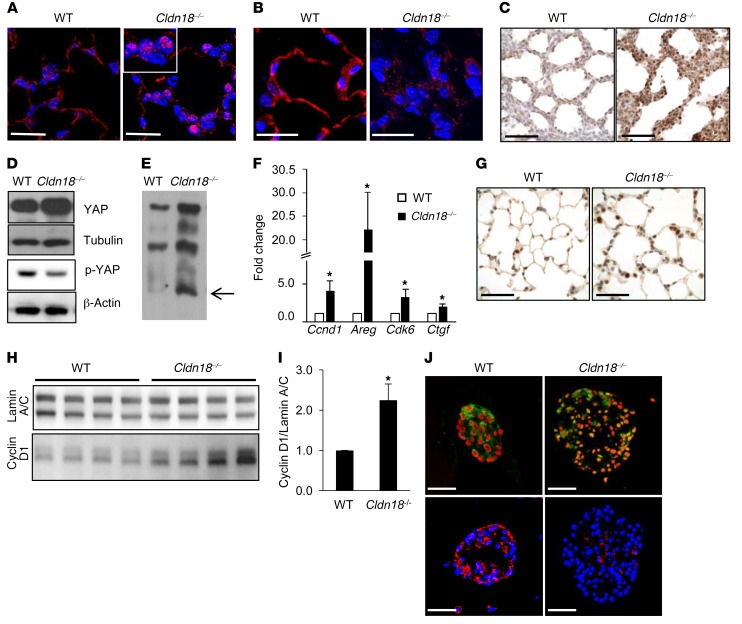
Consistent with its recognized role in regulating stem/progenitor cell proliferation and organ size (46), immunostaining revealed increased nuclear YAP (Figure 3A) and decreased cytoplasmic p-YAP (Figure 3B) in adult Cldn18–/– lungs, with increased nuclear YAP (brown) as early as E18 (Figure 3C). YAP protein (Figure 3D) and YAP mobility (Figure 3E) are increased, while p-YAP is decreased (Figure 3D) in Cldn18–/– AT2 cell lysates. Quantitative reverse transcription PCR (qRT-PCR) shows increased expression of YAP target genes including Ccnd1 (cyclin D1), Areg (amphiregulin), Ckd6 (cyclin-dependent kinase 6), and Ctgf (connective tissue growth factor) (Figure 3F), with increased cyclin D1 protein in adult Cldn18–/– mice (Figure 3, G–I). In 3D culture, Cldn18–/– AT2 cells also show increased nuclear YAP and decreased p-YAP (Figure 3J).
Figure 3. Activation of YAP signaling in whole lung and alveolar epithelial type II (AT2) cells in Cldn18–/– mice.
Representative immunofluorescence (IF) image shows increased nuclear YAP (A) and decreased cytoplasmic p-YAP (B) in lungs of adult Cldn18–/– mice (age ~2 months). n ≥ 3 mice of each genotype. DAPI (blue) is the nuclear counterstain. Scale bars: 20 μm. (C) Representative immunohistochemistry of lung sections at E18 shows increased nuclear YAP (brown) in Cldn18–/–mice. n = 3 for WT, 2 for Cldn18–/– mice. Hematoxylin is the nuclear counterstain (blue). Scale bars: 50 μm. (D) Western blot shows increased YAP and decreased p-YAP in Cldn18–/– AT2 cells (age ~2 months). Tubulin and β-actin are loading controls. n = 2 mice of each genotype. (E) Representative Phos-tag Western blot shows increased dephosphorylated YAP (arrow) in Cldn18–/– AT2 cells (age ~2 months). n = 3 independent experiments. (F) qRT-PCR shows increased expression of YAP target genes in Cldn18–/– versus WT lungs. n = 5 independent experiments. Z test. *P < 0.05. (G) Representative image shows increased expression of YAP target cyclin D1 (brown) in Cldn18–/– (age 2–9 months) mouse lung. Hematoxylin (blue) is the nuclear counterstain. n = 3 mice of each genotype. Scale bars: 50 μm. Western analysis (H) and quantification (I) of whole-lung samples demonstrate higher levels of cyclin D1 protein in Cldn18–/– compared with WT lungs. n = 4 mice of each genotype. Z test. *P < 0.05. (J) Representative image shows increased nuclear YAP (green) and decreased cytoplasmic p-YAP (red) in Cldn18–/– AT2 cells in 3D culture. PI (red) and DAPI (blue) are nuclear counterstains. Scale bars: 50 μm. n = 3 independent experiments. Bar graphs represent the mean ± SEM for F and I.
Inhibition of YAP signaling prevents increases in lung size and inhibits proliferation and CFE of AT2 cells from Cldn18–/– mice.
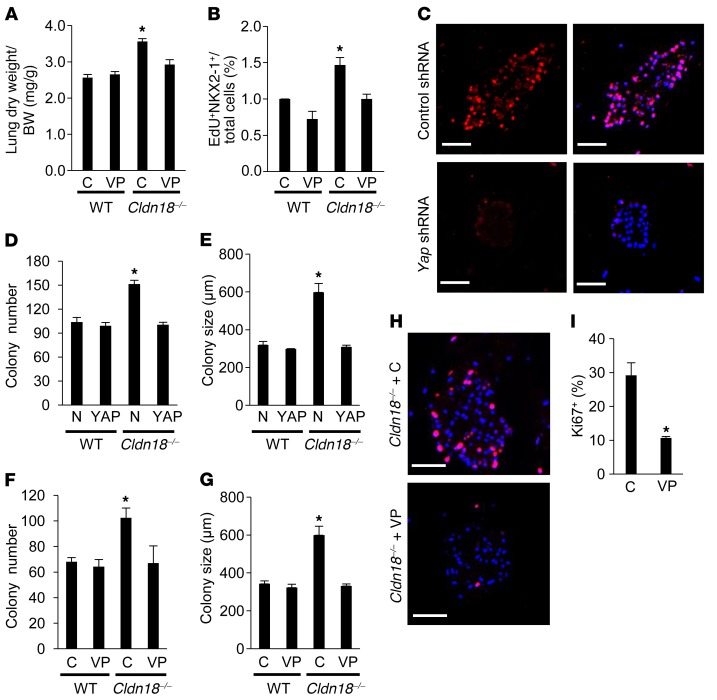
Verteporfin (VP), a pharmacologic inhibitor of YAP-TEAD association and YAP-induced liver overgrowth (48), administered from E13 through postnatal day 14 (P14) inhibits increases in lung weight relative to body weight at P16 (Figure 4A). Similarly, VP administered from P4–P12 inhibited increases in Cldn18–/– AT2 cell proliferation (Figure 4B) compared with Cldn18–/– mice treated with phosphate-buffered saline (PBS). Yap shRNA (Figure 4C) and VP prevent Cldn18-knockout-induced increases in colony number (Figure 4, D and F) and size (Figure 4, E and G). VP decreases Cldn18–/– AT2 cell proliferation (29.2% ± 3.7% control vs. 10.7% ± 0.4% VP) (Figure 4, H and I), confirming a role for YAP activation in regulation of lung stem/progenitor cell function downstream of CLDN18.
Figure 4. YAP inhibition decreases proliferation and colony-forming efficiency (CFE) of alveolar epithelial type II (AT2) cells from Cldn18–/– mice.
(A) Verteporfin (VP) prevents increases in lung dry weight/body weight (BW) ratio in Cldn18–/– lungs at postnatal day 16 (P16) following administration of VP from embryonic day 13 (E13) to P14. Control (C) = vehicle. n ≥ 9 mice for each group. Two-way ANOVA with Bonferroni’s correction. *P < 0.05 versus all others. (B) VP inhibits increases in EdU+NKX2-1+ cells in Cldn18–/– lungs following administration from P4 to P12. Control (C) = vehicle. Five fields (×40) were counted for each lung. n ≥ 3. Two-way ANOVA with Bonferroni’s correction. *P < 0.05 versus all others. (C) Representative image shows decreased YAP expression (red) in Cldn18–/– AT2 cells transduced with lentivirus expressing Yap shRNA in 3D culture. Scale bars: 50 μm. n = 2 independent experiments. Yap shRNA (MOI = 5) prevents increases in colony number (D) and size (E) in Cldn18–/– AT2 cells after 14 days of treatment. N is nonsilencing shRNA control. n = 3 independent experiments. Two-way ANOVA with Bonferroni’s correction. *P < 0.05 versus all others. VP (0.75 μM) prevents increases in colony number (F) and size (G) in Cldn18–/– AT2 cells after 14 days of treatment. n = 3 independent experiments. Two-way ANOVA with Bonferroni’s correction. *P < 0.05 versus all others. (H) Ki67 staining (red) and quantification (I) show VP (0.75 μM) inhibits Cldn18–/– AT2 cell proliferation. DAPI is the nuclear counterstain. n = 3 independent experiments. Unpaired 2-tailed t test. *P < 0.05. Scale bars: 50 μm. Bar graphs represent the mean ± SEM for A, B, D–G, and I.
