Abstract
While TLR-activated pathways are key regulators of B cell responses in mammals, their impact on teleost B cells are scarcely addressed. Here, the potential of Atlantic salmon B cells to respond to TLR ligands was shown by demonstrating a constitutive expression of nucleic-acid sensing TLRs in magnetic sorted IgM+ cells. Of the two receptors recognizing CpG in teleosts, tlr9 was the dominating receptor with over ten-fold higher expression than tlr21. Upon CpG-stimulation, IgM secretion increased for head kidney (HK) and splenic IgM+ cells, while blood B cells were marginally affected. The results suggest that CpG directly affects salmon B cells to differentiate into antibody secreting cells (ASCs). IgM secretion was also detected in the non-treated controls, again with the highest levels in the HK derived population, signifying that persisting ASCs are present in this tissue. In all tissues, the IgM+ cells expressed high MHCII levels, suggesting antigen-presenting functions. Upon CpG-treatment the co-stimulatory molecules cd83 and cd40 were upregulated, while cd86 was down-regulated under the same conditions. Finally, ifna1 was upregulated upon CpG-stimulation in all tissues, while a restricted upregulation was evident for ifnb, proposing that salmon IgM+ B cells exhibit a type I IFN-response.
Introduction
Adaptive immunity, present in all jawed vertebrates, is based on broad repertoires of antigen binding receptors expressed on B and T cells. In addition, B cells possess the capacity to directly sense and respond to pathogens through pattern recognition receptors (PRRs), including Toll-like receptors (TLRs). The development of different B cell subsets is determined in synergy between the B cell receptor, other receptor-derived signals and downstream signaling pathways1. These processes, as well as the various functionally distinct B cell subsets, are well studied in higher vertebrates, but less is known about the characteristics of teleosts B cells.
Teleosts and mammals diverged more than 350 million years ago; despite similarities between their immune systems, distinct differences are present. Lacking both bone marrow and lymph nodes, the anterior kidney (or head kidney; HK), and spleen are the organs involved in teleost B cell maturation and differentiation2. IgM, IgD and IgT are the only immunoglobulin (Ig) classes identified in fish3,4. IgM is regarded as a universal vertebrate Ig and was the first teleost Ig to be characterized5. Teleost IgM is a structural and functional homolog to mammalian IgM and the most prevalent isotype in fish serum present as a tetramer6. IgT (IgZ in cyprinids) is a unique teleost Ig7–9 involved in mucosal immunity analogous to mammalian IgA10–13. Compared to IgM and IgT, teleost IgD functions are less examined, however, in channel catfish (Ictalurus punctatus) IgD is suggested to be a mediator of innate immunity14. Two B cell populations were initially described in rainbow trout (Oncorhynchus mykiss); one subset expressing both IgM and IgD and one expressing only IgT13. These two B cell types are localized differently, with IgM+ cells being the main B cell population in spleen, kidney and blood15, while IgT+ cells dominate at mucosal sites12,13,16. Recently a third subset; expressing only IgD and homing mainly to the gills, was also identified in trout17.
Besides mediating humoral adaptive responses, mammalian B cells secrete cytokines and present antigen to T cells18,19. Naïve mammalian B cells are generally divided into three subsets; follicular B cells, B-1 B cells and marginal zone (MZ) B cells. Mature follicular B cells home to follicles in the secondary lymphoid organs (lymph nodes and spleen), where they present T cell dependent antigens (TD) to T cells. MZ B cells and B-1 B cells, respectively, reside in the marginal zone of the spleen or in body cavities and both have important roles in T cell independent antibody (Ab) responses. These cells occupy a niche between innate and adaptive immunity, secreting broadly cross-reactive IgM Abs20,21. Upon innate signaling, e.g. TLR-ligands, both MZ- and B1-B cells rapidly differentiate into Ab-secreting plasma cells, while follicular B cells only do this in the context of prior BCR activation22. Whether teleost B cells fall into several functionally distinct subpopulations is presently unknown. However, some of the features that characterize mammalian B-1 cells are described for teleost B cells; for example a high phagocytic activity23,24 and constitutive expression of PRRs25. Previous in vivo studies by our group showed that the combination of the TLR-ligand CpG and poly I:C, increased the neutralizing Ab response to a virus vaccine in Atlantic salmon (Salmo salar)26,27. This prompted us to use CpG as a model to investigate Atlantic salmon B cell biology, a topic of particular interest due to the high economic value of this species in the aquaculture industry and the urgent need for more efficient virus vaccines. A question of particular interest was whether the TLR-ligand CpG could directly stimulate B cells in vitro to secrete Abs and also to show enhanced antigen presentation functions stemming from increased expression of costimulatory molecules or MHC II molecules.
IgM+ B cells that were obtained by magnetic activated cell sorting (MACS) were found to constitutively express nucleic acid sensing TLRs, providing a foundation for TLR ligands to aid in shaping salmon B cell responses. Indeed, upon CpG stimulation, IgM secretion was increased in IgM+ cells; with the highest induction in HK compared to spleen and the lowest secretion in blood. In addition, gene expression analysis showed that the capacity of salmon IgM+ cells to trigger type I interferon (IFN-I) responses and present antigen appeared to be modulated by CpG stimulation. The results presented here provide a platform for further in-depth studies, dissecting different B cell subsets in teleost fish and their functional capacities related to humoral immunity, antigen presentation and regulatory functions.
Results
IgM+ B cells are the dominating B cell population in salmon kidney, blood and spleen
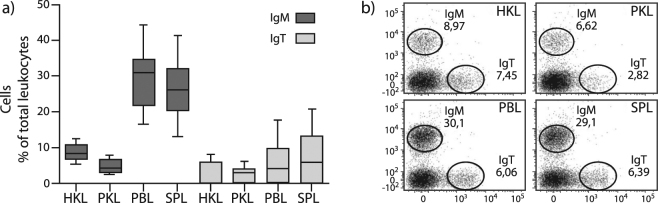
The percentage of IgM+ and IgT+ B cells in relation to total leukocytes in salmon HK, posterior kidney (PK), peripheral blood (PB) and spleen were analyzed by flow cytometry using trout anti-IgM and anti-IgT mAbs (Fig. 1). For all tissues, the most abundant B cell population was the IgM+ B cells (Fig. 1a,b). The IgM+ population constituted about 30% of all leukocytes. In PB and spleen, and had a higher abundance compared to HK and PK (~5–10%). Both IgM+ and IgT+ cells showed a larger individual variation in PB (17 to 44% and 0.1 to 18%, respectively) and spleen (13 to 41% and 0.1 to 21%, respectively), that was not seen in the HK or PK. In four to five of the individuals analyzed, there were less than 2% IgT+ cells, which was evident in all tissues.
Figure 1.
IgM+ cells are the dominating B cell population in Atlantic salmon systemic lymphoid tissues. Flow cytometry analysis of Atlantic salmon head kidney (HKL), posterior kidney (PKL), peripheral blood (PBL) and spleen (SPL) leukocytes stained with trout anti-IgM and IgT mAbs. (a) Median frequencies of IgM+ and IgT+ B cells of total leukocytes (n = 12). The box indicates 25th and 75th percentiles and the bars min and max values. (b) Representative flow cytometry dot plots showing the IgM and IgT percentages in the systemic lymphoid tissues.
Purity and viability of MACS sorted IgM+ B cells from HK, spleen and PB
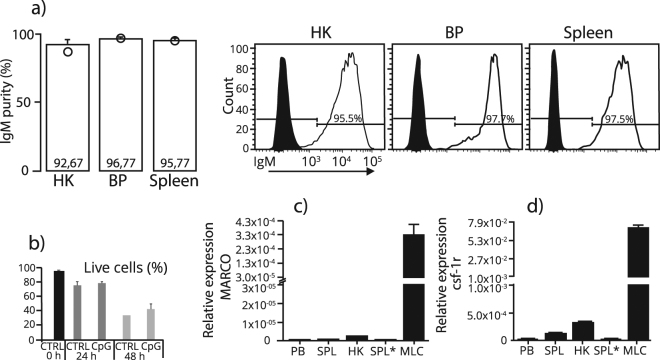
To study B cell biology of salmon, cultures of IgM+ cells were obtained by MACS. Before proceeding to further experiments, a basic characterization of these cells was done by purity and viability testing. As shown by flow cytometry, the purity of the IgM+ B cells was >95% for PB and SP and >92% for HK (Fig. 2a). Viability was 98% after MACS and decreased to 78 and 35% after 24 and 48 hours in culture, respectively. Viability in CpG stimulated IgM+ cells was in the same range as in unstimulated cells (Fig. 2b).
Figure 2.
Purity and viability of IgM+ B cells sorted by magnetic activated cell sorting (MACS). (a) Upon sorting, the mean percentages of IgM+ cells from HK, PB and spleen (n = 3 for each tissue) were analysed by flow cytometry. The circle (○) represents total percentage of viable cells before gating for IgM+ events. Histogram represents one representative individual for each tissue, where IgM+ events are presented by the transparent peak and non-stained events by the black peak. (b) Viability of IgM+ cells kept in culture with or without CpG for 0, 12 and 24 hours. (c and d) The relative expression of MARCO and csf-1r in MACS and FACS sorted IgM+ cells, and in macrophage-like cells (MLC).
