Abstract
The genome of the wild diploid strawberry species Fragaria vesca, an ideal model system of cultivated strawberry (Fragaria × ananassa, octoploid) and other Rosaceae family crops, was first published in 2011 and followed by a new assembly (Fvb). However, the annotation for Fvb mainly relied on ab initio predictions and included only predicted coding sequences, therefore an improved annotation is highly desirable. Here, a new annotation version named v2.0.a2 was created for the Fvb genome by a pipeline utilizing one PacBio library, 90 Illumina RNA-seq libraries, and 9 small RNA-seq libraries. Altogether, 18,641 genes (55.6% out of 33,538 genes) were augmented with information on the 5′ and/or 3′ UTRs, 13,168 (39.3%) protein-coding genes were modified or newly identified, and 7,370 genes were found to possess alternative isoforms. In addition, 1,938 long non-coding RNAs, 171 miRNAs, and 51,714 small RNA clusters were integrated into the annotation. This new annotation of F. vesca is substantially improved in both accuracy and integrity of gene predictions, beneficial to the gene functional studies in strawberry and to the comparative genomic analysis of other horticultural crops in Rosaceae family.
Keywords: genome annotation, PacBio-RNAseq, Illumina-RNAseq, strawberry, Fragaria vesca
1. Introduction
The diploid strawberry F. vesca, also known as Alpine or woodland strawberry, is the most widely distributed species naturally growing in the northern hemisphere. It also contributed to one set of the octoploid genome of the cultivated strawberry Fragaria × ananassa.1F. vesca genome (∼240 Mb) is relatively small and its genome sequence was published in 2011.2 Several attributes of F. vesca make it a good model species of fruit crops, such as short stature, short life cycle, recurrent flowering, and high efficiency of transformation. To facilitate the gene functional studies in strawberry, we have identified and profiled the expression of protein-coding genes, lncRNAs and miRNAs using extensive transcriptome datasets generated mainly from flower and fruit tissues at different developmental stages with >80 individual libraries in F. vesca.3–7 As an emerging model species, F. vesca is playing more and more important roles both in the fields of basic biological research and in horticultural and agricultural research.
The genome of F. vesca was first sequenced from a fourth-generation inbred line of Hawaii4 (F. vesca ssp. vesca) solely by second generation short read technologies, and its genome annotation v1.1 only contains protein-coding genes derived from ab initio gene predictions.2 Then, an updated annotation version (v1.1.a2) became available using 50 RNA-seq libraries generated from 25 different fruit tissue types to improve the annotation accuracy of protein-coding genes.8 In the meantime, a refined assembly of the F. vesca reference genome, named Fvb, was generated based on dense linkage maps of the North American diploid F. vesca ssp. bracteata.9 Compared with the original genome FvH4, Fvb features many translocations and inversions. Although Fvb still contains a large number of gaps that could be closed by utilizing long read sequencing technology, it has a much shorter unanchored pseudo-chromosome. The location of FvMYB10, a master regulator of anthocyanin production in strawberry, is moved from Chromosome 6 in FvH4 to Chromosome 1 in Fvb, where it is supposed to be according to genetic studies, illustrating that Fvb is of high quality.10 However, the gene annotation of Fvb (v2.0.a1) is not updated and merely obtained from the realignment of v1.1.
Previously, we analysed the transcriptomes of floral and fruit tissues in F. vesca.3,4 During data analysis and gene functional studies, we noticed that a considerable proportion of genes are mis-annotated. The RNA-seq datasets from F. vesca (90 individual libraries) provide a rich data resource to re-annotate the genome. In addition, the PacBio full-length transcripts obtained from the fruit receptacle in F. vesca are also available,7 which were proven to significantly increase the accuracy of genome annotation without any assembly.11,12
The annotation v2.0.a1 of Fvb contains only the coding regions from translation start site (ATG) to stop codon of protein-coding genes. However, recent studies highlight the importance of short ORFs (open reading frame) in 5′ UTRs in fine-tuning gene functions.13 The 3′ UTR sequences are also frequently needed for designing experiments. In addition, alternative splicing is an important regulatory mechanism at the post-transcriptional level, and up to 60% multi-exon protein-coding genes in plant genomes possess more than one isoform.7 Besides protein-coding genes, non-coding transcripts are pervasively expressed and constitute an integral part of the transcriptomes, including long non-coding RNAs (lncRNAs) and miRNAs.5,6 Collectively, an updated genome annotation for F. vesca that marries the Fvb assembly with the broader collection of accurately predicted genes, coding and noncoding, is timely and highly desirable.
Here, we optimized the genome annotation pipeline to improve the gene predictions in Fvb. We used a combination of MAKER2, AUGUSTUS, Program to Assemble Spliced Alignments (PASA), and manual curation to annotate protein-coding genes through integrating ab initio gene predictions, 90 RNA-seq transcriptome libraries and one PacBio full-length transcript library.14–17 The new annotation named V2.0.a2 not only updates the gene models of 13,168 loci, but also augments alternatively spliced isoforms of 7,370 genes, 5′ and 3′ UTRs of 18,641 genes, 1,938 lncRNAs, 171 miRNAs, and 51,714 small RNA clusters. Overall, V2.0.a2 provides a much more complete annotation of the F. vesca genome and will better serve gene functional studies in strawberry and other Rosaceae species.
2. Materials and methods
2.1. Transcriptome datasets used in this study
Both full-length transcripts generated by PacBio and RNAseq datasets from different F. vesca accessions were used as evidence for gene annotation. The PacBio reads were generated from pooled fruit receptacles at different developmental stages.7 A total of 90 Illumina RNA-seq libraries are respectively generated from fruits, flowers, meristems, and roots.3,4,18 In addition, nine small RNA-seq libraries generated from vegetative, flower, and fruit tissues were also used for small RNA identification in this study.5 Details of these datasets are described in Supplementary Table S1.