CLDN18 modulates AT2 cell proliferation and CFE and interacts with p-YAP to regulate YAP signaling.
CLDN18 overexpression in Cldn18–/– AT2 cells in 3D culture decreases CFE (Figure 5, A and B), nuclear YAP (Figure 5C), and proliferation (Figure 5D). CLDN18 overexpression in MLE-15 cells similarly inhibits proliferation and YAP nuclear localization (Supplemental Figure 14, A–C). CLDN18 overexpression decreases transcriptional activity of a YAP-responsive luciferase reporter in MLE-15 cells but not that of a nonphosphorylatable constitutively active YAP mutant, YAP5SA (Figure 5E), implicating Hippo kinases in regulation of YAP activity by CLDN18. Co-immunoprecipitation (co-IP) was performed to evaluate CLDN18/YAP interactions. p-YAP is identified in the CLDN18 Ab–precipitated protein complex in WT AT2 cell membrane lysates (Figure 5F). Additionally, the TJ-associated protein zonula occludens-1 (ZO-1) coprecipitates with CLDN18 and p-YAP in WT AT2 cell lysates (Figure 5F). An overexpressed CLDN18-green fluorescent protein (GFP) fusion protein associates with p-YAP and ZO-1 in MLE-15 cell membranes (Supplemental Figure 14D), consistent with p-YAP localization at TJs. Confocal images show that overexpressed DsRed-YAP and CLDN18-GFP colocalize at sites of cell-cell contact in MLE-15 cells (Figure 5G), suggesting that CLDN18 regulation of YAP signaling involves p-YAP sequestration at TJs. Mass spectrometry identified CLDN18 as a YAP binding partner in WT (but not Cldn18–/–) AT2 cells (Supplemental Figure 14, E and F). Co-IP using p-LATS1/2 Ab (Figure 5H) for pulldown followed by Western blotting for p-YAP, or the reciprocal using p-YAP antibody (Figure 5I) for pulldown followed by Western blotting for p-LATS1/2, reveals a reduction of p-LATS interaction with p-YAP as well as an overall reduction of p-LATS and p-YAP in AT2 cell membranes from Cldn18–/– mice. Furthermore, CLDN18 interacts with p-LATS in WT AT2 cell membranes (Figure 5H and Supplemental Figure 14G). These findings suggest that in addition to membrane sequestration, CLDN18 promotes interaction between p-LATS and p-YAP resulting in increased YAP phosphorylation and furthermore, that CLDN18 modulates Hippo kinases to regulate YAP phosphorylation and activity.
Figure 5. CLDN18 regulates AT2 cell proliferation and YAP activity, and CLDN18 and YAP interact.
CLDN18 overexpression prevents increases in colony number (A) and size (B). n = 3 independent experiments. Unpaired 2-tailed t test. *P < 0.05. Nuclear YAP (C) and EdU labeling (red) (D) decrease following transduction of Cldn18–/– AT2 cells with CLDN18-expressing lentivirus. GFP fluorescence (green) (D) indicates most cells were transduced with virus. n = 3. Scale bars: 50 μm. (E) CLDN18 overexpression inhibits activation of a YAP luciferase reporter, but not of YAP mutant, YAP5SA, that is resistant to phosphorylation by LATS1/2. YAP– and YAP+ indicate absence versus presence of overexpressed YAP, respectively. n = 3 independent experiments. One-way ANOVA with Bonferroni’s correction. *P <0.05. (F) Endogenous CLDN18 associates with p-YAP and ZO-1 in WT AT2 cell membranes. IgG = negative control. Input is cell lysate. (G) pDsRed-YAP was cotransfected with pCMV-CLDN18-GFP (upper panel) or control vector pCMV-GFP (lower panel) into MLE-15 cells. YAP colocalizes with CLDN18 at sites of cell-cell contact. n = 2. Scale bars: 5 μm. (H) IP with anti–p-LATS1/2 Ab shows decreased p-LATS1/2 and decreased association of p-LATS1/2 with p-YAP in Cldn18–/– (lane 4) compared with WT (lane 3) AT2 cell membranes. IgG = negative control IP with WT and Cldn18–/– lysates (lanes 1 and 2, respectively). n = 3 independent experiments. Input is lung tissue lysate; p-LATS1/2 cannot be detected in input. (I) IP with anti–p-YAP Ab shows decreased p-YAP and decreased association of p-YAP with p-LATS1/2 in Cldn18–/– (lanes 2 and 4) compared with WT (lanes 1 and 3) AT2 cell membranes. IgG = negative control. n = 3 independent experiments. Input is lung tissue lysate; p-LATS1/2 cannot be detected in input. Bar graphs represent the mean ± SEM for A, B, and E. Inputs were run in the same gel (F, H, and I) but were noncontiguous.
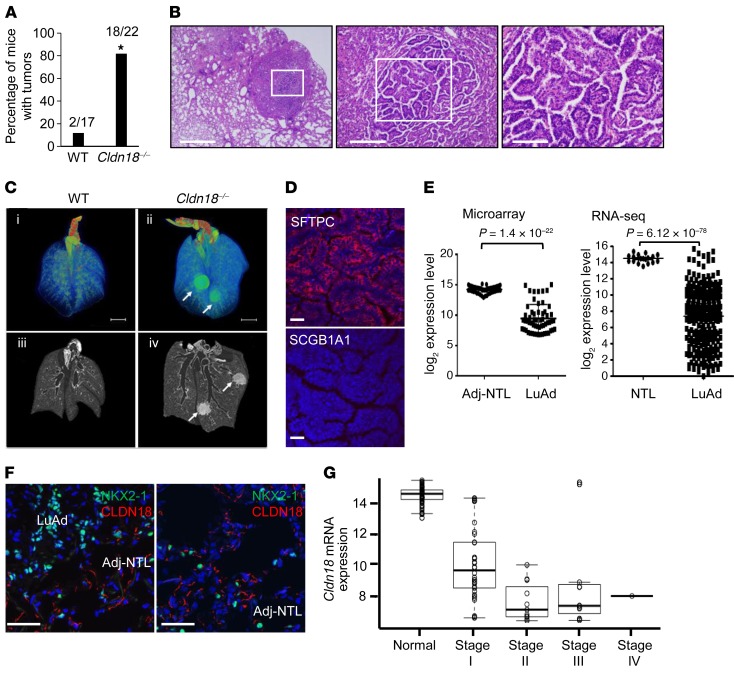
Loss of CLDN18 leads to adenocarcinoma development in aged Cldn18–/– mice and CLDN18 is downregulated in human LuAd.
Alterations of CLDN18 in LuAd (49, 50) and the known role of AT2 cells as the cell of origin of LuAd led us to evaluate lung tumor development in Cldn18–/– mice. Lungs of 9- to 10-month-old Cldn18–/– mice do not show obvious tumors. However, by 18–20 months of age, approximately 80% of Cldn18–/– mice compared with approximately 12% of WT mice develop tumors (Figure 6, A and B), generally in a subpleural location and typically adenocarcinomas with papillary features and fibrovascular cores (Figure 6B). Tumor cells show increased nuclear/cytoplasmic ratio and mitotic figures (Supplemental Figure 15A). Alveolar mononuclear cells (likely macrophages) are associated with nearly all tumors (Supplemental Figure 15B). Representative micro-CT images show that some Cldn18–/– mice have more than one lung tumor at 16 months of age (Figure 6C and Supplemental Table 2) without visible tumors in WT mice. Tumor cells express AT2 cell marker SFTPC, but not club cell marker SCGB1A1 (Figure 6D), suggesting an AT2 cell origin.
Figure 6. Increased tumor development in aged Cldn18–/– mice.
