Abstract
Rickettsia (R.) helvetica is the most prevalent rickettsia found in Ixodes ricinus ticks in Germany. Several studies reported antibodies against R. helvetica up to 12.5% in humans investigated, however, fulminant clinical cases are rare indicating a rather low pathogenicity compared to other rickettsiae. We investigated growth characteristics of R. helvetica isolate AS819 in two different eukaryotic cell lines with focus on ultra-structural changes of host cells during infection determined by confocal laser scanning microscopy. Further investigations included partially sequencing of rickA, sca4 and sca2 genes, which have been reported to encode proteins involved in cell-to-cell spread and virulence in some rickettsiae. R. helvetica grew constantly but slowly in both cell lines used. Confocal laser scanning microscopy revealed that the dissemination of R. helvetica AS819 in both cell lines was rather mediated by cell break-down and bacterial release than cell-to-cell spread. The cytoskeleton of both investigated eukaryotic cell lines was not altered. R. helvetica possesses rickA, but its expression is not sufficient to promote actin-based motility as demonstrated by confocal laser scanning microscopy. Hypothetical Sca2 and Sca4 proteins were deduced from nucleotide gene sequences but the predicted amino acid sequences were disrupted or truncated compared to other rickettsiae most likely resulting in non-functional proteins. Taken together, these results might give a first hint to the underlying causes of the reduced virulence and pathogenicity of R. helvetica.
Author summary
The pathogenicity of Rickettsia helvetica has not been investigated in depth to date. In humans, seroprevalences up to 12.5% against R. helvetica have been demonstrated with forest workers being predisposed to infection. However, fulminant clinical cases are rare indicating a rather low pathogenicity compared to other Rickettsia species. We therefore investigated growth characteristics of a R. helvetica tick isolate (AS819) in two different eukaryotic cell lines with focus on ultra-structural changes of host cells during infection as determined by confocal laser scanning microscopy. Further investigations included sequencing of rickA, sca4 and sca2 genes, which have been reported to encode proteins involved in cell-to-cell spread in some rickettsiae. In contrast to what is known from other rickettsiae, R. helvetica did not spread directly from cell to cell by actin-based motility presumably due to a deletion in the predicted Sca2 protein. As Sca2 is needed for virulence our results might indicate less virulence and pathogenicity of R. helvetica isolated from ixodid ticks in Germany.
Introduction
Rickettsia (R.) helvetica is the most prevalent rickettsia found in Ixodes (I.) ricinus ticks in Germany with varying prevalence up to 17% [1–4]. The organism has mainly been considered non-pathogenic and affected patients usually show a mild disease, manifesting in non-specific fever without erythema (so-called uneruptive fever), headache, and myalgia [5]. Hence, only a few laboratories in Germany focus on R. helvetica infection as a differential diagnosis and infections might be underdiagnosed due to mild symptoms in most cases. The latter might be the reason that, to the authors’ best knowledge, no clinical case in humans has been reported from Germany so far. However, more severe clinical cases have been demonstrated in Sweden including septicemia [6], myocarditis [7] and meningitis [8]. Only recently, complex phylogenetic studies showed that R. helvetica phylogenetically was misplaced in the spotted fever group (SFG) [9]. In contrast, R. conorii subsp. conorii, a typical representative of the SFG, causes Mediterranean spotted-fever, a disease that is characterized by fever, an eschar at the site of the tick bite, and a rash spreading to the palms and soles. The disease is endemic in southern Europe but single cases have also been reported from the central and northern European mainland [10].
As many obligate intracellular bacteria, rickettsiae proliferate in the cytoplasm as dispersed, individual bacteria but may also occasionally be found in clusters and in the nucleus [11]. The primary targets for rickettsiae during infection are endothelial cells of the middle and small vessels [12–13]. Disseminated infection of the endothelium and subsequent pathophysiological effects lead to most of the clinical characteristics described for rickettsial diseases [13]. Rapid spread within host tissues is a crucial step in many infectious diseases [14]. For some Rickettsia species it has been proven that rickettsiae harness the host cell actin cytoskeleton for intracellular movement and cell-to-cell spread. This phenomenon was first reported by Teysseire et al. [15] and Heinzen et al. [16] who observed associations between R. conorii and R. rickettsii and host F-actin. Most of the SFG rickettsiae (SFGR) as well as R. typhi assemble actin tails and undergo actin-based motility mediating cell-to-cell spread and enhancing virulence [17–18]. Two actin-polymerizing proteins have been identified in SFG rickettsiae: RickA, which activates the actin-related protein-2/3 (Arp2/3) complex of the host [19–20], and surface cell antigen 2 (Sca2) which has been suggested to mimic eukaryotic formin proteins [18,21]. Cardwell and Martinez [22] identified the minimal domain within the Sca2-protein of R. conorii that is sufficient for stimulating actin polymerization. Most recently, Sca4 was identified as a secreted effector of spread independent from actin-based motility in the SFG R. parkeri [23].
