Abstract
Extracellular RNAs are gaining clinical interest as biofluid-based noninvasive markers for diseases, especially cancer. In particular, derivatives of transfer RNA (tRNA) are emerging as a new class of small-noncoding RNAs with high biomarker potential. We and others previously reported alterations in serum levels of specific tRNA halves in disease states including cancer. Here, we explored seminal fluid for tRNA halves as potential markers of prostate cancer. We found that 5′ tRNA halves are abundant in seminal fluid and are elevated in prostate cancer relative to noncancer patients. Importantly, most of these tRNA halves are also detectable in prostatic tissues, and a subset were increased in malignant relative to adjacent normal tissue. These findings emphasize the potential of 5′ tRNA halves as noninvasive markers for prostate cancer screening and diagnosis and provide leads for future work to elucidate a putative role of the 5′ tRNA halves in carcinogenesis.
Keywords: Extracellular 5′ tRNA halves, prostate cancer biomarkers, seminal fluid
Introduction
Since their discovery in bodily fluids, extracellular RNAs, primarily microRNAs (miRNAs), are increasingly recognized as significant players in cell to cell communication.1–3 More importantly, as they can be detected with noninvasive methods, extracellular RNAs are gaining clinical interest as promising markers of physiologic and pathologic states.4–8
Recently, derivatives of small-noncoding RNAs (sncRNAs) have emerged as a new class of molecules with important functions and biomarker applications.9–12 One such novel molecule type, transfer RNA (tRNA) halves, has been detected in cell lines, tissues, serum, plasma, and other bodily fluids, in both normal and pathologic conditions.13–16 The tRNAs are cleaved in the anticodon loop by angiogenin to produce 5′- and 3′-tRNA halves (30-35 nucleotides) or in the D-stem or T-stem by Dicer or RNase Z to produce shorter fragments (13-20 nucleotides) called tRNA-derived RNA fragments (tRFs).10,17–23 The 5′ tRNA halves were first observed in stressed cultured cells19,24,25 where they associate with the translational repressor YB-1 and suppress protein synthesis to redirect cellular energy toward repairing the stress-induced damage.25–27 The tRNA halves also control other important cellular functions, including regulation of the small interfering RNA (siRNA) pathway by direct binding to Dicer-2.28 In support of non–stress-related functions, tRNA halves were detected under nonstress conditions in human cells19,29 and plants30; also, stresses such as amino acid and glucose starvation and UV irradiation failed to induce tRNA half accumulation.19,20,25 Furthermore, we have shown that 5′ tRNA halves are highly expressed in hematopoietic and lymphoid tissues of the mouse and in human leukocytes in the absence of stress,15 suggesting a role in hematopoietic and immune processes.31 The shorter tRFs also have a wide functional scope.32 They regulate gene expression by acting in a miRNA-like fashion,33–35 inhibit translation,36 and act as tumor suppressors through a mechanism that destabilizes pro-oncogenic transcripts.37
Using high-throughput small RNA sequencing in a series of studies, we have shown that 5′ tRNA halves circulate stably in serum and plasma.14–16 Later studies reported tRNA-derived small RNAs in human, rat, and monkey serum samples38,39 and in extracellular vesicles from human semen.40 We also presented evidence that associates changes in the abundance of circulating 5′ tRNA halves with physiologic states (ie, aging, calorie restriction, and dwarfism6,15), and disease states (ie, cancer14,16). Changes in the circulating levels of specific 5′ tRNA halves correlated with breast tumor characteristics, including estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) status.14 Such a correlation suggests a link between carcinogenesis and specific 5′ tRNA halves; in support of this concept, tRNA-derived sncRNAs were shown to influence cell proliferation and metastasis by regulating translation and messenger RNA (mRNA) stability.18,37,41
Taken together, the observations presented above link extracellular tRNA halves to pathophysiological states, including cancer, and highlight the potential utility of this small RNA species as a novel noninvasive marker of disease. To gain further evidence for this concept, we mined publicly available small RNA-Seq data sets generated from seminal fluid and prostate tumor tissues to explore 5′ tRNA halves as potential markers of prostate cancer.