Since macrophages bind IgM through their Fc-receptor, there might be a possibility of macrophage contamination within the IgM+ MACS purified cells. To test this, the expression levels of genes encoding the scavenger receptor MARCO and the csf-1r, known as markers of monocyte-macrophage lineages in mammals28 and fish29, were analyzed in freshly MACS-purified HK, spleen and PB B cell populations. For comparison, the expression of the same genes was tested in HK-derived untreated macrophage-like cells (MLC), and as expected high mRNA levels of both genes were seen (Fig. 2c,d). Notably, while MARCO expression was undetected in IgM+ cells derived from PB and spleen (Cq cut-off to 37.8), and at very low levels in the HK cells (Cq > 36), a modest csf-1r expression was apparent in cells from all three tissues (Cq = 30–34), and again, HK IgM+ cells yielded the highest expression (Supplementary Fig. S1). A comparison of the relative expression of MARCO and csf-1r between the IgM+ cells and the MLC are presented in Fig. 2c,d. A 324, 122, and 282 fold higher expression of MARCO was found in the MLC compared to PB, HK and spleen, respectively (Fig. 2c). In the same tissues, the csf-1r was 2690, 217 and 560 fold higher expressed in the MLC than in the IgM+ cells, respectively (Fig. 2d). In FACS-sorted splenic IgM+ cells (n = 5), both MARCO and csf-1r were expressed about the same level as in MACS-sorted cell, Cq > 37.8 and >32, respectively (Supplementary Fig. S1). Additionally, the expression of the T cell marker cd4–230 was explored and found to be undetectable across the tissues (Cq cut-off set to 38) (Supplementary Table S2). Our results indicate an absence of contaminating T cells in the sorted IgM+ populations, while traces of myeloid marker genes were detectable, most prominently in the HK derived cells.
Atlantic salmon IgM+ B cells express high levels of tlr9 and tlr8a1
Characterization of the basal TLR expression in salmon B cells from lymphoid organs is key to understanding how TLR signaling may affect salmon B cell responses. Here, we focused on a set of nucleic acid recognizing TLRs shown to respond to ss-RNA (TLR8a1), ds-ssRNA (TLR3 and 22)31 and CpG DNA (TLR9 and 21)32. tlr3, 8 and 9 are orthologues to mammalian TLRs, whereas tlr21 and 22 are absent in mammals. Figure 3 shows that all five tlrs were expressed by the IgM+ B cell populations and that tlr8a1 and tlr9 had the highest relative expression, representative across all tissues. tlr21 was the lowest expressed tlr-gene, and was respectively 18, 9 and 23-fold less expressed compared to tlr9 in HK (p < 0.001), PB (p < 0.05) and spleen (p < 0.01) IgM+ B cells. The individual variation of tlr8a1, one of several teleost tlr8 isotypes33, was low both within and across tissues. For the two tlrs recognizing dsRNA, the expression of the fish specific tlr22, was relatively stable across tissues and individuals, while the tlr3 expression displayed a higher individual variation.
Figure 3.
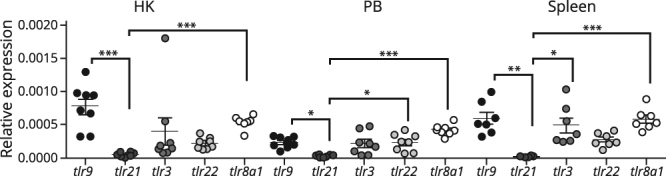
Relative expression of nucleic acid-sensing TLRs in Atlantic salmon IgM+ B cells derived from head kidney, peripheral blood (PB) and spleen. tlr expression was analyzed by RT-qPCR in IgM+ B cells sorted by MACS (n = 6 to 8). Data are presented as relative expression from individual fish (dots) where the bars indicate mean and s.e.m. Statistical significance between the tlr expression levels are indicated by brackets and the asterisks indicates the strength of significance: *p < 0.05 **p < 0.01 ***p < 0.001.
CpG-stimulation alters sigm transcript levels in IgM+ B cells
Constitutive expression levels of both tlr9 and 21 transcripts were evident when analyzing the MACS- sorted IgM+ B cells; hence, this was followed up by investigating if their ligand CpG would affect the expression pattern of selected immune genes. For all the assayed genes, the effect of non-CpG treatment was negligible or minimal (Supplementary Fig. S2). The basal expression level of secreted IgM (sigm) was higher across the three tissues compared to membrane bound IgM (migm) (Supplementary Table 2). Higher basal levels of sigm transcripts (about 16-fold) were observed in IgM+ HK (mean Cq = 18.2, SD = 0.3) and spleen (mean Cq = 18.3, SD = 0.1) compared to PB (Mean Cq = 22.5, SD = 0.2) B cells.
In general, the control supernatant, CAS, provided the weakest transcript induction, or even inhibited the expression of the B cell marker genes compared to the untreated control. Similarly, neither CpG nor PAS exhibited significant induction of migm in any of the tissues over time (Fig. 4b). For sigm, a gradual increase in induction was observed over time in B cells derived from PB and spleen (Fig. 4a). The CpG alone and the combination treatment, respectively, displayed fold inductions of 3.8 and 12.8 for PB, and 3.4 and 7.6 for spleen at 48 hours. Significant increase in sigm mRNA levels compared to the CAS treatment (p < 0.05) were observed in PB B cells stimulated with PAS or the combination at 12 hours, which was still significant in the combination treatment, at 24 and 48 hours (p < 0.05). The sigm transcript regulation was least affected by the stimulations in HK derived IgM+ B cells, although a significant (p < 0.05) increase was apparent in all the three stimulations compared to CAS at 24 hours. Of notice, while HK response was unaffected over time, for spleen B cells the PAS and combination stimulation yielded a significant induction of sigm (p < 0.05) compared to CAS at 48 hours.
Figure 4.
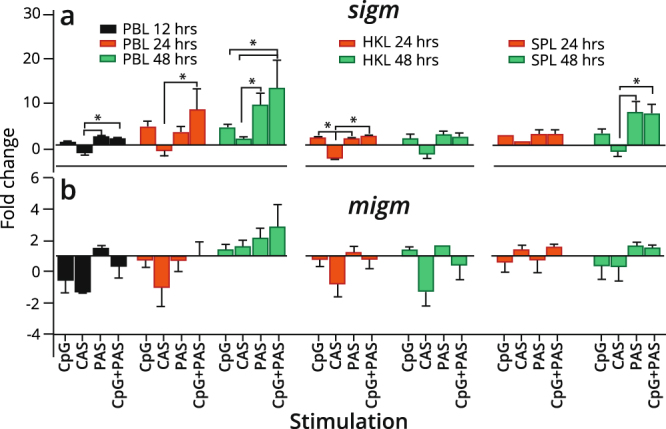
Expression of B cell markers genes in IgM+ B cells treated with CpG and/or condition supernatants. IgM+ B cells from peripheral blood (PB), head kidney (HK) and spleen leukocytes were MACS sorted and analyzed by RT-qPCR for expression of (a) secreted IgM (sigm) and (b) membrane IgM (migm). Gene expression data at each time point were normalized against the reference gene EF1aB and fold changes were calculated using the unstimulated sample at the same time point74. Data represent mean ± s.e.m. from at least three individuals (n = 3 to 6). The line intersecting the y-axis at 1 represents the unstimulated control that the fold change of the treatments are in relation to. Significant fold changes (p < 0.05) are indicated by *. CAS; Control adherent cell supernatant, PAS; Pulse adherent cell supernatant.
Induction of professional antigen presenting cell marker genes in IgM+ B cells by CpG
The capabilities of B cells to phagocytose and present antigens (Ag) have been described in trout34 and zebrafish19. Hence, we hypothesized that IgM+ B cells from Atlantic salmon may express distinct transcripts of selected Ag presentation and co-stimulatory genes upon CpG stimulation. Accordingly, we measured the relative expression of cd83, cd86, cd40 and mhcII in IgM+ B cells obtained from the three tissues. CpG alone and the combination treatments augmented the expression of cd83, a marker for mature dendritic cells (DCs) and activated B cells35. For the three tissues, significant increase in cd83 expression was evident in CpG and combination compared with CAS and PAS at 24 hours (Fig. 5a). However, in HK and spleen, cd83 transcript levels slightly declined after 24 hours for the CpG and combination treatments (Fig. 5a), while the expression was still significantly maintained after 48 hours in PB IgM+ B cells. cd86 is one of the best-defined co-stimulatory molecules in mammals and plays a major role in providing co-stimulation to T cells by both DCs and B cells36. Recently, the involvement of CD86 in B cell-initiated adaptive immunity in a zebrafish model was reported19. Interestingly, except for CpG treated spleen B cells, the expression of cd86 was downregulated compared to the control for all the treatment groups and time points in HK and PB (Fig. 5b). CD40 is a transmembrane glycoprotein and a member of the TNF receptor superfamily expressed by APCs37. Interactions between CD40 and its ligand, CD40L, holds a major role in the cross-talk between T cells and APCs. CpG and PAS, both as standalone stimulations, and in combination, induced a modest, although comparable expression of cd40 transcripts in sorted IgM+ B cells from the three tissues (Fig. 5c). Significantly (p < 0.05) higher induction of cd40 was observed in CpG and PAS stimulated HK B cells at 24 hours compared to CAS. mhcII was constitutively expressed in IgM+ B cells from all the assayed tissues (basal level expression of Cq = 20.7, SD = 0.4 for PB; Cq = 20.9, SD = 0.8 for HK; Cq = 20.9, SD = 0.9 for spleen). At transcript level (Fig. 5d), the stimulations did not markedly affect the regulation of mhcII although the CAS simulation resulted in a more pronounced, yet insignificant, downregulation.