2.2. High-fidelity gene models identified from SMRT reads
The RS_IsoSeq pipeline (v2.3) was employed to analyse the PacBio data. First, the reads longer than 500 bp with the minimum full pass number of 2 and a quality score above 90 were retained. Then, the full-length reads having 5′ primer, 3′ primer, and a poly-A tail were further selected for downstream analysis. Next, these full-length reads were collapsed into consensus transcripts by ICE and Quiver in the isoseq_cluster panel. The LoRDEC software was used to correct the sequencing errors in the consensus transcripts using Illumina reads as the reference (parameters: -k 19 -s 3).19 The corrected consensus transcripts were then mapped by GMAP with >85% alignment coverage and >90% alignment identity.20 At last, the aligned transcripts were used for the identification of the best gene models using Program to Assemble Spliced Alignments (PASA).14 The scripts gff3_to_SNAP_train.pl bundled with SNAP, gmes_petap.pl bundled with GeneMark, and optimize_augustus.pl bundled with AUGUSTUS were used for training of each tool by the high-fidelity gene models with default parameters.
2.3. A comprehensive transcriptome built from RNA-seq datasets and SMRT reads using PASA
The RNA-seq reads with more that 90% of their bases having a quality score higher than 28 were retained using fastq_quality_filter (-q 28 -p 90), and the bases with low quality score in each RNA-seq read were trimmed off using fastx_trim built in FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/ (5 September 2017, date last accessed)). Each library was mapped individually to Fvb9 downloaded from GDR (https://www.rosaceae.org (5 September 2017, date last accessed)) using Tophat221 with the following parameters: maximum intron size, 10,000bp; minimum intron size, 20 bp; fr-firststrand for strand-specific libraries.
Reference-based assembly: For flower and fruit tissues, the aligned BAM files of the two biological replicates were, respectively, merged and sorted using Samtools (version 1.2)22 due to relatively lower sequencing depth, resulting in a total of 37 BAM files. The analyses on other libraries were carried out individually. Next, transcripts were assembled individually for each library by Stringtie23 with default settings except that the minimum isoform fraction was set to 0.3 to remove weakly expressed isoforms.
De novo assembly: All the RNA-seq reads were pooled together for de novo assembly using Trinity24 with default parameters.
To build a comprehensive transcriptome, the ID accessions of full-length and de novo transcripts were first fed into PASA, and full-length, genome-guided and de novo transcripts generated in parallel were then collapsed, mapped back to Fvb, and reconstructed by PASA through its alignment assembly module. The comprehensive transcriptome was later used as the mRNA evidence for MAKER2 described in the following section.
2.4. MAKER2 annotation
MAKER2 was used for generating an initial gene annotation.15 First, RepeatModeler (http://www.repeatmasker.org/ (5 September 2017, date last accessed)) was employed to identify and classify repeats from the genome Fvb, and then RepeatMasker (Repbase version 20160829) was used to generate a masked genome. Plant protein sequences were downloaded from UniProt (http://www.uniprot.org/ (5 September 2017, date last accessed)) on 21 January 2017. MAKER2 was run on the masked Fvb with following evidence: Fvb annotation v2.0.a1, the comprehensive transcriptome, UniProt proteins, AUGUSTUS trained models, SNAP trained models, and GeneMark trained models.
2.5. Further improve the genome using Augustus and EVM
To use Augustus,25 the following evidence were provided: (i) intron hints converted from uniquely mapped RNA-seq reads using the script bet_to_gff.pl bundled with GeneMark; (ii) intron hints converted from PacBio full-length transcripts by GMAP (http://bioinf.uni-greifswald.de/bioinf/wiki/pmwiki.php?n=Augustus.PacBioGMAP (5 September 2017, date last accessed)); (iii) Protein hints generated from mapping UniPro proteins to masked genome by Exonerate;26 (iv) repeat hints from the RepeatMasker output (http://bioinf.uni-greifswald.de/bioinf/wiki/pmwiki.php?n=Augustus.IncorporateRepeats (5 September 2017, date last accessed)). These hints were fed into Augustus to generate the high confidence gene models.
Next, EVidenceModeler (EVM)27 was used to combine MAKER2 gene models, Augustus gene models, transcripts from SMRT and Illumina RNA-seq, and UniProt proteins with a nonstochastic weighted value into confident consensus gene models. The weight value for each part is: six for MAKER2 gene models, eight for Augustus gene models, 10 for SMRT transcripts, eight for Illumina transcripts, and three for homologous proteins. Finally, PASA was used to improve the EVM gene models by modifying gene structures and adding UTR annotations and alternatively spliced isoforms.
2.6. Manual curation of new annotation
All the gene models were inspected one by one and manually curated using a plug-in called Apollo28 in JBrowse Genome Viewer29 based on the mapped reads from different RNA-seq libraries.
2.7. Alternative splicing analysis
AStalavista30 was employed to classify the alternative splicing events in the v2.0.a2 annotation. It groups the events into four major types, namely IR, ES, AA, and AD. The event belonging to each type were extracted from the AStalavista output by following codes: IR (AS code: 1^2-,0), ES (AS code: 1- 2^, 0), AA (AS code: 1-,2-), and AD (AS code: 1^,2^).