Methods
Bioinformatics analysis of a small RNA-Seq data set generated from seminal fluid of men with and without prostate cancer
Fastq files of raw sequencing reads were downloaded from Gene Expression Omnibus (accession GSE5668642). The data set was generated by sequencing of small RNA extracted from 2 seminal fluid RNA pools, representing groups of 6 men with prostate cancer or 6 men without cancer. The RNA population sequenced in this data set was extracted from the nonsperm fraction of seminal fluid, which includes prostatic epithelial, urothelial, and inflammatory cells. The downloaded sequencing reads were trimmed by removing adaptor sequences with FASTX-Toolkit (hannonlab.cshl.edu) and aligned to the GRCh38/hg38 human genome with Bowtie v1.1.2.43 The alignment files were analyzed with Bedtools v2.25.044 to annotate and count reads mapped to noncoding RNA genes from Ensembl GRCh38 release 86, tRNA genes from Genomic tRNA Database,45,46 and YRNA from the UCSC GENCODE v24 track.47 The obtained read counts were compared with find differences in the abundance of tRNA halves between noncancer and cancer seminal fluid samples. Read counts were normalized by equalizing the total mapped reads between cancer and noncancer samples before comparison and calculation of fold change. However, the statistical significance of these findings is limited using pooled samples instead of biological replicates.
Bioinformatics and statistical analysis of a small RNA-Seq data set generated from prostrate primary tumor and matched normal tissues
Fastq files of raw sequencing reads were downloaded from the Gene Expression Omnibus accession GSE89193.48 This small RNA-Seq data set was generated from formalin-fixed paraffin-embedded (FFPE) tissue blocks isolated from prostatectomies performed on men diagnosed with prostate cancer. The study included 2 sequencing batches with read-length 37 and 50 nucleotides. In our analysis, we used only the batch with 50-nucleotide read-length consisting of 25 patients; tRNA halves are 30- to 34-nucleotide long and thus would not be found in the batch with 37 nucleotides read after adapter removal.
Sequencing reads from 25 tumor-normal pairs were aligned to the GRCh38/hg38 human genome using the same parameters used for the analysis of the small RNA-Seq from the seminal fluid samples. Aligned sequencing reads were pooled into 2 groups: normal and tumor. Pooled samples were analyzed to examine the length distribution of the reads, the pattern and size of peaks, and the types and proportions of small RNAs from which the reads originate; pooling is used only to assess the characteristics of the reads and not to measure the differential expression of small RNAs between control and cancer groups. The sequencing reads were processed and aligned to the GRCh38/hg38 human genome as described above for the seminal fluid samples.
To detect statistically significant differential expression of tRNA halves, read counts from 25 tumor-normal pairs were analyzed with the Bioconductor package EdgeR using a statistical test appropriate for paired designs.49 The test adjusts for baseline differences between patients using an additive linear model with “Patient” as the blocking factor. P values were adjusted for multiple testing using the Benjamini and Hochberg method to control the false discovery rate (FDR).
Results
To evaluate 5′ tRNA halves as potential biofluid-based cancer biomarkers, we interrogated a publicly available small RNA-Seq data set generated from seminal fluid samples collected from normal subjects and patients with prostate cancer. Men with cancer had a mean prostate-specific antigen (PSA) level of 6.6 (SD: 3.09, range: 3.2-11.9), and noncancer controls had a mean PSA level of 6.0 (SD: 3.21, range: 2.5-10.5). As both groups had similarly elevated levels of serum PSA, markers that can distinguish between the groups could be better diagnostic markers than PSA. Selth and colleagues42 used this data set to assess the potential of seminal fluid miRNAs as diagnostic biomarkers of prostate cancer. Here, we analyzed the same data set to determine whether 5′ tRNA halves are present in seminal fluid and whether their abundance is altered in association with prostate cancer.
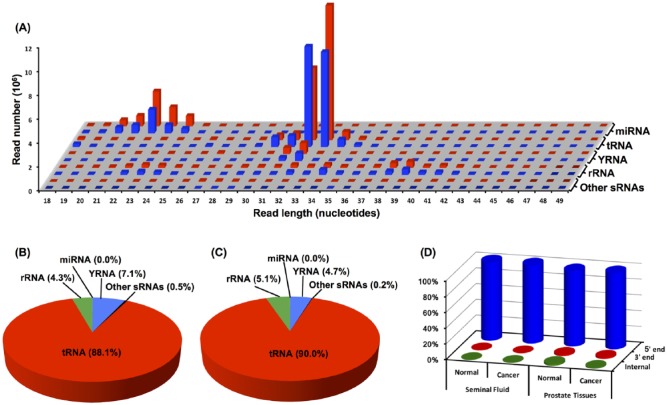
Alignment of sequencing reads to the human genome and annotation with noncoding RNA genes revealed that tRNAs map to a 30- to 34-nucleotide peak, whereas miRNAs map to a 20- to 24-nucleotide peak (Figure 1A). Comparable peaks are present in both cancer and noncancer seminal fluid samples (Figure 1A). We previously reported the same 2-peak pattern in human and mouse serum samples.14,15
Figure 1.