So far—to the authors’ knowledge—only a single study on growth characteristic of R. helvetica in eukaryotic cells exists [24] besides the original description of R. helvetica in 1993 [25]. Based on host cell decomposition and intranuclear growth of R. helvetica, the study by Elfving et al. [24] underlined the pathogenic ability of R. helvetica. Here, we report growth characteristics of R. helvetica isolate AS819 in two different eukaryotic cell lines with focus on ultra-structural changes of host cells during infection as determined by confocal laser scanning microscopy (LSM). Further investigations included sequencing of rickA, sca4 and sca2 genes, which have been reported to encode proteins involved in cell-to-cell spread in some rickettsiae.
Materials and methods
Rickettsia isolates and eukaryotic cell lines
Eukaryotic cell lines used in this study were obtained from LGC Standards, Wesel, Germany. L929 cells (murine fibroblasts from connective tissue; up to 25 passages from the original LGC Standards culture) and Vero E6 cells (African green monkey kidney cells; up to 39 passages from the original LGC Standards culture) were grown in Minimum Essential Medium (MEM) supplemented with Gibco GlutaMAX and 1x MEM non-essential amino acids (NEAA) solution (Life Technologies GmbH, Darmstadt, Germany) and 3% fetal calf serum (FCS) at 37°C, 5% CO2. The R. helvetica AS819 isolate (seventh passage from the original culture) used for the growth studies was isolated from I. ricinus. Species identity was confirmed based on 100% ompB-sequence identity (4,848 nt, accession number MF163037) to R. helvetica type strain C9P9 (accession number AF123725). R. conorii (Moroccan isolate VR141) and R. honei (VR1472) were obtained from ATCC, Manassas, USA. All rickettsiae were cultured on Vero E6 monolayers at 32°C in MEM prepared as described above. Flasks were checked daily for detrimental alteration of the monolayer. Rickettsiae were grown until cell layers revealed plaque formation (R. conorii, R. honei) or detached from the flasks surface (R. helvetica). The amount of R. helvetica in culture supernatants was determined by quantitative gltA real-time PCR as described below. A non-infected cell control (MOCK) was carried along with every infection for the evaluation of changes in the cell layers.
Cultivation and quantification of Rickettsia helvetica in two eukaryotic cell lines
L929 and Vero E6 cells (105 cells/well) seeded in 4-well BD Falcon CultureSlides (BD Biosciences, Heidelberg, Germany) were grown to confluent monolayers overnight at 37°C, 5% CO2. Prior to infection, monolayers were rinsed with FCS-free medium. Infection was performed with 100 μl of a seed culture containing 5 x 105 R. helvetica AS819-genome equivalents (corresponding to 5 genome equivalents per cell) determined by quantitative gltA real-time PCR as described below. After one hour of incubation at room temperature whilst constantly rocking, wells were filled up with 900 μl MEM supplemented with Gibco GlutaMAX NEAA and 3% FCS. Of every 4-well slide, three wells contained infected cells whilst the remaining served as a non-infected cell control (MOCK). Culture slides were incubated at 32°C, 5% CO2. Experiments were performed over a period of 20 days. A slide of each cell line was randomly selected on a daily basis and prepared for the quantification of rickettsia in the cell culture supernatant and the cell layer, respectively. Briefly, cell culture supernatants of each well were harvested individually and stored at -80°C. One ml of cell culture medium per well was added to the remaining cell layers and the slides were frozen at -80°C for one hour. Subsequently, freeze-thawed cells were scraped off, harvested and stored at -80°C until further processing. Nucleic acids were isolated from 200 μl of all samples applying the MagNA Pure LC Total Nucleic Acid Isolation Kit (Roche, Mannheim, Germany) and the MagNA Pure LC 2.0 system (Roche) according to the manufacturer’s instructions. Nucleic acids were eluted in a total volume of 50 μl. Quantification of rickettsiae was carried out by a real-time PCR targeting the single copy citrate synthase gene gltA in a Stratagene MX3000P Thermocycler (Agilent Technologies, München, Germany) as described earlier [26–27]. Five μl of nucleic acids were used as a template for the PCR. A commercially available DNA-standard (AmpTech GmbH, Hamburg, Germany) with a defined concentration (2.77 x 1010 copies/μl) was used for the quantification of Rickettsia genome equivalents. The number of R. helvetica copies/μl of template was calculated using the Stratagene Software by comparing the samples to a serial dilution of the DNA-standard (2.77 x 105 to 2.77 x 101 copies/μl).
