Abstract
It is well known that saturated fatty acids (SFAs) and unsaturated fatty acid, in particular omega-3 polyunsaturated fatty acids (n-3 PUFAs), have different effects on inflammatory signaling: SFAs are pro-inflammatory but n-3 PUFAs have strong anti-inflammatory properties. We have reported that palmitic acid (PA), a saturated fatty acid, robustly amplifies lipopolysaccharide (LPS) signaling to upregulate proinflammatory gene expression in macrophages. We also reported that the increased production of ceramide (CER) via sphingomyelin (SM) hydrolysis and CER de novo synthesis plays a key role in the synergistic effect of LPS and PA on proinflammatory gene expression. However, it remains unclear if n-3 PUFAs are capable of antagonizing the synergistic effect of LPS and PA on gene expression and CER production. In this study, we employed the above macrophage culture system and lipidomical analysis to assess the effect of n-3 PUFAs on proinflammatory gene expression and CER production stimulated by LPS and PA. Results showed that DHA strongly inhibited the synergistic effect of LPS and PA on proinflammatory gene expression by targeting nuclear factor kappa B (NFκB)-dependent gene transcription. Results also showed that DHA inhibited the cooperative effect of LPS and PA on CER production by targeting CER de novo synthesis, but not SM hydrolysis. Furthermore, results showed that myriocin, a specific inhibitor of serine palmitoyltransferase, strongly inhibited both LPS-PA-stimulated CER synthesis and proinflammatory gene expression, indicating that CER synthesis is associated with proinflammatory gene expression and that inhibition of CER synthesis contributes to DHA-inhibited proinflammatory gene expression. Taken together, this study demonstrates that DHA antagonizes the boosting effect of PA on LPS signaling on proinflammatory gene expression by targeting both NFκB-dependent transcription and CER de novo synthesis in macrophages.
Introduction
Metabolic endotoxemia is a pathological condition in which circulating lipopolysaccharide (LPS) is elevated in patients with obesity or diabetes as a result of high-fat diet (HFD)-increased intestinal permeability that facilitates the translocation of microbiome-derived LPS from the intestine to the bloodstream [1, 2]. It has been shown that bacterial LPS activity in human serum is associated with dyslipidemia, obesity and chronic inflammation [3]. In addition to LPS, plasma free saturated fatty acid (SFA) is also increased in patients with obesity or diabetes and promotes inflammation and cardiovascular disease [4–6]. When both LPS and SFA in circulation are elevated, they may act in concert to boost a strong proinflammatory response that cannot be generated by LPS or SFA alone. Indeed, our recent study supported the above notion as it showed a cooperative stimulation of atherogenesis by combining treatment of LPS with feeding SFA-rich HFD in animal models [7]. Our in vitro study also showed that palmitic acid (PA), the most abundant SFA in plasma [8], amplified LPS-triggered signaling for upregulating proinflammatory genes such as interleukin 6 (IL-6) in macrophages [9]. Furthermore, our study to explore the underlying mechanisms showed that the stimulation of ceramide (CER) production by PA and LPS plays a key role in the synergistic effect of PA and LPS on proinflammatory gene expression [9].
CER is mainly generated by the breakdown of sphingomyelin by sphingomyelinases (SMases) that include acid SMase (aSMase), neutral SMase (nSMase) or alkaline SMase [10]. Of these SMases, aSMase and nSMase are considered as the major SMases for the production of cellular CER. CER can be also generated by de novo synthesis catalyzed by enzymes such as serine palmitoyl transferase and CER synthase. CER plays key roles in a variety of cellular responses, regulating cell growth, differentiation, senescence, apoptosis and inflammation [11].
In contrast to the proinflammatory effects of SFAs, omega-3 (n-3) polyunsaturated fatty acids (PUFAs) have anti-inflammatory properties [12, 13]. A recent clinical study showed that supplementation with n-3 PUFAs in obese patients reduced proinflammatory cytokines and C-reactive protein, and increased adiponectin in circulation [14]. Another clinical study also showed that n-3 PUFAs reduced the circulating level and adipose tissue expression of proinflammatory cytokines in obese patients [15]. Although the beneficial effects of n-3 PUFAs on obesity or diabetes have been increasingly appreciated in recent years [16, 17], the underlying mechanisms whereby n-3 PUFAs inhibit inflammation have not been fully understood.
In this study, we have employed the cell model in which PA amplifies LPS signaling for proinflammatory molecule expression [9] to determine if n-3 PUFAs is capable of antagonizing PA-boosted LPS proinflammatory signaling. Moreover, since we have shown in this cell model that increased production of CER, a bioactive sphingolipid [18], is involved in the synergistic stimulation of proinflammatory cytokine expression by LPS and PA [9], we also determined if n-3 PUFAs antagonizes the stimulatory effect of LPS and PA on CER production.
Materials and methods
Cell culture
RAW264.7 cells, the murine macrophages used extensively in the investigations of the role of macrophages in inflammation [19], were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and grown in Dulbecco's modified Eagle's medium (DMEM) (ATCC, Manassas, VA) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (HyClone, Logan, UT). The cells were maintained in a 37°C, 90% relative humidity, 5% CO2 environment. The cells were seeded into 12-well plates at a density of 2 x 105/well and allowed to grow for 24 h before treatment. The cells at passage 5–8 were used and were 80% confluent at the treatment. For cell treatment, LPS from E. coli serotype 055:B5 (Sigma, St. Louis, MO) was used. The LPS was highly purified by phenol extraction and gel filtration chromatography, and was cell culture tested.
Fatty acid preparation
The PA, DHA, eicosapentaenoic acid (EPA), oleic acid (OA), linoleic acid (LA) and palmitoleic acid (POA) used in this study were purchased from Sigma (St. Louis, MO) and prepared as described previously [20, 21]. Briefly, fatty acids were dissolved in 0.1 N NaOH and 70% ethanol at 70°C to make concentration of 50 mM. The solution was kept at 55°C for 10 min, mixed, and brought to room temperature before treatment.
Enzyme-linked immunosorbent assay (ELISA)
IL-6 in medium was quantified using sandwich ELISA kits according to the protocol provided by the manufacturer (Biolegend, San Diego, CA).
Real-time polymerase chain reaction (PCR)
Total RNA was isolated from cells using RNeasy minikit (Qiagen, Santa Clarita, CA). First-strand complementary DNA (cDNA) was synthesized with the iScriptTM cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA) using 20 μl of reaction mixture containing 0.5 μg of total RNA, 4 μl of 5x iScript reaction mixture, and 1 μl of iScript reverse transcriptase. The complete reaction was cycled for 5 minutes at 25 oC, 30 minutes at 42 oC and 5 minutes at 85°C using a PTC-200 DNA Engine (MJ Research, Waltham, MA). The reverse transcription reaction mixture was then diluted 1:10 with nuclease-free water and used for PCR amplification in the presence of the primers [mouse serine palmitoyltransferase (SPT)1: 5’ primer sequence, AGTGGTGGGAGAGTC CCTTT; 3’ primer sequence, CAGTGACCACAACCCTGATG]. Primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). Real-time PCR was performed in duplicate using 25 μl of reaction mixture containing 1.0 μl of RT mixture, 0.2 μM of both primers, and 12.5 μl of iQTM SYBR Green Supermix (Bio-Rad Laboratories). Real-time PCR was run in the iCyclerTM real-time detection system (Bio-Rad Laboratories) with a two-step method. The hot-start enzyme was activated (95°C for 3 min) and cDNA was then amplified for 40 cycles consisting of denaturation at 95°C for 10 sec and annealing/extension at 60°C for 45 sec. A melt-curve assay was then performed (55°C for 1 min and then temperature was increased by 0.5°C every 10 sec) to detect the formation of primer-derived trimmers and dimmers. Mouse glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a control (5’ primer sequence, GDDTTCCGTGTTCCTACC; 3’ primer sequence, GCCTGCTTCACCACCTTC). Data were analyzed with the iCycler iQTM software. The average starting quantity (SQ) of fluorescence units was used for analysis. Quantification was calculated using the SQ of targeted cDNA relative to that of GAPDH cDNA in the same sample.
