Abstract
Integrated omics is becoming a new channel for investigating the complex molecular system in modern biological science and sets a foundation for systematic learning for precision medicine. The statistical/machine learning methods that have emerged in the past decade for integrated omics are not only innovative but also multidisciplinary with integrated knowledge in biology, medicine, statistics, machine learning, and artificial intelligence. Here, we review the nontrivial classes of learning methods from the statistical aspects and streamline these learning methods within the statistical learning framework. The intriguing findings from the review are that the methods used are generalizable to other disciplines with complex systematic structure, and the integrated omics is part of an integrated information science which has collated and integrated different types of information for inferences and decision making. We review the statistical learning methods of exploratory and supervised learning from 42 publications. We also discuss the strengths and limitations of the extended principal component analysis, cluster analysis, network analysis, and regression methods. Statistical techniques such as penalization for sparsity induction when there are fewer observations than the number of features and using Bayesian approach when there are prior knowledge to be integrated are also included in the commentary. For the completeness of the review, a table of currently available software and packages from 23 publications for omics are summarized in the appendix.
Keywords: Statistical learnings, integrated omics, exploratory learning, regression, network learning
Exploratory Methods
Dimension reduction
Dimension reduction is an important multivariate statistical approach; it is used to identify latent structure which is not observable but presented in the observations that are results of these structures. The number of dimensions or factors of the latent structure needs to be less than the number of variables, and the groupings of variables or weighted combinations of all variables are the statistical representations defined for the latent structure.
Principal component analysis (PCA) and factor analysis are 2 elementary statistical techniques for dimension reduction. In the literature for integrated omics, dimension reduction methods have presented several variations from PCA and factor analysis. These variations include multiple factor analysis (MFA),1 consensus PCA (CPCA), multiple-block PCA (MBPCA),2 and nonnegative matrix factorization (NMF).3
PCA and its variations
Hassani et al4 first introduced a CPCA method for multiple omics data sets, referred as “blocks,” and 3 validation tools in 2010. A block represents one type of omics measurement, and multiple blocks are collected from same biological samples. They used the genetic fingerprinting data and metabolite fingerprinting Fourier transform infrared spectra as an example which subdivides spectra into blocks of polysaccharide region, fingerprint region, protein region, and fatty acid region.
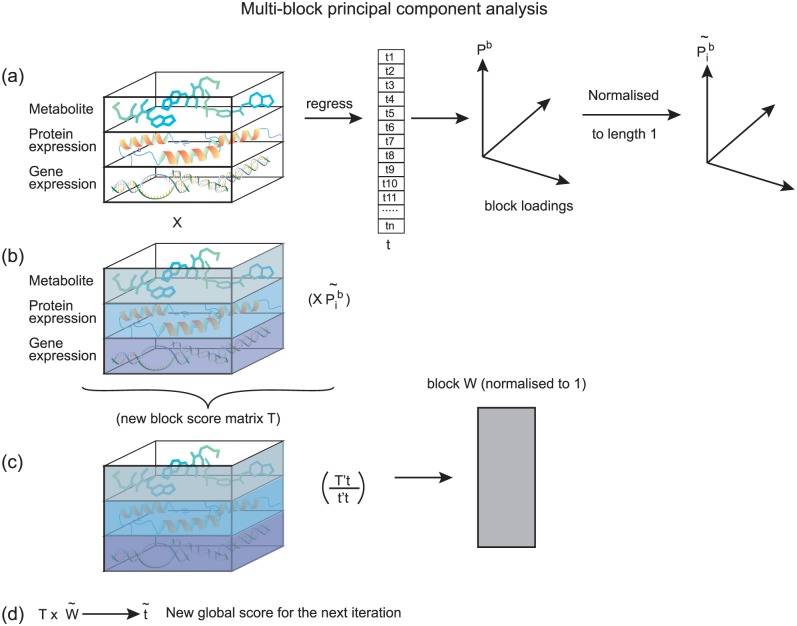
Consensus PCA uses an iterative algorithm (NIPALS [Nonlinear Iterative Partial Least Squares])5,6 to identify the latent bilinear structure from the combined measurement data. NIPALS can identify latent structure parameters including the block and global loading scores, block scores, and global scores iteratively (Figure 1). The authors described 3 methods for choosing components: root mean square error (RMSE), uncertainty t test, and stability plot. The RMSE is used to select the number of principal components through the RMSE plot. An uncertainty t test uses a t statistic estimated from loading coefficients to assess whether the measurement significantly contributes to the CPCA. The stability plot assesses any outlying observations. All 3 methods are used at both the block level and the global level.
Figure 1.
Multiblock principal component analysis (A, B, C). The multiblock principal component analysis starts from a random global score vector t (a randomly chosen starting scale for the principal component space). The blocks of data X (different omics measurements) are regressed via t and result in the principal loading Pb which represents the importance (weight) of each omics measurement variable contributing to the latent structure components. The loading Pb is normalized to , and a new block score is formed by multiplying with the data blocks X. The new block scores of vector ts are combined and become the block score matrix T. T is used to regress on the global score vector t resulting in weight vector w which is normalized to the length of 1. The new global score vector t for the next iteration is then calculated by multiplying the weight w and the new block score matrix T.
In 2013, the same group of authors2 compared 3 different deflation strategies for MBPCA. In iterative algorithms, such as NIPALS, latent components are extracted in a specific order. “Deflation” is the structure associated with each component subtracted off to reveal the next components; it corresponds to taking residuals in a regression. The choice of deflation strategy affects the interpretation of the structure by affecting which components of the estimated latent structure are forced to be orthogonal.
Conesa et al7 proposed a multiway approach to identify the underlying components that interconnect with different omics variables, with explicit modeling of 3-way latent structure. They use a dimension-reducing technique TUCKER3 for intra-omics analysis and the N-partial least squares (N-PLS) for inter-omics analysis. The different omics data sets comprise functional genomics measurements of transcriptomic, metabolomic, and physiological data sets. TUCKER3 is suggested to be an appealing data integration strategy because it can accommodate the structure of the data from a multifactorial design experiment (ie, time × treatment × protein expression), and N-PLS can infer the relationships between biomolecular measurements in multidimensional space.
Factor analysis and its variations
In contrast to PCA which projects the observations into the new latent structuralized space, factor analysis identifies latent structures that can be used to form (or explain) the observed data. Sanchez et al1 introduced MFA that can be used to reduce dimension and integrate supplementary information with the original omics data sets in a common space. Multiple factor analysis starts from a PCA on each block (type) of data and followed by jointly analyzing the singular-value normalized data using the global PCA. The normalized singular value represents the square root of the first eigenvalue. Sanchez et al1 suggested that using MFA is expected to avoid the problem and is suitable for different types of omics data sets.
NMF and others
As a dimension reduction technique, conventional NMF method decomposes the data matrix using a latent factor matrix W and a basic component data matrix.
Nonnegative matrix factorization is similar to PCA, but using nonnegative constraints instead of orthogonality constraints. Its solution is less uniquely defined but more interpretable for the nonnegative omics measurements, such as microRNA (miRNA) and gene expression. Yang and Michailidis3 introduced an integrated NMF (iNMF) algorithm to handle the heterogeneous multiple omics data sets and reduce the overall dimensions. The joint conventional NMF decomposes m multiple nonnegative data matrices by using the nonnegative common latent factor matrix W and m basic nonnegative component data matrix H, assuming that the m data sets have common latent structures. The “iNMF” adds the variation in the latent factor matrix W and uses a penalty term to control the variation for latent factor matrix across different data matrices. In contrast to the “orthogonality” constraints approach used in PCA, partial least squares (PLS), and canonical correlation analysis which maintains the center of mass, the “iNMF” uses constraint over “nonnegativity” for a better interpretation. Both approaches are to identify the best approximations for the original data sets. The iNMF has also been extended to cope with sparsity using a sparsity parameter in the penalty term. These methods were proposed for expression data sets with continuous measurements.