Preparation of cell lines for confocal laser scanning microscopy (LSM)
Cells were seeded in μ-Slide 8 well ibiTreat chambers (ibidi GmbH, Martinsried, Germany). Briefly, 2.5 x 105 cells/well were grown to near confluence overnight and were infected with 5 genome equivalents per cell as described above. After an initial incubation period of 1 h at room temperature, slides were transferred to 32°C, 5% CO2. Fixation and staining procedures were performed at room temperature. At 24 hours-intervals, culture supernatants were discarded and cells were washed using phosphate-buffered saline (PBS, pH 7.2) pre-heated to 37°C. Cells were fixed for 15 min in methanol-free formalin (3.7%). Following a wash with PBS, cells were permeabilized with 0.1% Triton X-100 in PBS for 4 min. To reduce nonspecific binding and background signal, blocking was performed in PBS with 1% bovine serum albumin for 60 min. Thereafter, cells were incubated with an anti-SFG Rickettsia antibody (1:2, Fuller Laboratories, Fullerton, USA) for 30 min. After washing with PBS, a mouse monoclonal anti-alpha-tubulin antibody (1:200, Molecular Probes, Life Technologies) was added for 30 min and F-actin was stained with Alexa Fluor 568 Phalloidin (1:100, Molecular Probes, Life Technologies) in parallel. Samples were washed four times and the secondary antibody for Rickettsia staining, an Alexa Fluor 647-labeled goat anti-human IgG (1:2000, Molecular Probes, Life Technologies), was added for 30 minutes. In addition, Alexa Fluor 488-labeled goat anti-mouse IgG (1:2000, Molecular Probes, Life Technologies) was added for staining of microtubuli. Washing another four times was followed by nuclei counterstaining with DAPI (4'.6-Diamidino-2-Phenylindole, Dilactate, 1:5000, Molecular Probes, Life Technologies). Confocal images were obtained with a Zeiss LSM 710 using an EC Plan-Neofluar 40x/1.30 Oil DIC M27 objective and using the ZEN software (Zeiss, Jena, Germany). For R. helvetica-infected cells, staining of slides was performed immediately after fixation each day during the incubation period of 20 d. R. conorii served as a control but the incubation period was limited to five days after infection due to the rapid cell spread of this rickettsia within cells. Images of L929 or Vero cells infected with R. helvetica were analyzed using the software Daime [28]. The detection of nuclei and rickettsiae by the software was optimized by comparing manual counts and software-based counts for three pictures each and the following parameters were chosen: Nuclei stained with DAPI were detected using threshold detection for objects larger than 500 pixels. Rickettsiae were detected using edge detection for objects larger than 20 pixels. As it was not possible to visually separate single rickettsiae in highly infected host cells at later time points, we decided to use the signal area instead of the number of objects in order to quantify the infection progress. The signal area corresponding to rickettsiae in at least 450 host cells was analyzed in images from five, ten, fifteen and twenty days post infectionem (p.i.) and the proportion of the area of rickettsiae to the area of cell nuclei was calculated for each time point.
Plaque assay
Confluent L929 and Vero E6 cells grown in 6-well plates were washed two times with FCS-free Medium. One ml of R. helvetica culture supernatant (undiluted and serially diluted 10−1–10−4) was added per well. One well contained an uninfected control. Plates were incubated at room temperature for 1 h while constantly rocking. Double concentrated M199 (Gibco Thermo Fisher Scientific, Waltham, USA) containing 10% FCS was mixed with an equal volume of pre-heated 2% Agarose (Agarose LE, Biozym, Hessisch Oldendorf, Germany) and each well was then filled with 4 ml of agarose overlay. Plates were incubated for 21 d at 32°C, 5% CO2. Formalin (3.7%) fixation (room temperature, 1 h) was done on days 7, 14, and 21 p.i.. After removal of the agarose plugs the cells were stained using crystal violet (1%) in 20% ethanol (30 min, room temperature). After staining, the plates were washed and examined. In parallel the BSL-2 Rickettsia R. honei was used as a positive (i.e. plaque-forming) control.