Figure 5.
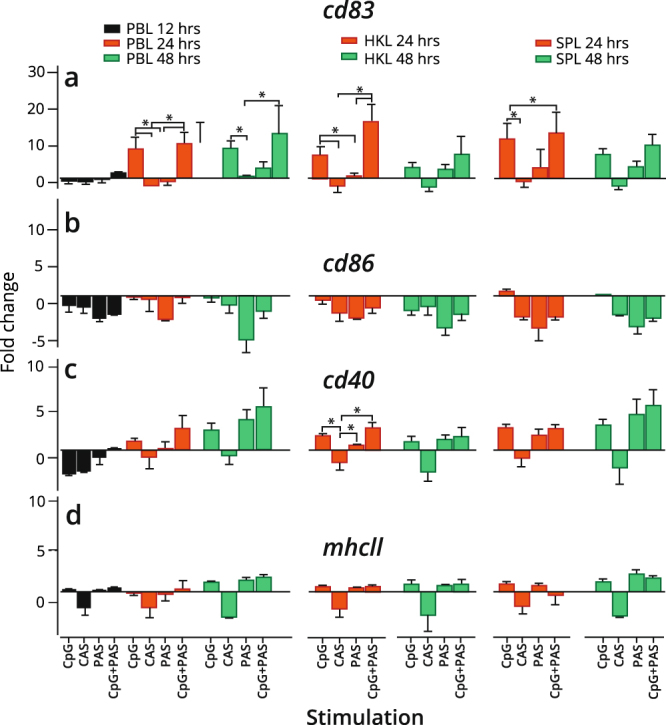
Expression of APC marker and co-stimulatory genes in IgM+ B cells treated with CpG and/or condition supernatants. IgM+ B cells from Atlantic salmon peripheral blood (PB), head kidney (HK) and spleen leukocytes were MACS purified, stimulated in vitro or left untreated and analyzed by RT-qPCR for expression of (a) cd83, (b) cd86, (c) cd40 and (d) mhcII. Gene expression data at each time point were normalized against the endogenous control gene EF1aB and fold changes were calculated using the unstimulated sample at the same time point74. Data represent mean ± s.e.m from at least three individuals (n = 3 to 6). The line intersecting the y-axis at 1 represents the unstimulated control that the fold change of the treatments are in relation to. Significant fold changes (p < 0.05) are indicated by *. CAS; Control adherent cell supernatant, PAS; Pulse adherent cell supernatant.
CpG-stimulation induce antibody secretion in HK and spleen IgM+ B cells
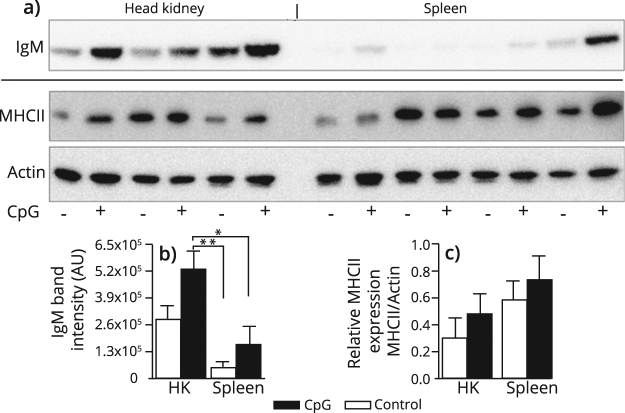
To validate the qPCR results, the IgM content in cell culture supernatants and the MHCII expression in cell lysates from MACS-purified B cells were analyzed by Western blot. In line with the transcript data, translational results revealed that IgM secretion was enhanced in CpG treated cells compared to untreated controls (Fig. 6a,b). A pronounced individual variation in IgM secretion was observed for both controls and stimulated cells. Of notice, significantly higher levels of secreted IgM were detected in CpG-stimulated HK B cells when compared to the spleen cells (Fig. 6b). The lowest levels of secreted IgM were detected in PB B cell culture supernatants (Supplementary Fig. S4). At protein level, CpG-treatment affected MHCII expression modestly, although not significantly different, compared with the control cells (Fig. 6c).
Figure 6.
Protein level analysis of IgM and MHCII expression by Western blot. MACS purified IgM+ B cells from head kidney (HK) and spleen were treated with CpG (+; 2 µM) or left untreated (−) for 72 hours before harvest of supernatants and cell lysates. Figures present results from two independent experiments. (a) Representative Western blot analysis showing IgM secretion in supernatants (upper panel), and MHCII expression in cell lysates (middle panel). An anti-actin Ab was applied as loading control (lower panel). MHCII and actin were run on the same gel; after MHCII detection, the gel was washed and reprobed with the anti-actin Ab. Bar graphs showing the mean band intensity (AU) of (b) secreted IgM and (c) MHCII relative expression in HK and spleen B cells (n = 6). Significantly higher IgM secretion is indicated by * where *p < 0.05 and **p < 0.01. The original full blot images can be found in Supplemental Fig. S5.
CpG-stimulation induces type I ifn gene expression in IgM+ B cells
To assess the capability of salmon IgM+ B cells to induce type I IFN responses, the expression of type I ifn genes were measured. CpG alone significantly (p < 0.05) up-regulated ifna1 transcripts in B cells derived from the systemic tissues. The combination of CpG and PAS, however, induced a more pronounced expression than the standalone stimulation, which suggests an additive effect (Fig. 7a). Over time, ifna1 expression patterns were similar for HK and spleen B cells and declined at 48 hours post stimulation, while the expression in PB IgM+ B cells was kept significantly higher at the same time point.
Figure 7.
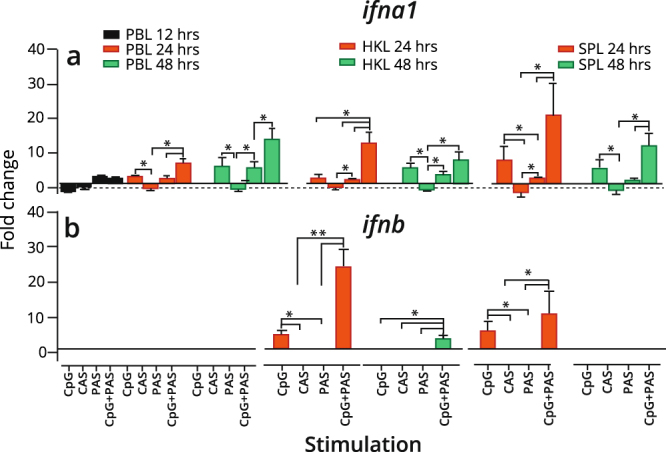
Expression of type I IFN genes in IgM+ B cells treated with CpG and/or condition supernatants. IgM+ B cells from Atlantic salmon peripheral blood (PB), head kidney (HK) and spleen (SP) leukocytes were MACS sorted, stimulated in vitro or left untreated and analyzed by RT-qPCR for expression of (a) ifna1 and (b) ifnb genes. Gene expression data at each time point were normalized against the endogenous control EF1aB and fold changes were calculated using unstimulated cells at the same time point74. Data represent mean ± s.e.m from at least three individuals (n = 3 to 6). The line intersecting the y-axis at 1 represents the unstimulated control that the fold change of the treatments are in relation to. Significant fold changes, p < 0.05 or p < 0.01, are indicated by * or **, respectively. CAS; Control adherent cell supernatant, PAS; Pulse adherent cell supernatant.
Upregulation of ifnb transcripts was observed only in IgM+ B cells obtained from HK and spleen at 24 hours for the two CpG treatments and was highly significant (p < 0.01) compared to CAS, while the expression was undetectable in PB IgM+ B cells (Fig. 7b). The basal expression of ifnb was undetected in IgM+ B cells obtained from all the tissues (Cq cut-off set to 36), while the expression of ifnc (Supplementary Table S2) was undetectable both in controls and stimulated IgM+ B cells.