(A) Frequency of lung tumors in mice (age 18–20 months) determined by H&E staining. Bar graph represents the mean ± SEM. Fisher’s exact test. n = 17–22 mice. *P < 0.05. (B) H&E staining of a representative lung adenocarcinoma (LuAd) in a Cldn18–/– mouse at 3 different magnifications, shows peripheral/subpleural location in lung (left panel) and papillary features (middle and right panels). n = 22. Scale bars (from left to right): 1 mm, 200 μm, and 100 μm. (C) Ex vivo micro-computed tomography (micro-CT) lung images. Lung specimens from age-paired (15–16 months) WT and Cldn18–/– mice (n = 3) were scanned. Upper panel (i and ii): 3D reconstruction of representative WT and Cldn18–/– mouse lungs. Scale bar: 5 mm. Lower panel (iii and iv): Representative coronal sections of micro-CT images corresponding to lungs in upper panel. Tumors are indicated by arrows. (D) Cells in tumors of Cldn18–/– mice are SFTPC+ (red) but SCGB1A1–. DAPI (blue) is the nuclear counterstain. n = 3 biological replicates. Scale bar: 20 μm. (E) Cldn18 mRNA expression is reduced based on data from the Illumina Human WG-6v3.0 Beadarray in 58 matched microdissected LuAd and adjacent nontumor lung (Adj-NTL) (left panel) (Cldn18.1) and RNA-seq data from 287 LuAd and 19 NTL based on data generated by The Cancer Genome Atlas (TCGA) Research Network (Cldn18.1 and -18.2) (right panel). Paired t test for Beadarray data and unpaired t test for TCGA data. (F) Representative confocal images of CLDN18 (red) and NKX2-1 (green) show decreased CLDN18 protein expression in human LuAd compared with Adj-NTL. n = 3. Scale bars: 50 μm. DAPI is the nuclear counterstain. (G) Stage-dependent decreases in CLDN18 mRNA expression using microarray data from 58 matched human LuAd and adjacent NTL samples fit with an exponential regression model. P = 1.75 × 10–32.
We analyzed CLDN18 expression in human LuAd using microarray data from matched human LuAd and adjacent nontumor lung samples (51) and RNA sequencing (RNA-seq) data generated by The Cancer Genome Atlas (TCGA) Research Network (52). Analysis of gene expression data shows significant reduction of CLDN18 mRNA in human LuAd compared with nontumor lung tissue (Figure 6E). Double immunofluorescence staining for CLDN18 (red) and NKX2-1 (green) shows decreased CLDN18 protein expression in human LuAd compared with adjacent nontumor lung (Figure 6F and Supplemental Figure 16), with stage-dependent reductions in human LuAd (Figure 6G). Tumors in both Cldn18–/– mice (Figure 7A) and human LuAd (Figure 7B) show strong nuclear YAP expression.
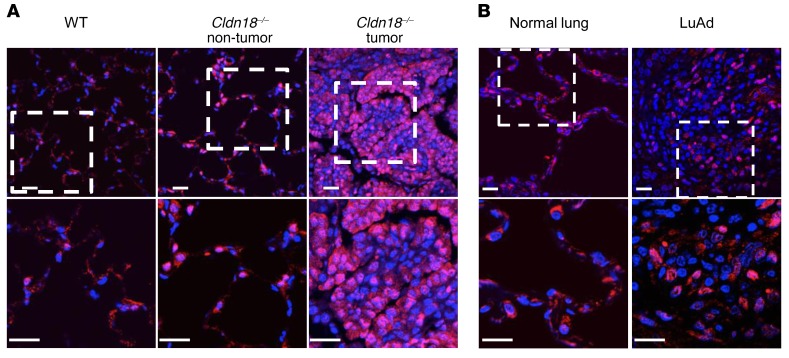
Figure 7. Nuclear YAP expression in lung adenocarcinoma (LuAd).
(A) Nuclear YAP (red) is present in Cldn18–/– mouse lungs in areas both with and without tumor. Lower panel shows higher magnification views of rectangle in upper panel. DAPI is the nuclear counterstain. n = 2. Scale bars: 20 μm. (B) Nuclear YAP is increased in human LuAd compared with normal lung. Lower panel shows higher magnification views of rectangle in upper panel. DAPI is the nuclear counterstain. n = 2. Scale bars: 20 μm.
Discussion
Located at sites of cell-cell contact, AJC proteins are well suited to transmit extracellular signals to regulate normal tissue maintenance and restore homeostasis following disruption of the epithelial barrier (53). We previously showed a critical nonredundant role for CLDN18 in regulating alveolar epithelial TJ composition and permeability properties in Cldn18–/– mice (24). Unexpectedly, we found an increase in size of lung, stomach, and kidney, all major sites of CLDN18 expression. We focused on the lung to further explore mechanisms underlying increased organ size. We demonstrate that increased lung size is mainly the result of parenchymal expansion accompanied by increased abundance and proliferation of AT2 cells, progenitors of distal lung epithelium. Mice with deletion of Cldn18 only in AT2 cells similarly showed increases in AT2 cell number and the proportion of AT2 cells in S and G2/M phase, indicating epithelial cell specificity. These findings suggest a potentially novel role for alveolar epithelial CLDN18 in restricting stem/progenitor cell proliferation (and thereby organ size). Consistent with in vivo results, Cldn18–/– AT2 cells in 3D culture showed increased CFE, which was maintained after passaging. Lungs of Cldn18–/– mice showed increased nuclear YAP with upregulation of several YAP target genes. Cldn18–/– AT2 cells in 3D culture also showed increased nuclear YAP, while inhibition of YAP signaling decreased Cldn18–/– AT2 cell proliferation in vivo and proliferation and CFE in vitro. Conversely, overexpression of CLDN18 decreased YAP nuclear localization and CFE of Cldn18–/– AT2 cells and decreased YAP transcriptional activity. Endogenous CLDN18 interacted with both ZO-1 and p-YAP, suggesting complex formation among these proteins localized to TJs, while overexpressed CLDN18 was similarly associated with both ZO-1 and p-YAP. Colocalization of YAP and CLDN18 at sites of cell-cell contact further suggests that CLDN18 plays a role in sequestering p-YAP at TJs. Cldn18–/– mice show increased propensity to develop LuAd with age, while human LuAd show stage-dependent reduction of CLDN18 and increased nuclear YAP. These findings identify a potentially novel role for integral TJ protein CLDN18 in regulating organ size and restricting distal lung epithelial progenitor capacity and proliferation, and indicate a role for YAP activation in regulating distal lung epithelial progenitor function.
A possible role for claudins in regulation of organ size and/or cell proliferation was suggested by the phenotype of Cldn15–/– mice that showed upper small intestine enlargement with expansion of proliferating crypt cells and an increased number of crypts per villus (14), although underlying mechanisms were not explored. In the current study, we demonstrate an increase in lung size with expansion and increased proliferation of AT2 cells, uncovering a role for CLDN18 in regulation of organ size and progenitor function. In the stomach, which was also enlarged, we observed expansion of the gastric mucosa (Supplemental Figure 4) with increased proliferation (data not shown). These results differ from previous studies in Cldn18.2–/– mice in which deletion of the stomach-specific isoform led to paracellular H+ leakage, upregulation of interleukin-1β, and atrophic gastritis (23) without organ enlargement or tumorigenesis, perhaps as a result of compensatory increases in Cldn18.1. The paradigm of CLDN18 restricting progenitor cell proliferation can therefore be extended to other sites of CLDN18 expression.
Hippo/YAP signaling regulates stem/progenitor cell proliferation and differentiation in many organs (26, 46, 54). In the lung, YAP is required for correct proximal-distal patterning of the airway and regulation of airway epithelial cell fate (55, 56). Conditional deletion of YAP led to disruption of branching morphogenesis, with expansion of distal epithelial progenitors and reciprocal reduction of proximal Sox2-expressing progenitors and impaired adult airway epithelial cell differentiation (56). Nuclear YAP promoted airway basal cell identity, whereas YAP overexpression blocked basal cell terminal differentiation, and YAP overexpression in differentiated secretory cells promoted a stem cell–like identity (55). Activation of YAP in lung airway epithelium following deletion of Mst1/2 under control of the Scgb1a1 promoter inhibited lung sacculation and maturation and resulted in club cell hyperplasia with increased bronchiolar epithelial cell proliferation and decreased differentiation (57). Additionally, increased nuclear YAP and decreased p-YAP were observed in airway epithelial cells following naphthalene injury or club cell ablation, suggesting a role for YAP signaling in bronchiolar epithelial repair following injury (57).