Sequence analyses of rickA, sca4 and sca2 genes and deduced amino acids
Amplification of the partial rickA gene was conducted by PCR using primers described by Balraj et al. [29]. PCR conditions were established using DNA from R. conorii VR141. Direct sequencing of PCR products was carried out by GATC Biotech AG sequencing service (Konstanz, Germany). Determination of open reading frames and subsequent amino acid (aa) alignments with corresponding sequences retrieved from the GenBank database (http://www.ncbi.nlm.nih.gov/) were performed using the software package BioEdit v. 7.2.5 [30]. The BioEdit Sequence Alignment Editor, Version 7.2.5 and the implemented ClustalW, Version 1.4 [31] were applied for sequence analyses. In addition, sequencing results of rickA were complemented by data obtained from whole genome pyrosequencing of R. helvetica AS819 using the Roche 454 GS-FLX platform (data not yet published). 377,211 shotgun reads (135,929,920 bases) were assembled using GS De Novo Assembler version 2.3, GS Reference Mapper version 2.3, and DNASTAR SeqMan Ngen version 10.1.0. On average, the coverage of the R. helvetica AS819 plasmid was 146-fold and the coverage of the genome was 34-fold. We predicted CDSs using the RAST prokaryotic genome annotation server (http://rast.nmpdr.org/rast). RNAs were identified using tRNAscan-SE v. 1.23 (tRNA), aragorn v. 1.2.34 (tRNA, tmRNA), and RNammer v.2.1 (rRNA). Database searches were done using BLAST and infernal. Hereby sca2 gene sequences were obtained.
Results
Growth kinetics of R. helvetica AS819 in eukaryotic cell lines
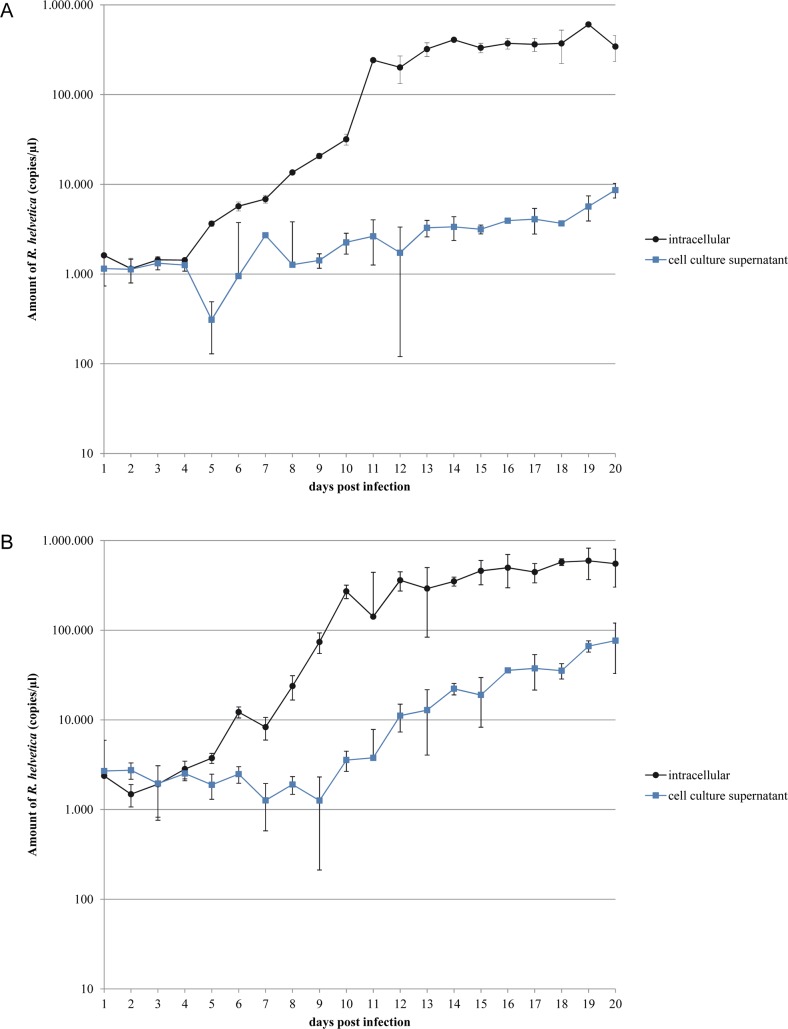
The growth of R. helvetica AS819 in two eukaryotic cell lines is summarised in Fig 1A and 1B. Results were obtained from triplicates of infected cultures per cell line and time point. After two to four days of lag, a continuous increase of R. helvetica genome equivalents (given as copies/μl) was seen in both cell lines resulting in a maximum of 6 x 105 copies/μl at day 19. Propagation in Vero E6 cells (Fig 1A) revealed a slight increase of intracellular R. helvetica DNA copies from day four to eleven after infection. A 100-fold increase was observed from day eleven to day twelve after infection. In L929 cells (Fig 1B), DNA copies of intracellular R. helvetica tripled from day five to day six and a sharp increase of DNA was seen from day seven to day ten resulting in an approximately 1,000-fold rise. A maximum of 100-fold difference between intracellular and extracellular DNA copies/μl was measured in Vero E6 cells (Fig 1A). In contrast, less than 10-fold difference was found using L929 cell lines (Fig 1B).
Fig 1.