Discussion
A better understanding of basic B cell biology, such as responsiveness to potential vaccine adjuvants and the mechanisms mediating their activity, is crucial for the development of new vaccines for farmed fish. For Atlantic salmon, many aspects of B cell functionality are unknown, partly due to the lack of population specific mAbs. Here we have used mAbs available against trout IgM and IgT combined with flow cytometry to separate and quantify B cell subpopulations in systemic immunological organs of non-immunized salmon. The monoclonal IgF1–18 (6-1-18) anti-trout IgM Ab is earlier reported to recognize both salmon IgM isotypes38, while the cross-reactivity of the anti-trout IgT mAb to salmon IgT is reported herein (Supplementary Fig. S3)
Single IgM and IgT positive cells were identified in total leukocytes from PB, spleen, HK and PK at percentages varying from three to 44% for IgM and between 0.1 to 21% for IgT. The data are in accordance with a similar study in trout13 demonstrating markedly higher abundances of IgM+ B cells compared to IgT+ B cells in systemic lymphoid tissues. As reported previously in several other teleost fish species13,39,40 and also in salmon38, PB and spleen here harbored the highest number of IgM+ cells, ~30% of total leukocytes. The percentages of IgT+ cells were in general low, and across the tissues investigated, several individuals had less than two percent of IgT+ B cells. The trout anti-IgT mAb is directed against the constant heavy chain domains two to four of IgT subclass 113 and was recently shown to cross-react with the other two trout IgT subclasses41. Three IgT subclasses are present in salmon as well, showing ~80% identity both to each other and to trout IgT9. Western blot analysis of a Flag-tagged CH4-domain of salmon IgT overexpressed in HEK293 cells verified cross-reactivity of the trout IgT Ab to salmon IgT (Supplementary Fig. S3). Whether the trout Ab is able to recognize all three salmon IgT+ B cell subclasses is presently unknown, thus we cannot conclude if one or all three salmon IgT subclasses are present in the investigated tissues.
Induction of protective Ab responses is a major goal for the development of prophylactic vaccines for farmed fish. Studies by us and other groups have reported the potential of CpG-based adjuvants for fish vaccines26,27,42. A combination of the TLR-ligands CpG and poly I:C formulated in a whole-virus vaccine for salmon induced high titer-protective Abs after vaccination26,27. However, how these TLR ligands potentiate Ab responses in bony fish remain poorly understood. One important scope of this study was to investigate the direct effects of TLR engagement on salmon B cell functions. In this context, we used purified IgM+ B cells obtained by MACS purification. While these cells were devoid of T cell marker genes, traces of well-known monocyte-macrophage lineage marker genes were detected. However, since their numbers were very low, their contribution to the overall mRNA levels expressed in the cultivated cells is most likely modest. Of notice, macrophage marker-gene expression were comparable between MACS and FACS purified IgM+ cells, suggesting that the levels of B cell purity by the two methods are comparable. Since bony fish B cells are suggested to have a myeloid origin34, one could speculate on whether myeloid restricted proteins like CSF-1R are totally absent in B cells.
To date, the expression pattern of TLRs in fish has mainly been monitored in whole tissues43–45, while knowledge regarding their expression in defined leukocyte subsets are scarce. Here, transcripts of the five nucleic-acid binding TLRs were detected in the purified IgM+ B cells from the three tissues. This suggests that salmon B cells have the potential to respond directly to CpG DNA and ds/ssRNA. The relative expression levels for each TLR showed little variability across the different tissues; slightly elevated levels of tlr9 and tlr8a1 were detected compared to the other TLRs, while tlr21 was the least expressed TLR. A similar study in trout reported higher constitutive levels of tlr9 and tlr8a2 compared to tlr3 in IgM+ positive B cell populations from blood, kidney and spleen25, although such differences were less apparent in our data. Variations in fish species, age and cell sorting procedures may explain the dissimilarities between these results. As in salmon, the fish specific receptor, tlr22, was abundant in trout B cells from all three tissues25. The most profound difference reported here is between tlr9 and 21, where tlr9 had 10 to 30-fold higher basal expression levels compared to tlr21. TLR21 is a non-mammalian TLR, first identified in chicken, with functions resembling TLR946,47. Later, tlr21 genes have been identified in several teleost species including salmonids48,49. In zebrafish, both TLR9 and 21 respond to CpG-ODNs and slight differences in their ligand recognition profiles are shown with TLR9 possessing a broader sequence specific range compared to TLR2132. Whether this holds true for other fish species, including salmonids, warrants further investigations.
An important question for this study was whether CpG stimulation could activate salmon B cells to become Ab secreting cells (ASCs). Secreted IgM protein was detected in non-stimulated IgM+ cells from spleen and HK, with markedly higher levels in HK derived B cells compared to spleen (Fig. 5a,b). In contrast, IgM secretion from PB IgM+ cells were hardly detected (Supplementary Fig. S4). RT-qPCR correspondingly demonstrated that non-treated IgM+ cells from PB expressed the lowest levels of secreted igm transcripts (mean Cq of 22.5), while basic levels in HK and spleen derived cells were about 16-fold higher (mean Cq of 18.2 and 18.3, respectively). The presence of Ig-secreting cells in the absence of immune activation suggest that plasmablasts/plasma cells might be present in these organs, and probably at a much higher abundance in HK compared to spleen and blood. This observation is in agreement with previous reports for other teleosts50,51 and suggest that ASCs may persist from previous antigenic exposures or that these cells spontaneously secrete IgM, thus resembling the mammalian B-1 cells. In our study, the levels of secreted IgM protein increased upon in vitro CpG stimulation, especially for HK derived cells. The levels of sigm transcripts in the IgM+ cells from the three tissues interrelated with the levels of secreted IgM in cell supernatants, where only secreted Ig transcripts derived from HK cells were significantly higher than the controls. The lack of salmon plasma cell markers impeded further characterization of the ASCs. For B cells activated through the BCR prior to cell harvest, CpG may contribute to terminal differentiation towards plasma cells. Whether CpG alone exerts an effect on naïve B cells, or, alternatively, that the combination of BCR triggering by the anti-IgM Ab and TLR signaling elicits differentiation into ASCs, is still an open question.
In addition to promote Ab responses, TLR signaling is involved in B cell cytokine secretion and antigen presentation52. In our data, the mhcII gene-expression pattern in IgM+ cells was similar across all the three tissues, and its levels were non-significantly upregulated (~2-fold) by the stimulations (Fig. 4d). The detection of MHCII protein by Western blotting in lysates from IgM+ cells validated the finding (Fig. 5b). In general, information about the role of teleost B cells as APCs is lacking. It has been reported that a population of IgM+/MHC-II+ cells from salmon HK has the ability to accumulate ovalbumin antigen in vitro53, thus, supporting a role of HK-derived B cells in the antigen-presenting process. In this study, cd83 and cd40 transcript levels were slightly upregulated in CpG-treated HK-derived IgM+ cells (Fig. 5a,c), while at the same time cd86 levels decreased (Fig. 5b). These results were observed for all tissues. However, in contrast to the earlier study53, where the B cells were cultivated and stimulated within a heterogeneous leukocyte population and subsequently FACS-sorted, the purified B cells were here directly stimulated. Our current data show that CpG alone, or possibly in combination with BCR stimulation, is sufficient to affect the gene expression pattern of central antigen presenting markers on salmon B cells. The rapid up-regulation of cd83 transcripts in the B cells described herein is also observed in murine B cells after TLR-engagement or BCR ligation54. While cd83 mRNA expression was solely upregulated in CpG-treated B cells, both CpG alone and PAS induced elevated cd40 mRNA levels in all the tissues when compared to the CAS, although significantly different only for HK at 24 hours. This is similar to the situation in mammals where both TLR signaling and cytokines derived from myeloid cells regulate cd40 expression in B cells55. Results based on micro-array data, have shown that a suite of genes encoding secreted proteins, which includes cytokines, chemokines and TNF receptor superfamily members, are expressed in salmon adherent leukocytes stimulated with the same CpG ODN as used here56. It is therefore likely that PAS contains molecules with a potential to selectively upregulate different B cell transcripts.
The high phagocytic capacity of teleost B cells24,57, combined with the upregulation of different APC markers reported here and by others19,53,58, suggest that TLR-mediated activated B cells are capable of modulating T cell responses. As in our study, high expression of surface MHCII levels and cd83 transcripts for LPS-stimulated trout splenic B cells has been reported elsewhere58, underscoring their role as APCs. However, the down-regulation of cd86 in our study contradicts the upregulation of this gene obtained in the previous study58. This may suggest that B cells show both similar and contrasting responses to the two microbial molecules CpG and LPS, which may influence their functions. Another important aspect is that the LPS-stimulation in the previous work58 was not performed on purified B cells, and hence, the activation by other cell types and their secretory products may directly or indirectly influence B cell responses. The enhanced effect of the PAS supernatant when used with CpG, as reported herein, underscores that molecules derived from other cell types significantly shape the B cell responses to TLR ligation.