AT2 cells serve as progenitors of adult distal lung, undergoing both self-renewal and transdifferentiation to give rise to AT1 cells during normal turnover and following injury (44, 45, 58). Knowledge of signaling pathways that regulate stem cell proliferation and/or differentiation during alveolar repair and regeneration is limited. A recent study implicated YAP activation in response to mechanical tension in distal lung progenitor proliferation and expansion after pneumonectomy, suggesting a role for YAP signaling in alveolar regeneration in this context (59). In the current study, we show that loss of CLDN18 leads to YAP activation and progenitor expansion in distal lung in the absence of injury, uncovering a role for YAP signaling in regulating distal lung epithelial progenitor maintenance and proliferation downstream of CLDN18 and suggesting that CLDN18 regulates AT2 cell quiescence by restricting YAP activity. Cldn18–/– AT2 cells were still able to give rise to AT1 cells in vivo and in both 2D and 3D culture in vitro, indicating that YAP activation as a result of loss of CLDN18 did not significantly affect distal lung epithelial cell fate (55). Outcome of YAP activation may thus be context dependent and distinct in conducting airways and distal lung.
The Hippo pathway is the major regulator of YAP activity (60, 61). Upstream signals that activate Hippo (and inactivate YAP) include extracellular soluble factors via G protein–coupled receptors, mechanotransduction, cell-cell contact, and polarity (28, 62), including apical polarity complexes such as Crb3 (63). There is also accumulating evidence for Hippo kinase–independent modulation of YAP activity through direct interactions with cell membrane- and cytoskeleton-associated proteins that regulate apical-basolateral polarity and cell-cell contact (26). In this regard, AJ proteins E-cadherin and α-catenin inhibit YAP signaling by both Hippo-independent and -dependent mechanisms (39, 40). Similarly, AMOTs, TJ-associated scaffolding proteins that maintain TJ integrity and epithelial polarity, regulate YAP activity leading to sequestration in cytoplasm and at TJs (64). AMOTs also promote LATS activation by serving as a scaffold that brings Hippo kinases and YAP together at TJs (65). We have demonstrated in the current study a role for integral TJ protein CLDN18 in regulating YAP activity. Co-IP demonstrates interaction among p-YAP, CLDN18, ZO-1, and p-LATS1/2 in WT AT2 cells. Furthermore, when overexpressed, CLDN18 and YAP colocalize at sites of cell-cell contact, suggesting sequestration of p-YAP at TJs. Decreased p-YAP in Cldn18–/– lungs and AT2 cells and decreased p-LATS/p-YAP interaction in membranes of Cldn18–/– AT2 cells suggest that CLDN18 is required for Hippo signaling, and furthermore that CLDN18 serves a scaffolding function that promotes interaction between the Hippo kinases and p-YAP at TJs. Loss of CLDN18 likely disrupts a membrane protein complex that includes YAP and Hippo kinases leading to increased nuclear translocation of dephosphorylated YAP. Elucidation of specific protein-protein interactions that mediate CLDN18 interaction with p-YAP/YAP will require further investigation.
Claudin expression is frequently altered in cancer (18, 66, 67), with variable up- and downregulation depending on tissue type and disease stage (15, 18, 68–70). There is thus no unifying concept as to how claudin dysregulation contributes mechanistically to carcinogenesis. CLDN18 expression is altered in both gastric cancer and LuAd, although most studies have not distinguished between expression of Cldn18.1 versus Cldn18.2 isoforms. CLDN18 was significantly downregulated in gastric cancer compared with surrounding normal gastric mucosa and its expression correlated inversely with proliferative potential at the invasive front and invasive properties of a gastric cancer cell line (71), while downregulation of CLDN18 correlated with poor survival (72). In contrast, there are reports of sustained membranous CLDN18.2 expression in gastric cancer, as well as ectopic upregulation in tumors of lung, ovary, and esophagus (73). Based on these studies, Ab-mediated therapeutic targeting of CLDN18.2 in cancers with upregulation of CLDN18.2 has been suggested (74). However, CLDN18 expression was detected in only approximately 20% of LuAd (50), with overall expression being lower in tumors than in normal lung tissue, while CLDN18.2 was expressed in only 3.7% of LuAd (74). Thus, targeting of CLDN18.2 would not be helpful for the majority of LuAd. Our analysis (Figure 6E) indicates that the CLDN18.1 isoform is downregulated in LuAd. Consistent with these findings, overexpression of CLDN18.1 was found to suppress abnormal proliferation and motility of A549 cells, a human LuAd cell line, via inhibition of PI3K/PDK1/Akt signaling (75). Therapeutic targeting of CLDN18 should therefore take into account the specific isoform that is altered in a particular subtype of LuAd.
We show that loss of CLDN18 activates YAP signaling and promotes proliferation of AT2 cells, recognized cells of origin for LuAd. Furthermore, loss of CLDN18 leads to development of LuAd in aged mice, while TCGA data show stage-specific downregulation of CLDN18.1 in LuAd. Mechanisms underlying CLDN18.1 downregulation in LuAd remain to be determined. YAP expression correlates with poor prognosis in non–small cell lung cancer (49) and YAP activity drives tumor progression in mouse models of LuAd (34). Although YAP hyperactivation regulates cellular properties important for cancer development (e.g., promotion of stemness, proliferation and dedifferentiation, and resistance to apoptosis), precise mechanisms leading to tumorigenesis as a result of YAP activation remain unclear (33). The number of stem cell divisions within a tissue correlates strongly with risk of carcinogenesis, suggesting accumulation of random mutations and genomic instability due to uncontrolled AT2 cell proliferation in Cldn18–/– mice as a potential mechanism underlying tumorigenesis (76). Our results establish an important regulatory role for TJs in general and CLDN18 in particular in restricting YAP activity to prevent uncontrolled progenitor cell proliferation and resulting tumorigenesis, while also suggesting that transient modulation of CLDN18 and/or YAP function may be of benefit by harnessing progenitor cells to promote regeneration following lung injury.
Methods
Constitutive and epithelial cell–specific Cldn18-knockout mice.
Generation of Cldn18–/– mice has recently been reported (24). Sftpc+/creERT2 (Harold A. Chapman, UCSF) mice (77) crossed to Cldn18fl/fl mice were further crossed with ROSATm/Tm reporter mice (78), yielding mice with the genotype Sftpc+/creERT2;Cldn18fl/fl;ROSA+/Tm. Cldn18 knockout was induced by administration of Tmx at a dose of 100 mg/kg i.p. for 2 consecutive days. Control mice with the genotype Sftpc+/creERT2;ROSA+/Tm received the same dose of Tmx to label AT2 cells.
Tissue preparation and morphological analysis.
Tissues harvested from WT and Cldn18–/– mice were prepared as previously described (24). To quantify tumors, H&E–stained lung sections from WT and Cldn18–/– mice were examined by light microscopy in a blinded fashion.
Lung volume measurement.
Lung volume was measured as previously described (79). Briefly, mice were mechanically ventilated for 10 minutes with 100% oxygen. The tracheal tube was then clamped for 10 minutes to allow circulating blood to absorb oxygen and collapse airspaces. Total volume at pressures of 0, 5, 10, 15, 20, 25, and 30 cmH2O was measured using a suspension technique after inflation with PBS.
Micro-CT imaging.
Lungs were fixed and inflated with 4% paraformaldehyde (PFA) at 20 cmH2O pressure overnight and then incubated through a serial ethanol gradient (50%, 70%, 80%, 90%, and 100%) followed by incubation with 100% hexamethyldisilazane overnight before air drying (80). Lung specimens were scanned at an isotropic resolution of 10 microns at 45 kVp, 200 mAs (81). CT image raw data were analyzed using AMIRA software (FEI) to create volume renderings (82). 3D segmentation to compartmentalize the lung into tissue and conducting airway was performed based on threshold of gray value difference between tissue and air. Small sections (~1 mm3) were cut from the distal portion of the lung and scanned at an isotropic resolution of 0.7 microns. Volumes of whole lung (VTlung) and conducting airway (VCairway) were measured based on CT scan contrast data of whole lung. Alveolar airspace and parenchymal fractions (Falv and Fpar) of 1-mm3 lung sections were calculated based on CT scan contrast data of the small section, assuming that the 2 fractions could be applied to the remainder of distal lung. Alveolar airspace (Valv) and parenchymal (Vpar) volume of the whole lung were calculated by multiplying (VTlung – VCairway) by Falv and Fpar, respectively. The relationship among different compartments shown in Supplemental Table 1 is presented as the following equation: VTlung = VCairway + Valv + Vpar. For radiographic measurement of lung tumors, regions of high density on acquired CT images were automatically detected, counted, and volumetrically quantified. A CT threshold (320 Hounsfield units [HU]) was used to segment lung tumor from normal lung areas.
Isolation of mouse AT2 cells.