PCR arrays
RNA isolated from duplicate samples was pooled for the reverse transcription to make cDNA. The first-strand cDNA was synthesized from RNA using RT2 First Strand Kit (SuperArray Bioscience Corp., Frederick, MD). Mouse Toll-like receptor (TLR) pathway-focused PCR Arrays (SuperArray Bioscience Corp.) were performed using 2X SuperArray RT2 qPCR master mix and the first strand cDNA by following the instruction from the manufacturer.
Transfection and luciferase activity assay
To study IL-6 transcription, RAW 264.7 cells were transiently transfected with p1168huIL6P-luc+ plasmids (Belgian Co-ordinated Collections of Micro-organisms, BCCMTM, Brussels, Belgium) (1 μg/106 cells) with transfection reagent FuGENE HD (Promega Corporation, Madison, WI). Cells were cotransfected with the Renilla luciferase reporter plasmid pRL-TK (Promega) (50 ng/106 cells) as an internal control. Twenty-four hours after transfection, the cells were stimulated with LPS, PA or both LPS and PA. To study nuclear factor kappa B (NFκB)-mediated transcription, RAW264.7 cells were transfected with 1 μg of NFκB Cignal Reporter (Qiagen Inc., Valencia, CA) using FuGENE® HD as transfection reagent for 24 h. The renilla luciferase constructs were used as control. The cells were then treated with fresh medium containing 1 ng/ml of LPS, 100 μM of PA or LPS plus PA for 12 or 24 h. After the treatment, the cells were rinsed with cold PBS and lysed with the buffer from Dual-Luciferase Reporter Assay System (Promega). Both firefly and renilla luciferase levels were assayed in a luminometer using the dual-luciferase reporter assay reagents (Promega) according to the instruction from the manufacturer. The firefly luciferase levels were normalized to the renilla luciferase levels.
Lipidomics
RAW264.7 cells were collected, fortified with internal standards, extracted with ethyl acetate/isopropyl alcohol/water (60:30:10, v/v/v), evaporated to dryness, reconstituted in 100 μl of methanol and subjected to lipidomic analysis of CER and SM as described previously [22]. Simultaneous ESI/MS/MS analyses of CERs and SMs were performed on a Thermo Finnigan TSQ 7000 triple quadrupole mass spectrometer operating in a multiple reaction monitoring positive ionization mode. The phosphate contents of the lipid extracts were used to normalize the MS measurements of sphingolipids and measured with a standard curve analysis and a colorimetric assay of ashed phosphate [23].
aSMase activity assay
RAW264.7 cells were treated and cell lysate was then used for aSMase activity assay using an aSMase assay kit (Cayman Chemical, Ann Arbor, Michigan) according to the manufacture’s instructions.
Statistical analysis
The experiments were performed in duplicates for 3 times and the data were presented as Least Squares means ± SD. One-way ANOVA was performed to determine the statistical significance among different experimental groups. A value of P<0.05 was considered significant.
Results
DHA strongly inhibits the upregulation of proinflammatory genes by LPS or LPS plus PA
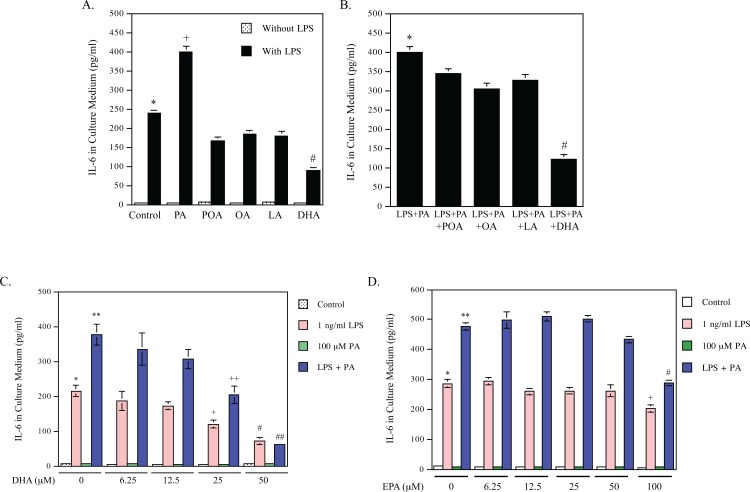
To compare the effects of different free fatty acids including SFA, monounsaturated fatty acid (MUFA) and PUFA on LPS-induced secretion of IL-6, a major proinflammatory cytokine [24], we used PA as a SFA, OA and POA as MUPAs, LA as a n-6 PUFA, DHA and EPA as n-3 PUFAs in this study. As shown in Fig 1A, while none of these fatty acids stimulated IL-6 secretion by itself, PA is the only fatty acid to have a synergy with LPS to stimulate IL-6 secretion. In contrast to PA, DHA inhibited LPS-induced IL-6 secretion by 62% and POA, OA and LA inhibited LPS-induced IL-6 secretion by 30%, 23%, 25%, respectively. Next, we determined the effect POA, OA, LA or DHA on the synergistic effect of LPS and PA on IL-6 secretion. As shown in Fig 1B, while POA, OA, LA reduced LPS-PA-stimulated IL-6 secretion by 14%, 24% and 19%, respectively, DHA markedly attenuated it by 70%. Our study further showed that DHA inhibited IL-6 secretion stimulated by LPS or LPS plus PA in a dose-dependent manner and DHA at 50 μM inhibited IL-6 secretion stimulated by LPS or LPS plus PA by 70% and 80%, respectively (Fig 1C). EPA, another ɷ-3 PUFA, was less potent than DHA in the inhibition of IL-6 secretion stimulated by LPS or LPS plus PA (Fig 1D).
Fig 1. DHA inhibits IL-6 expression stimulated by LPS or LPS plus PA.
A. RAW264.7 macrophages were treated with 100 μM of palmitic acid (PA), palmitoleic acid (POA), oleic acid (OA), linoleic acid (LA), or docosahexaenoic acid (DHA) in the absence or presence of 1 ng/ml of LPS for 24 h. After treatment, IL-6 in culture medium was quantified using ELISA. + vs. #, p<0.01; # vs. *, p<0.01. B. RAW264.7 macrophages were treated with both 1 ng/ml LPS and 100 μM of PA in the absence or presence of 100 μM of POA, OA, LA or DHA for 24 h. After treatment, IL-6 in culture medium was quantified using ELISA. # vs. *, p<0.01. C and D. RAW264.7 macrophages were treated with 1 ng/ml of LPS, 100 μM of PA or both 1 ng/ml LPS and 100 μM of PA in the absence or presence of different concentrations of DHA (C) or eicosapentaenoic acid (EPA) (D) for 24 h. After treatment, IL-6 in culture medium was quantified using ELISA. In panel C: ** vs. *, p<0.01; + vs. *, p<0.05; # vs. *, p<0.01; ++ vs. **, p<0.05; ## vs. **, p<0.01. In panel D: ** vs. *, p<0.01; + vs. *, p<0.05; # vs. **, p<0.01.
To further elucidate the anti-inflammatory effect of DHA on the gene expression in macrophages in response to LPS or LPS plus PA, we profiled the gene expression using a Toll-like receptor (TLR) pathway-based PCR array. Data (Table 1) showed the synergistic effect of LPS and PA on the expression of a number of genes including monocyte chemoattractant protein 1 (MCP-1), CD86, colony stimulating factor 3 (CSF3), IL-1α, IL-1β, IL-6 and cyclooxygenase-2 (COX-2). For example, while LPS and PA alone induced MCP-1 expression by 18- and 11-fold, respectively, the combination of LPS and PA increased MCP-1 expression by 48-fold. Strikingly, although DHA did not inhibit the baseline expression of these genes, it potently antagonized the combined stimulation by LPS and PA of the expression of the above proinflammatory genes by 70–98%. It is noteworthy that the percent inhibition by DHA on the combined effect of LPS and PA appears to be higher than that on the effect of LPS or PA alone (Table 1). Interestingly, DHA exerted a stimulatory effect on the expression of IL-10, an anti-inflammatory cytokine [25]. While LPS, PA and LPS plus PA increased IL-10 expression by 2-, 2- and 4-fold, respectively, DHA further augmented the increases to 7-, 7- and 14-fold.