The other streamlines in dimension reduction include Serra et al8 who combined dimension reduction and cluster analysis to multiple genomic data sets. The algorithm involves prototype extraction and ranking which aims to reduce dimension by filtering variables using variance and rank the prototype based on their abilities to separate classes. Su et al9 proposed an integrated framework, which applied different dimensional reduction and feature extraction techniques, and used image-omics and functional omics data for the classification of breast cancer staging. They demonstrate an improvement of 3% in classifications using the integrated data compared with using the image-omics data only.
Clustering Methods
In integrated omics, clustering methods appear to be the commonly used approaches for subjects or features partitioning. They are useful tools to provide exploratory view of the underlying clusters pattern. The data set from multi-omics may have a complex data topology; new strategies are required to identify the partitioning structure of the integrated information. Apart from the conventional clustering approach using different distance measures, newly proposed methods use maximum likelihood method and some include penalized terms to control for complexity in feature selections. Among these studies, Newman and Cooper10 and Aibar et al11 introduced and modified the stochastic clustering method: self-organizing map (SOM) that has been used in cartography in geography. Shen et al12 and Kim13 used the latent variable approach with penalty terms to optimize the likelihood for cluster memberships. Sharma et al14 used iterative maximized likelihood method to cluster both categorical and continuous variables.
Iterative maximum likelihood–based approaches
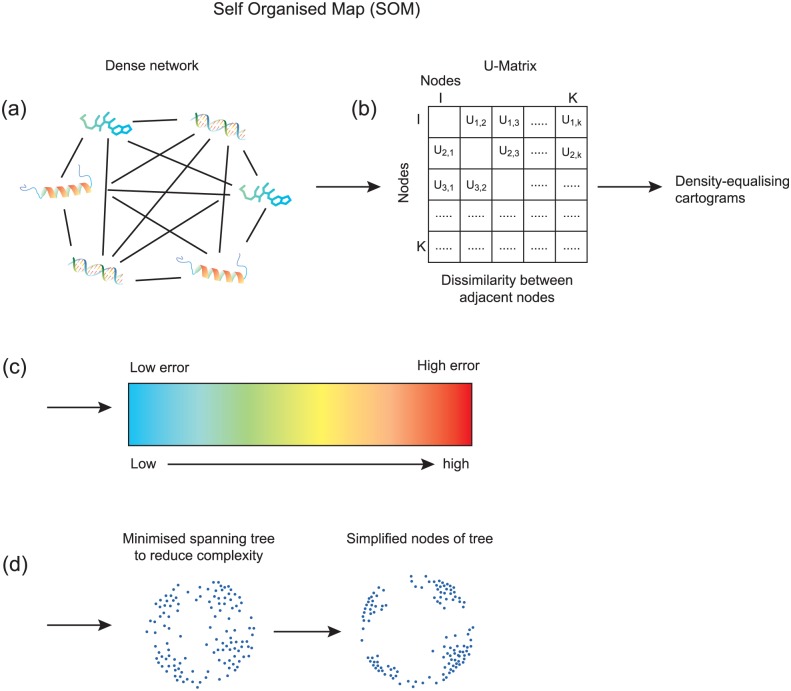
Newman and Cooper10 presented an unsupervised clustering technique which bases on the SOM (Figure 2), a stochastic clustering method to reduce the number of dimensions and preserve the local topology of gene expressions. Initial SOM measures the similarity of adjacent nodes and derives the dissimilarity surface (error matrix). The error matrix is used to identify borders of clusters and group similar data points and separate dissimilar data points iteratively. The AutoSOME method uses density equalization, which is a technique of cartography, to ensemble these graphical features output from SOM and to rescale the SOM output lattices. The density equalization treats nodes of high errors with high density and forces these nodes separating from each other; conversely, it treats nodes of low errors with low densities and aggregates them. A minimal spanning tree algorithm is then built from the rescaled nodes to identify the final clusters solution. Using the similar approach, Aibar et al11 applied the SOM in transcriptomics samples from 3 real data sets: myelodysplastic syndrome, Alzheimer disease, and colorectal cancer, to classify patients from different disease stages.
Figure 2.
AutoSOME (Automatic clustering using Self Organized Map) (A, B, C, D). Self-organizing map (SOM) is a stochastic clustering method to reduce the number of dimensions and preserve the local topology of gene expressions.
Sharma et al14 proposed a maximum likelihood–based clustering approach that can be applied to both categorical and continuous data. In system biology, this method can be applied to microarray expression and single-nucleotide polymorphism (SNP) data. It identifies the optimal solution that maximizes the likelihood for the n class clusters following the data topology. The iterative algorithm includes the following steps: initialize the cluster members, shift one sample from one cluster to another, and recalculate the total likelihood of m clusters based on the new mean and covariance matrices of each cluster. The proposed likelihood-based algorithm uses both the distance measures and variance components in the samples.
Regularization-based methods to control for complexity in feature selections
Regularization or penalty constraints are one common approach in statistical modeling for controlling complexity and achieve precision when the number of observations is far beyond the number of features or when the real associations between molecular features are known to be much smaller than all the possible associations.
Shen et al12 proposed a penalty-based clustering method (iCluster) to identify the number of clusters and membership of clusters for the integrated genetic and genomic features (copy number variation [CNV], DNA methylation, SNP). The main idea is to treat the latent variables of clusters as missing information and use expectation and maximization algorithm to estimate parameters of the penalized complete data likelihood. The penalty term induces sparsity in the weighting matrix for the latent variables and achieves simplicity of the clusters. Their paper introduces 3 types of penalty functions, namely, lasso, elastic net, and fused lasso to control the number of clusters.
Kim13 proposed group penalty method for group-structured and tight integrative clustering in which group lasso is presented as an updated version of iCluster.12 Under the penalized regression framework, the joint penalty complete log-likelihood was extended by adding a group lasso penalty term. Because it is possible that multiple feature modules share the same feature, for example, 2 miRNAs regulate the same gene. The group lasso regularization, which is based on multiple feature modules, contains overlapped features (ie, messenger RNA [mRNA], CNV, 2 methylations) and maintains the biological information in the model building.
Chi et al15 created a convex biclustering method to partition samples and features under a regulation penalty path. They use the distance-based measurements for clusters and iteratively shrink both the column (features) and samples (rows) simultaneously. The biclustering method is motivated by solving problems in the high-dimensional genome data and can be extended to use in the omics study for 2-dimensional partition problems.
Network Learning Methods
Network composing nodes and arcs provide an advanced tool to demonstrate the interactions between large numbers of variables (molecular features) in integrated omics. In network learning theories, variables are presented as nodes, causal relation or associations are presented as the arcs or edges between nodes. The graphical model and Bayesian network (BN) provide probabilistic estimates between nodes in these networks.16 The learning methods for causal and conditional-dependent networks can be used to investigate the multilayer associations and causal relations between omics features in integrated omics studies. When the causal relations are not the focus, matrix-based statistics are used to measure the associations between the linked data sets. The existing method for omics data sets includes canonical correlation and RV. Developments in matrix statistics for integrated omics blossomed in the past decade include the maximal first-order partial correlation coefficients (MF-PCcor) and adjusted RV.