For cell isolation for 3D culture, mouse lungs were digested with elastase (4 U/ml, LS002280, Worthington Biochemical). Cells were resuspended at 106 cells in 100 μl of HEPES-buffered salt solution (HBSS) in a mixture of Abs, including anti-CD45, anti-CD31, anti-CD34, anti–SCA-1, anti-EPCAM, anti-CD24, and relevant isotype controls. AT2 cells (CD45–CD31–CD34–SCA-1–CD24–) were sorted from the EPCAM+ lung epithelial population using a MoFlo XDP (Beckman Coulter) or FACSAria (BD Biosciences) sorter as previously described (83). Data were analyzed using FlowJo software (TreeStar). For passaging, colonies were dissociated using dispase (354235, BD Biosciences) followed by magnetic selection for EPCAM+ cells. For 2D culture and cell cycle analysis, AT2 cells were isolated from WT and Cldn18–/– mice as previously described (84). For cell cycle analysis of Tm+ AT2 cells, single-cell suspensions were generated by dissociation with dispase of lungs from Sftpc+/creERT2;Cldn18fl/fl;ROSA+/Tm and control Sftpc+/creERT2;ROSA+/Tm mice.
3D Matrigel culture.
Sorted AT2 cells (4,000–5,000 cells) were grown in 3D culture on Transwell filter inserts (353095, BD Biosciences) as previously described (83). Images were taken after 14–16 days in culture. For passaging, EPCAM+ cells (~5,000) were replated in Matrigel with fresh MLg cells (American Type Culture Collection). ImageJ software (NIH) was used for quantification of colony size. Filters were fixed in 4% PFA, embedded in Histogel (HG-4000, Thermo Fisher Scientific), and processed for paraffin embedding.
Culture of MLE-15, 293T, and AT2 cells.
MLE-15 cells (Jeffrey A. Whitsett, University of Cincinnati) were cultured in HITES medium as previously described (85). For 2D culture, mouse primary AT2 cells were seeded on Transwell polycarbonate filters (Corning) coated with rat laminin-5 (Millipore) at 750,000 cells/cm2 and cultured as previously described (84).
Abs.
Primary Abs used for Western blotting were CLDN18 (700178, Life Technologies), AQP5 (AQP005, Alomone Labs), T1α (A205002, Affinity Bioreagents/Thermo Fisher Scientific), RAGE (MAB11451, R&D Systems), caveolin-1 (ab2910, Cell Signaling Technology), pro-SFTPC (AB3786, EMD Millipore), eIF-4E (610270, BD Biosciences Pharmingen), lamin A/C (sc20681, Santa Cruz Biotechnology), ZO-1 (40-2200, Life Technologies), cyclin D1 (2978s, Cell Signaling Technology), p-YAP (4911, Cell Signaling Technology), YAP (4912, Cell Signaling Technology), and α-tubulin (6-11B-1, Sigma-Aldrich).
Primary Abs used for immunostaining were NKX2-1 (MS-699-P1, Leica-Novocastra), cyclin D1 (2978s, Cell Signaling Technology), CLDN18 (700178, Life Technologies), Ki67 (ab16667, Abcam), pro-SFTPC (WRAB-9337, Seven Hills Bioreagents), p-YAP (4911, Cell Signaling Technology), YAP (4912, Cell Signaling Technology), and AQP5 (sc-9890, Santa Cruz Biotechnologies).
Primary Abs used for FACS were CD45 (30-F11, eBiosciences), CD34 (13-0341-82, eBiosciences), CD31 (13-0311, eBisciences), SCA-1 (108109, Biolegend), EPCAM (118215, Biolegend), and CD24 (562789, BD Biosciences).
Abs used for co-IP were p-LATS1/2 (A8125, One World Lab), p-YAP (4911, Cell Signaling Technology), and CLDN18 (700178, Life Technologies).
Immunofluorescence.
Samples were incubated with primary Abs at 4°C overnight following microwave antigen retrieval (Antigen Unmasking Solution, Vector Laboratories). Slides were incubated with Alexa 488–labeled goat anti-mouse Ab (Life Technologies), FITC-conjugated donkey anti-goat IgG (sc2024, Santa Cruz Biotechnologies), or goat biotinylated anti-rabbit/-mouse IgG followed by Cy3-conjugated streptavidin (Jackson ImmunoResearch) or fluorescein-labeled Avidin DCS (Vector Laboratories). Nuclear staining was performed with 4′,6-diamidino-2-phenylindole (DAPI) or propidium iodide (PI) Vectashield mount (Vector Laboratories). Images were captured using a Nikon Eclipse 80i microscope with cooled CCD camera (QImaging). Confocal images were captured using a Zeiss LSM 510 confocal system (Carl Zeiss) or Leica TCS SP8 confocal microscope (Leica Microsystems).
Quantification of NKX2-1+ cells.
Lung sections from WT and Cldn18–/– mice were stained for NKX2-1 with DAPI as nuclear counterstain. Five random pictures were taken using the ×40 lens of a Nikon Eclipse 80i microscope. Numbers of NKX2-1+ and total cells (DAPI+) were counted in each of 5 lung fields for each genotype. Approximately 4,000 cells from each group were counted.
Quantification of SFTPC+ and Tm+ cells.
Lung sections stained with SFTPC with DAPI as nuclear counterstain were scanned with an Axio Scan.Z1 (Carl Zeiss Microscopy). The number of SFTPC+ and DAPI+ cells were counted in 2 randomly chosen regions for each genotype using Imaris software (Bitplane). Approximately 50,000 DAPI+ cells from each group were counted. For Tm+ cell counting, frozen lung sections were stained with DAPI, scanned, and counted.
EdU labeling.
5-Ethynyl-2′-deoxyuridine (EdU) was injected i.p. at 50 mg/kg body weight 24 hours prior to harvest. EdU incorporation was detected with a Click-iT Plus EdU Imaging Kit (10337, Life Technologies) following immunostaining for NKX2-1. E18 lung sections were scanned (Axio Scan.Z1) and a total of approximately 13,000 and approximately 17,000 NKX2-1+ cells were counted from each genotype using Imaris software. To determine the percentage of EdU+NKX2-1+ cells postnatally, a total of approximately 1,000 NKX2-1+ cells from 5 random fields were counted. To label proliferating MLE-15 cells, EdU (10 μM) was added to culture medium 48 hours after transfection. To label proliferating AT2 cells in 3D culture, EdU (10 μM) was added to medium 3 hours before harvesting cells.
Nuclear protein extraction.
Nuclear protein was extracted from MLE-15 cells using a Panomics Nuclear Extraction Kit (AY2002).
Western analysis.
Preparation of protein lysates from AT2 cells or whole lung and subsequent Western analysis were performed as previously described (86). Antigen-Ab complexes were visualized and quantified by enhanced chemiluminescence (Pierce) using a Fluor-Chem Imaging System (Model 8900, Alpha Innotech) or an Azure Imager c300 (Azure Systems). See complete unedited blots in the supplemental material.
RNA isolation, RT-PCR, and qRT-PCR.
RNA extraction and cDNA synthesis were performed as previously described (86). qRT-PCR was performed with SYBR-Green reagent (Applied Biosystems) in a 7900HT Fast Real-Time PCR System (Applied Biosystems). Sequences of primers used are as follows: Cldn18 forward 5′-GACCGTTCAGACCAGGTACA-3′ and reverse 5′-GCGATGCACATCATCACTC-3′; Ccnd1 forward 5′-gcgtaccctgacaccaatct-3′ and reverse 5′-cacaacttctcggcagtcaa-3′; Cdk6 forward 5′-gcctatgggaaggtgttcaa-3′ and reverse 5′-gggctctggaactttatcca-3′; Ctgf forward 5′-CCACCCCAAACCAGTCATAA-3′ and reverse 5′-TGCTGTGCAGGTGATAAAGC-3′; and Areg forward 5′-CATCGGCATCGTTATCACAG-3′ and reverse 5′-ACAGTCCCGTTTTCTTGTCG-3′.
Cell cycle analysis.
Cell cycle analysis for AT2 cells was performed using PI as previously described (87). Tm+ AT2 cells were stained with Hoechst 33323 (H3570, Thermo Fisher Scientific). Flow cytometry analysis was performed using a FACSAria or LSRII flow cytometer (BD Biosciences) and ModFit LT Version 4 or 5 software.
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay.
TUNEL assay was performed using the In Situ Cell Death Detection Kit (11684795910, Roche Diagnostics).
Virus generation.