Table 1. The effect of DHA on gene expression stimulated by LPS, PA or LPS plus PA.
| Genes | Fold increase by: | % inhibition by DHA | Fold increase by: | % inhibition by HDA | Fold increase by: | % inhibition by HDA | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| DHA | LPS | LPS+DHA | PA | PA+DHA | LPS+PA | LPS+PA+DHA | ||||
| MCP-1 | 11 | 18 | 11 | 36 | 11 | 10 | 9 | 48 | 6 | 87 |
| CD86 | 2 | 5 | 2 | 61 | 6 | 3 | 55 | 10 | 3 | 70 |
| CSF3 | 3 | 3 | 4 | - | 7 | 2 | 73 | 95 | 3 | 97 |
| IL-1α | 3 | 10 | 1 | 87 | 6 | 2 | 74 | 30 | 5 | 84 |
| IL-1β | 1 | 10 | 3 | 71 | 3 | 4 | - | 29 | 4 | 87 |
| IL-6 | 3 | 39 | 2 | 96 | 10 | 2 | 77 | 196 | 3 | 98 |
| COX-2 | 1 | 3 | 2 | 33 | 5 | 2 | 67 | 11 | 2 | 81 |
| IL-10 | 5 | 2 | 7 | (up 288%) | 2 | 7 | (up 337%) | 4 | 14 | (up 339%) |
RAW264.7 cells were treated with 1 ng/ml of LPS, 100 μM of palmitic acid (PA) or LPS plus PA in the absence or presence of 100 μM of DHA for 24 h and RNA was isolated from duplicate wells, combined and subjected to gene expression analysis using a Toll-like receptor (TLR) pathway-focused PCR array as described in the Methods. Full names for the abbreviations of the genes: MCP-1, monocyte chemoattractant protein 1; CSF3, colony stimulating factor 3; COX-2, cyclooxygenase-2.
DHA and other unsaturated fatty acids inhibit the effect of LPS and PA on CER synthesis
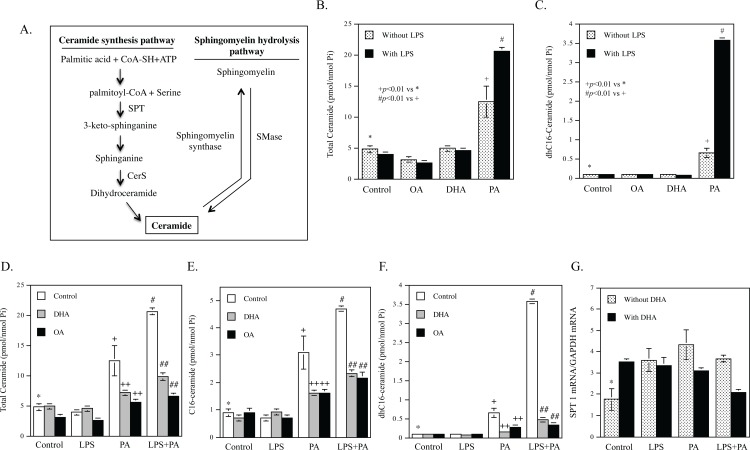
CER is generated by either CER synthesis or SM hydrolysis [26] (Fig 2A). In addition, it is also generated by reacylation of sphingosine through the salvage pathway [27]. We have shown in our previous study that LPS and PA synergistically increase CER generation by stimulating both CER de novo synthesis and SM hydrolysis, and the increased CER in turn plays an important role in the upregulation of proinflammatory molecules [9]. In this study, we examined the effect of DHA on the stimulation of CER generation by LPS and PA. To determine if the effect is DHA specific, we compared the effect of DHA with that of OA.
Fig 2. DHA inhibits the CER production increased by LPS and PA.
A. Diagram for CER de novo synthesis and sphingomyelin hydrolysis pathways. SPT, serine palmitoyltransferase; CerS, ceramide synthase; SMase, sphingomyelinase. B and C. LPS and PA synergistically stimulate total (B) and dhC16-CER production (C). RAW264.7 macrophages were treated with 100 μM of OA, DHA or PA in the absence or presence of 1 ng/ml of LPS for 12 h. After treatment, total and dhC16-CER were quantified using lipidomics. + vs. *, p<0.01; # vs. +, p<0.01. D-F. Either DHA or OA inhibited PA- or LPS plus PA-stimulated CER production. RAW264.7 macrophages were treated with 1 ng/ml LPS, 100 μM PA or both LPS and PA in the absence or presence of 100 μM DHA or OA for 12 h. After treatment, total CER (D), C16-CER (E) and dhC16-CER (F) were quantified using lipidomics. + vs. *, p<0.01; # vs. +, p<0.01; ++ vs. +, p<0.01; ## vs. #, p<0.01. G. DHA inhibits SPT1 mRNA expression stimulated by LPS and PA. RAW264.7 macrophages were treated with 1 ng/ml LPS, 100 μM PA or both LPS and PA in the absence or presence of 100 μM of DHA for 12 h. After treatment, SPT1 mRNA was quantified using real-time PCR. * vs. +, p<0.05; * vs. #, p<0.05; * vs. ^, p<0.05; ^^ vs. ^, p<0.01.
Data from lipidomic analysis showed that DHA had no effect on total CER, OA reduced total CER by 35%, but PA increased total CER by 2.59-fold (Fig 2B and Table 2). Furthermore, LPS had no effect on total CER in control cells and cells treated with OA or DHA, but augmented PA-increased total CER by 44% (Fig 2B and Table 2). Similarly, PA increased dihydro (dh)C16-CER and LPS further augmented PA-increased dhC16-CER production by 5.5-fold (Fig 2C and Table 2). DHA had no effect on the baseline level of CER, but significantly inhibited the stimulatory effect of PA and LPS plus PA on total CER content by 46% and 52%, respectively (Fig 2D and Table 2). Interestingly, OA also exerted inhibition on total CER content increased by PA and PA plus LPS by 55% and 68%, respectively (Fig 2D). DHA or OA also inhibited C16-CER (Fig 2E) and dhC16-CER generation (Fig 2F) stimulated by PA or PA plus LPS. Given that dhC16-CER is an intermediate during CER de novo synthesis [28], these results indicate that DHA or OA inhibits PA- or LPS plus PA-stimulated CER de novo synthesis. In addition to dhC16-CER and C16-CER, DHA or OA also attenuated the stimulation of C18-, C20-, C22-CER production by PA or LPS plus PA (Table 2). Results from our lipidomic analysis also showed that POA and LA had similar inhibitory effect as DHA and OA on CER de novo synthesis (data not shown).
Table 2. The effect of DHA on CER content regulated by LPS, PA or LPS plus PA.
| dhC16-CER | C16-CER | C18-CER | C20-CER | C22-CER | C24-CER | C24:1-CER | Total CER | ||
|---|---|---|---|---|---|---|---|---|---|
| Control | Control | 0.10±0.01 g1 | 0.91±0.13 g2 | 0.10±0.02 | 0.11±0.01 | 0.51±0.05 | 1.61±0.17 | 1.31±0.14 | 4.80±0.56 g |
| LPS | 0.12±0.01 | 0.72±0.05 | 0.11±0.01 | 0.11±0.01 | 0.42±0.02 | 1.52±0.08 | 1.10±0.06 | 3.90±0.26 | |
| PA | 0.65±0.12 a1 | 3.09±0.60 a2 | 0.82±0.19 | 0.47±0.14 | 2.86±0.60 | 2.73±0.49 | 1.51±0.32 | 12.43±2.53 a | |
| LPS+PA | 3.58±0.06 b1 | 4.70±0.08 b2 | 1.60±0.22 | 0.88±0.01 | 4.29±0.09 | 3.23±0.01 | 1.87±0.04 | 20.58±0.56 b | |
| DHA | Control | 0.10±0.01 | 1.10±0.02 | 0.11±0.01 | 0.10±0.01 | 0.52±0.04 | 1.60±0.03 | 1.40±0.05 | 4.90±0.16 |
| LPS | 0.08±0.01 | 0.91±0.01 | 0.10±0.01 | 0.04±0.01 | 0.40±0.02 | 1.52±0.01 | 1.30±0.03 | 4.52±0.10 | |
| PA | 0.16±0.01 c1 | 1.62±0.08 c2 | 0.22±0.02 | 0.15±0.03 | 1.07±0.04 | 2.30±0.18 | 1.39±0.09 | 7.13±0.47 c | |
| LPS+PA | 0.47±0.02 d1 | 2.33±0.12 d2 | 0.42±0.01 | 0.26±0.03 | 1.64±0.17 | 2.81±0.06 | 1.65±0.23 | 9.81±0.66 d | |
| OA | Control | 0.10±0.01 | 0.90±0.15 | 0.10±0.01 | 0.10±0.01 | 0.2±0.04 | 0.30±0.04 | 1.30±0.11 | 3.10±0.43 |
| LPS | 0.10±0.01 | 0.70±0.03 | 0.10±0.01 | 0.1±0.01 | 0.2±0.01 | 0.3±0.02 | 0.9±0.09 | 2.60±0.18 | |
| PA | 0.27±0.01 e1 | 1.62±0.12 e2 | 0.39±0.02 | 0.14±0.03 | 0.67±0.02 | 0.72±0.07 | 1.59±0.06 | 5.61±0.34 e | |
| LPS+PA | 0.33±0.02 f1 | 2.15±0.23 f2 | 0.43±0.06 | 0.15±0.01 | 0.74±0.04 | 0.83±0.05 | 1.74±0.11 | 6.59±0.51 f | |
a, p<0.01 vs g
b, p<0.01 vs a and g
c, p<0.01 vs a
d, p<0.01 vs b
e, p<0.01 vs a
f, p<0.01 vs b.