Estimating associations between omics data sets
Kayano et al17 introduced ranking-based MF-PCcor to estimate the associations within the metabolite network and cope with outlying samples. The partial correlation coefficient bases on the normalized rank of the expression data and the maximal first-order partial correlation estimates the edges between metabolites.
Mayer et al presented an unbiased estimate of matrix correlation–adjusted RV coefficient.18 RV was originally used as a similarity index for 2 matrics; it is a generalization of the correlation from 2 variables to 2 data sets. It is the ratio between trace of cross-correlation matrices’ product and trace of squared correlation matrices’ product:
The adjusted RV, an unbiased estimate of the matrix correlation, replaces the squared correlation with the conventional adjusted R squares in linear regressions. The adjusted RV is applied to multiple system biology data sets for the identification of biologically meaningful subgroups and can be used as the input for clustering and multiscaling analysis.
Estimating structure of multilayer networks formed by integrated omics data sets
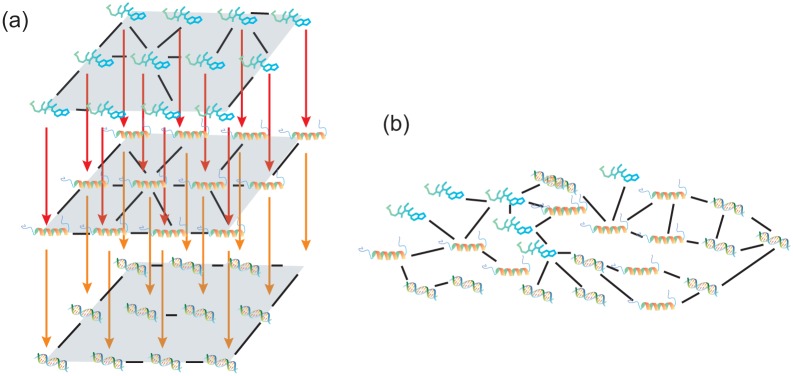
Angione et al19 introduced the multilayer network (multiplex) method for the integrated omics data. It is known that iCluster and similarity network fusion are not designed for the analysis of cross-omics data.19 iCluster does not scale all measurements and needs preselection of genes. Similarity network fusion only creates aggregated layers from genes. Angione proposes a method to model the linkage between genotypes and phenotypes. It constitutes multiplex networks of transcriptomics and fluxomics (a duplex) and fuses the 2 networks into one using a weighting network fusion approach. The proposed method uses a linear program to map the gene expression onto the metabolite model. Network with 2 layers is constructed with nodes representing environmental condition and edges representing similarity between nodes regarding gene or metabolite expression. The final derived single network is used to identify clusters of conditions with similarities. The weighted fusion approach of multiplex networks uses the weight to reflect the importance of gene or metabolite to the nodes (environmental conditions). Figure 3 provides the visualization map of the multiplex fusion algorithm.
Figure 3.
A multilayer network (A, B). Multiplex fusion algorithm.19
Mosca and Milanesi20 presented a network analysis method similar to Angione et al19 to integrate biological components and their interactions from multiple omics data sets. They propose to use molecular interactions and multiple objectives (MOs) for the simultaneous optimization, basing on statistical criteria at the network level and component level. Different statistical criteria are set for different objective functions in the MO optimizations. Of these criteria, hypervolume indicator, which presents the volume of the dominated portion (suboptimal points) of the objective space, is used as the quality measure of MO optimization process. The introduced algorithm integrates a weighted network from multiple omics data sets and optimizes the weighted networks.
Cun and Frohlich21,22 presented netClass algorithm of joining networks using smoothing approach. It uses smoothing method (kernel-based smoothing network diffusion) on the feature-wise marginal statistics over the structure of a joint protein-protein and miRNA-target gene interaction graph. Random walk kernel is used for smoothing and a permutation test is used to select features of each data set. The package provides an analytical tool to integrate miRNA and mRNA expression data, with protein-protein interactions and miRNA-target gene information.
Apart from developing new learning methods, some studies applied the existing methods into integrated omics. One typical study of these applications is the work by Peñagaricano et al23 who applied BN (R package bnlearn) to explore the causal networks underlying fat deposition and muscularity in pigs, using genotype, transcriptomic, and phenotype data sets. The study group introduces an integrated analysis using marginal associations between genotypic and phenotypic traits (genotype and phenotype data) via pQTL, marginal associations between genotypic and expression traits (genotype and transcript expression mRNA data) via eQTL, and identifies the colocalized joint significant eQTL and pQTL from the mapping analysis. They provide a summary of several methods to infer the causal genotype-phenotype network. One of the causal structure learning techniques is the inductive causation (IC) algorithm and its extended version, Incremental Association Markov Blanket (IAMB). The IC algorithm starts with determining conditional associations of a pair of variables (A and B) given all other variables, by searching any possible subsets of other variables as the dependency set. It follows by the second set of conditional independent tests including the adjacent variable C of A and B. The resultant partially directed graph is then filled with undirected edges as many as possible so long as that there is no new V structure and new directed edge formed. The extended version of the IC-IAMB algorithm includes a screening process to identify the Markov blanket of every variable X. The IAMB involves a set of conditional independent test for a pair of variable X and Y given subset W; it reduces the computation complexity without compromising accuracy. PARADIGM24 is another BN tool that is developed for the integrated omics expression data; it is a factor BN graph method that requires a differentiating state for each variable and their pathways.
There are study groups only providing tools for building a network and visualizing these networks. Appendix 1 includes a summary of these tools. One example is the BisoGenet,25 which is a network building tool assigning biological functional relations of protein and protein, protein and genes, based on a local in-house database “SysBiomics.” This server provides network building and visualization functions, given input entities nodes and edges.
Regression-Based Methods
In the integrated omics literature, the regression equations are set for explaining inter- or intrasystem relation and interactions. The strategies of parallel or sequential regressions are sometimes used with constraints. Parallel regressions are chosen to model causal relations between multiple molecular responses (ie, metabolites and genes) on continuous or categorical scale and their interacting effects as well as factors of interests, ie, pathway membership. Multivariate responses technique is not suitable due to the necessity of including interresponse relations in the explanatory factors of these models. One example of these interresponse relations is as follows: an active pathway membership of gene affects metabolites involved in the same pathway.
Parallel regressions
The parallel regressions are used in different omics responses to explain intersystem responses simultaneously. One example is the model proposed by Jauhiainen et al26 to integrate transcriptomic and metabolomic data to make an informed pathway-level decision. They proposed 2 linear models to describe responses of the gene and metabolite expression on pathway memberships. The fixed and random effect metabolite linear models include the pathway membership of gene presented by the regression coefficients from its parallel linear model; the mixed model includes random effects on the metabolite level. The random term allows the effects from unselected genes in the pathway being measured as these genes could post effects on the metabolite even if they are not selected at the gene level. The model selection occurs at 2 levels: firstly to select differentially expressed genes and subsequently which genes are allowed to influence the metabolite expression, and secondly, on the global pathway level to pick out the active pathways.
Poisson et al27 presented 2 joint tests for gene expression and metabolite information using 2 parallel logistics regressions. The gene expression and metabolite information are fitted in separate logistic regression, both of which predict the probability being in the interested gene or metabolite set S. The first test involves a 2-degree-of-freedom Wald test on the resultant regression coefficients. The second test is an enrichment test statistics using the sum of square statistics for gene and metabolite which are constructed as a 2-dimensional vector by permutation. A similar enrichment strategy was given by Pey et al28 who used an optimized pathway analysis model enriched by the classification based on upregulated or downregulated gene/protein expression. The optimization is divided into 3 stages to minimize the associations between flux and reactions in the classes. Results of gene expression measured by transcriptomics and protein data measured by proteomics are used to infer the forming of pathways.