Mouse Yap shRNA-expressing vector (TRCN0000300325) was obtained from Sigma-Aldrich. CLDN18.1 cDNA (NCBI reference sequence: NM_016369.3) cloned into lentiviral vector pLenti-C-turboGFP (pLenti-C-turboGFP-C18) was from Origene. Lentiviral expression plasmids were cotransfected into 293T cells together with helper plasmids pCMVΔR8.91 and pMDG as previously described (85). Viral titers were determined by p24 Elisa Assay Kit (Cell Biolabs).
Lentiviral YAP knockdown and CLDN18 overexpression in AT2 cells.
Freshly isolated AT2 cells (100,000 cells) were mixed with virus expressing Yap shRNA (MOI = 5) (nonsilencing shRNA used as control) or CLDN18 (pLenti-C-turboGFP-CLDN18) (pLenti-C-turboGFP as control) (MOI = 3) and incubated overnight at 4°C. The next day, virus-transduced cells were washed, mixed with MLg fibroblasts, and seeded for 3D culture. 3D cultures were processed for CFE analysis and harvested at day 14 for H&E staining and immunostaining.
VP treatment.
VP, brand name Visudyne (1786, Selleckchem) was dissolved in dimethyl sulfoxide (DMSO) at 100 mg/ml and diluted in PBS to 15 mg/ml. VP was administered i.p. at 100 mg/kg body weight daily from P4 to P11, and lungs were harvested at P12 for evaluation of cell proliferation. To examine dry weight/body weight ratios, pregnant females were injected (100 mg/kg body weight) at E13, E15, E17, and E19 followed by postnatal injection into pups from P4 and then every other day up to P14 at the same dose of VP. Lungs were harvested on P16. VP was added to AT2 cells in 3D culture from day 2 at a concentration of 0.75 μM. 3D cultures were imaged and harvested for staining at day 14.
Transient transfections in MLE-15 cells.
Twenty-four hours after seeding, 0.5 μg 5xUAS-Luc, 0.3 μg GAL4-TEAD (from Jiandie Lin, University of Michigan), and 30 ng pEGFP-C3-Yap (from Marius Sudol, National University of Singapore) or pCMV-Flag-YAP5SA were cotransfected into MLE-15 cells together with 0.25 μg pCMV6-AC-CLDN18-GFP or control pCMV6-AC-CLDN18-GFP (Origene). Luciferase assay was performed 48 hours following transfection and normalized to protein concentration.
Phos-tag Western blotting.
Phos-tag gels were prepared according to the manufacturer’s instructions (300-93523, Wako Chemicals). Protein separation and immunoblotting were performed using standard protocols for Western blotting.
Cell membrane isolation.
Cells were resuspended in 2 ml mannitol buffer (300 mM mannitol, 10 mM HEPES, pH 7.4, 3 mM EGTA, and 1 mM EDTA), followed by sonication. Cell membrane fractions were obtained by ultracentrifugation for 30 minutes at 60,000 g at 4°C. Membrane pellets were resuspended in RIPA buffer (150 mM NaCl, 1.0% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0) and incubated on ice for 2 hours, followed by ultracentrifugation for 30 minutes at 60,000 g. Supernatants were collected and protein concentration determined by Bio-Rad Protein assay (500-0006, Bio-Rad).
Cross-linking Ab to protein A/G beads.
Cross-linking of protein A/G Sepharose beads (Santa Cruz Biotechnology) with CLDN18, p-LATS1/2, p-YAP, or YAP Ab or IgG were performed as previously described (85).
Co-IP.
Thirty microliters of cross-linked CLDN18, p-LATS1/2, or p-YAP Ab or IgG was added to each sample (70 μg in 200 μl RIPA buffer) and incubated overnight at 4°C. Immunoprecipitated proteins were eluted in Laemmli sample loading buffer.
Mass spectrometry.
Membrane protein lysate (70 μg in RIPA buffer) from mouse AT2 cells was immunoprecipitated with mouse monoclonal anti-YAP Ab (SC101199, Santa Cruz Biotechnology) cross-linked with protein A/G beads. Immunoprecipitates were loaded onto SDS gels and stained with colloidal Coomassie blue (24590, Thermo Fisher Scientific). Mass spectrometric analysis of bands of interest was performed at the Proteomics Core Facility in the Liver Center at the University of Southern California (USC).
Microarray and RNA-seq analysis of CLDN18 expression in LuAd.
The relationship between CLDN18 expression and tumor/nontumor status and/or stage was examined using CLDN18 mRNA expression microarray data from Illumina Human WG-6 v3.0 Expression BeadChips, processed as described previously (51). Level 3 CLDN18 RNA-seq data were directly downloaded from TCGA data portal website (52) (https://tcga-data.nci.nih.gov/tcga/) and log2 transformed. To evaluate the relationship between tumor stage and CLDN18 expression, we built an exponential regression model in R (version 3.1.1) examining expression versus stage, defining normal lung as stage 0, tumor stage 1 as 1, and similarly for the other stages.
Statistics.
Values are the mean ± SEM. Significance (P < 0.05) for 3 or more group means with 1 and 2 factors was determined by 1-way and 2-way ANOVA, respectively. Post hoc analyses were performed with Bonferroni’s corrections. Two group means and a 2 × 2 contingency table were compared for significance using 2-sided t tests and Fisher’s exact test, respectively. Z tests were used to determine if ratiometric (i.e., normalized) data were different from control. Statistical analyses were performed using SPSS version 19 (IBM) and Microsoft Office Excel 2013.
Study approval.
De-identified human LuAd and nontumor lung samples were collected at Keck Hospital of USC (USC IRB protocol HS-06-00447). Paraffin-embedded normal lung tissue was obtained from remnant human transplant lungs under an approved protocol (USC IRB protocol HS-07-00660). All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at USC.
Author contributions
BZ conceived and directed experiments, analyzed and interpreted experimental data, and wrote the manuscript. PF conceived and directed experiments, analyzed and interpreted data, and edited the manuscript. JL performed experiments in cell lines and tumor tissue and micro-CT analysis. DRC performed 3D cultures and staining of lung tissues. YL performed co-IP, Western blotting, immunostaining, and 3D culture. FXY ran Phos-tag gels and Western blots. AM performed mouse AT2 cell isolation and 3D cultures. BV performed micro-CT analysis. GL performed experiments in primary AT2 cells and lung tissues. NOC performed cell cycle analysis and 3D culture. MS performed lung volume measurements. MNK and WE performed mouse tumor pathological analysis. PC performed micro-CT analysis and edited the manuscript. JML, IALO, PM, and EDC conceived experiments and edited the manuscript. CY and CNM performed RNA-seq and microarray analysis. KG conceived experiments, provided plasmids, and edited the manuscript. BRS conceived experiments and provided expertise for 3D cultures, interpreted experimental data, and edited the manuscript. ZB conceived the study, conceived and directed experiments, interpreted experimental data, and wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the Hastings Foundation, Whittier Foundation, and research grants ES017034 (to E.D. Crandall), HL108634 (to E.D. Crandall), R35HL135747 (to Z. Borok), HL112638 (to Z. Borok), HL126877 (to Z. Borok), HL114094 (to Z. Borok and I.A. Laird-Offringa), and HL114959 (to B. Zhou) from the NIH and LA1-06915 (to B.R. Stripp) from the California Institute of Regenerative Medicine. E.D. Crandall is Hastings Professor and Kenneth T. Norris Jr. Chair of Medicine. Z. Borok is Edgington Chair in Medicine. Critical reading of the manuscript and statistical analysis by Kwang-Jin Kim is appreciated. Histology and microscopy services were provided by the Cell and Tissue Imaging Core of the USC Research Center for Liver Diseases (P30 DK048522 and S10 RR022508) and Norris Comprehensive Cancer Center Core (P30 CA014089). Micro-CT services were performed in the USC Molecular Imaging Center. Flow cytometry was performed in Core Facilities at USC (P30CA014089 and the USC Office of the Provost, Dean’s Development Funds, Keck School of Medicine of USC), Cedars-Sinai Medical Center and Children’s Hospital Los Angeles.
Version 1. 02/05/2018
Electronic publication
Version 2. 03/01/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(3):970–984.https://doi.org/10.1172/JCI90429.
See the related Commentary at Claudin-18: unexpected regulator of lung alveolar epithelial cell proliferation.
Contributor Information
Beiyun Zhou, Email: bzhou@usc.edu.
Per Flodby, Email: flodby@usc.edu.
Jiao Luo, Email: jiaoluo68@gmail.com.
Yixin Liu, Email: yixinliu@usc.edu.