Trout peritoneal IgM+ cells, upon in vivo exposure to E. coli or VHSV, are shown to differentiate towards IgM-secreting cells, and at the same time these cells show decreased levels of MHCII surface expression compared to non-treated controls59. As the terminal differentiation of mammalian plasma cells is shown to be accompanied by the loss of MHCII expression29, these authors argue that decreased MHCII surface levels may be a coincident indicator for trout plasma cell differentiation. This somewhat differs from our data where CpG treatment of IgM+ cells did not significantly affect MHCII levels, while increased IgM secretion clearly indicated CpG-induced differentiation towards plasma cells. However, the specific stage of differentiation of these cells towards the terminal plasma cell was not identified. Most of these cells may be at the plasmablast stage and might not have accomplished MHCII downregulation after 72 hours in vitro CpG treatment.
In our data, high individual variations in IgM secretion and MHCII proteins levels were apparent for both controls and stimulated IgM+ cells. This heterogeneity in responses suggest variations in the B cell composition between the individual donor fish. As a result, the secreted IgM may reflect both the presence of previously activated plasmablast/plasma cells and naïve B cells responding to the TLR-ligation. The impact of CpG treatment on MHCII levels may be different on naïve versus previously activated B cells. This proposed heterogeneity in the B cell populations restricts the usage of our data to evaluate if any association between plasma cell development and MHCII levels exists. Anyhow, whether active suppression of class II genes is an underlying mechanism for bony fish plasma cell differentiation or not, is an interesting question that calls for future investigation only attainable using subpopulation specific mAbs.
In general, information on CD40 signaling in teleost B cells is limited. However, studies in both zebrafish60 and salmon61 have reported that structural characteristics of CD40 and its ligand (CD40L or CD154) are conserved between mammals and fish. Salmon cd40 was upregulated in cultivated HK leucocytes after exposure to different PAMPs including CpG61, which is in accordance with the present data. In zebrafish, CD40/IgM double positive lymphocytes were reported60, suggesting that CD40 is present in teleost B cells. The present work further support this, showing that salmon HK, spleen and PB B cells express considerable levels of cd40 (Supplementary Table S2). Opposite to cd40 and cd83, decreased levels of cd86 were apparent upon CpG treatment across the tissues, which may suggest a mechanism to avoid prolonged T cell activation62.
Teleost fish possess a well-developed type I IFN system encoded by multiple IFN genes. Six IFN-I subtypes are identified in the salmon genome, where the subtypes ifna, ifnb and ifnc are the best-defined representatives63. Here, CpG-stimulated B cells derived from HK and spleen showed increased transcript levels of ifnb, while elevated ifna1 mRNA levels were detected in B cells from all three tissues (Fig. 7). Together, these results propose that salmon B cells exhibit a type I IFN-response. In line with this, rainbow trout IgM+ PB and spleen cells have been shown to transcribe ifn1 upon poly I:C and VHSV infection64 and type I IFN responses have been reported for CpG B treated murine B cells65. In general, the basal levels of IFN transcripts in the purified salmon B cells were low (ifna1 mean Cq 31.8–33.7) or non-detectable for ifnb and ifnc (Supplementary Table 2). Interestingly, elevated ifna1 mRNA levels were detected both for CpG alone and for CpG + PAS during the course of time, while ifnb levels peaked at 24 hours and then declined to its basal levels at 48 hours. A possible explanation for this is that the initial ifna1 burst triggers the transcription of interferon regulatory factors, which mediate a positive feedback loop that lead to the induction of a second wave of ifna1 transcription66,67. The cytosolic receptors RIG-I/MDA5 that recognize viral RNA are suggested to be the main receptors to activate pathways leading to ifna1 expression in salmon68. Our results imply that other receptors specific for DNA induce ifna1 gene transcription in salmon B cells. The inverted (GpC) ODN control was non-stimulatory (Supplementary Fig. S2), proposing that the ifna1 induction in salmon IgM+ cells is mediated through a CpG-specific TLR. Unlike ifna1, ifnb is reported to show modest basal expression in most salmon organs, while a rapid and potent induction appear in salmon HK and spleen upon treatment with R848, a ligand for TLR7/TLR868. This observation, together with the rapid increase in ifnb levels in the purified HK and spleen B cells upon TLR9 ligation, suggest that signaling through the TLR7 family activates pathways important for ifnb expression in salmon leukocytes. The significant increase in ifnb expression when combining PAS and CpG, suggest that other signals provided by the supernatant are necessary for optimal ifnb production in B cells.
B cells have important protective roles against many viral infections, mainly through the production of neutralizing Abs. In mammals type I IFN promotes B cell activation and Ab responses, including class switch during viral infections69,70. It is thus possible that type I IFNs produced by salmon B cells, affect the regulation and functional activities of the B cells themselves, in addition to that of other immune cells. Interestingly, by using a DNA vaccine model, adjuvant effects of Atlantic salmon type I IFNs were demonstrated and shown to increase the virus specific IgM response71. These results establish a link between type I IFNs and the adaptive immune system in salmon, although the mechanisms behind are currently unknown. In support, our group has earlier reported that CpG and poly I:C significantly enhance virus-specific humoral responses in vivo, where a substantial increase in ifna1 transcripts in HK and spleen were detected at 12 and 48 hours post injection26. In the previous study26, however, whether the IFNs was produced by B cells or by other cell types was not investigated. The implications of direct interactions between type I IFNs and B cells in salmon has not been elucidated and is a topic for future studies.
Materials and Methods
Fish
Atlantic salmon (Salmo salar L.) roe from Aqua Gen (Aqua Gen, Kyrksæterøra, Norway) was hatched and smoltified at Tromsø Aquaculture Research Station (Tromsø, Norway). The fish were kept in seawater at natural temperature and photoperiod and fed with dry commercial feed (Skretting, Stavanger, Norway) before transfer to the research facility of the Arctic University of Norway, Tromsø, where they were kept at similar conditions until sacrifice. Fish were acclimated for at least two weeks before sampling. Two batches of fish were used; 400 g fish for FACS analysis and 1000–1500 g fish for MACS sorting and immune gene expression analysis of IgM+ B cells. We confirm that the experimental protocols used for the live fish experiments were based on the Animal Welfare Act (https://www.regjeringen.no/en/dokumenter/animal-welfare-act/id571188/) and performed in accordance with relevant guidelines and regulations given by the Norwegian Animal Research Authority.
Reagents
Phosphorothioate-modified B class CpG 2006 ODN (5′-T*CG T*CG T*TT T*GT C*GT T*TT G*TC G*T*T-3′, * indicates phosphothioate bonds) and non-CpG 2007 ODN, inverting all CGs to GCs, were purchased from Integrated DNA technologies. The monoclonal Abs IgF1-18 (6-1-18) anti-trout IgM38 and anti-trout IgT13 are described earlier. Isotype specific secondary Abs; IgG1-RPE (for IgM) and IgG2a-APC (for IgT) were both from Jackson ImmunoResearch. Fixable Viability Dye 780 (FVD780) was purchased from eBioscience. MACS MS columns and anti-Mouse IgG1 MicroBeads were purchased from Miltenyi Biotec. Anti-actin Ab was purchased from Sigma, while the preparation of MHCIIβ antiserum is described previously56.
Cell isolation
Leukocytes from HK, PK, spleen and PB were isolated on Percoll (GE Healthcare) gradients72,73. Briefly, HK and spleen were sampled aseptically and kept on ice-cold transport medium (L-15 medium with 10 U/ml penicillin, 10 μg/ml streptomycin, 2% fetal bovine serum (FBS), 20 U/ml heparin) until homogenization on 100-μm cells strainers (Falcon). The resulting cell suspensions were layered on 25/54% discontinuous Percoll gradients and centrifuged at 400 × g for 40 min at 4 °C. Cells at the interface were collected and washed twice in L-15 medium. PB was collected from the caudal vein using heparinized vacutainer tube and diluted immediately at least two fold in ice-cold transport medium. The resulting suspension was layered on a 54% Percoll gradient, centrifuged, and harvested as above. Cells were counted using an automatic cell counter (NucleoCounter, YC-100). The isolation of HK monocytes/macrophages were performed as described elsewhere56 and the adherent cells were cultivated in L-15+ (L-15 supplemented with 5% FBS and penicillin/streptomycin) in 24 well plates (Nunclon Delta Surface, Thermo Scientific) for 3 days before being analyzed.