2.8. Prediction of lncRNAs
Two approaches were used to identify lncRNAs. The transcripts combined from the assemblies of Illumina RNA-seq and PacBio full-length reads were used for screening lncRNAs. The mono-exonic transcripts were removed to reduce false positives due to transcriptional noise and ambiguous alignment. In the first approach, the transcripts with class_code ‘u’ (unknown intergenic transcript), ‘o’ (generic exonic overlap with a reference transcript), ‘x’ (natural antisense transcript, NAT), and ‘i’ (intronic transcript) were selected. Then, the transcripts with length > 200nt and ORF < 300nt were retained. Next, the left transcripts were blasted against the Swiss-Protein database by BLASTX (2.6.0)31 with E-values < 1e-3 to remove potential protein-coding transcripts. At last, we used Coding Potential Calculator32 to calculate the CPC score of each transcript, and only those with a CPC score < 0 were considered as lncRNA candidates. In the second approach, lncRNAs were identified from the same set of transcripts by FEELnc33 using the specificity threshold 0.97. The transcripts overlapping protein-coding genes in v2.0.a2 and < 200 nt in length were eliminated. FEELnc used the ORF coverage (i.e. length of the longest ORF/length of the lncRNA transcript), k-mer frequency, nucleotide frequency, and codon usage to distinguish between mRNAs and lncRNAs. Finally, the consensus transcripts identified by both methods were designated as lncRNAs. These lncRNAs were grouped into two major types (genic and intergenic) and six subtypes based on the localization and the direction of transcription relative to the proximal protein-coding genes by the FEELnc classifier module called FEELncclassifier.
2.9. Alignment of small RNAs
The raw reads from 9 small RNA-seq libraries generated from tissues of F. vesca5 were used for the identification of small RNAs. First, the 3′ adapters were trimmed off using Cutadapt.34 Then, the resulting .fasta file was mapped against the Fvb genome using bowtie (version 0.12.8)35 with following parameters: -q -v 0 -p 30 -S -a -m 50. ShortStack36 was subsequently employed to identify small RNA clusters, and only the dominant 20- to 24- nt in length and DCL-derived small RNA loci (more possibly related to RNAi) called out by ShortStack were retained. The expression level of each small RNA cluster was normalized using the count mode in ShortStack with default settings.
2.10. Alignment of miRNAs
The stem-loop sequences of 171 miRNAs in F. vesca predicted by a previous report5 were mapped to the new F. vesca genome assembly Fvb by bowtie2 with default parameters, then the SAM file was converted into the GFF format by an in-house python script and integrated into the v2.0.a2 annotation file.
2.11. Experimental verification of new gene models
Total RNA was isolated from the fruit receptacle of YW5AF7 (the 7th generation inbred line of Yellow Wonder, an accession of F. vesca) using the Plant Total RNA Isolation Kit (Sangon Biotech, Shanghai, China, No. SK8631) following manufacturer’s instructions. cDNA was synthesized from 1 μg total RNA in 20 μl solution using the PrimeScriptTM RT reagent kit (TaKaRa, Shiga, Japan, Cat# RR047A). KOD DNA polymerase (TOYOBO Bio-Tech, Cat# F0934K) was used to amplify the coding regions of selected genes for Sanger sequencing with primers listed in Supplementary Table S5. The sequencing results were aligned with the sequences obtained from new annotations by Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/ (5 September 2017, date last accessed)).
3. Results and discussion
3.1. Update of gene models of protein-coding genes in Fvb based on SMRT and Illumina RNA-seq datasets
In this study, a new Fvb annotation named version 2.0.a2 (v2.0.a2) was created that includes protein-coding genes, alternatively spliced isoforms of multiexon genes, small RNAs, and lncRNAs. An optimized workflow to annotate protein-coding genes and their isoforms is shown in Figure 1. To take advantage of the extensive RNA-seq datasets, MAKER2, specifically designed for second-generation genome projects, was first used to generate an initial protein-coding gene annotation.15 Input data for MAKER2 include (i) repeats masked genome Fvb, (ii) ab initio gene predictors, including SNAP, GeneMaker, and Augustus, each trained with high-fidelity gene models, (iii) plant protein sequences downloaded from UniProt databases, (iv) transcripts assembled from a total of 90 Illumina RNA-seq libraries, (v) full length transcripts obtained from one PacBio transcriptome library (Supplementary Table S1). The Illumina RNA-Seq datasets were generated from a series of tissue types in F. vesca, including leaf, seedling, root, shoot apical meristem, flower meristem, anther and carpel from flowers at different developmental stages and different fruit tissues at five stages with an average of 30 million and a range from 12 million to 89 million reads per library (see details in Supplementary Table S1). In total, there are 2.5 billion RNA-Seq short reads. The PacBio data was generated from fruit receptacles pooled from small green to mature stages in F. vesca, containing 82,360 full-length consensus transcripts derived from 442,601ReadofInserts.
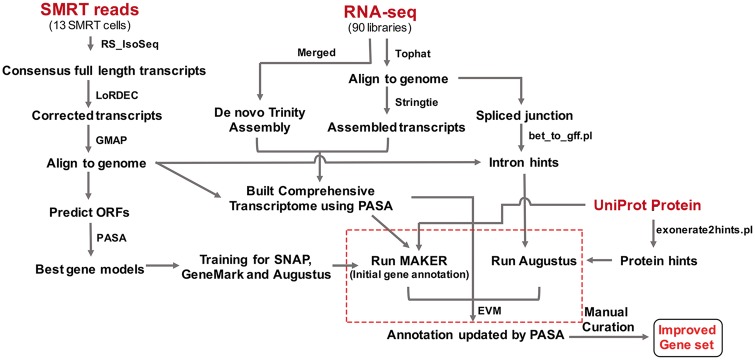
Figure 1.