Growth of Rickettsia helvetica AS819 in Vero E6 (A) and L929 (B) cell cultures. Genome equivalents measured by quantitative gltA PCR of infected cell cultures are given at several time points during infection. The values from triplicate infected cultures were averaged; standard deviation error bars are given. Growth studies using Vero E6 (A) and L929 (B) were performed simultaneously. ● indicate gltA-copies/μl template-DNA purified from cell layers, and ■ represent gltA-copies/μl template-DNA obtained from culture supernatants.
Growth characteristics determined by laser scanning confocal microscopy
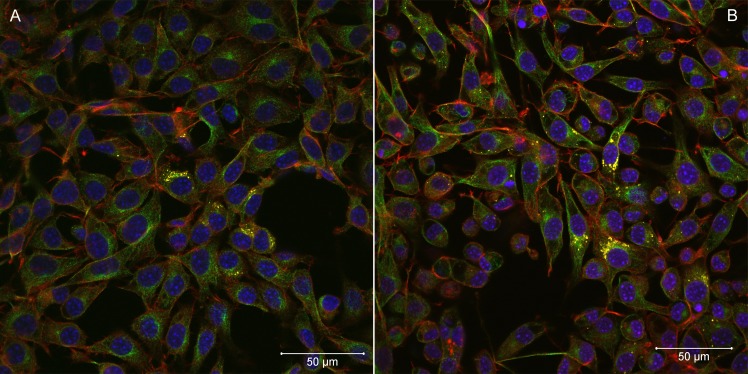
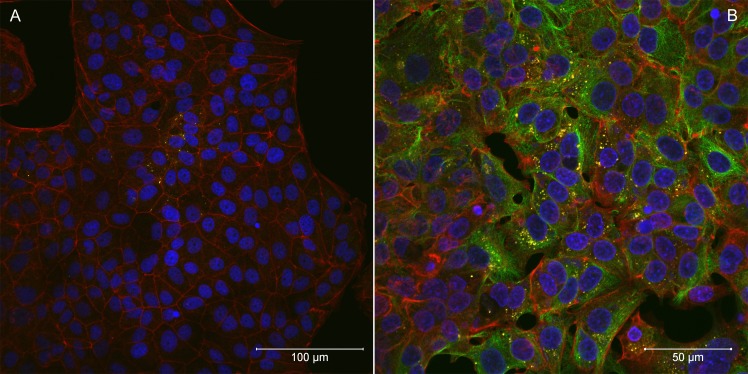
Rapid cell-to-cell spread was not seen in R. helvetica (isolate AS819)-infected cell monolayers. LSM analyses revealed that the percentage of infected cells was rather constant over a period of several days indicating little cell-to-cell spread. Five to ten days p.i., infected cells were scattered throughout the monolayers (Figs 2 and 3). At that time, up to ten rickettsiae were countable within the cytoplasm of a single cell. From day fifteen until the end of the experiment, the number of infected host cells increased. However, infected cells remained in clusters indicating that R. helvetica initiated mainly new infection of adjacent cells. R. helvetica build up in large numbers in the cytoplasm of single cells were visible. No intranuclear bacteria were seen.
Fig 2.
Laser scanning confocal microscopy of Rickettsia helvetica AS819-infected L929 cells five days (A) and ten days (B) after infection. F-actin was stained with Alexa Fluor 568 Phalloidin (red), α-tubulin appears green (Alexa Fluor 488), nuclei were stained with DAPI (blue), and rickettsiae are coloured yellow (magnification 40x). The bar indicates 50 μm.
Fig 3.
Laser scanning confocal microscopy of Rickettsia helvetica AS819-infected Vero E6 cells five days (A) and 20 days (B) after infection. F-actin was stained with Alexa Fluor 568 Phalloidin (red), nuclei were stained with DAPI (blue), and rickettsiae are coloured yellow (magnification 40x). The bar indicates 100 μm (a) and 50 μm (b).
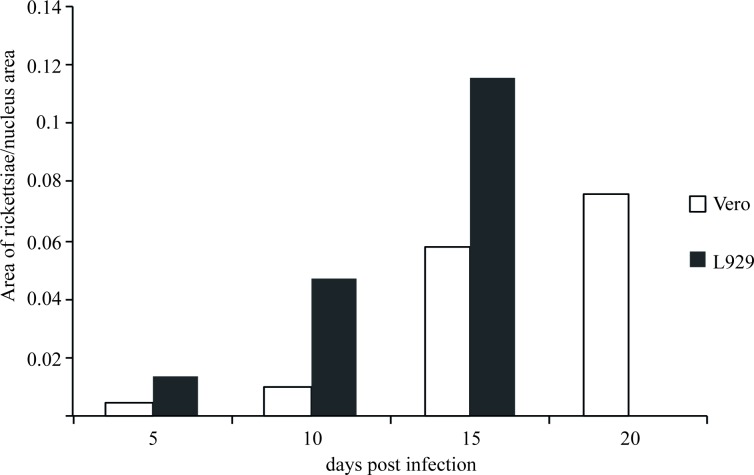
A quantification of rickettsiae per nucleus area over time revealed higher numbers of R. helvetica AS819 within L929 cells compared to Vero E6 cells during the whole infection experiment (Fig 4). In both cell lines the number of rickettsiae per cell nucleus doubled from time point to time point until 15 days after infection. The experiment could be pursued until day 20 p.i. using Vero E6 cells and Fig 4 shows an additional slight increase from day fifteen to day 20 after infection. The detachment of L929 cells increased after day fifteen p.i., hence, due to the low number of attached cells LSM analyses were impossible after that point in time.