Sequential regressions
Acharjee et al29 presented a sequential analytical approach starting from using the random forest to screen variables from individual “omic” data set, followed by further selection of the redundant variables via eQTL (quantity trait linkage). One advantage of the study is using the well-known regulatory genetic and metabolic pathways to validate the method. The method in the study is applied to transcriptomic (mRNA), proteomic (2D gel), and metabolomic (liquid chromatography-mass spectrometry and gas chromatography-mass spectrometry) data. First, the analysis starting with a random forest algorithm is implemented using R package randomForest, and a permutation test is proposed by the author to determine the metabolite/protein/RNA significance for predicting the trait. Second, the integrated linkage map is used and implemented via R package metanetwork. Finally, the final selected gene, protein, and metabolites including the trait are used to construct the network. The network’s nodes are formed by the aforementioned molecules and traits, and the edges are representing the strength of the interactions measured by regularized partial correlations.
Partial least squares
Partial least squares is a multivariate technique used to identify latent structures of both predictors and responses by maximizing the covariance between them. It is widely used in the integrated omics study. Since Wold introduced the NIPALS algorithm for PCA and PLS in chemometrics in the 1980s, NIPALS became the popular computer algorithm for PLS. Lê Cao et al30 proposed a sparse PLS (SPLS) using lasso penalization for integrated omics. Sparse PLS optimizes the square error terms with a penalty term of loading vectors of response matrix Y and predictor matrix X.
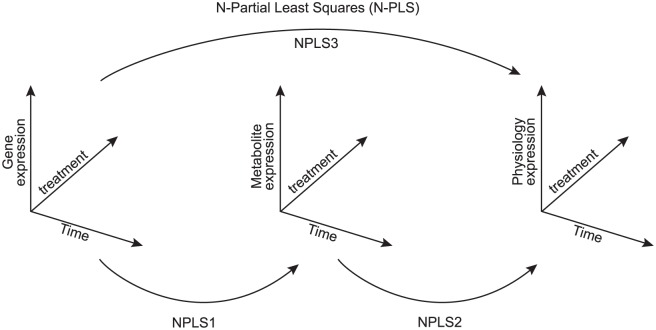
Fonville et al31 introduced the extended orthogonal signal correction (OSC) PLS (O-PLS) in 2010. It weights the predictor variables using the orthogonal components in the covariance matrix between the response (Y) and the predictor variables (X). The O-PLS filters out the “structure noise” basing on the covariance matrix for Y and X. It becomes one of the popular approaches in metabolomics due to its easy interpretation. A similar idea named N-PLS was also given by Conesa et al,7 N-PLS construct data array responses (Y) from multiple omics platforms and the predictor data blocks (X) curated from another type of omics platform in the multifactorial (N) spaces. It finds latent spaces that can maximize the covariance between X and Y and decomposing X from the improved version. The authors proposed a gene selection procedure using a gene-associated parameter p that reflects the contribution of each gene (Figure 4).
Figure 4.
N-partial least squares (N-PLS) construct data array with responses (Y) from different omics platforms7. The predictor data blocks (X) curated from another type of omics platform in the multifactorial (N) spaces.
Chen and Li32 presented 3 stochastic discrete dynamic equations to describe the relations among genes, proteins, miRNAs, and DNA methylations. These stochastic dynamic equations provide quantitative predictions of measurements of mRNA, miRNA, and protein expression at a specific time point. The quantitative measurements involve their expression levels at time t, interactions, respectively, for miRNA-mRNA, protein-protein, and the degradation of mRNA, as well as rate of miRNA-mRNA coupling. These stochastic equations describe the intermolecular relations included in protein-protein interaction, miRNA and gene regulatory network, and the measurement errors. In addition to the 3 stochastic equations, an extra equation for path gene protein is added to construct the integrated genetic and epigenetic cellular network. The regulatory and interactive parameters included in these 4 dynamic equations are evaluated using temporal data and solved by the constrained least square parameter estimation problem.
Another example is given by Pavel et al33 who integrated 3 types of molecular data: mutation, CNV, and gene expression via a fuzzy system score for each gene and sample. Biological rules are created based on the defined categories of these 3 molecular data sets. A fuzzy logic modeling is used to cluster and subtype discovery and to recover many known suppressor genes and oncogenes and subtypes in colorectal cancer cells.
Biological Knowledge Enrichment Learning
As defined in machine learning literature, supervised learning method uses response variable and training data or prior knowledge to provide a prediction for response variable. In statistical learning, the prior knowledge can either be used to set prior in the Bayesian model or inform the model selection. Bayesian modeling provides the essential framework to incorporate known information in analysis. It is called supervised learning in the context of prediction because the “true” value Y is part of the training data. In the context of estimating causal relationships between omics variables, however, the value of Y is not the goal of the analysis and these do not involve knowing the true causal relations.
Pavel et al33 gave examples of using the biological knowledge for forming biological rules in the cluster. Poisson et al27 introduced enrichment tests learned by biological knowledge to jointly evaluate gene expression and metabolite abundance. Nguyen and Hob34 proposed a semisupervised machine learning method to identify disease-related genes via the publicly available database. The method starts with identified disease-related proteins from the public databases which provide known biological information for the proteins to be analyzed. The included databases are UniProt, Gene Ontology, Pfam, InterDom, Reactome, and gene expression. The proteins of interests are divided into disease-related group or not related group according to the integrated information from these databases. After the division, data are extracted and preprocessed according to the feature functions, namely, the protein sequence length, keywords appeared in the database related to each protein, enzyme function, protein interaction with disease, protein pathways involvements, protein domain involvements, and domain-domain interaction (interDom) involvements. The final procedure uses the Gaussian random field and harmonic functions to learn a new set of the disease gene. Gomez-Cabrero et al35 used rank statistics to identify the most correlated gene markers of the comorbidities of the patients with chronic obstructive pulmonary disease via public data and then introduced the relative risk and correlations for binary variables to cluster the disease. Kamburov et al36 presented a Web-based tool IMPaLA for joint pathway analysis of transcriptomic, proteomic, and metabolomic data from multiple data sets. Joint P value is given for multiple data sets comprising metabolites and genes/proteins in the learning.
Meta-analysis
Meta-analysis is used for data integration on the summarized statistics level, and it is also used as a tool to combine analysis integrated from different studies on the level of individual observation which is called the mixed approach.1 In the latter application, meta-analysis is used to provide information to refine the analysis of a new study.37 The semisupervised method proposed by Nguyen and Ho34 is an example of the mixed approach which uses meta-analysis to provide prior biological information from multiple publicly available databases to update results in the later analysis. These meta-analyses use both statistical and biological inputs in the integration. Kim13 extended a meta-analytical framework for PCA. The aim of using the meta-analytical framework of PCA is to use multiple data sets to provide the common PCs (Principal components). Two methods are presented to summarize the common PCs: (1) decomposition of sum of variance decomposition and (2) minimization of sum of squared cosine (SSC) maximization. Sum of variance decomposition uses the weighted sum of covariance matrices from m data sets to find the common eigenvector matrix. Minimization of SSC maximization uses m eigenvectors derived from the multiple data sets (studies) to form an eigenvector matrix.
The publicly available software packages for meta-analysis include CNAmet,38 Rtopper,39 iClusterPlus,40 and the STATegra41 Bioconductor package.