Annotation workflow for strawberry protein-coding genes. The SMRT transcripts were used for the training of SNAP, GeneMark and Augustus. RNA-Seq libraries were used to build comprehensive transcriptomes by a combination of de novo and genome-guided assemblies. UniProt protein sequences were also provided for accurate gene annotation. The red dotted box indicates the core tools used for the annotation. In the end, manual curation was carried out to assure the accuracy.
The ambiguity of transcript reconstruction will affect the accuracy of gene annotation when using MAKER2. To avoid assembly errors in gene prediction, it is advantageous to use the transcript information in unassembled mapped reads.37 Augustus is a gene finder that can incorporate data from RNA-seq reads (junctions reads) directly into weighted predictions, join exons into a single gene and identify alternative transcription events.25 Therefore, Augustus was employed to further improve gene predictions through incorporating the intron hints from RNA-seq reads and protein hints from UniProt databases. Then, the gene structures predicted by both MAKER2 and Augustus were combined into consensus gene models using EVidenceModeler (EVM),27 and the gene models generated from EVM were updated by PASA.14 At last, the new annotation was inspected across the entire genome in IGV38 to evaluate the prediction and select the optimal gene models by comparing with the aligned RNA-seq reads, and about 1,000 (3%) genes were manually curated in this way.
Annotation v2.0.a2 of the genome Fvb contains a final set of 33,538 protein-coding loci with 50,738 transcripts (Table 1 and Supplementary Table S2). The Locus IDs for 30,012 identical or modified genes stay the same between the older annotation v2.0a1 and this annotation (v2.0.a2), while 3,525 genes (novel or split) were newly numbered using the same nomenclature as previous annotations (i.e. GeneXXXXX). The Locus IDs in both versions for the newly modified genes are listed in Supplementary Table S3. One improvement of v2.0.a2 is that a total of 18,641 genes were augmented by inclusion of 5′ UTRs and/or 3′ UTRs, representing nearly 55.6% of all protein-coding genes (Fig. 2a and b). Moreover, the gene models of 13,168 protein-coding genes were modified and updated (Fig. 2a and c). More specifically, 2,692 genes were derived from splitting longer genes (previously fused genes), and 213 genes were obtained by fusing neighbouring genes (Fig. 2a, d, and e). In addition, 2,031 genes were removed, and 631 genes were added (Fig. 2a). To further evaluate the quality of the annotation, the BUSCO genes39 were compared between v2.0.a1 and v2.0.a2. The results show a significant increase of complete BUSCOs from 88.9% for v2.0.a1 to 95.7% for v2.0.a2, while the fragmented and missing BUSCOs were reduced in v2.0.a2 (Table 1), indicating a higher quality annotation. These updates will better serve gene structure and functional studies.
Table 1.
Summary of the v2.0.a2 annotation
| Type | V2.0.a1 | V2.0.a2 |
|---|---|---|
| Protein-coding genes | ||
| Number of genes | 33,673 | 33,538 |
| Mean length of genomic loci | 2,863 | 2,856 |
| Mean exon number | 5.1 | 4.62 |
| Mean CDS length | 1,188 | 1,123 |
| Mean length of introns | 408 | 320 |
| Genes with 5′ UTR | – | 17,616 |
| Genes with 3′ UTR | – | 17,971 |
| Genes with both 5′ and 3′ UTR | – | 16,946 |
| Mean 5′ UTR length (bp) | – | 320 |
| Mean 3′ UTR length (bp) | – | 513 |
| Number of genes with isoforms | – | 7,370 |
| Mean isoform number per gene | 1.00 | 1.51 |
| Genes with functional annotations | 27,875 | 28,798 |
| Genes with GO terms | 17,156 | 22,106 |
| Complete BUSCOs | 88.9% | 95.7% |
| Fragmented BUSCOs | 5.6% | 1.9% |
| Missing BUSCOs | 5.5% | 2.4% |
| Non-coding genes | ||
| microRNAs (miRNAs) | – | 171 |
| Small RNA clusters | – | 51,714 |
| Long non-coding RNAs (lncRNAs) | – | 1,938 |
| Total loci | 33,673 | 87,361 |
Figure 2.
Illustration of the modifications to protein-coding genes in v2.0.a2. (a) Summary of the major types of modifications. The examples of four types are shown in the subfigures b to e. ‘Number of genes’ indicates the number of genes with each type of modifications in v2.0.a2. (b) 5′ and 3′ UTRs were added for gene20640. (c) Gene12679 has different and fewer exons than the v2.0.a1 equivalent. (d) Gene20642 in v2.0.a1 was split to create two loci, gene36495 and gene36496. (e) Gene12833 and gene12832 in v2.0.a1 were fused to create a single locus, gene12832. Exons are in blue, untranslated regions (UTRs) are in dark red, and introns are indicated by the thin black lines. The modified regions were highlighted by a dotted rectangle.
3.2. Functional annotation of protein-coding genes
To update the functional annotation of novel and pre-existing protein-coding genes in v2.0.a2, all the protein sequences were blasted against known proteins in the InterPro databases using InterProScan.40 In addition, each gene was also blasted against the NCBI non-redundant (nr) protein database to gain the Gene ontology (GO) term by Blast2go.41 As a result, a total of 28,798 and 22,106 genes in v2.0.a2 gained functional annotations and GO terms, respectively (Table 1). Especially, there is a great increase in the percentage of genes with GO terms, from 51% in v2.0.a1 to 66% in v2.0.a2 (Table 1). For novel genes, 527 of the 631 loci (83.5%) were assigned with functional annotations.