Length distribution and annotation of reads obtained by sequencing small RNAs extracted from seminal fluid. (A) Length of mapped reads from seminal fluid of noncancer (blue bars) or patients with cancer (red bars). The length of small RNA sequencing reads is plotted against their abundance. Shown here are only sequencing reads that map to the GRCh38/hg38 human genome and that are annotated as miRNA, tRNA, YRNA, rRNA, or other sRNAs (snRNA, snoRNA, scRNA, scaRNA). (B, C) Annotation of reads located in the 30- to 34-nucleotide peaks. Pie charts showing the percent of reads mapping to the indicated specific types of small RNAs in data sets obtained by sequencing of small RNAs in seminal fluid samples from (B) noncancer patients or (C) patients with cancer. (D) Relative abundance of reads that map to the 5′ versus 3′ ends of tRNA genes. Pooled reads from seminal fluid from patients with or without cancer and from prostate tumor and normal tissue pairs were analyzed to determine the number of reads that align with 5′ or 3′ ends of tRNA genes. Reads that map beyond the initial 5 nucleotides on either the 5′ or the 3′ ends are labeled “Internal.” The y-axis represents the percentage of the total reads that map to 5′ end, 3′ end, or internally to tRNA genes.
In subsequent analyses, we only considered reads mapping to tRNA genes in the 30- to 34-nucleotide peak as this is the size range of 5′ tRNA halves. The total number of reads in the 30- to 34-nucleotide peaks was highly comparable between the noncancer (36 242 600) and cancer (36 481 628) samples. Annotation analysis (Figure 1B and C) showed that the reads in the 30- to 34-nucleotide peak map mainly to tRNA: 88.1% in noncancer and 90.0% in cancer samples. The remaining reads map to sequences annotated as encoding YRNA, ribosomal RNA (rRNA), or other small RNAs (scRNA, scaRNA, snRNA, snoRNA, vaultRNA).
The length of reads that map to tRNA (30-34 nucleotides) is roughly half of the size of the full-length tRNAs. Therefore, we classified the reads based on their overlap with 5′ or 3′ ends of tRNA genes. We found more than 99.6% of the tRNA-derived reads align with the 5′ end of a tRNA in both noncancer and cancer samples (Figure 1D). Thus, only reads mapping to the 5′ ends of tRNA genes are considered for subsequent differential expression analysis.
5′ tRNA halves are increased in seminal fluid of patients with prostate cancer
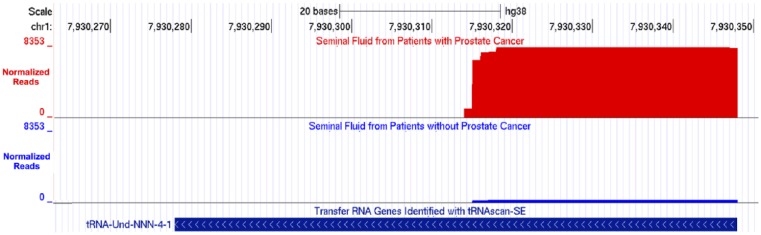
To identify changes in seminal fluid 5′ tRNA halves that may associate with a prostate cancer diagnosis, we compared their abundance in noncancer and cancer seminal fluid samples. We found that the seminal fluid from patients with cancer had increased levels of 71 types of 5′ tRNA halves derived from the isodecoders of tRNA-Ala, -Arg, -Cys, -Gln, -Leu, -Lys, -Met, -Ser, -Thr, -Trp, and the pseudogene tRNA-Und-NNN-4-1 (Table S1). Notably, the 5′ tRNA half derived from the pseudogene tRNA-Und-NNN-4-1 is the most differentially expressed; its levels are 25-fold higher in cancer than in noncancer seminal fluid (Table S1 and Figure 2).
Figure 2.
UCSC genome browser screenshots illustrating alignment of reads to the tRNA-Und-NNN-4-1 pseudogene. The alignment (number of reads, y-axis) shows that the numbers of reads mapping to the 5′ end of tRNA-Und-NNN-4-1 gene are significantly higher in seminal fluid from patients with (red) than without (blue) prostate cancer.