Fig 4. Quantification of rickettsiae per nucleus area over time.
The infection progress over time is shown as the increase of rickettsiae per cell nucleus area. For both cell lines a continuous increase of rickettsiae can be observed until day 15 p.i. After this point in time, L929 cells detached and could not be further investigated while a slight increase was still observable in Vero E6 cells from day 15 to day 20 p.i. Images with at least 450 host cells were analysed for each time point and cell line.
In neither cell line actin polymerization due to R. helvetica AS819 was detected in contrast to the R. conorii–infected cell lines (Fig 5). Further, in the latter a prominent plaque formation was visible on day four p.i. indicating rapid cell-to-cell spread. The analysis of images of the MOCK-infected controls did not yield any signals corresponding to rickettsiae.
Fig 5. Laser scanning confocal microscopy of Rickettsia conorii VR141-infected Vero E6 cells four days after infection.
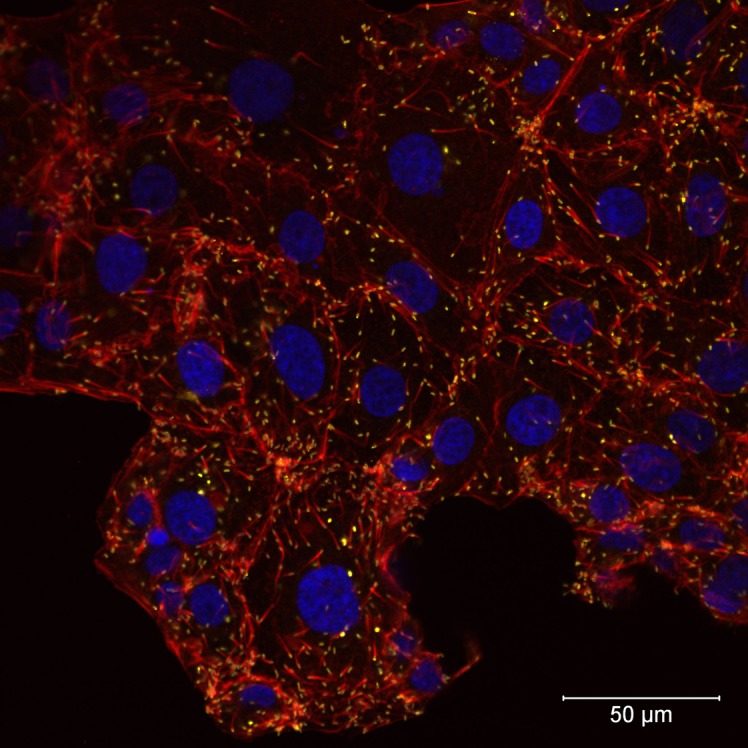
Actin tails at one pole of R. conorii VR141 can be seen. F-actin was stained with Alexa Fluor 568 Phalloidin (red), nuclei were stained with DAPI (blue), and R. conorii VR141 is presented in yellow (magnification 40x). The bar indicates 50 μm.
Plaque formation
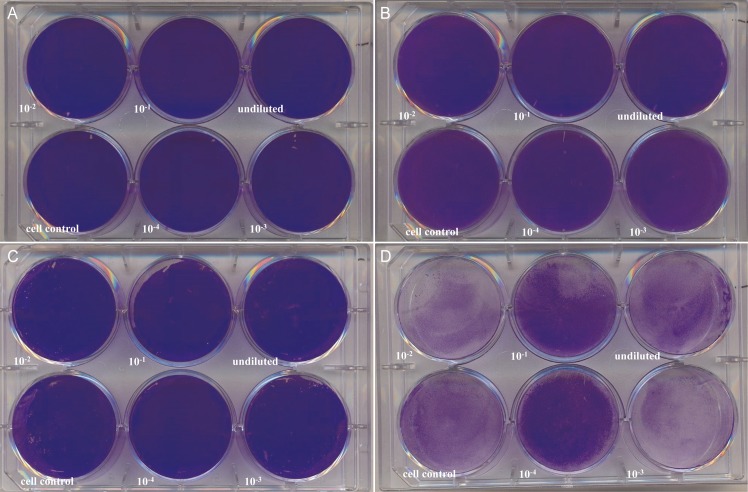
No plaque formation was seen in R. helvetica-infected cell lines at 7 d p.i. (Fig 6A and 6B), 14 p.i. and 21 d p.i. (Fig 6C and 6D). As noticed before, the detachment of L929 cells increased with prolonged incubation time (Fig 6D). In contrast, plaque formation was readily observed in both cell lines infected with R. honei at 7 d p.i. (Fig 7A and 7B).