a1, p<0.01 vs g1
b1, p<0.01 vs a1 and g1
c1, p<0.01 vs a1
d1, p<0.01 vs b1
e1, p<0.01 vs a1
f1, p<0.01 vs b1.
a2, p<0.01 vs g2
b2, p<0.01 vs a2 and g2
c2, p<0.01 vs a2
d2, p<0.01 vs b2
e2, p<0.01 vs a2
f2, p<0.01 vs b2.
The concentration unit of the values in the table: pmole/nmol phosphate.
Since it has been shown that LPS upregulated the mRNA expression of SPT, a rate-limiting enzyme in CER de novo synthesis [29–31], we tested our hypothesis that DHA inhibits SPT1 mRNA expression. Results (Fig 2G) showed that LPS, PA or LPS plus PA increased SPT1 mRNA expression in the absence of DHA. Interestingly, DHA significantly attenuated LPS plus PA-stimulated SPT mRNA expression.
DHA has no effect on the synergistic stimulation of SM hydrolysis by LPS and PA
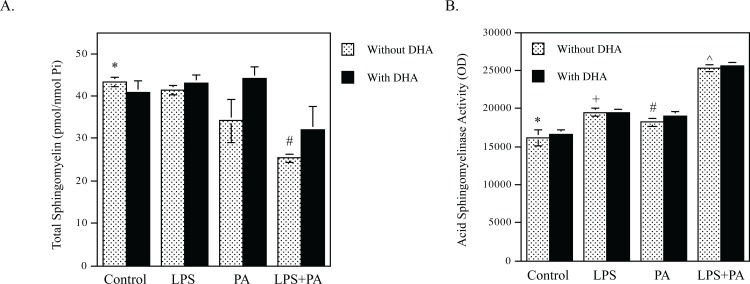
Since we have shown that LPS and PA synergistically stimulate CER production by increasing not only CER de novo synthesis, but also SM hydrolysis [9], we determined the effect of DHA on LPS-PA-stimulated SM hydrolysis. Data showed that while LPS and PA synergistically reduced SM content, DHA did not change SM content significantly (Fig 3A and Table 3). Furthermore, since we have shown that aSMase is responsible for increased SM hydrolysis in response to LPS and PA [9], we determined the effect of DHA on aSMase activity stimulated by LPS and PA. Results showed that while LPS and PA cooperatively stimulated aSMase activity, DHA did not attenuate LPS-PA-stimulated aSMase activity (Fig 3B).
Fig 3. DHA has no effect on SM hydrolysis and ASMase activity stimulated by LPS and PA.
A. RAW264.7 macrophages were treated with 1 ng/ml of LPS, 100 μM of PA or both 1 ng/ml LPS and 100 μM of PA in the absence or presence of 100 μM of DHA for 12 h. After treatment, cellular sphingomyelin was quantified using lipidomics. * vs. #, p<0.01. B. RAW264.7 macrophages were treated with 1 ng/ml of LPS, 100 μM of PA or both 1 ng/ml LPS and 100 μM of PA in the absence or presence of 100 μM of DHA for 2 h. After treatment, cellular ASMase activity was determined as described in the Methods. * vs. +, p<0.05; * vs. #, p<0.05; * vs. ^, p<0.05.
Table 3. The effect of DHA on sphingomyelin content regulated by LPS, PA or LPS plus PA.
| C14-SM | C16-SM | C18-SM | C20-SM | C22-SM | C22:1-SM | C24-SM | C24:1-SM | Total SM | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Without DHA | Control | 0.86±0.21 | 24.87±4.87a1 | 2.18±0.87 | 0.66±0.19 | 3.76±0.73 | 0.70±0.18 | 3.00±1.09 a2 | 6.98±2.13 a3 | 43.30±0.20 a |
| LPS | 0.95±0.18 | 24.5±4.79 | 1.84±0.79 | 0.59±0.15 | 3.35±0.71 | 0.68±0.12 | 3.08±0.97 | 5.80±2.08 | 41.40±0.06 | |
| PA | 0.59±0.01 | 20.56±1.51 | 2.36±1.08 | 0.66±0.20 | 2.98±1.05 | 0.60±0.11 | 1.73±1.09 | 4.44±1.95 | 34.16±7.08 | |
| LPS+PA | 0.46±0.05 | 16.81±2.23b1 | 1.52±0.48 | 0.38±0.09 | 1.75±0.41 | 0.37±0.07 | 1.01±0.54 b2 | 2.85±1.09 b3 | 25.31±0.44 b | |
| With DHA | Control | 1.05±0.15 | 25.15±0.69 | 1.95±0.28 | 0.58±0.06 | 3.24±0.49 | 0.67±0.12 | 2.25±0.71 | 5.60±2.03 | 40.80±2.91 |
| LPS | 1.22±0.26 | 28.49±3.79 | 1.78±0.14 | 0.53±0.01 | 2.83±0.19 | 0.63±0.06 | 2.22±0.29 | 4.89±1.37 | 42.91±1.96 | |
| PA | 0.75±0.26 | 28.00±3.26 | 2.44±0.05 | 0.72±0.06 | 4.01±0.51 | 0.57±0.01 | 2.57±0.08 | 4.80±1.18 | 44.10±2.80 | |
| LPS+PA | 0.67±0.31 | 21.34±5.04 | 1.73±0.35 | 0.45±0.09 | 2.66±0.74 | 0.43±0.08 | 1.42±0.01 | 3.25±0.30 | 32.16±6.38 | |
b, p<0.01 vs a
b1, p<0.01 vs a1
b2, p<0.01 vs a2
b3, p<0.01 vs a3.
The concentration unit of the values in the table: pmole/nmol phosphate.