Transcription factors and regulators and protein kinases usually play crucial roles during signal transduction. In the iTAK pipeline42 that we used for the classification of these genes, transcription factors are defined as proteins that regulate the expression of target genes by binding to specific cis-elements in promoter regions, while transcriptional regulators operate indirectly via interaction with the basal transcription apparatus (e.g. transcription factors) or by altering the accessibility of DNA to TFs via chromatin remodelling to regulate the expression of target genes. As a result, we identified 1,542 transcription factors from 68 families, and 390 transcriptional regulators in the v2.0.a2 annotation by using the iTAK pipeline (Supplementary Table S4).42 The total number of transcription factors is comparable to that of v2.0.a1, but a few gene families have gained relatively more members, such as the B3 family from 67 to 78, MYB from 112 to 118, and bHLH from 90 to 101 (Supplementary Table S4). There are 1,055 protein kinase encoding genes in v2.0.a2, which are 90 more in the older annotation (Supplementary Table S4).
Disease resistance is an important agricultural trait for strawberry as well as other fruit crops, thus we also used the RGAugury pipeline43 to analyse R-genes and pattern-recognition receptors (PRRs), collectively called resistance gene analogs (RGAs), in the two annotation versions of Fvb. The RGAs are classified into four major families based on conserved domains and motifs in this pipeline, namely NBS encoding, membrane associated RLP (receptor-like protein) and RLK (receptor-like protein kinase), and TM-CC (transmembrane-coiled-coil).43 The first three families were further grouped into a few subfamilies. A total of 1,190 RGAs were identified, including 340 NBS encoding genes, 122 RLPs, 590 RLKs, and 138 TM-CCs in v2.0.a2 (Supplementary Table S5). Although the gene number in the NBS encoding family is comparable between the two annotations, the TNL subfamily has much more members (from 17 to 79), while the TX subfamily has fewer members (from 124 to 59) (Supplementary Table S5). The size of the RLP and RLK families doesn’t change much. However, the gene number in the TM-CC family is significantly decreased, from 192 genes in v2.0.a1 to 138 genes in v2.0.a2.
With the advantage of strand-specific RNA-seq reads, we were able to distinguish the sense and antisense transcripts from one locus. One example of such a locus having overlapping transcripts derived from opposite strands is gene02467, which codes for a tRNA ligase. This gene is located in one intron of an extremely long gene called gene02466 that encodes the ATP-binding cassette transporter A1 (Fig. 3a). By contrast, the same locus was previously annotated to have four genes in v2.0.a1 (Fig. 3a). FvSOC1, the homolog of AtSOC1 (SUPPRESSOR OF THE OVEREXPRESSION OF CONSTANS), is a key negative regulator of flowering and a positive regulator of runnering in F. vesca.44FvSOC1 does not exist in v2.0.a1. However, the modified gene19424 with a brand new gene model in v2.0.a2 has exactly the same sequence as the EST of FvSOC1 (accession number in NCBI: JF806634) (Fig. 3b). In addition, some gene models were fine-tuned in v2.0.a2 to have a higher annotation quality. For instance, FvTFL1, another crucial floral repressor in F.vesca,45,46 has a new translation start site producing a shorter protein, which becomes consistent with the published sequence in NCBI (accession number: JF806631) (Fig. 3c). A few other mis-annotated genes encoding glutathione S-transferases (GSTs), the red/far-red photoreceptor phytochrome B, and a bHLH transcription factor were PCR amplified, cloned and sequenced to confirm the v2.0.a2 annotation (Supplementary Fig. S1 and Table S6). Taken together, not only were a large proportion of pre-existing genes corrected, but also novel genes with potentially important roles were discovered and incorporated into the new annotation.
Figure 3.
Examples of three loci with improved annotations. (a) IGV view of the RNA-seq reads for the overlapping genes (gene02466 and gene02467) with opposite transcription directions. (b) The gene model of gene19424 (FvSOC1) becomes dramatically different in new annotation. (c) The gene model of gene30276 (FvTFL1) is modified in new annotation. Grey peaks indicate the read coverage. Pink bars indicate reverse strand reads. Light blue bars indicate forward strand reads. Blue bars indicate exons in v2.0.a1. Orange bars indicate exons in the reverse strand in v2.0.a2. Green bars indicate exons in the forward strand in v2.0.a2.
3.3. Alternatively spliced genes in v2.0.a2
Alternative splicing (AS) is an important regulatory mechanism at post-transcriptional level and is prevalent in animals as well as plants.47,48 However, the v2.0.a1 annotation does not provide any information about the isoforms alternatively spliced from the pre-mRNAs. The v2.0.a2 annotation takes into account and provides the specific information on the alternatively spliced transcripts predicted by our analysis pipeline (Fig. 1). To increase the prediction accuracy, weakly expressed transcripts in the RNA-seq datasets were removed during the assembly using Stringtie.23 A total of 16,705 alternative transcripts were detected genome-wide from 7,370 multiexon genes, accounting for 21.98% of the 33,538 protein-coding genes. The average transcript number per gene is 1.51. As we used a more stringent filtering strategy, much fewer isoforms were presented in v2.0.a2 than that of our previous study (47,000 isoforms from 13,591 alternatively spliced genes, accounting for 41.6% of all the protein coding genes).7
Among the AS genes, about half of them (46.85%) possess only two isoforms, 23.19% genes possess three isoforms, and 1.28% genes have >10 isoforms (Fig. 4a). 7,370 AS genes showed enriched GO terms in all kinds of metabolic processes (Supplementary Table S7), consistent with our previous studies.7,49 To better characterize alternative splicing (AS) in the new annotation, AStalavista was used to group the AS events into a few major types.30 3,434 events are of alternative 3′ splice sites (A3), 1,351 events are of alternative 5′ splice sites (A5), 4,245 events are due to intron retention (IR), and 1,029 events result from exon skipping (ES) (Fig. 4b). Consistent with other studies,7,49 the percentage of IR (32.39%) is the greatest, while the percentage of ES (7.85%) is the least.