Expression of tRNA halves in prostate cancer tissues corroborates their presence in the seminal fluid
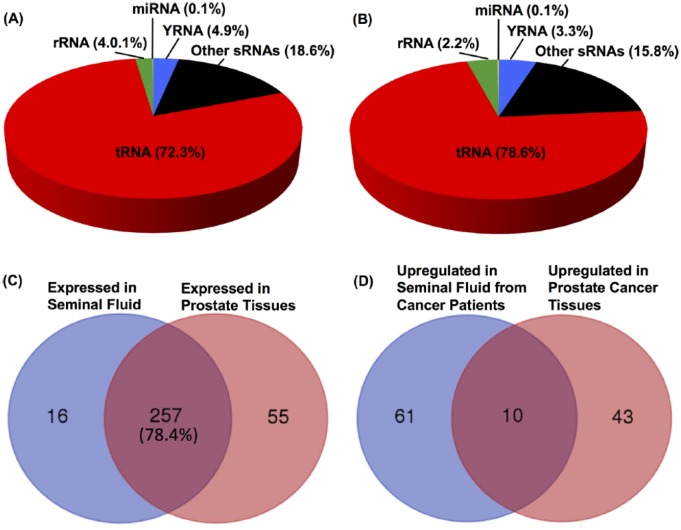
To investigate whether tRNA halves are also present in solid prostate cancer tissue besides seminal fluid, we downloaded and analyzed a small RNA-Seq generated from FFPE prostate cancer samples.48 In this data set, prostate primary tumor and matched normal tissues were collected from men diagnosed with prostate cancer. Aligned sequencing reads from 25 tumor-normal pairs were pooled into 2 groups (normal and cancer) to examine the general characteristics of the reads including length distribution and types and proportions of small RNAs from which the reads originated. Similar to the seminal fluid samples, high proportions of the reads in the 30- to 34-nucleotide peak map to tRNA: 72.3% in the normal and 78.6% in the tumor samples (Figure 3A and B). In both prostate cancer and matched normal tissues, most of the tRNA-derived reads (>95%) align with the 5′ end of tRNA (Figure 1D). A comparison between the tRNA halves identified in the 2 data sets indicates that 94.1% of tRNA halves found in seminal fluid were also detectable in prostate tissues (Figure 3C), suggesting a prostatic origin.
Figure 3.
Analysis of the reads peak from prostate tumor and adjacent normal tissue and comparison with seminal fluid. (A and B) Annotation of reads located in the 30- to 34-nucleotide peak. Pie charts showing the percent of reads mapping to the indicated specific types of small RNAs in pooled data sets from (A) normal and (B) tumor tissue samples. Pooling is used only for qualitative analysis of the reads and not to measure the differential expression of small RNAs between control and cancer groups. (C) Comparison of tRNA halves expressed in seminal fluid with those expressed in prostate solid tissues. tRNA halves are considered expressed in prostate tissues if CPM >25. CPM is the average tRNA read counts-per-million computed over all libraries; it represents a measure of the overall expression level of the tRNA halves. tRNA halves are considered expressed in seminal fluid samples if the sum of normalized read counts from cancer and noncancer samples is higher than 25. (D) Comparison of 5′ tRNA halves that are upregulated in seminal fluid from prostate cancer relative to noncancer patients and 5′ tRNA halves that are upregulated in prostate cancer tissue relative to adjacent normal tissue. The Venn diagram shows ten 5′ tRNA halves (see Table 1) that were upregulated in solid prostate tumor tissue and seminal fluid from patients with prostate cancer.
A prostate cancer diagnosis is associated with increased expression of 5′ tRNA halves in prostate tumor tissues and in seminal fluid
To identify potential tumor-associated changes in the levels of 5′ tRNA halves, we compared read counts from 25 tumor-normal pairs using a statistical test designed for paired experiments. We detected fifty-five 5′ tRNA halves differentially expressed between normal and cancer tissues using an FDR <0.05 (Table S2). Furthermore, we compared the 5′ tRNA halves differentially expressed in solid prostate tumor tissues (Table S2) to the 5′ tRNA halves that are increased in seminal fluid from patients with cancer (Table S1). As illustrated in Figure 3D and Table 1, 10 of the 5′ tRNA halves increased in seminal fluid from patients with prostate cancer were simultaneously significantly upregulated in prostate cancer tissue relative to adjacent normal tissue.
Table 1.