Fa-Xing Yu, Email: fxyu@fudan.edu.cn.
Alicia McConnell, Email: amcconnell@enders.tch.harvard.edu.
Bino Varghese, Email: binoav@gmail.com.
Guanglei Li, Email: 2096931323@qq.com.
Nyam-Osor Chimge, Email: chimgee@usc.edu.
Mitsuhiro Sunohara, Email: mitsuhiro.sunohara@nifty.ne.jp.
Wafaa Elatre, Email: welatre@gmail.com.
Peter Conti, Email: Peter.Conti@med.usc.edu.
Chenchen Yang, Email: yang529@usc.edu.
Parviz Minoo, Email: minoo@usc.edu.
Barry R. Stripp, Email: barry.stripp@cshs.org.
Edward D. Crandall, Email: ecrandal@usc.edu.
Zea Borok, Email: zborok@usc.edu.
References
- 1.Farquhar MG, Palade GE. Junctional complexes in various epithelia. J Cell Biol. 1963;17:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Meer G, Gumbiner B, Simons K. The tight junction does not allow lipid molecules to diffuse from one epithelial cell to the next. Nature. 1986;322(6080):639–641. doi: 10.1038/322639a0. [DOI] [PubMed] [Google Scholar]
- 3.Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol. 1998;141(7):1539–1550. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morita K, Furuse M, Fujimoto K, Tsukita S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci U S A. 1999;96(2):511–516. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cereijido M, Contreras RG, Shoshani L, Flores-Benitez D, Larre I. Tight junction and polarity interaction in the transporting epithelial phenotype. Biochim Biophys Acta. 2008;1778(3):770–793. doi: 10.1016/j.bbamem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Furuse M, Sasaki H, Fujimoto K, Tsukita S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J Cell Biol. 1998;143(2):391–401. doi: 10.1083/jcb.143.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cording J, et al. In tight junctions, claudins regulate the interactions between occludin, tricellulin and marvelD3, which, inversely, modulate claudin oligomerization. J Cell Sci. 2013;126(Pt 2):554–564. doi: 10.1242/jcs.114306. [DOI] [PubMed] [Google Scholar]
- 8.Van Itallie CM, Anderson JM. Claudin interactions in and out of the tight junction. Tissue Barriers. 2013;1(3):e25247. doi: 10.4161/tisb.25247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tatum R, et al. Renal salt wasting and chronic dehydration in claudin-7-deficient mice. Am J Physiol Renal Physiol. 2010;298(1):F24–F34. doi: 10.1152/ajprenal.00450.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konrad M, et al. Mutations in the tight-junction gene claudin 19 (CLDN19) are associated with renal magnesium wasting, renal failure, and severe ocular involvement. Am J Hum Genet. 2006;79(5):949–957. doi: 10.1086/508617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furuse M, et al. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: a lesson from claudin-1-deficient mice. J Cell Biol. 2002;156(6):1099–1111. doi: 10.1083/jcb.200110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben-Yosef T, et al. Claudin 14 knockout mice, a model for autosomal recessive deafness DFNB29, are deaf due to cochlear hair cell degeneration. Hum Mol Genet. 2003;12(16):2049–2061. doi: 10.1093/hmg/ddg210. [DOI] [PubMed] [Google Scholar]
- 13.Miyamoto T, et al. Tight junctions in Schwann cells of peripheral myelinated axons: a lesson from claudin-19-deficient mice. J Cell Biol. 2005;169(3):527–538. doi: 10.1083/jcb.200501154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamura A, et al. Megaintestine in claudin-15-deficient mice. Gastroenterology. 2008;134(2):523–534. doi: 10.1053/j.gastro.2007.11.040. [DOI] [PubMed] [Google Scholar]
- 15.Singh AB, Sharma A, Dhawan P. Claudin family of proteins and cancer: an overview. J Oncol. 2010;2010:541957. doi: 10.1155/2010/541957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soini Y. Tight junctions in lung cancer and lung metastasis: a review. Int J Clin Exp Pathol. 2012;5(2):126–136. [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuda Y, et al. Gastric and intestinal claudin expression at the invasive front of gastric carcinoma. Cancer Sci. 2007;98(7):1014–1019. doi: 10.1111/j.1349-7006.2007.00490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ouban A, Ahmed AA. Claudins in human cancer: a review. Histol Histopathol. 2010;25(1):83–90. doi: 10.14670/HH-25.83. [DOI] [PubMed] [Google Scholar]
- 19.Koval M. Claudin heterogeneity and control of lung tight junctions. Annu Rev Physiol. 2013;75:551–567. doi: 10.1146/annurev-physiol-030212-183809. [DOI] [PubMed] [Google Scholar]
- 20.Schlingmann B, Molina SA, Koval M. Claudins: gatekeepers of lung epithelial function. Semin Cell Dev Biol. 2015;42:47–57. doi: 10.1016/j.semcdb.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohta H, Chiba S, Ebina M, Furuse M, Nukiwa T. Altered expression of tight junction molecules in alveolar septa in lung injury and fibrosis. Am J Physiol Lung Cell Mol Physiol. 2012;302(2):L193–L205. doi: 10.1152/ajplung.00349.2010. [DOI] [PubMed] [Google Scholar]
- 22.Türeci O, et al. Claudin-18 gene structure, regulation, and expression is evolutionary conserved in mammals. Gene. 2011;481(2):83–92. doi: 10.1016/j.gene.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi D, et al. Deficiency of claudin-18 causes paracellular H+ leakage, up-regulation of interleukin-1β, and atrophic gastritis in mice. Gastroenterology. 2012;142(2):292–304. doi: 10.1053/j.gastro.2011.10.040. [DOI] [PubMed] [Google Scholar]
- 24.Li G, et al. Knockout mice reveal key roles for claudin 18 in alveolar barrier properties and fluid homeostasis. Am J Respir Cell Mol Biol. 2014;51(2):210–222. doi: 10.1165/rcmb.2013-0353OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaFemina MJ, et al. Claudin-18 deficiency results in alveolar barrier dysfunction and impaired alveologenesis in mice. Am J Respir Cell Mol Biol. 2014;51(4):550–558. doi: 10.1165/rcmb.2013-0456OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13(8):877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piccolo S, Dupont S, Cordenonsi M. The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev. 2014;94(4):1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- 28.Harvey KF, Hariharan IK. The hippo pathway. Cold Spring Harb Perspect Biol. 2012;4(8):a011288. doi: 10.1101/cshperspect.a011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21(21):2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cherrett C, Furutani-Seiki M, Bagby S. The Hippo pathway: key interaction and catalytic domains in organ growth control, stem cell self-renewal and tissue regeneration. Essays Biochem. 2012;53:111–127. doi: 10.1042/bse0530111. [DOI] [PubMed] [Google Scholar]
- 31.Barry ER, et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493(7430):106–110. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tremblay AM, Camargo FD. Hippo signaling in mammalian stem cells. Semin Cell Dev Biol. 2012;23(7):818–826. doi: 10.1016/j.semcdb.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15(2):73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lau AN, et al. Tumor-propagating cells and Yap/Taz activity contribute to lung tumor progression and metastasis. EMBO J. 2014;33(5):468–481. doi: 10.1002/embj.201386082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szymaniak AD, Mahoney JE, Cardoso WV, Varelas X. Crumbs3-mediated polarity directs airway epithelial cell fate through the Hippo pathway effector Yap. Dev Cell. 2015;34(3):283–296. doi: 10.1016/j.devcel.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao B, et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25(1):51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem. 2011;286(9):7018–7026. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim NG, Koh E, Chen X, Gumbiner BM. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc Natl Acad Sci U S A. 2011;108(29):11930–11935. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silvis MR, et al. α-catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci Signal. 2011;4(174):ra33. doi: 10.1126/scisignal.2001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlegelmilch K, et al. Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell. 2011;144(5):782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asselin-Labat ML, Filby CE. Adult lung stem cells and their contribution to lung tumourigenesis. Open Biol. 2012;2(8):120094. doi: 10.1098/rsob.120094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rock J, Königshoff M. Endogenous lung regeneration: potential and limitations. Am J Respir Crit Care Med. 2012;186(12):1213–1219. doi: 10.1164/rccm.201207-1151PP. [DOI] [PubMed] [Google Scholar]
- 43.Rock JR, Hogan BL. Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu Rev Cell Dev Biol. 2011;27:493–512. doi: 10.1146/annurev-cellbio-100109-104040. [DOI] [PubMed] [Google Scholar]