Fig 6. Rickettsia helvetica AS819 plaque-forming assay.
Vero E6 and L929 monolayers were infected for 21 days with serial dilutions of R. helvetica AS819. After crystal violet staining, plaques were absent at 7 d p.i. in Vero E6 (A) and L929 (B) cells as well as after 21 d p.i. (C, Vero E6; D, L929). Detachment of L929 cells (D) increased with incubation time in agreement with earlier observations.
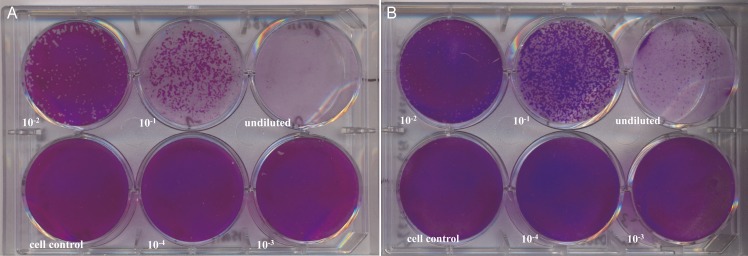
Fig 7. Plaque formation by Rickettsia honei in different cell lines.
Vero E6 and L929 monolayers were infected for 7 days with serial dilutions of R. honei. After crystal violet staining plaques were noticed in Vero E6 (A) and L929 (B) cells.
Hypothetical RickA, Sca4 and Sca2 in R. helvetica AS819
The PCR targeting rickA and subsequent pyrosequencing resulted in a nucleotide sequence of 1,677 nt (accession number MF163038) with highest similarity (i.e. 90%; 1,528 nt/1,694 nt, 40 gaps) to the complete coding sequence of the rickA gene of R. raoultii strain DnS14 (accession number EU340900). The open reading frame encoded a deduced 559 aa sequence. Using the RAST annotation server, this sequence was identified as hypothetical Wiscott-Aldrich Syndrome Protein (WASP)-like protein.
A protein BLAST search revealed 100% identity (559/559 aa) to a hypothetical protein of the R. helvetica type strain C9P9 (accession number WP010421970) followed by 83% similarity (465/560 aa) to the Arp2/3 complex-activating protein RickA of R. felis (accession number WP039594871). Hypothetical RickA in R. helvetica AS819 revealed an N-terminal domain for binding monomeric actin (G-actin binding site) and several proline (P)-rich repeats were counted within the protein sequence (S1 Fig). In addition, the commonly called WCA region composed of a WASP-homology 2 (WH2) region, a central (C region), and an acidic domain was identified in R. helvetica AS819. As described for partial RickA of different other Rickettsia species, human WASP and N-WASP, a conserved motif (ФXXФXXФXXXRXXФ) was found in the C region (S1 Fig), with Ф representing an aliphatic amino acid, X any residue, and R an arginine [32]. In addition, R. helvetica AS819 possesses two WH2 regions (S1 Fig) which has also been described for several other rickettsiae.