Figure 4.
Features of the alternative splicing events in v2.0.a2. (a) Pie chart showing the percentage of genes with different number of isoforms among the 7,370 alternatively spliced genes. Different colour indicates genes with different number of isoforms per gene. (b) Summary of the different types of alternative splicing events that generate alternative transcripts in v2.0.a2.
3.4. Non-coding RNAs
Non-coding RNAs are the integral part of the genome. To further improve the annotation of F.vesca genome, the loci producing small RNAs and lncRNAs were both integrated into the new annotation v2.0.a2.
3.4.1. Small RNAs
Thus far, most of the small RNAs with known functions are 21–24nt long, such as small interfering RNAs (siRNAs), miRNAs, phasiRNAs, and tasiRNAs. These small RNAs regulate the expression of target genes through transcriptional gene silencing (TGS) or post-transcriptional gene silencing (PTGS). Among them, 171 miRNAs, including 44 novel miRNAs and 127 conserved miRNAs, have been previously identified in F. vesca,5 but their locations in the genome are lacking. In this study, the stem-loop precursor sequences of these miRNAs were all uniquely aligned to Fvb, and the coordinates for the stem-loop and mature miRNA sequences were integrated into v2.0.a2 (Supplementary Table S8).
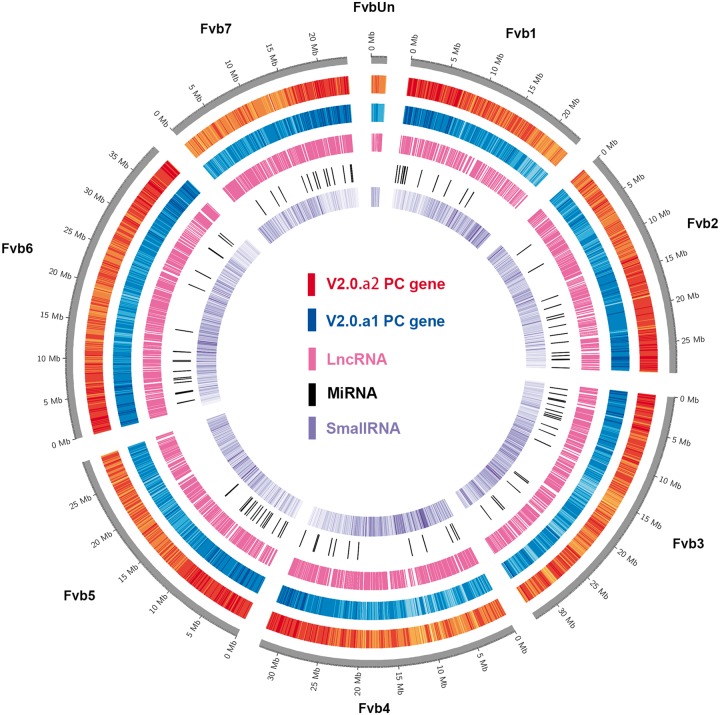
Besides, siRNAs of 24nt are most widespread in the genome.6 Here, we analyzed a total of 9 small RNA-seq libraries generated from open flower, 4 DPA (days post anthesis)-seed, 10 DPA-seed, 4 DPA-carpel wall, 10 DPA-carpel wall, flower bud, young leaf, young seedling, and 10 DPA-fruit receptacle in F. vesca from a previous study.5 ShortStack,36 a popular pipeline used for small RNA annotation in Arabidopsis and other species,49–51 was employed to identify small RNA clusters with 20–24nt small RNAs in Fvb. Finally, 51,714 small RNA clusters were identified with 24nt small RNAs as the most abundant type, accounting for 98.46% out of all small RNA clusters (Supplementary Table S9). Furthermore, 15,705 (30.4%) and 1,544 (2.99%) of these clusters overlap with protein-coding genes and lncRNAs, respectively. When the gene density of small RNA clusters was plotted across the seven chromosomes, it roughly negatively correlated with that of protein-coding genes in both v2.0.a1 and v2.0.a2 (Fig. 5).
Figure 5.
Circular representation of genome-wide distribution of the genomic features. Tracks from outer to inner circles indicate: protein-coding (PC) gene models in v2.0.a2, v2.0.a1, lncRNA, miRNA, and smallRNA loci, respectively. Colour density represents gene density. Darker colour indicates a higher density.