Discussion
We sought to give an overview of the existing methods in integrated omics presented in the past decade. The following discussion provides insights in their implementations and limitations, in particular, for those variations extended from conventional methods. Table 1 summarizes the statistical distributions of different omics platform measurements for the discussion.
Table 1.
Similarities and differences across different platforms of omics.
| Transcriptomics Gene expression |
Transcriptomics Gene expression |
Proteomics Protein abundance |
Metabolomics Concentration of small molecules |
Common single-nucleotide polymorphism genotypes | MicroRNA expression | DNA methylation | |
|---|---|---|---|---|---|---|---|
| Technology | RNA sequencing | Microarray | Mass spec. | Mass spec. | Microarray | Microarray | Microarray |
| Statistical distributions used | Log-normal distributed/Poisson distributed | Normal or log-normal distributed | Log-normal–distributed peptide intensity to form hierarchical protein abundance | Log-normal–distributed peptide intensity to form hierarchical protein abundance | Binomial distributed | Log-normal distributed/Poisson distributed | Binomial distributed |
CPCA, MBPCA Versus MFA
Omics variables and study questions determine whether the analytical technique to be chosen is CPCA, MBPCA, or MFA. Intra- and interplatform variabilities also have their influences in the method selection.
Consensus PCA, operating on combined measurement, is for more uniformly curated data from the same or different platforms. Multiblock PCA can handle data with larger interplatform variability, potentially curated from complex experiments. Multiple factor analysis, an extension of factor analysis, is beneficial to studies when there is known biological knowledge to interpret latent common factors; it provides a technique for projecting supplementary variables (representing prior knowledge) on to the estimated factors. MFA has been used in research to investigate the insulin resistance when there are clinical data, DNA banding, and expression arrays that need to be integrated.1
Sanchez et al1 provided cross-validated estimates to determine number of relevant dimensions in CPCA, whereas MFA requires prespecified dimensions.
Cluster Analysis Methods
To compare these clustering methods, a published matrix42 for comparing cluster method is used as our reference. We condense the matrix to focus on the 5 statistical and implementation performances: (1) outliers detection, (2) providing number of clusters objectively, (3) providing uncertainty measures (ie, confidence interval), (4) handling mixed types of data variables, and (5) speed and memory use.
An enhanced version of the SOM, the AutoSOME cluster method, does not require prior knowledge of number of cluster and is less sensitive to outlying observations. Starting with a dissimilarity measure matrix in the SOM, the later adding processes include the density equalization algorithm and a graph-theory/minimum-spanning-tree algorithm to identify the objective number of clusters based on a threshold of P value. The limitation is that it can only be used in platforms producing continuous data, but with strength in its ability to handle both clean and noisy gene expression data and its stability in using the resampling method to derive the averaged cluster solution and confidence interval. Empirically, Newman and Cooper10 showed that applied transformation on the euclidean distance such as cubic operation achieves better separations and clusters using AutoSOME.
Comparatively, the regularization-based approach iCluster can be used for both categorical and continuous integrated omics data. Similar to the bicluster and group-regularized methods, iCluster allows faster estimation even in high-dimensional data sets.
Network Learning Methods
The proposed criteria used to compare these network learning methods in integrated omics are as follows: (1) purpose of the network learning, (2) handling complex network, (3) providing uncertainty measures, (4) speed and memory, and (5) providing prediction accuracy information.
Multiplex similarity networks are designed to handle complex networks with multiple layers; it provides a weighted similarity measures to account for the importance of each layer. The version at the time of this review does not provide prediction accuracy for model comparison and uncertainty measures.
The widely used BN is established with a longer history in other areas; it provides structure for modeling causal relations among variables. It has been used and developed in integrated omics recently; although it is not designed to handle multiple complex layer networks, it can be used for a limited number of mixed types of variables (such as phenotypes and expression measurements) using the hybrid Bayesian computing BN. It provides uncertainty measures for the marginal or conditional probabilities and uses information criteria such as BIC (Bayesian information criterion) and BDe (Bayesian Dirichlet equivalent uniform posterior probability) to assess goodness of fit in the structure.
Kernel-based smoothing approach in the netClass package uses a kernel-smoothed Support Vector Machine algorithm based on gene-wise t statistics to select the significant signatures from continuous expression data (miRNA and mRNA); it provides cross-validation to assess goodness of fit.
Among these methods, BN and its extended algorithm for omics data sets are designed for directed acyclic graphs: these require a known or hypothesis structure. Multiplex similarity networks are designed for multiple networks, and they can handle different types of variables (scales, counts, and binary variable) without requirement of a known structure.
Parallel Versus Sequential Regression Versus Multivariate PLS
Using a parallel or sequential approach to regression needs to be decided on the study purposes and complexities of the omics data sets. Parallel methods allow estimation of relations between different omics responses and their explanatory variables simultaneously. They are useful for pathway-level analysis, especially when data sets have different types of omics variables involved in the same pathway. A sequential approach is used to facilitate biological enrichment analysis following the feature reduction when each platform has large number of variables. The sequential approach allows selected gene, proteins, or metabolites to be included in the network construction at the final step.
Multivariate PLS is useful when the study requires extrapolating relations between multidimensional responses and explanatory variables because it takes account of the multiway structure of the data (eg, samples by platforms by time). Multivariate PLS variants include SPLS, O-PLS, and N-PLS that attempt to simplify the latent structure in different ways.
Both parallel and sequential regressions integrate the hierarchical structure of biological regulation in the models. The parallel approach requires a global fitness measure such as a pathway-level weighted combined for model selection. When there are a large number of omics variables from different platforms to be integrated, the sequential regression approach or a penalized PLS will be beneficial to cope with the large numbers of dimensions in regression, although the parallel regression approach can also use penalized approach for each model.
Bayesian versus non-Bayesian computation
Bayesian and frequentist approaches are not contradictory in integrated omics. In machine learning literature, supervised learning includes using Bayesian statistical approaches to integrating prior knowledge in the current observations. Sharma et al14 used the prior probability of cluster belongings in their iterative maximal likelihood algorithm for estimating the posterior probability of clusters membership. Although their computation method does not use the classic posterior samplings (ie, Markov chain Monte Carlo approach), they have employed the Newton-Raphson gradient ascending method to find the optimal estimates which have integrated the priors information. iCluster is another example of using Bayesian approach for identifying cluster membership, but using expectation-maximization (EM) algorithm in the computation, iCluster12 requires prior knowledge of the number of clusters. Multiple Dataset Integration43 uses the multinomial mixture model which requires prior knowledge of mixture probability and uses the Gibb samplings. PARADIGM24 and CONEXIC44 are 2 algorithms that use BN-based methods: the former uses EM algorithm in the computation of the unknown factor graph parameters and the latter is specifically designed for combining gene expression and copy numbers to construct a regression tree.
Bayesian method is preferred when the analysis requires integrating known knowledge (ie, pathway or network structure)45 but it requires larger computer memories and can be time-consuming to achieve better precisions in the estimation.
Closing Remarks
The presented methods for integrated omics are not only innovative but also diverse. The selections of analytical techniques are primarily determined by the research questions sought to answer. New methods are created for providing better strategies to integrate different omics measurements from different technology platforms that have both inter- and intraplatforms variabilities. Streamlining of these methods gives us a clear vision of how the statistical framework has been built to agree with other sciences. Future research requires more uniformed structure and methods in networks estimation and prediction for mixed types of measurements and more applications in precision medicines.