Flow cytometry analyses
For the Ig-frequency analysis, 3 × 105 HK, PK, PB and spleen leukocytes from twelve individuals were each pelleted at 500 × g directly after isolation, further washed in PBS+ (PBS with 0.5% BSA) and stained with anti-trout IgM (1:200 dilution) and anti-trout IgT (2 μg/mL) mAbs for 30 minutes. After washing with PBS+, the samples were incubated with isotype specific secondary Abs; IgG1-RPE and IgG2a-APC (1:400 dilution), respectively, and FVD780 (1 μl/ml) in PBS+ for 20 minutes. All staining and centrifugation steps were done at 4 °C in 96-well U-bottom plates (Nunclon Delta Surface). Ig-frequencies were analyzed on Aria II flow cytometer (Becton Dickinson), while data processing was done in FlowJo (Tree Star). Dead cells (FVD780+) and doublets (SSC-A vs SSC-H) were excluded before the frequency of IgM and IgT were determined
Magnetic activated cell sorting (MACS) of IgM+ B cells
The sorting procedure was performed as per the company’s recommendation (Miltenyi Biotec). All washes and incubations were done using MACS sorting buffer (SB; PBS with 0.5% bovine serum albumin and 2 mM EDTA) and spun at 450 × g 4 °C for 5 minutes. Briefly, after one wash with SB, 7–8 × 107 cells were incubated with a 1:200 dilution of anti-trout IgM mAb for 30 minutes at 4 °C. After two washes, cells were incubated in 80 μl SB and 20 μl anti-mouse IgG1 microbeads for 15 minutes at 4 °C. Sorted IgM+ B cells were pelleted immediately and resuspended in L-15+. Zero time-point samples were obtained before initiation of stimulations. A time course study of viability of cultured IgM+ MACS purified cells from blood was performed by flow cytometry using the FVD780 stain as described above (n = 2). The cells were seeded and stimulated with CpG (2 μM) as described below. Purity of sorted IgM+ B cells was determined by flow cytometry and RT-qPCR of cd4–2 (T cell marker), macrophage colony stimulating factor receptor (csf-1r) and the scavenger receptor MARCO genes transcripts. For comparison, the expression of the same genes was measured in HK-derived untreated MLC and FACS sorted IgM+ splenic cells, where MLC were excluded by gating.
Stimulation of sorted IgM+ B cells for gene expression study
MACS sorted IgM+ B cells, 1 × 106, were seeded in 100 μl L-15+ in 96-well U-bottom plates (Nunclon Delta Surface, Thermo Scientific). For stimulation, either PAS or CAS (see below) in a 1:2 dilution or 2 μM CpG B ODN was applied to the cells. In addition, some cells were stimulated with PAS (diluted 1:2) combined with CpG (2 μM). Controls received only L-15+. A non-CpG control (2 µM) was also included. Cells were incubated at 14 °C for 12, 24 or 48 hours before harvested for RNA isolation. Due to variations in cell yield at each isolation, the number of individuals for each treatment varied between 3 to 6.
Preparation of conditioned supernatant
Six million HKLs were pulsed with 2 μM CpG 2006 for 8 hours (pulse adherent cell supernatant; PAS) or left unstimulated (control adherent cell supernatant; CAS) at 14 °C. After the pulse treatment, culture media and non-adherent cells were removed, while the adherent cells were washed and further incubated at 14 °C for 72 hours. After centrifugation at 450 × g at 4 °C for five minutes, PAS and CAS, respectively, were harvested, pooled (three individuals) and stored at −80 °C until used.
RNA extraction and transcript analyses
Total RNA from sorted IgM+ B cells and HK MLC was isolated using the RNeasy Mini Kit (Qiagen) following the manufacturer’s recommendation. On-column DNase digestion was performed using RNase-free DNase set (Qiagen), and RNA was quantified using NanoDrop (ND 1000 Spectrophotometer). Twenty-five microliter cDNA reactions with 150 ng total RNA were synthesized using TaqMan reverse transcription reagents (Applied Biosystems) under the following conditions: 25 °C for 10 min, 48 °C for 30 min and 95 °C for 5 min. cDNA samples were diluted 1:5 and stored at −20 °C until use.
qPCR was run as 20 μl duplicate reactions with 6 ng cDNA/reaction on a 7500 Fast Real-Time PCR Systems (Applied Biosystems) according to their standard protocol. Primer and probe sequences are listed in Supplementary Table 1. A negative control (no template) reaction was performed for each primer pair. The Cq-threshold was set to 0.20 for both reference gene EF1aB and target genes. A melt curve analysis was also performed to ensure that a single product had been amplified. For the stimulated cells, fold change was calculated using the non-treated cells from each tissue and time point as a control74. Relative expression (zero hour samples) of tlr3, 22, 9, 21 and 8a1 were calculated using the 2−ΔCq method75 where ΔCq was calculated by subtracting the EF1aB Cq value from the target gene Cq value.
Western blot analysis of MHC II and secreted IgM
Supernatants and cell lysates from non-treated or CpG (2 µM) treated HK (n = 6) and spleen (n = 6) sorted IgM+ B cells were harvested after 72 hours culture at 14 °C. Supernatants were up-concentrated by 0.5 ml centrifugal filter columns (Millipore) and then diluted 32-fold before denaturation in 2× LDS buffer at 70 °C for 10 minutes. Cells were sampled in 2× LDS buffer and diluted to 1.62 × 105 cells/sample before denaturation. All samples were run on precasted 4–12% gradient NuPAGE Novex Bis-Tris gels and subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) with 1× MOPS buffer (Invitrogen) for 50 min at 200 V and 120 mA. MagicMark™ XP and SeeBlue Plus 2 pre-stained (both from Invitrogen), were loaded for molecular weight estimation. The proteins were blotted onto a polyvinylidene difluoride membrane, blocked with 5% BSA (Sigma) and incubated overnight with either anti-trout IgM Ab (1:200 dilution), anti-salmon MHCII Ab (1:1000)56 or anti-actin rabbit Ab (1:600). The respective blots were incubated for 1 hour with goat anti-mouse-HRP Ab or goat anti-rabbit-HRP Ab (1:8000 dilution; Santa Cruz Biotechnology) in 5% BSA. The blots were developed using SuperSignal West Femto Trial Kit (Thermo) and a KODAK Image Station 4000 MM Digital Imaging System. For IgM, band intensities (arbitrary units, AU) were determined by subtracting the background noise from the visualized bands, while the relative expression of MHCII was calculated by:
Statistical analysis
RT-qPCR data were based on duplicate measurements of three to six individuals and were analyzed in GraphPad Prism 5.04 or SPSS version 24. Statistical evaluation were performed using two-tailed non-parametric Mann-Whitney U or Dunn’s multiple comparison test following a significant Kruskal-Wallis test on the fold change and relative expression of the transcript data, respectively. The IgM secretion and MHCII protein expression data were analyzed by Mann-Whitney U. For all analyses, differences were considered significant at p < 0.05.
Electronic supplementary material
Acknowledgements
This study was supported by the Aquaculture program of The Research Council of Norway (grant no. 237315/E40 ViVaFish), the RDA-project: “Forskerskole i marin bioteknologi”, UiT the Arctic University of Norway and the National Institutes of Health Grant 2R01GM085207-05 (to J.O.S.) and by the National Science Foundation Grant NSF-IOS-1457282 (to J.O.S.). The authors thank Dr. Søren Grove, for providing the Western blot analysis demonstrating the cross-reactivity of the anti-trout IgT Ab to the CH4-domain of salmon IgT. We also thank Linn G. Tollersrud, Kjersti Julin and Guro Strandskog for their excellent technical assistance. Frøydis Strand is also acknowledged for the excellent technical help with the figures.
Author Contributions
S.J. and H.L.T. processed the samples used in this study and analyzed the data. S.J. performed cell stimulation, MACS sorting and RT-qPCR while H.L.T. performed FACS and Western blot analysis. S.J., H.L.T., I.J. and J.B.J. prepared and reviewed the manuscript. O.S. and K.S. provided the anti-trout IgT and IgM Abs, respectively, and reviewed the manuscript. I.J. and J.B.J. designed and supervised the experiments, collaborated in obtaining funding and J.B.J. directed the project.
Competing Interests
The authors declare no competing interests.
Footnotes
Shiferaw Jenberie and Hanna L. Thim contributed equally to this work.