3.4.2. LncRNAs
LncRNAs are defined as non-coding RNAs with a length >200 bp. To annotate lncRNAs in F. vesca, we used two independent methods to identify them and only retained the consensus ones (see Methods). Overall, we identified a total of 1,938 lncRNA loci with more than one exon in the F. vesca genome, corresponding to a total of 4,042 transcripts (2.1 isoforms per locus on average). The exon number distribution of lncRNAs showed that lncRNAs have fewer exons than protein coding genes. 53% of lncRNAs possess two or three exons, while only 32% of the protein-coding genes possess two or three exons (Fig. 6a). The gene length of lncRNAs varies from 201 to 12,722 bp, and the mean length is 1,479 bp. In this pipeline, the lncRNAs were first classified into two major groups, intergenic and genic, based on the location relative to protein-coding genes (Supplementary Fig. S2). Intergenic lncRNAs can be further classified into three subgroups: same_strand, located in the same strand with the neighbouring protein-coding gene; convergent, convergent transcription with the neighbouring protein-coding gene in different strands; divergent, divergent transcription with the neighbour protein-coding gene in different strands (Supplementary Fig. S2). Genic lncRNAs can also be classified into three subgroups: overlapping, lncRNA is longer at one end and shorter at the other end than the overlapped protein-coding gene; containing, lncRNA is longer than the overlapped protein-coding gene at both ends; nested, lncRNA is shorter than the overlapped protein-coding gene at both ends.33 Among the 4,042 lncRNA transcripts, the intergenic type contains 1,413 same_strand, 873 convergent, and 786 divergent lncRNA transcripts; the genic type contains 689 overlapping, 74 containing, and 207 nested transcripts (Fig. 6b;Supplementary Fig. S2 and Table S10).
Figure 6.

Identification and characterization of long noncoding RNAs in Fvb. Number of exons harboured by lncRNAs and protein-coding genes. (b) Classification of lncRNAs based on the location and transcription direction relative to adjacent protein-coding genes. The lncRNAs were grouped into two types: genic and intergenic. Each type was further grouped into three subtypes. The number above each bar indicates the number of genes in each subtype. The diagrams of each subtype are shown in Supplementary Fig. S2.
4. Conclusions
The F. vesca genome was first released with annotations of merely protein coding genes that were mainly derived from ab initio predictions.2 Then, an updated genome of high quality called Fvb was re-assembled based on genetic linkage mapping with the same annotation.9 RNA-seq datasets generated from strawberry fruits have been previously utilized for F. vesca genome reannotation,8 however, it was based on limited number and type of tissue samples, short Illumina reads, and was again restricted to protein-coding genes. With the recent advancement in genome sequencing, especially the availability of third generation long read sequencing data, and accumulation of large number of RNA-seq datasets from diverse tissues, creating a new and improved re-annotation for the most recent genome assembly is timely. Here, we developed an optimized pipeline to re-annotate the strawberry Fvb genome taking advantage of PacBio full-length transcripts and an extensive dataset of RNA-seq libraries. This new annotation, v2.0.a2, updated the F. vesca gene models and includes new information ranging from 5′ and 3′ UTRs to alternatively spliced transcripts. In addition, non-coding genes, including small RNAs and lncRNAs, were also integrated into this new annotation. To conclude, our new annotation is much more complete than the older annotation and will provide a valuable resource for the strawberry and the Rosaceae research community as well as for comparative and functional studies in flowering plants.
5. Data availability
Supplementary Table S2 is the gtf file of the new Fvb annotation v2.0.a2, which is also available in GDR (https://www.rosaceae.org/ (5 September 2017, date last accessed)) and SGR (http://bioinformatics.towson.edu/strawberry/ (5 September 2017, date last accessed)).
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Guogui Ning for the help on establishing the analysis pipeline. This work was supported by the National Natural Science Foundation of China (31572098 to C.K.) and the Scientific and Technological Self-innovation Foundation of Huazhong Agricultural University (2014RC005 to Z.L. and 2014RC017 to C.K.).
Author contributions
Conceived and designed the experiments: Y.L. and C.K. Analysed the data: Y.L., W.W., and M.P. Performed the experiments: J.F. and H.L. Wrote the paper: C.K., Y.L., and Z.L.
Supplementary data
Supplementary data are available at DNARES online.
Conflict of interest
None declared.
References
- 1. Liston A., Cronn R., Ashman T. L.. 2014, Fragaria: a genus with deep historical roots and ripe for evolutionary and ecological insights. Am. J. Bot., 101, 1686–99. [DOI] [PubMed] [Google Scholar]
- 2. Shulaev V., Sargent D. J., Crowhurst R. N., et al. 2011, The genome of woodland strawberry (Fragaria vesca). Nat. Genet., 43, 109–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kang C., Darwish O., Geretz A., Shahan R., Alkharouf N., Liu Z.. 2013, Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. Plant Cell, 25, 1960–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hollender C. A., Kang C., Darwish O., et al. 2014, Floral transcriptomes in woodland strawberry uncover developing receptacle and anther gene networks. Plant Physiol., 165, 1062–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xia R., Ye S., Liu Z., Meyers B., Liu Z.. 2015, Novel and recently evolved miRNA clusters regulate expansive F-box gene networks through phasiRNAs in wild diploid strawberry. Plant Physiol., 169, 594–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kang C., Liu Z.. 2015, Global identification and analysis of long non-coding RNAs in diploid strawberry Fragaria vesca during flower and fruit development. BMC Genomics, 16, 815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Y., Dai C., Hu C., Liu Z., Kang C.. 2017, Global identification of alternative splicing via comparative analysis of SMRT- and Illumina-based RNA-seq in strawberry. Plant J., 90, 164–76. [DOI] [PubMed] [Google Scholar]