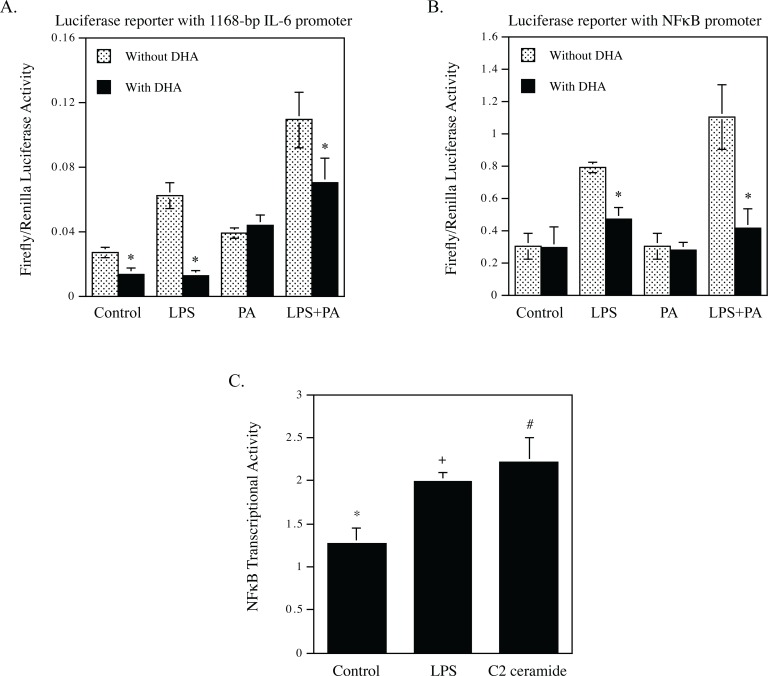
DHA inhibits NFκB-mediated transcriptional activation by LPS and PA
The results presented in Fig 1 demonstrated that DHA was more potent than other unsaturated fatty acids such as OA in the inhibition of proinflammatory gene upregulation by LPS or LPS plus PA, but the effect of DHA on CER generation is similar to that of OA (Fig 2), suggesting that a mechanism other than the inhibition of CER generation plays a key role in the inhibition of proinflammatory gene upregulation by LPS or LPS plus PA. To elucidate the underlying mechanism, we tested our hypothesis that DHA inhibits the upregulation of proinflammatory genes by targeting the transcription of proinflammatory genes. We transfected macrophages with plasmids containing 1168-bp IL-6 promoter sequence with the luciferase reporter gene prior to the exposure of cells to LPS, PA or LPS plus PA in the absence or presence of DHA. Luciferase activity assay indicated that LPS and PA had a synergy on IL-6 transcriptional activity, but DHA inhibited the effect of LPS or LPS plus PA on IL-6 transcription (Fig 4A). Since it is known that NFκB plays a key role in the expression of proinflammatory cytokines including IL-6 [32], we further determine the effect of DHA on NFκB transcriptional activity. Results showed that DHA significantly inhibited NFκB transcriptional activity stimulated by LPS or LPS plus PA (Fig 4B). To understand the linkage between CER production and proinflammatory gene expression, we determined if CER stimulates NFκB transcriptional activity. Results showed that C2-CER, a cell membrane permeable CER, exerted a similar stimulation as LPS on NFκB transcriptional activity (Fig 4C).
Fig 4. DHA inhibits IL-6 transcription and NFκB transcriptional activity.
A. RAW264.7 macrophages were transfected with plasmids containing an 1168-bp IL-6 promoter sequence and luciferase reporter gene for 24 h. After the transfection, the macrophages were treated with 1 ng/ml of LPS, 100 μM of PA or both LPS and PA for 24 h and cellular firefly and renilla luciferase activities were assayed. The ratios of firefly vs. renilla luciferase were calculated. * vs. cells with the same treatment in the absence of DHA, p<0.01. B. RAW264.7 macrophages were transfected with DNA construct containing the tandem repeats of NFκB transcriptional response element in the promoter and luciferase reporter gene for 24 h. After the transfection, the macrophages were treated with 1 ng/ml of LPS, 100 μM of PA or both LPS and PA for 24 h and cellular firefly and renilla luciferase activities were assayed. The ratios of firefly vs. renilla luciferase were calculated. The ratios of firefly vs. renilla luciferase activity were calculated. * vs. cells with the same treatment in the absence of DHA, p<0.01. C. RAW264.7 macrophages were transfected with DNA construct containing the tandem repeats of NFκB transcriptional response element in the promoter and luciferase reporter gene for 24 h. After the transfection, the macrophages were treated with 1 ng/ml of LPS or 50 μM of C2-CER for 24 h and cellular firefly and renilla luciferase activities were then assayed. The ratios of firefly vs. renilla luciferase activity were calculated. * vs. +, p<0.05; * vs. #, p<0.05.
CER de novo synthesis is involved in the upregulation of proinflammatory genes by LPS and PA
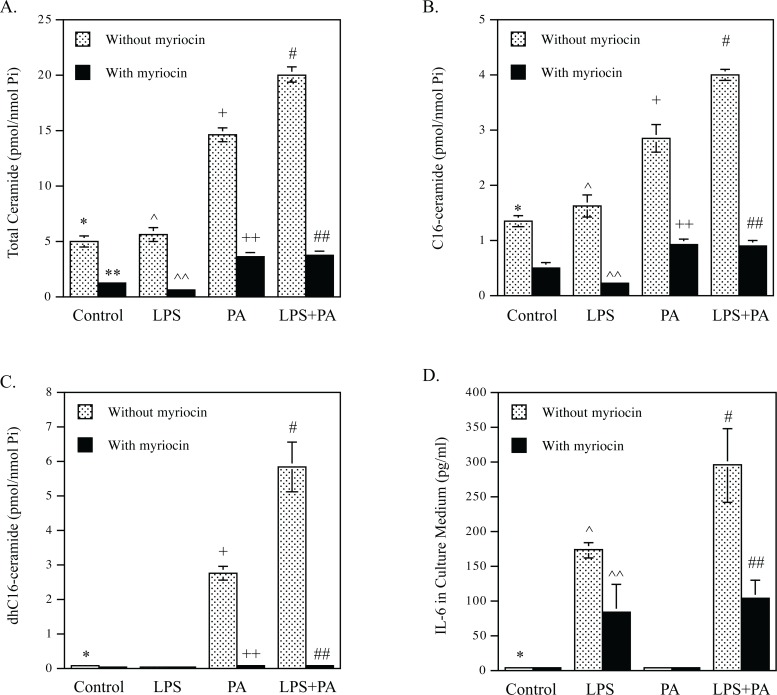
The above studies showed that DHA inhibited CER de novo synthesis in macrophages. To determine if the inhibition by DHA of CER de novo synthesis contributes to DHA-suppressed proinflammatory gene expression, we employed myriocin, a specific SPT inhibitor [33], to assess the impact of CER synthesis inhibition on proinflammatory gene expression. Since SPT is a rate-limiting enzyme in the CER de novo synthesis pathway [29–31], we expected that the treatment with myriocin would reduce CER de novo synthesis stimulated by LPS and PA. Indeed, lipidomics analysis showed that myriocin potently reduced the baseline level of total CER by 75% and diminished the stimulatory effect of PA and LPS plus PA on CER synthesis by 76% and 84%, respectively (Fig 5A and Table 4). Myriocin also potently inhibited the synthesis of C16-CER (Fig 5B and Table 4). Strikingly, myriocin nearly abolished dhC16-CER stimulated by PA or LPS plus PA (Fig 5C and Table 4). After confirming that myriocin markedly inhibits CER de novo synthesis stimulated by LPS and PA, we determined how myriocin affects upregulation of proinflammatory molecule expression by LPS and PA. Results showed that myriocin inhibited IL-6 secretion stimulated by LPS and LPS plus PA by 52% and 65%, respectively (Fig 5D). In addition to IL-6, data from PCR array showed that myriocin inhibited the stimulatory effect of LPS and PA on proinflammatory genes including MCP-1, CSF3, IL-1α, IL-1β and COX-2 (data not shown).
Fig 5. Myriocin inhibits IL-6 secretion stimulated by LPS or LPS plus PA.
A-C. RAW264.7 macrophages were treated with 1 ng/ml of LPS, 100 μM of PA or both LPS and PA in the absence or presence of 10 μM of myriocin for 12 h. After the incubation, total (A), C16-CER (B) and dhC16-CER (C) were quantified using lipidomics. D. RAW264.7 macrophages were treated with 1 ng/ml of LPS, 100 μM of PA or both LPS and PA in the absence or presence of 10 μM of myriocin for 24 h. After treatment, IL-6 in culture medium was quantified using ELISA. * vs. +, p<0.01; * vs. #, p<0.01; ** vs. *, p<0.01; ^^ vs. ^, p<0.01; ++ vs. +, p<0.01; ## vs. #, p<0.01.