5′ tRNA halves upregulated in solid prostate tumor tissue and simultaneously increased in seminal fluid.
| tRNA gene namea | Genomic coordinatesb | Prostate tissues | Seminal fluid | |||||
|---|---|---|---|---|---|---|---|---|
| CPMc | FCd | P valued | FDRd, % | Normale | Cancere | FCf | ||
| tRNA-Ala-CGC-2-1 | chr6:28673835-28673907 | 571 | 2.5 | .000 | 0.04 | 3 | 56 | 18.9 |
| tRNA-Leu-TAA-1-1 | chr6:144216546-144216629 | 981 | 2.2 | .000 | 0.11 | 28 | 74 | 2.7 |
| tRNA-Ser-CGA-2-1 | chr6:27209848-27209930 | 186 | 2.1 | .000 | 0.56 | 2 | 25 | 12.7 |
| tRNA-Ala-CGC-1-1 | chr6:26553502-26553574 | 1277 | 2.0 | .000 | 0.39 | 25 | 123 | 5.0 |
| tRNA-Gln-CTG-1-4 | chr15:65869061-65869133 | 627 | 2.0 | .001 | 1.63 | 39 | 169 | 4.3 |
| tRNA-Ala-AGC-8-2 | chr8:66114188-66114261 | 656 | 2.0 | .000 | 0.69 | 62 | 194 | 3.1 |
| tRNA-Ala-TGC-2-1 | chr6:28643444-28643516 | 419 | 1.9 | .001 | 1.65 | 24 | 142 | 6.0 |
| tRNA-Ala-AGC-8-1 | chr2:27051213-27051286 | 546 | 1.8 | .001 | 1.65 | 54 | 209 | 3.9 |
| tRNA-Gln-CTG-2-1 | chr6:27547751-27547823 | 504 | 1.8 | .002 | 2.56 | 30 | 168 | 5.7 |
| tRNA-Ala-TGC-3-2 | chr12:124921754-124921826 | 432 | 1.7 | .003 | 3.60 | 7 | 48 | 7.0 |
Abbreviations: CPM, counts per million; FC, fold change; FDR, false discovery rate; tRNA, transfer RNA.
tRNA gene name from genomic tRNA database (gtrnadb.ucsc.edu).
Genomic coordinates of the tRNA gene in the human GRCh38/hg38 genome.
Average tRNA read counts-per-million computed over all libraries and taking into account the estimated dispersions and the libraries sizes. It represents a measure of the overall expression level of the tRNA fragments.
Fold change, P value, and FDR (<5%) for differential abundance computed by EdgeR.
Read count of each tRNA half in the indicated sample. Read counts were normalized by equalizing the total mapped reads between cancer and noncancer samples.
Fold change represents read counts ratio of normal to cancer samples.
Discussion
The easy access and remarkable stability of sncRNAs circulating in body fluids have raised clinical interest in developing extracellular sncRNAs as minimally invasive disease markers. Particularly, the differential abundance of circulating miRNAs has proven useful in diagnosing various types of cancer, including prostate cancer.5,50 More recently, attention has shifted to a new class of circulating sncRNAs also with great diagnostic potential; these are smaller derivatives of known sncRNAs including tRNA. We have previously reported that serum abundance of 5′ tRNA halves changes in association with a cancer diagnosis14,16 and with physiologic states, ie, aging and calorie restriction.15 In the present report, we show that 5′ tRNA halves are expressed in prostate tissue, enriched in seminal fluid, and their abundance changes in association with a prostate cancer diagnosis.
We found that tRNA halves represent the bulk of the sequencing reads in both seminal fluid and prostate tumor tissues. Also, as in previous studies,15,51 we found that 5′ tRNA halves are significantly more abundant than their corresponding 3′-end derivatives. Others confirmed the predominance of the 5′-end derivatives of tRNA not only in body fluids (serum from healthy mice52 and cattle53) but also in solid tissues.54 Specific 5′ tRNA halves were also observed in breast cancer cells under hypoxic stress37 and in the media of breast epithelial cell lines.55 Thus, there may be a specific association between certain pathophysiologic states and the biogenesis and/or stability of 5′ tRNA halves. Factors responsible for the differences in RNA stability between the 5′-end and 3′-end fragments are yet to be investigated. However, a recent report underlined the importance of factors such as sex, race, and disease type in influencing the length, start and end points, and relative abundance of human tRNA fragments.56
Levels of 5′ tRNA halves were increased in seminal fluid from prostate cancer relative to noncancer patients. Changes in extracellular and intracellular levels of 5′ tRNA halves have been observed in cancer and other diseases even though their functional roles have not yet been elucidated.13,14,16,38 Thus, the presence of 5′ tRNA halves in seminal fluid and alterations of their abundance in association with prostate cancer should instigate studies to clarify the potential role of 5′ tRNA halves in prostate tumorigenesis and also may form the basis for developing cancer markers. Seminal fluid is gaining importance as a clinically relevant “liquid biopsy” of the prostate because it is more enriched in prostatic constituents than blood and thus presents a better source of prostate cancer–specific biomarkers.57
Interestingly, the most differentially expressed (ie, 25-fold higher in cancer than in noncancer samples) 5′ tRNA half is derived from the pseudogene, tRNA-Und-NNN-4-1; this tRNA pseudogene is predicted with tRNAscan-SE58 and its anticodon remains undetermined. This observation suggests that tRNA genes annotated as pseudogenes are not only highly expressed but also specifically processed into sncRNAs that can be secreted into body fluids in a manner that is affected by pathophysiological states. The tRNA pseudogenes have also been shown to be functional, eg, they regulate mRNA stability59; based on this and our findings, pseudo-tRNAs should be elevated to the status of tRNA genes and their potential functions investigated.