- 44.Barkauskas CE, et al. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest. 2013;123(7):3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adamson IY, Bowden DH. The type 2 cell as progenitor of alveolar epithelial regeneration. A cytodynamic study in mice after exposure to oxygen. Lab Invest. 1974;30(1):35–42. [PubMed] [Google Scholar]
- 46.Camargo FD, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17(23):2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 47.Alder JK, et al. Telomere dysfunction causes alveolar stem cell failure. Proc Natl Acad Sci U S A. 2015;112(16):5099–5104. doi: 10.1073/pnas.1504780112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu-Chittenden Y, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26(12):1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Dong Q, Zhang Q, Li Z, Wang E, Qiu X. Overexpression of yes-associated protein contributes to progression and poor prognosis of non-small-cell lung cancer. Cancer Sci. 2010;101(5):1279–1285. doi: 10.1111/j.1349-7006.2010.01511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merikallio H, Pääkkö P, Harju T, Soini Y. Claudins 10 and 18 are predominantly expressed in lung adenocarcinomas and in tumors of nonsmokers. Int J Clin Exp Pathol. 2011;4(7):667–673. [PMC free article] [PubMed] [Google Scholar]
- 51.Selamat SA, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22(7):1197–1211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gumbiner BM, Kim NG. The Hippo-YAP signaling pathway and contact inhibition of growth. J Cell Sci. 2014;127(Pt 4):709–717. doi: 10.1242/jcs.140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hiemer SE, Varelas X. Stem cell regulation by the Hippo pathway. Biochim Biophys Acta. 2013;1830(2):2323–2334. doi: 10.1016/j.bbagen.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 55.Zhao R, et al. Yap tunes airway epithelial size and architecture by regulating the identity, maintenance, and self-renewal of stem cells. Dev Cell. 2014;30(2):151–165. doi: 10.1016/j.devcel.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahoney JE, Mori M, Szymaniak AD, Varelas X, Cardoso WV. The hippo pathway effector Yap controls patterning and differentiation of airway epithelial progenitors. Dev Cell. 2014;30(2):137–150. doi: 10.1016/j.devcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lange AW, Sridharan A, Xu Y, Stripp BR, Perl AK, Whitsett JA. Hippo/Yap signaling controls epithelial progenitor cell proliferation and differentiation in the embryonic and adult lung. J Mol Cell Biol. 2015;7(1):35–47. doi: 10.1093/jmcb/mju046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rock JR, et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci USA. 2011;108(52):E1475–E1483. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Z, et al. MAPK-mediated YAP activation controls mechanical-tension-induced pulmonary alveolar regeneration. Cell Rep. 2016;16(7):1810–1819. doi: 10.1016/j.celrep.2016.07.020. [DOI] [PubMed] [Google Scholar]
- 60.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30(1):1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yimlamai D, Fowl BH, Camargo FD. Emerging evidence on the role of the Hippo/YAP pathway in liver physiology and cancer. J Hepatol. 2015;63(6):1491–1501. doi: 10.1016/j.jhep.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27(4):355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ling C, et al. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci USA. 2010;107(23):10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moleirinho S, Guerrant W, Kissil JL. The Angiomotins--from discovery to function. FEBS Lett. 2014;588(16):2693–2703. doi: 10.1016/j.febslet.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paramasivam M, Sarkeshik A, Yates JR, Fernandes MJ, McCollum D. Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol Biol Cell. 2011;22(19):3725–3733. doi: 10.1091/mbc.E11-04-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soini Y. Expression of claudins 1, 2, 3, 4, 5 and 7 in various types of tumours. Histopathology. 2005;46(5):551–560. doi: 10.1111/j.1365-2559.2005.02127.x. [DOI] [PubMed] [Google Scholar]
- 67.Chao YC, et al. Claudin-1 is a metastasis suppressor and correlates with clinical outcome in lung adenocarcinoma. Am J Respir Crit Care Med. 2009;179(2):123–133. doi: 10.1164/rccm.200803-456OC. [DOI] [PubMed] [Google Scholar]
- 68.Kwon MJ. Emerging roles of claudins in human cancer. Int J Mol Sci. 2013;14(9):18148–18180. doi: 10.3390/ijms140918148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh AB, Dhawan P. Claudins and cancer: Fall of the soldiers entrusted to protect the gate and keep the barrier intact. Semin Cell Dev Biol. 2015;42:58–65. doi: 10.1016/j.semcdb.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 70.Lee JW, et al. Increased expressions of claudin-1 and claudin-7 during the progression of cervical neoplasia. Gynecol Oncol. 2005;97(1):53–59. doi: 10.1016/j.ygyno.2004.11.058. [DOI] [PubMed] [Google Scholar]
- 71.Oshima T, et al. Down-regulation of claudin-18 is associated with the proliferative and invasive potential of gastric cancer at the invasive front. PLoS ONE. 2013;8(9):e74757. doi: 10.1371/journal.pone.0074757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanada Y, Oue N, Mitani Y, Yoshida K, Nakayama H, Yasui W. Down-regulation of the claudin-18 gene, identified through serial analysis of gene expression data analysis, in gastric cancer with an intestinal phenotype. J Pathol. 2006;208(5):633–642. doi: 10.1002/path.1922. [DOI] [PubMed] [Google Scholar]
- 73.Sahin U, et al. Claudin-18 splice variant 2 is a pan-cancer target suitable for therapeutic antibody development. Clin Cancer Res. 2008;14(23):7624–7634. doi: 10.1158/1078-0432.CCR-08-1547. [DOI] [PubMed] [Google Scholar]
- 74.Micke P, et al. Aberrantly activated claudin 6 and 18.2 as potential therapy targets in non-small-cell lung cancer. Int J Cancer. 2014;135(9):2206–2214. doi: 10.1002/ijc.28857. [DOI] [PubMed] [Google Scholar]
- 75.Shimobaba S, et al. Claudin-18 inhibits cell proliferation and motility mediated by inhibition of phosphorylation of PDK1 and Akt in human lung adenocarcinoma A549 cells. Biochim Biophys Acta. 2016;1863(6 Pt A):1170–1178. doi: 10.1016/j.bbamcr.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 76.Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347(6217):78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chapman HA, et al. Integrin α6β4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest. 2011;121(7):2855–2862. doi: 10.1172/JCI57673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arenkiel BR, et al. Activity-induced remodeling of olfactory bulb microcircuits revealed by monosynaptic tracing. PLoS ONE. 2011;6(12):e29423. doi: 10.1371/journal.pone.0029423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zheng T, et al. Inducible targeting of IL-13 to the adult lung causes matrix metalloproteinase- and cathepsin-dependent emphysema. J Clin Invest. 2000;106(9):1081–1093. doi: 10.1172/JCI10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scotton CJ, et al. Ex vivo micro-computed tomography analysis of bleomycin-induced lung fibrosis for preclinical drug evaluation. Eur Respir J. 2013;42(6):1633–1645. doi: 10.1183/09031936.00182412. [DOI] [PubMed] [Google Scholar]
- 81.Johnson KA. Imaging techniques for small animal imaging models of pulmonary disease: micro-CT. Toxicol Pathol. 2007;35(1):59–64. doi: 10.1080/01926230601184262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thiesse J, et al. Lung structure phenotype variation in inbred mouse strains revealed through in vivo micro-CT imaging. J Appl Physiol. 2010;109(6):1960–1968. doi: 10.1152/japplphysiol.01322.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen H, et al. Airway epithelial progenitors are region specific and show differential responses to bleomycin-induced lung injury. Stem Cells. 2012;30(9):1948–1960. doi: 10.1002/stem.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Demaio L, et al. Characterization of mouse alveolar epithelial cell monolayers. Am J Physiol Lung Cell Mol Physiol. 2009;296(6):L1051–L1058. doi: 10.1152/ajplung.00021.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Flodby P, et al. Cell-specific expression of aquaporin-5 (Aqp5) in alveolar epithelium is directed by GATA6/Sp1 via histone acetylation. Sci Rep. 2017;7(1):3473. doi: 10.1038/s41598-017-03152-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rieger ME, et al. p300/β-Catenin interactions regulate adult progenitor cell differentiation downstream of WNT5a/protein kinase C (PKC) J Biol Chem. 2016;291(12):6569–6582. doi: 10.1074/jbc.M115.706416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Darzynkiewicz Z, Bedner E, Smolewski P. Flow cytometry in analysis of cell cycle and apoptosis. Semin Hematol. 2001;38(2):179–193. doi: 10.1016/s0037-1963(01)90051-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.