Acknowledgments
The authors would like to express their gratitude to Ms Vivian Ward who helped to visualize the analytical processes for the review.
Appendix
Appendix 1.
Computing and analytical software (packages) for omics data sets.
| Names | Language used | Analytical functions | Include visualization | Provide public databases | Omics techniques | Designed for integrative omics analysisa | Involved statistical models | Designed for human study | Web-based open source |
|---|---|---|---|---|---|---|---|---|---|
| OmicKriging1,b | R | It is designed for predicting complex traits (quantitative and qualitative) by leveraging and integrating similarity in genetic and large-scale omics | No | NA | miRNA, mRNA, T, SNP, and other large-scale omics | Yes (subject level) | Yes. It uses an algorithm to optimize the composited similarity matrix which integrates different omics correlation matrices | Yes | No |
| TranSMART + Galaxy + MINERVA: a combination pipeline2,b | NA | TranSMART repository provides integration of low dimensional clinical data and high-dimensional molecular data sets, with built-in data mining and analysis applications Galaxy provides analytical pipeline, and MINERVAR provides conceptualized and visualization for molecular interaction networks |
Yes | eTRIKS, www.etriks.org | GE, T, P, M (not specified) | Yes (subject level) | Yes. Galaxy uses the R Bioconductor packages limma | Yes | Yes. Galaxy is a Web server and cloud bench |
| OmicsAnalyzer3 | JAVA | As a plug-in for cystoscope, it has the functions of mapping different data sets, estimating associations, and visualizations | Yes | No | NA | Yes (molecular ID level) | Yes | Not specified | NA |
| VANTED4 | JAVA | VANTED is a framework providing essential functions for system biology. It has 7 tasks including data integration, visualization and data analysis for correlation, clustering, differential analysis, and enrichment analysis. It also computes some topological features for the network | Yes | Connect to network database: MetaCrop, KEGG, RIMAS | Not specified | Yes (molecular ID level) | Yes | Yes | No |
| The DNA Microarray Inter-omics Analysis Platform5 | R Bioconductor packages and custom Java solutions It includes packages for lipidomics (ie, mixOMICs/integomics) |
It provides data process function and focuses on the integration of these 2 types: 1. Lipidomics and transcriptomics; 2. miRNA and mRNA. The integration analysis includes miRNA and mRNA interaction site recognition. It also provides low-level GE analysis including background adjustment and normalization, differential analysis, data mining functions which include gene expression-phenotype relationships (clustering, multidimensional scaling [MDS], artificial neural network), and biological pathway analysis MDS includes sparse PLS, regularized canonical correlation coefficient |
Yes | Murine nutrigenomics data set; Normal Human Dermal Fibroblasts (NHDF) | GE, miRNA, T, L, miRNA-mRNA interaction | Yes (subject level) | Yes | Yes | Yes |
| Lemon-Tree6 | JAVA | It is a modular network software. It provides a function (ganesh) for model-based Gibbs sampler to infer coexpression modules and condition clusters within each modular It also provides regulator program that forms decision trees with nodes of the regulator at the expression level. Function tight-cluster will build clusters formed by consensus modules of genes |
Yes | TCGA glioblastoma expression and copy number data | T, miRNA, GE, CNA, eQTL, any others Gene expression and candidate regulator types |
Yes (subject level) | Yes | Yes | No |
| integrOmics7,b | R | It provides regularized canonical correlation analysis, sparse partial least squares regression | Yes | No | M, L, C | Yes (subject level) | Yes | Yes | No |
| Mayday SeaSight8 | JAVA with an built-in R terminal | Mayday has the daily used methods for array analysis. It includes cluster, differentiation analysis, and machine learning methods. It also has a terminal connection with R which facilitates usage of R functions | Yes | KEGG, MetaCyc | GE, T | SeaSight provides the integrative function for GE and next generation sequence data (at the experiment level) | Yes | Yes | Yes |
| DASS-GUI9 | C++ | It provides 2 modes: 1. Calculation mode: DASS.cs which identifies the closed set and DASS.pv which calculates the statistical significance of the derived closed sets 2. Analytical mode: pattern analysis includes pattern hierarchy, enrichment analysis, and module validation |
No | No | NA | No | Yes. It uses biclustering and other data mining method | Yes | No |
| GeneTrail210 | Optimized C++ library based on Boost, Eigen 3, and GMP | It provides differential expression tests at the identifier level and set level. It also provides multiple tests corrections. Its gene set and phenotype strategies use an optimal permutation method to reduce computing time | No | No | T, M, P, GE, miRNA | No | Yes | Yes | Yes |
| OmixAnalyzer11 | Java, R, Perl | It includes differential analysis (t test and ANOVA), cluster, and functional enrichment, provides figures and reports. It targets mid-sized systems biology project | Yes | No | GE, EX, P (on its way) | No | Yes | Yes | Yes |
| Specmine12 | R | Identification of metabolites, univariate (corr, regression, ANOVA), multivariate (robust PCA, cluster), machine learning, and feature selection (classification and regression, validation) | Yes | No | M, S | No | Yes | Yes | No |
| imDEV13 | R and Visual Basic | It provides functions to execute multivariate R functions from Excel. It includes MDS methods (Cluster, PCA, PLS) and 2/3 dimensional visualizations | Yes | No | M,C | No | Yes | Yes | No |
| XMRF14 | R | Fitting Markov networks to a wide range of high-throughput genomics data | Yes | No | GE | No | Yes | Not specified | No |
| PathVisioRPC15 | Allowed access from R, Perl, Python, Java, C, C++, PHP | A Remote Procedure Call for PathVisio, provides a link/communicating between the interface (PathVisio) and the statistical analytical tools (scripts). PathVisioRPC wraps PathVisio functionality into XMLRPC functions which can be implemented in many languages for execution. The R package of PathVisio is RPathVisio |
Yes, it is provided from PathVisio | No | GE, M, T | No | No. PathVisio provides pathway analysis and data visualization software. It provides Z score for the pathway overrepresentation analysis using the input statistics (ie, fold changes) | Yes | Yes |
| COBRApy16 | Python, MATLAB | It uses constrained modeling to represent the complex biological process of metabolism and gene expression in a pathway. Constrained-based modeling includes a biological system constraint which is defined by the objective function and usually linear programming is used as the analytical method | Yes | No | GE, M | No | No. It applied linear programming (machine learning) | Yes | No |
| 3Omics17 | Perl and PHP scripts and running on a Linux-based Apache Web server | Correlation networking, coexpression, phenotyping, pathway enrichment, and GO (Gene Ontology) enrichment | Yes | PubMed database, KEGG, Human Cyc, iHOP, DAVID, Entrez Gene, OMIM, and UniProt | T, P, M | Yes (molecular ID level) | No | Yes | Yes |
| PaintOmics18 | Perl & Python scripts running on an Apache Web server | A joint visualization tool for transcriptomics and metabolomics | Yes | KEGG | GE, M | Yes (subject level) | No | Yes | Yes |
| COEUS19 | Jena, Java | It is a data integration software, a new sematic Web framework | No | Unipro, OMIM | Not specified | No | No | Yes | Yes |
| Cytoscape20 | JAVA | A popular tool for biological network visualization and data integration | Yes | No | All data types for biological network | No | No | Yes | Yes |
| Plug-in for Pathway Tools | Providing an add-on function for the pathway tools, a plug-in API for its GUI: 1. Expanded CLUSTAL alignment and to compare orthologs of selected gene at DNA level 2. IS element annotation and analysis 3. Map probes to PGDB genomes by probe sequences and facilitate preprocessing of gene expression data before visualization |
No | Pathway/Genome Databases (PGDBs) | GE | No | No | Yes | No | |
| MGV (Mayday Graph Viewer)21 | JAVA, it is an extension of the platform Mayday | It provides visualizations for cluster comparison between studies, cross data sets biological pathway, gene models, and probe centric view | Yes, mainly for visualization | No | T, M, P, GE | No | No | Yes | No |
| Omix22 | OVL script | A customized visualization tool for metabolic network | Yes | KEGG | T, M, F | No | No | No | No |
| MVBioDataSim23 | R | It is a multiview genomic data simulator | No | No | GE | No | No | No | No |
| ATHENA24,b | Implemented in C++ and uses the libGE (version 0.206) and GAlib (version 2.4.7) genetic algorithm library | Grammatical evaluation neural network is used to analyze associations between single, multiple level genetic interactions and clinical outcomes. It includes (1) variable/feature selection, (2) model main and interactions effects predicting clinical outcomes, and (3) interpretation prepared for further bioinformatics | No | (TCGA) data portal ovarian cancer | CNA, GM, miRNA, GE, C | Yes (subject level) | Machine learning method: extension of artificial neural network | Yes | No |
Abbreviations: C, clinical data/outcomes; CNA, copy number alteration; EX, exon arrays; GE, gene expression (microarray); GM, gene methylation; L, lipidomics; M, metabolomics; NA, not available; P, proteomics; S, spectral data; T, transcriptomics.