Table 4. The effect of myricin on CER synthesis stimulated by PA and LPS plus PA.
| dhC16-CER | C16-CER | C18-CER | C20-CER | C22-CER | C24-CER | C24:1- CER | Total CER | ||
|---|---|---|---|---|---|---|---|---|---|
| Without myriocin | Control | 0.06±0.006 e1 | 1.34±0.102 e2 | 0.06±0.005 e3 | 0.06±0.001 | 0.38±0.013 | 1.64±0.032 | 1.22±0.070 | 4.90±0.010 e |
| LPS | 0.04±0.004 | 1.62±0.213 | 0.09±0.006 | 0.07±0.011 | 0.39±0.039 | 1.77±0.205 | 1.39±0.137 | 5.52±0.625 | |
| PA | 2.75±0.100 a1 | 3.54±0.254 a2 | 3.40±0.069 a3 | 2.24±0.008 | 1.64±0.015 | 1.94±0.004 | 1.35±0.021 | 14.55±0.308 a | |
| LPS+PA | 5.83±0.721 b1 | 3.98±0.016 b2 | 7.48±0.082 b3 | 4.06±0.026 | 1.20±0.069 | 1.59±0.087 | 1.08±0.028 | 19.99±0.108 b | |
| With myriocin | Control | 0.03±0.005 | 0.48±0.059 | 0.04±0.008 | 0.03±0.003 | 0.09±0.011 | 0.15±0.030 | 0.39±0.023 | 1.23±0.136 |
| LPS | 0.02±0.002 | 0.21±0.010 | 0.02±0.000 | 0.01±0.001 | 0.04±0.003 | 0.13±0.036 | 0.19±0.017 | 0.62±0.069 | |
| PA | 0.07±0.003 c1 | 0.91±0.045 c2 | 0.30±0.001 c3 | 0.20±0.009 | 0.39±0.001 | 0.87±0.018 | 0.60±0.015 | 3.55±0.092 c | |
| LPS+PA | 0.08±0.002 d1 | 0.89±0.023 d2 | 0.49±0.047 d3 | 0.29±0.049 | 0.31±0.012 | 0.88±0.096 | 0.58±0.012 | 3.67±0.172 d | |
a, p<0.01 vs e
b, p<0.01 vs a and e
c, p<0.01 vs a
d, p<0.01 vs b.
a1, p<0.01 vs e1
b1, p<0.01 vs a1 and e1
c1, p<0.01 vs a1
d1, p<0.01 vs b1.
a2, p<0.01 vs e2
b2, p<0.01 vs a2 and e2
c2, p<0.01 vs a2
d2, p<0.01 vs b2.
a3, p<0.01 vs e3
b3, p<0.01 vs a3 and e3
c3, p<0.01 vs a3
d3, p<0.01 vs b3.
The concentration unit of the values in the table: pmole/nmol phosphate.
Discussion
A number of clinical studies have shown that supplementation with n-3 PUFAs in obese patients reduced proinflammatory cytokines [14, 15]. Increasing reports in recent years have also demonstrated the beneficial effects of n-3 PUFAs on obesity or diabetes [16, 17]. However, our understanding of the mechanisms whereby n-3 PUFAs inhibit inflammation remains incomplete. In the present study, we utilized a cell model that we had established to demonstrate that PA potently amplifies LPS inflammatory signaling in macrophages. We determined if n-3 PUFAs are capable of antagonizing the boosting effect of PA on LPS inflammatory signaling. Since we have shown in this cell model that LPS and PA stimulate gene expression by increasing CER production via both CER de novo synthesis and SM hydrolysis [9], we also determined how DHA affects CER production. The results clearly demonstrated that DHA was much more potent than OA, POA and LA in the inhibition of the boosting effect of PA on LPS signaling and that DHA inhibited the stimulation of CER production by LPS and PA by targeting de novo synthesis, but not SM hydrolysis.
An intriguing finding from this study is that unsaturated fatty acids such as OA, POA and LA have the similar effect as DHA in the inhibition of PA-increased CER de novo synthesis, indicating that the inhibition of PA-increased CER de novo synthesis is not specific for DHA. Given that DHA is more potent than OA, POA and LA in the inhibition of LPS plus PA-stimulated proinflammatory gene expression, it is suggested that a mechanism other than the inhibition of CER de novo synthesis is involved in the robust inhibition by DHA of the gene expression stimulated by LPS and PA.
In our previous study, we have shown that LPS and PA synergistically stimulate signaling activation that ultimately leads to an increase in NFκB transcriptional activity [9]. It has been well established that NFκB as a transcription factor plays a pivotal role in the induction and maintenance of the state of inflammation that underlies metabolic diseases such as obesity and type 2 diabetes [34]. We thus hypothesized that DHA antagonizes the stimulation by LPS and PA of NFκB-dependent transcription. Indeed, the finding from the present study supports our hypothesis as our results showed that DHA potently inhibited the effects of LPS or LPS plus PA on NFκB transcriptional activity. Remarkably, we found that while PA strongly enhanced LPS-induced NFκB activity, DHA inhibited NFκB activity stimulated by LPS and LPS plus PA by 47% and 87%, respectively (Fig 4B). The potential mechanisms by which n-3 PUFAs inhibit LPS-stimulated NFκB activity have been explored previously by Xue et al., who showed that n-3 PUFAs inhibited NFκB activation by stimulating AMP-activated protein kinase (AMPK) pathway [35]. Additionally, Li et al. also reported that n-3 PUFAs inhibited LPS-stimulated NFκB activity by activating peroxisome proliferator-activated receptor gamma [36].
While it is clear that the inhibition of NFκB transcriptional activity plays a key role in the suppression by DHA of proinflammatory gene expression, several lines of evidence from our study and others suggest that the inhibition of CER de novo synthesis also contributes to the powerful inhibition by DHA of proinflammatory gene expression.
First, several studies have demonstrated that PA or LPS increases cellular CER content by stimulating CER synthesis and the increased CER synthesis is responsible for the upregulation of proinflammatory gene expression by PA or LPS [30, 37, 38]. PA is known to augment CER synthesis mainly by its conversion to palmitoyl-CoA, a substrate for SPT to synthesize 3-keto-sphinganine that is a precursor of CER [37, 38]. On the other hand, LPS increases CER synthesis by activating TLR4 signaling that upregulates the expression or activities of the enzymes involved in CER de novo synthesis [37]. To demonstrate linkage between CER synthesis and the proinflammatory gene expression, these studies showed that treatment of cells with pharmacological inhibitors such as SPT inhibitor myriocin or CER synthase inhibitor fumonisin B1 [39] dampens the stimulatory effect of LPS or PA on proinflammatory gene expression. Our present study also showed that myriocin strongly inhibited IL-6 secretion stimulated by LPS or LPS plus PA. Second, our previous study [9] showed that myriocin or fumonisin B1 significantly reduced nuclear content of p65, a major subunit of NFκB [40], indicating that the elevated CER de novo synthesis results in the increased nuclear translocation of NFκB. Moreover, the role of CER in NFκB transcriptional activity is also confirmed by our finding from the present study that C2-CER stimulated NFκB transcriptional activity. Demarchi et al. also demonstrated that C2-CER activated NFκB subunits p50 and p65 in NIH 3T3 cells [41]. However, since C2-CER has a very short fatty acid chain that is quite different from the major intracellular CER such as C16-CER, it may not have all the biological properties of those CERs generated by CER de novo synthesis or SM hydrolysis. Third, our PCR analysis showed that although the combination of LPS and PA is much stronger than LPS alone in stimulation of gene expression, the degree of the inhibition by DHA on LPS plus PA-stimulated gene expression was similar to or even stronger than that on LPS-stimulated gene expression (Table 1). For example, while LPS and LPS plus PA increased IL-6 expression by 39- and 196-fold, respectively, DHA inhibited LPS- and LPS plus PA-stimulated IL-6 expression by 96% and 98%, respectively. Similarly, while LPS and LPS plus PA increased MCP-1 expression by 18- and 48-fold, respectively, DHA inhibited LPS- and LPS plus PA-stimulated MCP-1 expression by 36% and 87%, respectively. Obviously, in addition to targeting the LPS-induced inflammatory signaling, DHA also targets a PA-activated mechanism that contributes to proinflammatory gene expression and the mechanism is likely to be CER de novo synthesis.
It is noteworthy that, in contrary to myriocin that targets the baseline of CER de novo synthesis, DHA antagonizes the PA- or LPS plus PA-stimulated CER de novo synthesis. Our present study showed that myriocin potently reduced the baseline level of the CER content by 75% (Table 4), suggesting that it inhibits the fundamental CER synthesis. By inhibiting the fundamental CER synthesis, myriocin drastically inhibited PA- or LPS-PA-increased CER production by 76% and 84%, respectively. Contrarily, DHA had no effect on the baseline level of CER production, but reduced the increased CER production in response to PA or LPS plus PA (Table 2), suggesting that DHA specifically inhibits the mechanisms by which PA or LPS plus PA increases CER de novo synthesis. In fact, our findings on the inhibition of SPT1 mRNA expression by DHA (Fig 2G) support the above notion. Results showed that DHA did not inhibit the baseline level of SPT1 mRNA expression, but abolished the upregulated SPT1 mRNA expression in response to LPS plus PA.