The prostatic origin of the tRNA halves found in seminal fluid is suggested by their significant coexpression in prostate tissues: more than 94.1% of tRNA halves present in seminal fluid are also expressed in prostate solid tissues. We found a significant differential expression of 5′ tRNA halves in prostate tumor relative to adjacent normal tissues. Even though expression changes in the 5′ tRNA halves have not been previously reported, and shorter tRFs have been associated with prostate cancer.60,61 Particularly, Olvedy and colleagues identified a small subset of tRFs that are differentially expressed across increasing grades of prostate cancer with potential as prognostic markers.60 However, they did not detect the longer 5′ tRNA halves because the sequenced read-length is 35 nucleotides; 5′ tRNA halves are 30- to 34-nucleotide long and will not be present after adapter removal from 35-nucleotide reads.
Comparison of expression changes showed that ten 5′ tRNA halves are significantly upregulated in solid prostate tumor relative to adjacent normal tissue and simultaneously increased in seminal fluid from prostate cancer relative to noncancer patients. Forty-three 5′ tRNA halves were upregulated in prostate cancer tissues but did not increase in seminal fluid from cancer relative to noncancer. Sixty-one 5′ tRNA halves increased in seminal fluid from cancer relative to noncancer but did not change expression in prostate cancer tissues relative to adjacent normal tissue. Functional interpretation of these different classes of 5′ tRNA halves in relation to prostate cancer is challenging because a defined role of 5′ tRNA halves in tumorigenesis has not been determined yet. However, possible involvement of 5′ tRNA halves in prostate cancer is suggested by requirement of the 5′ half of tRNA-Asp-GUC for the proliferation of prostate carcinoma cells62; siRNA knockdown of this 5′ tRNA half, but not its 3′-counterpart, inhibited prostate carcinoma cell proliferation. It was also shown that tRNA halves were significantly more abundant in AR+ than AR− prostate cancer cells and in ER+ than ER− breast cancer cells, suggesting that the expression of tRNA halves is sex hormone dependent.62 As sex hormones and their receptors are crucial to the development and progression of prostate and breast cancers,63,64 the dependence of tRNA halves’ expression on sex hormones and their importance in proliferation strongly suggest they play a significant role in tumorigenesis. Furthermore, we previously reported that specific changes in the circulating levels of specific 5′ tRNA halves were associated with clinicopathologic characteristics of breast cancer including ER, PR, HER2, inflammation, and subsequent relapse.14 Other types of tRNA derivatives have been linked to prostate carcinogenesis18; knocking down a particular tRF causes loss of cell viability and inhibition of prostate cancer cells’ proliferation. Taken together, these observations imply potential involvement of tRNA-derived sncRNAs in tumorigenesis, but the underlying mechanisms need to be investigated to establish any clinical utility of 5′ tRNA halves in cancer diagnosis and prognosis.
Conclusions
Data mining of publicly available small RNA-Seq data sets revealed that 5′ tRNA halves are not only present in prostate tissues and seminal fluid but also that their abundance changes in association with a prostate cancer diagnosis. We found that the levels of a subset of 5′ tRNA halves increased in seminal fluid from prostate cancer relative to noncancer patients and simultaneously significantly upregulated in solid prostate tumor relative to adjacent normal tissue. These findings underscore the potential of 5′ tRNA halves as noninvasive markers for prostate cancer screening and diagnosis and provide leads for future work to elucidate the putative carcinogenic mechanisms of the 5′ tRNA halves. Although our findings do not establish causality, they suggest that 5′ tRNA halves may be involved in some aspect of prostate tumorigenesis. Thus, additional studies are warranted to explore the potential of 5′ tRNA halves as agents of carcinogenesis and as novel markers and therapeutic targets for cancer.