A correction to this article is available online at https://doi.org/10.1038/s41598-018-24843-9.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21895-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pone EJ, et al. Toll-like Receptors and B-cell Receptors Synergize to Induce Immunoglobulin Class Switch DNA Recombination: Relevance to Microbial Antibody Responses. Crit Rev Immunol. 2010;30:1–29. doi: 10.1615/CritRevImmunol.v30.i1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zwollo P, Cole S, Bromage E, Kaattari S. B Cell Heterogeneity in the Teleost Kidney: Evidence for a Maturation Gradient from Anterior to Posterior Kidney. J. Immunol. 2005;174:6608–6616. doi: 10.4049/jimmunol.174.11.6608. [DOI] [PubMed] [Google Scholar]
- 3.Fillatreau S, et al. The astonishing diversity of Ig classes and B cell repertoires in teleost fish. Front. Immunol. 2013;4:28. doi: 10.3389/fimmu.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hordvik I. Immunoglobulin isotypes in Atlantic salmon, Salmo salar. Biomolecules. 2015;5:166–177. doi: 10.3390/biom5010166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bengten E, Clem LW, Miller NW, Warr GW, Wilson M. Channel catfish immunoglobulins: repertoire and expression. Dev. Comp. Immunol. 2006;30:77–92. doi: 10.1016/j.dci.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 6.Mashoof, S. & Criscitiello, M. F. Fish Immunoglobulins. Biology (Basel) 5 (2016). [DOI] [PMC free article] [PubMed]
- 7.Danilova, N., Bussmann, J., Jekosch, K. & Steiner, L. A. The immunoglobulin heavy-chain locus in zebrafish: identification and expression of a previously unknown isotype, immunoglobulin Z. Nat. Immunol. 6 (2005). [DOI] [PubMed]
- 8.Hansen JD, Landis EDa, Phillips RB. Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: Implications for a distinctive B cell developmental pathway in teleost fish. PNAS. 2005;102:6919–6924. doi: 10.1073/pnas.0500027102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tadiso TM, Lie KK, Hordvik I. Molecular cloning of IgT from Atlantic salmon, and analysis of the relative expression of tau, mu, and delta in different tissues. Vet. Immunol. Immunopathol. 2011;139:17–26. doi: 10.1016/j.vetimm.2010.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Sunyer JO. Fishing for mammalian paradigms in the teleost immune system. Nat. Immunol. 2013;14:320–326. doi: 10.1038/ni.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Z, et al. Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. Proc. Natl. Acad. Sci. USA. 2013;110:13097–13102. doi: 10.1073/pnas.1304319110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Z, et al. Mucosal immunoglobulins at respiratory surfaces mark an ancient association that predates the emergence of tetrapods. Nat Commun. 2016;7:10728. doi: 10.1038/ncomms10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang YA, et al. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat. Immunol. 2010;11:827–835. doi: 10.1038/ni.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edholm ES, et al. Identification of two IgD + B cell populations in channel catfish, Ictalurus punctatus. J. Immunol. 2010;185:4082–4094. doi: 10.4049/jimmunol.1000631. [DOI] [PubMed] [Google Scholar]
- 15.Parra D, Takizawa F, Sunyer JO. Evolution of B cell immunity. Annu Rev Anim Biosci. 2013;1:65–97. doi: 10.1146/annurev-animal-031412-103651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tacchi L, et al. Nasal immunity is an ancient arm of the mucosal immune system of vertebrates. Nat Commun. 2014;5:5205. doi: 10.1038/ncomms6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castro R, et al. CCR7 is mainly expressed in teleost gills, where it defines an IgD + IgM- B lymphocyte subset. J. Immunol. 2014;192:1257–1266. doi: 10.4049/jimmunol.1302471. [DOI] [PubMed] [Google Scholar]
- 18.Lund FE. Cytokine-producing B lymphocytes – key regulators of immunity. Curr. Opin. Immunol. 2008;20:332–338. doi: 10.1016/j.coi.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu LY, et al. B cells in teleost fish act as pivotal initiating APCs in priming adaptive immunity: an evolutionary perspective on the origin of the B-1 cell subset and B7 molecules. J. Immunol. 2014;192:2699–2714. doi: 10.4049/jimmunol.1301312. [DOI] [PubMed] [Google Scholar]
- 20.Allman D, Pillai S. Peripheral B cell subsets. Curr. Opin. Immunol. 2008;20:149–157. doi: 10.1016/j.coi.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X. Regulatory functions of innate-like B cells. Cell. Mol. Immunol. 2013;10:113–121. doi: 10.1038/cmi.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genestier L, et al. TLR Agonists Selectively Promote Terminal Plasma Cell Differentiation of B Cell Subsets Specialized in Thymus-Independent Responses. J. Immunol. 2007;178:7779–7786. doi: 10.4049/jimmunol.178.12.7779. [DOI] [PubMed] [Google Scholar]
- 23.Haugland GT, Jordal AE, Wergeland HI. Characterization of small, mononuclear blood cells from salmon having high phagocytic capacity and ability to differentiate into dendritic like cells. PLoS One. 2012;7:e49260. doi: 10.1371/journal.pone.0049260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, et al. B lymphocytes from early vertebrates have potent phagocytic and microbicidal abilities. Nat. Immunol. 2006;7:1116–1124. doi: 10.1038/ni1389. [DOI] [PubMed] [Google Scholar]
- 25.Abos B, et al. Transcriptional heterogeneity of IgM + cells in rainbow trout (Oncorhynchus mykiss) tissues. PLoS One. 2013;8:e82737. doi: 10.1371/journal.pone.0082737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thim HL, et al. Immunoprotective activity of a Salmonid Alphavirus Vaccine: comparison of the immune responses induced by inactivated whole virus antigen formulations based on CpG class B oligonucleotides and poly I:C alone or combined with an oil adjuvant. Vaccine. 2012;30:4828–4834. doi: 10.1016/j.vaccine.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 27.Thim HL, et al. Vaccines (Basel) 2014. Vaccine Adjuvants in Fish Vaccines Make a Difference: Comparing Three Adjuvants (Montanide ISA763A Oil, CpG/Poly I:C Combo and VHSV Glycoprotein) Alone or in Combination Formulated with an Inactivated Whole Salmonid Alphavirus Antigen; pp. 228–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy JE, Tedbury PR, Homer-Vanniasinkam S, Walker JH, Ponnambalam S. Biochemistry and cell biology of mammalian scavenger receptors. Atherosclerosis. 2005;182:1–15. doi: 10.1016/j.atherosclerosis.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 29.Roca FJ, Sepulcre MA, Lopez-Castejon G, Meseguer J, Mulero V. The colony-stimulating factor-1 receptor is a specific marker of macrophages from the bony fish gilthead seabream. Mol. Immunol. 2006;43:1418–1423. doi: 10.1016/j.molimm.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 30.Maisey K, et al. Isolation and Characterization of Salmonid CD4 + T Cells. J. Immunol. 2016;196:4150–4163. doi: 10.4049/jimmunol.1500439. [DOI] [PubMed] [Google Scholar]
- 31.Matsuo A, et al. Teleost TLR22 Recognizes RNA Duplex to Induce IFN and Protect Cells from Birnaviruses. J. Immunol. 2008;181:3474–3485. doi: 10.4049/jimmunol.181.5.3474. [DOI] [PubMed] [Google Scholar]
- 32.Yeh DW, et al. Toll-like receptor 9 and 21 have different ligand recognition profiles and cooperatively mediate activity of CpG-oligodeoxynucleotides in zebrafish. Proc. Natl. Acad. Sci. USA. 2013;110:20711–20716. doi: 10.1073/pnas.1305273110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poynter S, Lisser G, Monjo A, DeWitte-Orr S. Biology (Basel) 2015. Sensors of Infection: Viral Nucleic Acid PRRs in Fish; pp. 460–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sunyer OJ. Evolutionary and Functional Relationships of B Cells from Fish and Mammals: Insights into their Novel Roles in Phagocytosis and Presentation of Particulate Antigen. Infect. Disord. Drug. Targets. 2012;12:200–212. doi: 10.2174/187152612800564419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prazma CM, Yazawa N, Fujimoto Y, Fujimoto M, Tedder TF. CD83 Expression Is a Sensitive Marker of Activation Required for B Cell and CD4 + T Cell Longevity In Vivo. Journal Immunol. 2007;179:4550–4562. doi: 10.4049/jimmunol.179.7.4550. [DOI] [PubMed] [Google Scholar]
- 36.Lim TS, et al. CD80 and CD86 differentially regulate mechanical interactions of T-cells with antigen-presenting dendritic cells and B-cells. PLoS One. 2012;7:e45185. doi: 10.1371/journal.pone.0045185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma DY, Clark EA. The role of CD40 and CD154/CD40L in dendritic cells. Semin. Immunol. 2009;21:265–272. doi: 10.1016/j.smim.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hedfors IA, Bakke H, Skjodt K, Grimholt U. Antibodies recognizing both IgM isotypes in Atlantic salmon. Fish Shellfish Immunol. 2012;33:1199–1206. doi: 10.1016/j.fsi.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 39.Köllner B, Wasserrab B, Kotterba GA, Fischer U. Evaluation of immune functions of rainbow trout (Oncorhynchus mykiss)—how can environmental influences be detected? Toxicol. Lett. 2002;131:83–95. doi: 10.1016/S0378-4274(02)00044-9. [DOI] [PubMed] [Google Scholar]