In our previous study, we reported that PA increased the percentage of CERs with shorter carbon chains such as C16-, C18-, C20-, and C22-CER and decreased the percentage of CERs with longer chains such as C24- and C24:1-CER [9]. Interestingly, the combination of PA and LPS further increased the percentage of CERs with the shorter carbon chains but decreased the percentage of the longer carbon chains, suggesting that the CERs with shorter carbon chains may play an important role in the upregulation of proinflammatory cytokines by LPS and PA. From the current study, it is interesting to find that DHA exerted its stronger inhibition on the CERs with shorter carbon chains than the CER with longer carbon chains. For example, DHA inhibited PA- and PA plus LPS-increased C16-CER by 48% and 50%, respectively, but only inhibited PA- and PA plus LPS-increased C24-CER by 16% and 13%, respectively (Table 2).
While we have focused on the role of CER in the inhibition by DHA of proinflammatory gene expression stimulated by LPS and PA, it is important to understand that DHA also exerts actions that are sphingolipid-independent to inhibit proinflammatory gene expression in macrophages. For example, De Boer et al. have reported that DHA inhibits the secretion of MCP-1 and IL-6 by reducing the degree of M1 polarization of macrophages [42]. Komatsu et al. have also shown that DHA suppresses nitric oxide production in interferon-gamma plus LPS-stimulated macrophage by inhibiting the oxidative stress [43]. Therefore, it appears that the regulation of sphingolipid pathway is one of the multiple mechanisms by which DHA exerts anti-inflammatory effects on macrophages.
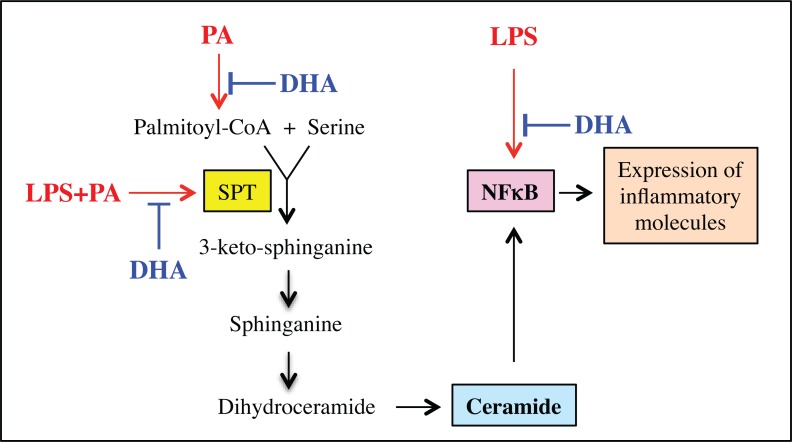
Taken together, we have demonstrated in this study that DHA antagonizes the boosting effect of PA on LPS inflammatory signaling in macrophages by targeting NFκB-dependent gene transcription and CER de novo synthesis (Fig 6). These findings provide novel insight into the molecular and signaling mechanisms involved in the inhibition of inflammatory response in macrophages by DHA.
Fig 6. Schematic diagram to show the proposed mechanism involved in the inhibition by DHA of proinflammatory gene expression stimulated by LPS or LPS plus PA in RAW264.7 macrophages.
Acknowledgments
We are thankful to the Lipidomics Shared Resource, Hollings Cancer Center, Medical University of South Carolina, for sphingolipid analysis. We are also grateful to Dr. Juan Loor and two anonymous referees who helped improving our manuscript during the reviews.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by a Merit Review grant BX000854 from the Biomedical Laboratory Research and Development Program of the Department of Veterans Affairs (Yan Huang) and NIH grant DE016353 (Yan Huang). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gomes JM, Costa JA, Alfenas RC. Metabolic endotoxemia and diabetes mellitus: A systematic review. Metabolism. 2017;68:133–44. doi: 10.1016/j.metabol.2016.12.009 [DOI] [PubMed] [Google Scholar]
- 2.Cani PD, Osto M, Geurts L, Everard A. Involvement of gut microbiota in the development of low-grade inflammation and type 2 diabetes associated with obesity. Gut microbes. 2012;3:279–88. doi: 10.4161/gmic.19625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lassenius MI, Pietilainen KH, Kaartinen K, Pussinen PJ, Syrjanen J, Forsblom C, et al. Bacterial endotoxin activity in human serum is associated with dyslipidemia, insulin resistance, obesity, and chronic inflammation. Diabetes Care. 2011;34:1809–15. doi: 10.2337/dc10-2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruiz-Nunez B, Dijck-Brouwer DA, Muskiet FA. The relation of saturated fatty acids with low-grade inflammation and cardiovascular disease. J Nutr Biochem. 2016;36:1–20. doi: 10.1016/j.jnutbio.2015.12.007 [DOI] [PubMed] [Google Scholar]
- 5.Das M, Pal S, Ghosh A. Association of metabolic syndrome with obesity measures, metabolic profiles, and intake of dietary fatty acids in people of Asian Indian origin. J Cardiovasc Dis Res. 2010;1:130–5. doi: 10.4103/0975-3583.70911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hellmann J, Zhang MJ, Tang Y, Rane M, Bhatnagar A, Spite M. Increased saturated fatty acids in obesity alter resolution of inflammation in part by stimulating prostaglandin production. J Immunol. 2013;191:1383–92. doi: 10.4049/jimmunol.1203369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu Z, Li Y, Brinson CW, Lopes-Virella MF, Huang Y. Cooperative stimulation of atherogenesis by lipopolysaccharide and palmitic acid-rich high fat diet in low-density lipoprotein receptor-deficient mice. Atherosclerosis. 2017;265:231–41. doi: 10.1016/j.atherosclerosis.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weigert C, Brodbeck K, Staiger H, Kausch C, Machicao F, Haring HU, et al. Palmitate, but not unsaturated fatty acids, induces the expression of interleukin-6 in human myotubes through proteasome-dependent activation of nuclear factor-kappaB. J Biol Chem. 2004;279:23942–52. doi: 10.1074/jbc.M312692200 [DOI] [PubMed] [Google Scholar]
- 9.Jin J, Zhang X, Lu Z, Perry DM, Li Y, Russo SB, et al. Acid sphingomyelinase plays a key role in palmitic acid-amplified inflammatory signaling triggered by lipopolysaccharide at low concentrations in macrophages. Am J Physiol Endocrinol Metab. 2013;305:E853–67. Epub 2013/08/08. doi: 10.1152/ajpendo.00251.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeidan YH, Hannun YA. The acid sphingomyelinase/ceramide pathway: biomedical significance and mechanisms of regulation. Curr Mol Med. 2010;10:454–66. [DOI] [PubMed] [Google Scholar]
- 11.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–50. doi: 10.1038/nrm2329 [DOI] [PubMed] [Google Scholar]
- 12.Li H, Ruan XZ, Powis SH, Fernando R, Mon WY, Wheeler DC, et al. EPA and DHA reduce LPS-induced inflammation responses in HK-2 cells: evidence for a PPAR-gamma-dependent mechanism. Kidney Int. 2005;67:867–74. doi: 10.1111/j.1523-1755.2005.00151.x [DOI] [PubMed] [Google Scholar]
- 13.Mullen A, Loscher CE, Roche HM. Anti-inflammatory effects of EPA and DHA are dependent upon time and dose-response elements associated with LPS stimulation in THP-1-derived macrophages. J Nutr Biochem. 2010;21:444–50. doi: 10.1016/j.jnutbio.2009.02.008 [DOI] [PubMed] [Google Scholar]
- 14.Allaire J, Couture P, Leclerc M, Charest A, Marin J, Lepine MC, et al. A randomized, crossover, head-to-head comparison of eicosapentaenoic acid and docosahexaenoic acid supplementation to reduce inflammation markers in men and women: the Comparing EPA to DHA (ComparED) Study. Am J Clin Nutr. 2016;104:280–7. doi: 10.3945/ajcn.116.131896 [DOI] [PubMed] [Google Scholar]
- 15.Itariu BK, Zeyda M, Hochbrugger EE, Neuhofer A, Prager G, Schindler K, et al. Long-chain n-3 PUFAs reduce adipose tissue and systemic inflammation in severely obese nondiabetic patients: a randomized controlled trial. Am J Clin Nutr. 2012;96:1137–49. doi: 10.3945/ajcn.112.037432 [DOI] [PubMed] [Google Scholar]