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: JMD conceived and designed the experiments, analyzed the data, and wrote the first draft of the manuscript. JMD, HA, and LAS agree with manuscript results and conclusions and made revisions and approved final version. All authors reviewed and approved the final manuscript.
Disclosures and Ethics: The authors declare that there is no conflict of interest. As a requirement of publication, authors have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality, and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
References
- 1. Turchinovich A, Samatov TR, Tonevitsky AG, Burwinkel B. Circulating miRNAs: cell-cell communication function? Front Genet. 2013;4:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hergenreider E, Heydt S, Tréguer K, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14:249–156. [DOI] [PubMed] [Google Scholar]
- 3. Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhao Y, Song Y, Yao L, Song G, Teng C, et al. Circulating microRNAs: promising biomarkers involved in several cancers and other diseases. DNA Cell Biol. 2017;36:77–94. [DOI] [PubMed] [Google Scholar]
- 5. Selth LA, Tilley WD, Butler LM. Circulating microRNAs: macro-utility as markers of prostate cancer? Endocr Relat Cancer. 2012;19:R99–R113. [DOI] [PubMed] [Google Scholar]
- 6. Victoria B, Dhahbi JM, Nunez Lopez YO, et al. Circulating microRNA signature of genotype-by-age interactions in the long-lived Ames dwarf mouse. Aging Cell. 2015;14:1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dhahbi JM, Atamna H, Li R, et al. MicroRNAs circulate in the hemolymph of Drosophila and accumulate relative to tissue microRNAs in an age-dependent manner. Genomics Insights. 2016;9:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dhahbi JM, Spindler SR, Atamna H, et al. Deep sequencing identifies circulating mouse miRNAs that are functionally implicated in manifestations of aging and responsive to calorie restriction. Aging (Albany NY). 2013;5:130–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rother S, Meister G. Small RNAs derived from longer non-coding RNAs. Biochimie. 2011;93:1905–1115. [DOI] [PubMed] [Google Scholar]
- 10. Sobala A, Hutvagner G. Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip Rev RNA. 2011;2:853–862. [DOI] [PubMed] [Google Scholar]
- 11. Rutjes SA, van der Heijden A, Utz PJ, van Venrooij WJ, Pruijn GJ. Rapid nucleolytic degradation of the small cytoplasmic Y RNAs during apoptosis. J Biol Chem. 1999;274:24799–24807. [DOI] [PubMed] [Google Scholar]
- 12. Vickers KC, Roteta LA, Hucheson-Dilks H, Han L, Guo Y. Mining diverse small RNA species in the deep transcriptome. Trends Biochem Sci. 2015;40:4–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dhahbi JM, Spindler SR, Atamna H, Boffelli D, Martin DI. Deep sequencing of serum small RNAs identifies patterns of 5′ tRNA half and YRNA fragment expression associated with breast cancer. Biomark Cancer. 2014;6:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dhahbi JM, Spindler SR, Atamna H, et al. 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics. 2013;14:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Victoria Martinez B, Dhahbi JM, Nunez Lopez YO, et al. Circulating small non-coding RNA signature in head and neck squamous cell carcinoma. Oncotarget. 2015;6:19246–19263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pederson T. Regulatory RNAs derived from transfer RNA? RNA. 2010;16:1865–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (TRFs). Genes Dev. 2009;23:2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fu H, Feng J, Liu Q, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. [DOI] [PubMed] [Google Scholar]
- 20. Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cole C, Sobala A, Lu C, et al. Filtering of deep sequencing data reveals the existence of abundant dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–229. [DOI] [PubMed] [Google Scholar]
- 23. Diebel KW, Zhou K, Clarke AB, Bemis LT. Beyond the ribosome: extra-translational functions of tRNA fragments. Biomark Insights. 2016;11:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol. 2009;185:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Emara MM, Ivanov P, Hickman T, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285:10959–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Durdevic Z, Mobin MB, Hanna K, Lyko F, Schaefer M. The RNA methyltransferase Dnmt2 is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila. Cell Rep. 2013;4:931–937. [DOI] [PubMed] [Google Scholar]