Integration occurs at the molecular level: the input data are IDs of gene, protein, and metabolite and merged by these ID; results are derived using public databases (ie, pathway enrichment analysis via information of KEGG). Integration occurs at the subject level: the input data are an original expression or sequence variables from the same subject, data are merged by subject ID.
Software package that has functions to integrate clinical data and omics data and provides advanced statistical techniques for integrated data analysis.
References
- 1. Wheeler HE, Aquino-Michaels K, Gamazon ER, et al. Poly-omic prediction of complex traits: omicKriging. Genet Epidemiol. 2013. 38:402–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Satagopam V, Gu W, Eifes S, et al. Integration and visualization of translational medicine data for better understanding of human diseases. Big Data. 2016;4:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stoltmann T, Zimmermann K, Koschmieder A, Leser U. OmixAnalyzer: a web based system for management and analysis of high-throughput omics data sets. Lecture Notes Comp Sci. 2013;7970:46–53. [Google Scholar]
- 4. Rohn H, Junker A, Hartmann A, et al. VANTED V2: a framework for systems biology applications. BMC Syst Biol. 2012;6:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Waller T, Gubała T, Sarapata K, Piwowar M, Jurkowski W. DNA microarray integromics analysis platform. BioData Mining. 2015;8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bonnet E, Calzone L, Michoel T. Integrative multi-omics module network inference with lemon-tree. PLoS Comp Biol. 2015;11:e1003983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lê Cao KA, Ignacio González I, Déjean S. IntegrOmics: an R package to unravel relationships between two omics datasets. Bioinformatics. 2009;25:2855–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Battke F, Nieselt K. Mayday seaSight: combined analysis of deep sequencing and microarray data. PLoS ONE. 2011;6:e16345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hollunder J, Friedel M, Kuiper M, Wilhelm T. DASS-GUI: a user interface for identification and analysis of significant patterns in non-sequential data. Bioinformatics. 2010;26:987–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stöckel D, Kehl T, Trampert P, et al. Multi-omics enrichment analysis using the GeneTrail 2 web service. Bioinformatics. 2016;32:1502–1508. [DOI] [PubMed] [Google Scholar]
- 11. Xia T, Hemert JV, Dickerson JA. OmicsAnalyzer: a cytoscape plug-in suite for modeling omics data. Bioinformatics. 2010;26:2995–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. specmine [computer program]. Version 1.0: R; 2015 https://github.com/cran/specmine
- 13. Grapov D, Newman JW. imDEV: a graphical user interface to R multivariate analysis tools in Microsoft Excel. Bioinformatics. 2012;28:2288–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. XMRF [computer program]. Version 1.0: R; 2015 https://cran.r-project.org/web/packages/XMRF
- 15. Bohler A, Eijssen LMT, van Iersel MP, et al. Automatically visualise and analyse data on pathways using pathVisioRPC from any programming environment. BMC Bioinformatics. 2015;16:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ebrahim A, Lerman JA, Palsson BO, Hyduke DR. COBRApy: Constraints-based reconstruction and analysis for Python. BMC Syst Biol. 2013;7:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kuo TC, Tian TF, Tseng YJ. 3Omics: a web-based systems biology tool for analysis, integration and visualization of human transcriptomic, proteomic and metabolomic data. BMC Syst Biol. 2013;7:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. García-Alcalde F, García-López F, Dopazo J, Conesa A. Paintomics: a web based tool for the joint visualization of transcriptomics and metabolomics data. Bioinformatics. 2011;27:137–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lopes P, Luís Oliveira J. COEUS: “semantic web in a box” for biomedical applications. J Biomed Semant. 2012;3:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smoot M, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Symons S, Nieselt K. MGV: a generic graph viewer for comparative omics data. Bioinformatics. 2011;27:2248–2255. [DOI] [PubMed] [Google Scholar]
- 22. Droste P, Miebach S, Niedenführ S, Wiechert W, Nöh K. Visualizing multi-omics data in metabolic networks with the software Omix: a case study. BioSystems. 2011;105:154–161. [DOI] [PubMed] [Google Scholar]
- 23. Fratello M, Serra A, Fortino V, Raiconi G, Tagliaferri R, Greco D. A multi-view genomic data simulator. BMC Bioinformatics. 2015;16:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim D, Li R, Dudek SM, Ritchie MD. ATHENA: identifying interactions between different levels of genomic data associated with cancer clinical outcomes using grammatical evolution neural network. BioData Mining. 2013;6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: IZ conducted the review and writing of the first draft of the paper. TL provided insightful suggestions to the structure and edited the paper. Both reviewed and approved the final manuscript.