- 40.Miyadai T, Ootani M, Tahara D, Aoki M, Saitoh K. Monoclonal antibodies recognising serum immunoglobulins and surface immunoglobulin-positive cells of puffer fish, torafugu (Takifugu rubripes) Fish Shellfish Immunol. 2004;17:211–222. doi: 10.1016/j.fsi.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 41.Zhang N, Zhang XJ, Chen DD, Oriol Sunyer J, Zhang YA. Molecular characterization and expression analysis of three subclasses of IgT in rainbow trout (Oncorhynchus mykiss) Dev. Comp. Immunol. 2017;70:94–105. doi: 10.1016/j.dci.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu CS, Sun Y, Hu YH, Sun L. Identification and analysis of a CpG motif that protects turbot (Scophthalmus maximus) against bacterial challenge and enhances vaccine-induced specific immunity. Vaccine. 2010;28:4153–4161. doi: 10.1016/j.vaccine.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 43.Altmann S, et al. Toll-like receptors in maraena whitefish: Evolutionary relationship among salmonid fishes and patterns of response to Aeromonas salmonicida. Fish Shellfish Immunol. 2016;54:391–401. doi: 10.1016/j.fsi.2016.04.125. [DOI] [PubMed] [Google Scholar]
- 44.Arnemo M, Kavaliauskis A, Gjoen T. Effects of TLR agonists and viral infection on cytokine and TLR expression in Atlantic salmon (Salmo salar) Dev. Comp. Immunol. 2014;46:139–145. doi: 10.1016/j.dci.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 45.Rebl A, Siegl E, Kollner B, Fischer U, Seyfert HM. Characterization of twin toll-like receptors from rainbow trout (Oncorhynchus mykiss): evolutionary relationship and induced expression by Aeromonas salmonicida salmonicida. Dev. Comp. Immunol. 2007;31:499–510. doi: 10.1016/j.dci.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 46.Brownlie R, et al. Chicken TLR21 acts as a functional homologue to mammalian TLR9 in the recognition of CpG oligodeoxynucleotides. Mol. Immunol. 2009;46:3163–3170. doi: 10.1016/j.molimm.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 47.Keestra AM, de Zoete MR, Bouwman LI, van Putten JP. Chicken TLR21 is an innate CpG DNA receptor distinct from mammalian TLR9. J. Immunol. 2010;185:460–467. doi: 10.4049/jimmunol.0901921. [DOI] [PubMed] [Google Scholar]
- 48.Lee PT, et al. Identification and characterisation of TLR18-21 genes in Atlantic salmon (Salmo salar) Fish Shellfish Immunol. 2014;41:549–559. doi: 10.1016/j.fsi.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 49.Rebl A, Goldammer T, Seyfert HM. Toll-like receptor signaling in bony fish. Vet. Immunol. Immunopathol. 2010;134:139–150. doi: 10.1016/j.vetimm.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 50.Bromage ES, Kaattari IM, Zwollo P, Kaattari SL. Plasmablast and Plasma Cell Production and Distribution in Trout Immune Tissues. J. Immunol. 2004;173:7317–7323. doi: 10.4049/jimmunol.173.12.7317. [DOI] [PubMed] [Google Scholar]
- 51.Davidson GA, Lin SH, Secombes CJa, Ellis EA. Detection of specific and ‘constitutive’ antibody secreting cells in the gills, head kidney and peripheral blood leucocytes of dab (Limanda limanda) Vet. Immunol. Immunopathol. 1997;58:363–374. doi: 10.1016/S0165-2427(97)00017-2. [DOI] [PubMed] [Google Scholar]
- 52.Jiang W, et al. TLR9 stimulation drives naive B cells to proliferate and to attain enhanced antigen presenting function. Eur. J. Immunol. 2007;37:2205–2213. doi: 10.1002/eji.200636984. [DOI] [PubMed] [Google Scholar]
- 53.Iliev DB, Thim H, Lagos L, Olsen R, Jorgensen JB. Homing of Antigen-Presenting Cells in Head Kidney and Spleen - Salmon Head Kidney Hosts Diverse APC Types. Front. Immunol. 2013;4:137. doi: 10.3389/fimmu.2013.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kretschmer B, et al. CD83 modulates B cell function in vitro: increased IL-10 and reduced Ig secretion by CD83Tg B cells. PLoS One. 2007;2:e755. doi: 10.1371/journal.pone.0000755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bjorck P, Axelsson B, Paulie S. Expression of CD40 and CD43 during activation of human B lymphocytes. Scand. J. Immunol. 1991;33:211–218. doi: 10.1111/j.1365-3083.1991.tb03751.x. [DOI] [PubMed] [Google Scholar]
- 56.Iliev DB, et al. CpG-induced secretion of MHCIIbeta and exosomes from salmon (Salmo salar) APCs. Dev. Comp. Immunol. 2010;34:29–41. doi: 10.1016/j.dci.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 57.Overland HS, Pettersen EF, Ronneseth A, Wergeland HI. Phagocytosis by B-cells and neutrophils in Atlantic salmon (Salmo salar L.) and Atlantic cod (Gadus morhua L.) Fish Shellfish Immunol. 2010;28:193–204. doi: 10.1016/j.fsi.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 58.Abos B, et al. Distinct Differentiation Programs Triggered by IL-6 and LPS in Teleost IgM(+) B Cells in The Absence of Germinal Centers. Sci. Rep. 2016;6:30004. doi: 10.1038/srep30004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Castro R, Abos B, Gonzalez L, Granja AG, Tafalla C. Expansion and differentiation of IgM(+) B cells in the rainbow trout peritoneal cavity in response to different antigens. Dev. Comp. Immunol. 2017;70:119–127. doi: 10.1016/j.dci.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 60.Gong YF, Xiang LX, Shao JZ. CD154-CD40 interactions are essential for thymus-dependent antibody production in zebrafish: insights into the origin of costimulatory pathway in helper T cell-regulated adaptive immunity in early vertebrates. J. Immunol. 2009;182:7749–7762. doi: 10.4049/jimmunol.0804370. [DOI] [PubMed] [Google Scholar]
- 61.Lagos LX, Iliev DB, Helland R, Rosemblatt M, Jorgensen JB. CD40L–a costimulatory molecule involved in the maturation of antigen presenting cells in Atlantic salmon (Salmo salar) Dev. Comp. Immunol. 2012;38:416–430. doi: 10.1016/j.dci.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 62.Zheng Y, et al. CD86 and CD80 Differentially Modulate the Suppressive Function of Human Regulatory T Cells. J. Immunol. 2004;172:2778–2784. doi: 10.4049/jimmunol.172.5.2778. [DOI] [PubMed] [Google Scholar]
- 63.Robertsen B. The role of type I interferons in innate and adaptive immunity against viruses in Atlantic salmon. Dev. Comp. Immunol. 2018;80:41–52. doi: 10.1016/j.dci.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 64.Abos B, et al. Early activation of teleost B cells in response to rhabdovirus infection. J. Virol. 2015;89:1768–1780. doi: 10.1128/JVI.03080-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oganesyan G, et al. IRF3-dependent type I interferon response in B cells regulates CpG-mediated antibody production. J. Biol. Chem. 2008;283:802–808. doi: 10.1074/jbc.M704755200. [DOI] [PubMed] [Google Scholar]
- 66.Bergan V, Kileng O, Sun B, Robertsen B. Regulation and function of interferon regulatory factors of Atlantic salmon. Mol. Immunol. 2010;47:2005–2014. doi: 10.1016/j.molimm.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 67.Iliev DB, Sobhkhez M, Fremmerlid K, Jorgensen JB. MyD88 interacts with interferon regulatory factor (IRF) 3 and IRF7 in Atlantic salmon (Salmo salar): transgenic SsMyD88 modulates the IRF-induced type I interferon response and accumulates in aggresomes. J. Biol. Chem. 2011;286:42715–42724. doi: 10.1074/jbc.M111.293969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Svingerud T, et al. Atlantic salmon type I IFN subtypes show differences in antiviral activity and cell-dependent expression: evidence for high IFNb/IFNc-producing cells in fish lymphoid tissues. J. Immunol. 2012;189:5912–5923. doi: 10.4049/jimmunol.1201188. [DOI] [PubMed] [Google Scholar]
- 69.Bon AL, et al. Type I Interferons Potently Enhance Humoral Immunity and Can Promote Isotype Switching by Stimulating Dendritic Cells In Vivo. Immunity. 2001;14:461–470. doi: 10.1016/S1074-7613(01)00126-1. [DOI] [PubMed] [Google Scholar]
- 70.Coro ES, Chang WLW, Baumgarth N. Type I IFN Receptor Signals Directly Stimulate Local B Cells Early following Influenza Virus Infection. J. Immunol. 2006;176:4343–4351. doi: 10.4049/jimmunol.176.7.4343. [DOI] [PubMed] [Google Scholar]
- 71.Chang CJ, Sun B, Robertsen B. Adjuvant activity of fish type I interferon shown in a virus DNA vaccination model. Vaccine. 2015;33:2442–2448. doi: 10.1016/j.vaccine.2015.03.093. [DOI] [PubMed] [Google Scholar]
- 72.Jørgensen BJ, Johansen A, Stenersen B, Ann-Inger Sommer IA. CpG oligodeoxynucleotides and plasmid DNA stimulate Atlantic salmon (Salmo salar L.) leucocytes to produce supernatants with antiviral activity. Dev. Comp. Immunol. 2001;25:313–321. doi: 10.1016/S0145-305X(00)00068-9. [DOI] [PubMed] [Google Scholar]
- 73.Reitan LJ, Thuvander A. In vitro stimulation of salmonid leucocytes with mitogens and with Aeromonas salmonicida. Fish Shellfish Immunol. 1991;1:297–307. doi: 10.1016/S1050-4648(05)80067-1. [DOI] [Google Scholar]
- 74.Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.