- 29. Kawaji H, Nakamura M, Takahashi Y. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nowacka M, Strozycki PM, Jackowiak P, Hojka-Osinska A, Szymanski M, Figlerowicz M. Identification of stable, high copy number, medium-sized RNA degradation intermediates that accumulate in plants under non-stress conditions. Plant Mol Biol. 2013;83:191–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dhahbi JM. 5′ tRNA halves: the next generation of immune signaling molecules. Front Immunol. 2015;6:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10:1798–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yeung ML, Bennasser Y, Watashi K, Le SY, Houzet L, Jeang KT. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 2009;37:6575–6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Burroughs AM, Ando Y, de Hoon MJ, et al. Deep-sequencing of human argonaute-associated small RNAs provides insight into miRNA sorting and reveals argonaute association with RNA fragments of diverse origin. RNA Biol. 2011;8:158–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sobala A, Hutvagner G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013;10:553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goodarzi H, Liu X, Nguyen HCB, Zhang S, Fish L, Tavazhoie SF. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Y, Zhang C, Shi J, et al. Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J Mol Cell Biol. 2014;6:172–174. [DOI] [PubMed] [Google Scholar]
- 39. Akat KM, Moore-McGriff D, Morozov P, et al. Comparative RNA-sequencing analysis of myocardial and circulating small RNAs in human heart failure and their utility as biomarkers. Proc Natl Acad Sci U S A. 2014;111:11151–11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vojtech L, Woo, Hughes S, et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014;42:7290–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Maute RL, Schneider C, Sumazin P, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1404–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Selth LA, Roberts MJ, Chow CW, et al. Human seminal fluid as a source of prostate cancer-specific microRNA biomarkers. Endocr Relat Cancer. 2014;21:L17–L21. [DOI] [PubMed] [Google Scholar]
- 43. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:P841–P842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chan PP, Lowe TM. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016;44:D184–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res. 2012;22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ding Y, Wu H, Warden C, et al. Gene expression differences in prostate cancers between young and old men. PLoS Genet. 2016;12:e1006477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–888. [DOI] [PubMed] [Google Scholar]
- 51. Dhahbi JM, Spindler SR, Atamna H, Boffelli D, Mote P, Martin DI. 5 ′ YRNA fragments derived by processing of transcripts from specific YRNA genes and pseudogenes are abundant in human serum and plasma. Physiol Genomics. 2013;45:990–998. [DOI] [PubMed] [Google Scholar]
- 52. Van Goethem A, Yigit N, Everaert C, et al. Depletion of tRNA-halves enables effective small RNA sequencing of low-input murine serum samples. Sci Rep. 2016;6:37876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Casas E, Cai G, Neill JD. Characterization of circulating transfer RNA-derived RNA fragments in cattle. Front Genet. 2015;6:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Selitsky SR, Baran-Gale J, Honda M, et al. Small tRNA-derived RNAs are increased and more abundant than microRNAs in chronic hepatitis B and C. Sci Rep. 2015;5:7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tosar JP, Gámbaro F, Sanguinetti J, Bonilla B, Witwer KW, Cayota A. Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res. 2015;43:5601–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Telonis AG, Loher P, Honda S, et al. Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies. Oncotarget. 2015;6:24797–24822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roberts MJ, Richards RS, Gardiner RA, Selth LA. Seminal fluid: a useful source of prostate cancer biomarkers? Biomark Med. 2015;9:77–80. [DOI] [PubMed] [Google Scholar]
- 58. Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:W54–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rudinger-Thirion J, Lescure A, Paulus C, Frugier M. Misfolded human tRNA isodecoder binds and neutralizes a 3′ UTR-embedded Alu element. Proc Natl Acad Sci U S A. 2011;108:E794–E802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Olvedy M, Scaravilli M, Hoogstrate Y, Visakorpi T, Jenster G, Martens-Uzunova ES. A comprehensive repertoire of tRNA-derived fragments in prostate cancer. Oncotarget. 2016;7:24766–24777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Martens-Uzunova ES, Jalava SE, Dits NF, et al. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene. 2012;31:978–991. [DOI] [PubMed] [Google Scholar]
- 62. Honda S, Loher P, Shigematsu M, et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci U S A. 2015;112:E3816–E3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lonergan PE, Tindall DJ. Androgen receptor signaling in prostate cancer development and progression. J Carcinog. 2011;10:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Haldosen LA, Zhao C, Dahlman-Wright K. Estrogen receptor beta in breast cancer. Mol Cell Endocrinol. 2014;382:665–672. [DOI] [PubMed] [Google Scholar]