- 8. Darwish O., Shahan R., Liu Z., Slovin J. P., Alkharouf N. W.. 2015, Re-annotation of the woodland strawberry (Fragaria vesca) genome. BMC Genomics, 16, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tennessen J. A., Govindarajulu R., Ashman T. L., Liston A.. 2014, Evolutionary origins and dynamics of octoploid strawberry subgenomes revealed by dense targeted capture linkage maps. Genome Biol. Evol., 6, 3295–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hawkins C., Caruana J., Schiksnis E., Liu Z.. 2016, Genome-scale DNA variant analysis and functional validation of a SNP underlying yellow fruit color in wild strawberry. Sci. Rep., 6, 29017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Minoche A. E., Dohm J. C., Schneider J., et al. 2015, Exploiting single-molecule transcript sequencing for eukaryotic gene prediction. Genome Biol., 16, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dong L., Liu H., Zhang J., et al. 2015, Single-molecule real-time transcript sequencing facilitates common wheat genome annotation and grain transcriptome research. BMC Genomics, 16, 1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu G., Yuan M., Ai C., et al. 2017, uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature, 545, 491–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haas B. J., Delcher A. L., Mount S. M., et al. 2003, Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res., 31, 5654–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Holt C., Yandell M.. 2011, MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics, 12, 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stanke M., Diekhans M., Baertsch R., Haussler D.. 2008, Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics, 24, 637–44. [DOI] [PubMed] [Google Scholar]
- 17. Stanke M., Waack S.. 2003, Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics, 19, Ii215–25. [DOI] [PubMed] [Google Scholar]
- 18. Toljamo A., Blande D., Karenlampi S., Kokko H.. 2016, Reprogramming of strawberry (Fragaria vesca) root transcriptome in response to phytophthora cactorum. PLoS One, 11, e0161078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Salmela L., Rivals E.. 2014, LoRDEC: accurate and efficient long read error correction. Bioinformatics, 30, 3506–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu T. D., Watanabe C. K.. 2005, GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics, 21, 1859–75. [DOI] [PubMed] [Google Scholar]
- 21. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L.. 2013, TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol., 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li H., Handsaker B., Wysoker A., et al. 2009, The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25, 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pertea M., Pertea G. M., Antonescu C. M., Chang T. C., Mendell J. T., Salzberg S. L.. 2015, StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol., 33, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grabherr M. G., Haas B. J., Yassour M., et al. 2011, Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol., 29, 644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stanke M., Schoffmann O., Morgenstern B., Waack S.. 2006, Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics, 7, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Slater G. S., Birney E.. 2005, Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics, 6, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haas B. J., Salzberg S. L., Zhu W., et al. 2008, Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol., 9, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee E., Helt G. A., Reese J. T., et al. 2013, Web Apollo: a web-based genomic annotation editing platform. Genome Biol., 14, R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stein L. D., Mungall C., Shu S. Q., et al. 2002, The generic genome browser: a building block for a model organism system database. Genome Res., 12, 1599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Foissac S., Sammeth M.. 2007, ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res., 35, W297–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J.. 1990, Basic local alignment search tool. J. Mol. Biol., 215, 403–10. [DOI] [PubMed] [Google Scholar]
- 32. Kong L., Zhang Y., Ye Z.-Q., et al. 2007, CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res., 35, W345–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wucher V., Legeai F., Hedan B., et al. 2017, FEELnc: a tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res., 45, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martin M. 2011, Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J., 17, 10–2. [Google Scholar]
- 35. Langmead B. 2010, Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics, doi: 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Axtell M. J. 2013, ShortStack: comprehensive annotation and quantification of small RNA genes. RNA, 19, 740–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Steijger T., Abril J. F., Engström P. G., et al. 2013, Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods, 10, 1177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robinson J. T., Thorvaldsdottir H., Winckler W., et al. 2011, Integrative genomics viewer. Nat. Biotechnol., 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Simao F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M.. 2015, BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics, 31, 3210–2. [DOI] [PubMed] [Google Scholar]
- 40. Jones P., Binns D., Chang H.-Y., et al. 2014, InterProScan 5: genome-scale protein function classification. Bioinformatics, 30, 1236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Conesa A., Götz S., García-Gómez J. M., Terol J., Talón M., Robles M.. 2005, Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21, 3674–6. [DOI] [PubMed] [Google Scholar]
- 42. Zheng Y., Jiao C., Sun H., et al. 2016, iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol Plant, 9, 1667–70. [DOI] [PubMed] [Google Scholar]
- 43. Li P., Quan X., Jia G., Xiao J., Cloutier S., You F. M.. 2016, RGAugury: a pipeline for genome-wide prediction of resistance gene analogs (RGAs) in plants. BMC Genomics, 17, 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mouhu K., Kurokura T., Koskela E. A., Albert V. A., Elomaa P., Hytönen T.. 2013, The Fragaria vesca homolog of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 represses flowering and promotes vegetative growth. Plant Cell, 25, 3296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Iwata H., Gaston A., Remay A., et al. 2012, The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant J., 69, 116–25. [DOI] [PubMed] [Google Scholar]
- 46. Koskela E. A., Mouhu K., Albani M. C., et al. 2012, Mutation in TERMINAL FLOWER1 reverses the photoperiodic requirement for flowering in the wild strawberry Fragaria vesca. Plant Physiol., 159, 1043–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee Y., Rio D. C.. 2015, Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem., 84, 291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reddy A. S., Marquez Y., Kalyna M., Barta A.. 2013, Complexity of the alternative splicing landscape in plants. Plant Cell, 25, 3657–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cheng C. Y., Krishnakumar V., Chan A. P., Thibaud-Nissen F., Schobel S., Town C. D.. 2017, Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J., 89, 789–804. [DOI] [PubMed] [Google Scholar]
- 50. Jex A. R., Nejsum P., Schwarz E. M., et al. 2014, Genome and transcriptome of the porcine whipworm Trichuris suis. Nat. Genet., 46, 701–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Coruh C., Cho S. H., Shahid S., Liu Q., Wierzbicki A., Axtell M. J.. 2015, Comprehensive annotation of Physcomitrella patens small RNA loci reveals that the heterochromatic short interfering RNA pathway is largely conserved in land plants. Plant Cell, 27, 2148–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.