References
- 1. Sanchez A, Fernandez-Real J, Vegas E, et al. Multivariate methods for the integration and visualization of omics data. Paper presented at: Spanish Symposium on Bioinformatics; October 27-29, 2012; Torremolinos, Spain. [Google Scholar]
- 2. Hassani S, Hanafi M, Qannari E, Kohler A. Deflation strategies for multi-block principal component analysis revisited. Chemomet Intel Lab Syst. 2013;120:154–168. [Google Scholar]
- 3. Yang Z, Michailidis G. A non-negative matrix factorization method for detecting modules in heterogeneous omics multi-modal data. Bioinformatics. 2015;32:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hassani S, Martens H, Qannari E, Hanafi M, Borge G, Kohler A. Analysis of omics data: graphical interpretation- and validation tools in multi-block methods. Chemomet Intel Lab Syst. 2010;104:140–153. [Google Scholar]
- 5. Wold H. Estimation of principal components and related models by iterative least squares. In: Krishnaiah PR, ed. Multivariate Analysis. New York, NY: Academic Press; 1966:391–420. [Google Scholar]
- 6. Miyashita Y, Itozawa T, Katsumi H, Sasaki S. Comments on the NIPALS algorithm. J Chemometrics. 1990;4:97–100. [Google Scholar]
- 7. Conesa A, Prats-Montalbán J, Tarazona S, Nueda MJ, Ferrer A. A multiway approach to data integration in systems biology based on Tucker3 and N-PLS. Chemomet Intel Lab Syst. 2010;104:101–111. [Google Scholar]
- 8. Serra A, Fratello M, Fortino V, Raiconi G, Tagliaferri R, Greco D. MVDA: a multi-view genomic data integration methodology. BMC Bioinformatics. 2015;16:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Su H, Shen Y, Xing F, et al. Robust automatic breast cancer staging using a combination of functional genomics and image-omics. Paper presented at: 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); August 25-29, 2015; Milan, Italy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Newman A, Cooper J. AutoSOME: a clustering method for identifying gene expression modules without prior knowledge of cluster number. BMC Bioinformatics. 2010;11:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aibar S, Abaigar M, Campos-Laborie F, Sánchez-Santos J, Hernandez-Rivas J, Las Rivas J. Identification of expression patterns in the progression of disease stages by integration of transcriptomic data. Paper presented at: Statistical Methods for Omics Data Integration and Analysis; September 7-11, 2015; Valencia, Spain. [Google Scholar]
- 12. Shen R, Wang S, Mo Q. Sparse integrative clustering of multiple omics dataset. Ann Appl Statist. 2013;7:269–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim S. Statistical Learning Methods for Omics Data Integration in Dimension Supervised and Unsupervised Machine Learning. Pittsburgh, PA: The Department of Biostatistics, University of Pittsburgh; 2015. [Google Scholar]
- 14. Sharma A, Shigemizu D, Boroevich K, et al. Stepwise iterative maximum likelihood clustering approach. BMC Bioinformatics. 2016;17:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chi CE, Allen GI, Baraniuk RG. Convex biclustering. Biometrics. 2017;73:10–19. [DOI] [PubMed] [Google Scholar]
- 16. Alpaydin E. Introduction to Machine Learning. Cambridge, MA: The MIT Press; 2010. [Google Scholar]
- 17. Kayano M, Imoto S, Yamaguchi R, Miyan S. Multi-omics approach for estimating metabolic networks using low-order partial correlations. J Computat Biol. 2013;20:571–582. [DOI] [PubMed] [Google Scholar]
- 18. Mayer C, Lorent J, Horgan G. Exploratory analysis of multiple omics datasets using the adjusted RV coefficient. Stat Appl Genet Molec Biol. 2011;10:14. [DOI] [PubMed] [Google Scholar]
- 19. Angione C, Conway M, Lió P. Multiplex methods provide effective integration of multi-omic data in genome-scale models. BMC Bioinformatics. 2016;17:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mosca E, Milanesi L. Network-based analysis of omics with multi-objective optimization. Mol BioSyst. 2013;9:2971–2980. [DOI] [PubMed] [Google Scholar]
- 21. Cun Y, Frohlich H. netClass: an R-package for network based, integrative biomarker signature discovery. Bioinformatics. 2014;30:1326–1326. [DOI] [PubMed] [Google Scholar]
- 22. Cun Y, Frohlich H. Network and data integration for biomarker signature discovery via network smoothed T-statistics. PLoS ONE. 2013;8:e73074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peñagaricano F, Valente BD, Steibel JP, et al. Exploring causal networks underlying fat deposition and muscularity in pigs through the integration of phenotypic, genotypic and transcriptomic data. BMC Syst Biol. 2015;9:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vaske C, Benz S, Sanborn J, Earl D, Szeto C, Zhu J. Inference of specific pathway activities from multi-dimensional cancer genomics data using paradigm. Bioinformatics. 2010;26:i237–i245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martin A, Ochagavia M, Rabasa L, Miranda J, Fernandez-de-Cossio J, Bringas R. BisoGenet: a new tool for gene network building, visualization and analysis. BMC Syst Biol. 2015;11:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jauhiainen A, Nerman O, Michailidis G, Jornsten R. Transcriptional and metabolic data integration and modeling for identification of active pathways. Biostatistics. 2012;13:748–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Poisson LM, Taylor JM, Ghosh D. Integrative set enrichment testing for multiple omics platforms. BMC Bioinformatics. 2011;12:459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pey J, Valgepea K, Rubio A, Beasley JE, Planes FJ. Integrating gene and protein expression data with genome-scale metabolic networks to infer functional pathways. BMC Systems Biology. 2013;7:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Acharjee A, Bjorn Kloosterman B, Visser R, Maliepaard C. Integration of multi-omics data for prediction of phenotypic traits using random forest. Paper presented at: Statistical Methods for Omics Data Integration and Analysis; November 10-12, 2014; Heraklion, Greece. [Google Scholar]
- 30. Lê Cao KA, Rossouw D, Robert-Granié C, Besse P. A sparse PLS for variable selection when integrating omics data. Stat Appl Genet Molec Biol. 2008;7:35. [DOI] [PubMed] [Google Scholar]
- 31. Fonville JM, Richards SE, Barton RH, et al. The evolution of partial least squares models and related chemometric approaches in metabonomics and metabolic phenotyping. J Chemometrics. 2010;24:636–649. [Google Scholar]
- 32. Chen BS, Li CW. Constructing an integrated genetic and epigenetic cellular network for whole cellular mechanism using high-throughput next-generation sequencing data. BMC Syst Biol. 2016;10:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pavel AB, Sonkin D, Reddy A. Integrative modeling of multi-omics data to identify cancer drivers and infer patient-specific gene activity. BMC Syst Biol. 2016;10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nguyen TP, Hob TB. Detecting disease genes based on semi-supervised learning and protein-protein interaction networks. Artif Intell Med. 2012;54:63–71. [DOI] [PubMed] [Google Scholar]
- 35. Gomez-Cabrero D, Menche J, Vargas C, et al. From comorbidities of chronic obstructive pulmonary disease to identification of shared molecular mechanisms by data integration. Paper presented at: Statistical Methods for Omics Data Integration and Analysis; September 7-11, 2015; Valencia, Spain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kamburov A, Cavill R, Ebbels TR, Keun HC. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics. 2011;27:2917–2918. [DOI] [PubMed] [Google Scholar]
- 37. Kannan L, Ramos M, Re A, et al. Public data and open source tools for multi-assay genomic investigation of disease. Bioinformatics. 2016;17:603–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Louhimo R, Hautaniemi S. CNAmet: an R package for integration of copy number, expression and methylation data. Bioinformatics. 2011;27:887–888. [DOI] [PubMed] [Google Scholar]
- 39. Tyekucheva S, Marchionni L, Karchin R, Parmigiani G. Integrating diverse genomic data using gene sets. Genome Biol. 2011;12:R105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. iClusterPlus: integrative clustering of type genomic data [computer program]. R package version 1.12.1; 2016. https://bioconductor.org/packages/release/bioc/html/iClusterPlus.html
- 41. STATegRa: classes and methods for multi-omics data integration [computer program]. R package version 1.10.0; 2017. http://bioconductor.org/packages/release/bioc/html/STATegRa.html
- 42. Andreopoulos B, An A, Wang X, Schroeder M. A roadmap of clustering algorithms: finding a match for a biomedical application. Brief Bioinformatics. 2008;10:297–314. [DOI] [PubMed] [Google Scholar]
- 43. Kirk P, Griffin J, Savage R, Ghahramani Z, Wild D. Bayesian correlated clustering to integrate multiple datasets. Bioinformatics. 2012;28:3290–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Akavia UD, Litvin O, Kim J, Sanchez-Garcia F, Kotliar D, Causton HC. An integrated approach to uncover drivers of cancer. Cell. 2010;143:1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ahmad A, Fröhlich H. Integrating heterogeneous omics data via statistical inference and learning techniques. Genom Comput Biol. 2016;2:e32. [Google Scholar]