- 16.Pinel A, Morio-Liondore B, Capel F. n-3 Polyunsaturated fatty acids modulate metabolism of insulin-sensitive tissues: implication for the prevention of type 2 diabetes. J Physiol Biochem. 2014;70:647–58. doi: 10.1007/s13105-013-0303-2 [DOI] [PubMed] [Google Scholar]
- 17.Robinson LE, Mazurak VC. N-3 polyunsaturated fatty acids: relationship to inflammation in healthy adults and adults exhibiting features of metabolic syndrome. Lipids. 2013;48:319–32. doi: 10.1007/s11745-013-3774-6 [DOI] [PubMed] [Google Scholar]
- 18.Bartke N, Hannun YA. Bioactive sphingolipids: metabolism and function. J Lipid Res. 2009;50 Suppl:S91–6. doi: 10.1194/jlr.R800080-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi HY, Daniels MP, Liu Y, Chen LY, Alsaaty S, Levine SJ, et al. A cytosolic phospholipase A2-initiated lipid mediator pathway induces autophagy in macrophages. J Immunol. 2011;187:5286–92. doi: 10.4049/jimmunol.1004004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green CD, Olson LK. Modulation of palmitate-induced endoplasmic reticulum stress and apoptosis in pancreatic beta-cells by stearoyl-CoA desaturase and Elovl6. Am J Physiol Endocrinol Metab. 2011;300:E640–9. doi: 10.1152/ajpendo.00544.2010 [DOI] [PubMed] [Google Scholar]
- 21.Schwartz EA, Zhang WY, Karnik SK, Borwege S, Anand VR, Laine PS, et al. Nutrient modification of the innate immune response: a novel mechanism by which saturated fatty acids greatly amplify monocyte inflammation. Arterioscler Thromb Vasc Biol. 2010;30:802–8. doi: 10.1161/ATVBAHA.109.201681 [DOI] [PubMed] [Google Scholar]
- 22.Shaner RL, Allegood JC, Park H, Wang E, Kelly S, Haynes CA, et al. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J Lipid Res. 2009;50:1692–707. doi: 10.1194/jlr.D800051-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Veldhoven PP, Bell RM. Effect of harvesting methods, growth conditions and growth phase on diacylglycerol levels in cultured human adherent cells. Biochim Biophys Acta. 1988;959:185–96. [DOI] [PubMed] [Google Scholar]
- 24.Yudkin JS, Kumari M, Humphries SE, Mohamed-Ali V. Inflammation, obesity, stress and coronary heart disease: is interleukin-6 the link? Atherosclerosis. 2000;148:209–14. [DOI] [PubMed] [Google Scholar]
- 25.Couper KN, Blount DG, Riley EM. IL-10: the master regulator of immunity to infection. J Immunol. 2008;180:5771–7. [DOI] [PubMed] [Google Scholar]
- 26.Mullen TD, Hannun YA, Obeid LM. Ceramide synthases at the centre of sphingolipid metabolism and biology. Biochem J. 2012;441:789–802. doi: 10.1042/BJ20111626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitatani K, Idkowiak-Baldys J, Hannun YA. The sphingolipid salvage pathway in ceramide metabolism and signaling. Cell Signal. 2008;20:1010–8. doi: 10.1016/j.cellsig.2007.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stiban J, Fistere D, Colombini M. Dihydroceramide hinders ceramide channel formation: Implications on apoptosis. Apoptosis. 2006;11:773–80. doi: 10.1007/s10495-006-5882-8 [DOI] [PubMed] [Google Scholar]
- 29.Memon RA, Holleran WM, Uchida Y, Moser AH, Grunfeld C, Feingold KR. Regulation of sphingolipid and glycosphingolipid metabolism in extrahepatic tissues by endotoxin. J Lipid Res. 2001;42:452–9. [PubMed] [Google Scholar]
- 30.Chang ZQ, Lee SY, Kim HJ, Kim JR, Kim SJ, Hong IK, et al. Endotoxin activates de novo sphingolipid biosynthesis via nuclear factor kappa B-mediated upregulation of Sptlc2. Prostaglandins Other Lipid Mediat. 2011;94:44–52. doi: 10.1016/j.prostaglandins.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasset L, Zhang Y, Dunn TM, Di Lorenzo A. Sphingolipid De Novo Biosynthesis: A Rheostat of Cardiovascular Homeostasis. Trends Endocrinol Metab. 2016;27:807–19. doi: 10.1016/j.tem.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novotny NM, Markel TA, Crisostomo PR, Meldrum DR. Differential IL-6 and VEGF secretion in adult and neonatal mesenchymal stem cells: role of NFkB. Cytokine. 2008;43:215–9. doi: 10.1016/j.cyto.2008.05.015 [DOI] [PubMed] [Google Scholar]
- 33.Hojjati MR, Li Z, Zhou H, Tang S, Huan C, Ooi E, et al. Effect of myriocin on plasma sphingolipid metabolism and atherosclerosis in apoE-deficient mice. J Biol Chem. 2005;280:10284–9. doi: 10.1074/jbc.M412348200 [DOI] [PubMed] [Google Scholar]
- 34.Tornatore L, Thotakura AK, Bennett J, Moretti M, Franzoso G. The nuclear factor kappa B signaling pathway: integrating metabolism with inflammation. Trends Cell Biol. 2012;22:557–66. doi: 10.1016/j.tcb.2012.08.001 [DOI] [PubMed] [Google Scholar]
- 35.Xue B, Yang Z, Wang X, Shi H. Omega-3 polyunsaturated fatty acids antagonize macrophage inflammation via activation of AMPK/SIRT1 pathway. PloS one. 2012;7:e45990 doi: 10.1371/journal.pone.0045990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamblin M, Chang L, Fan Y, Zhang J, Chen YE. PPARs and the cardiovascular system. Antioxid Redox Signal. 2009;11(6):1415–52. doi: 10.1089/ARS.2008.2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schilling JD, Machkovech HM, He L, Sidhu R, Fujiwara H, Weber K, et al. Palmitate and lipopolysaccharide trigger synergistic ceramide production in primary macrophages. J Biol Chem. 2013;288(5):2923–32. doi: 10.1074/jbc.M112.419978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haversen L, Danielsson KN, Fogelstrand L, Wiklund O. Induction of proinflammatory cytokines by long-chain saturated fatty acids in human macrophages. Atherosclerosis. 2009;202:382–93. doi: 10.1016/j.atherosclerosis.2008.05.033 [DOI] [PubMed] [Google Scholar]
- 39.Mullen TD, Jenkins RW, Clarke CJ, Bielawski J, Hannun YA, Obeid LM. Ceramide synthase-dependent ceramide generation and programmed cell death: involvement of salvage pathway in regulating postmitochondrial events. J Biol Chem. 2011;286:15929–42. doi: 10.1074/jbc.M111.230870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vermeulen L, De Wilde G, Notebaert S, Vanden Berghe W, Haegeman G. Regulation of the transcriptional activity of the nuclear factor-kappaB p65 subunit. Biochem Pharmacol. 2002;64:963–70. [DOI] [PubMed] [Google Scholar]
- 41.Demarchi F, Bertoli C, Greer PA, Schneider C. Ceramide triggers an NF-kappaB-dependent survival pathway through calpain. Cell Death Differ. 2005;12:512–22. doi: 10.1038/sj.cdd.4401592 [DOI] [PubMed] [Google Scholar]
- 42.De Boer AA, Monk JM, Robinson LE. Docosahexaenoic acid decreases pro-inflammatory mediators in an in vitro murine adipocyte macrophage co-culture model. PloS one. 2014;9:e85037 doi: 10.1371/journal.pone.0085037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Komatsu W, Ishihara K, Murata M, Saito H, Shinohara K. Docosahexaenoic acid suppresses nitric oxide production and inducible nitric oxide synthase expression in interferon-gamma plus lipopolysaccharide-stimulated murine macrophages by inhibiting the oxidative stress. Free Radic Biol Med. 2003;34:1006–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.