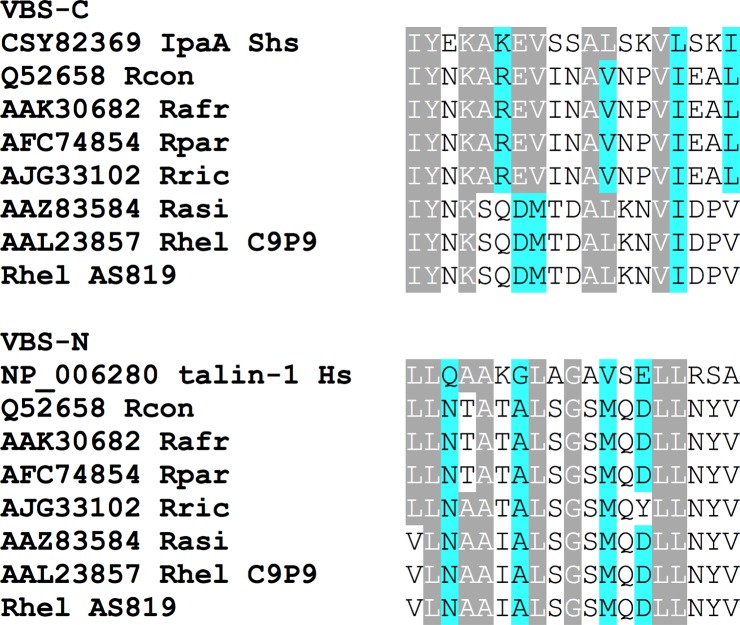
Analyses of nucleotide fragments obtained by pyrosequencing also resulted in 1,917 nt (accession number MF163040) that revealed 97% (1,863 nt/1,918 nt, 13 gaps) similarity to the partial protein PS 120 (D) gene sequence of R. helvetica type strain C9P9 (accession number AF163009) but 99% (1,901/1,918 nt, 13 gaps) to the partial sca4 nucleotide sequence of R. asiatica strain IO-1 (accession number DQ110869). Four partial R. helvetica sca4 gene sequences found in GenBank were identical (767/767 nt, accession number KR150775) or revealed 99% similarity (accession numbers KT825971, KT825970, FJ358501) to the sequence of R. helvetica AS819. The open reading frame resulted in a 639 aa sequence that shared 95% (599/632 aa, 4 gaps) similarity to the partial protein PS 120 sequence of R. helvetica type strain C9P9 (accession number AAL23857) and revealed 99% identity (630/637 aa, 4 gaps) to the partial Sca 4 sequence of R. asiatica strain IO-1 (accession number AAZ83584). Within the aa sequence two stretches were recognized that resemble vinculin-binding sites (Fig 8).
Fig 8. Amino acid sequence alignments of the indicated vinculin-binding-sites.
Sca4 of various Rickettsia shares significant similarity with vinculin-binding-site (VBS) sequences in mammalian talin-1 or Shigella invasion protein IpaA. Gray: aa residues identical to IpaA2 or talin-1; blue: aa residues similar to IpaA2 or talin-1. Abbreviations: Rcon—R. conorii, Rafr—R. africae, Rpar—R. parkeri, Rric—R. rickettsii, Rasi—R. asiatica, Rhel—R. helvetica, Shs—Shigella sonnei, Hs—Homo sapiens.
Furthermore, a 4,884-nucleic acid-stretch (accession number MF163039) with highest identity (99%, 4,858/4,888 nt, 7 gaps) to the R. helvetica C9P9 complete sca2 gene sequence (accession number AY355375) was obtained. This sequence contains also multiple stop codons and therefore seems to be a pseudogene. Therefore, the predicted aa sequence is disrupted compared to other rickettsiae (S2 Fig) most likely resulting in a non-functional protein.
Discussion
The pathogenicity of R. helvetica has not been investigated in depth to date. Its main vector, I. ricinus, is considered as a generalist with an extraordinarily broad host spectrum including mammals, birds, and reptiles [33]. Hence, potential transmission of R. helvetica to humans seems likely. In humans, seroprevalences up to 12.5% against R. helvetica have been demonstrated with forest workers being predisposed to infection [34–37]. However, fulminant clinical cases are rare indicating a rather low pathogenicity compared to other rickettsiae.
There has been only one study describing the life cycle, growth characteristics and host cell response of R. helvetica in a Vero cell line [24] besides the first description of this species [25]. Concurrent to the results by Elfving et al. [24] we observed a short lag phase of up to four days after infection of the monolayers. However, in contrast to that study [24] R. helvetica AS819 grew rather slowly. One possible explanation might be that host cell fragments in our seed culture competed with intact host cells for rickettsial attachment thereby decreasing uptake efficiency as has been described for R. prowazeki seeds [38]. Elfving et al. [24] also reported a lag phase but used suspensions of lysed cells. Moreover, the culture supernatant of our seed culture may have contained an unknown amount of dead or late-growth-phase rickettsia that were no longer in an active growth state [38]. The latter would lead to a lag phase in the intracellular growth [38] as was noticed in our experiments. The percentage of viable R. helvetica AS819 was not assessed hence, the copy numbers calculated for the seed may have included a large amount of dead organisms. The differences between the Swedish and our study might also be attributed to the different culture techniques used in the respective study: conventional culture in the study at hand versus shell vial centrifugation technique used by the Swedish colleagues [24]. The latter technique has been described to increase infectivity [39] but does not resemble the host-pathogen-interaction during natural infection. No intranuclear R. helvetica AS819 was detected in the first description [25] and in our experiments, which is in addition in contrast to the study by Elfving et al. [24] and might also result from the centrifugation step used in their study. We showed that R. helvetica AS819 grew constantly but not rapidly after a phase of adaptation in both tested Vero E6 and L929cell lines, which was confirmed by the LSM investigations. LSM revealed that the dissemination of R. helvetica AS819 in both cell lines was rather mediated by cell break-down and bacterial release than cell-to-cell spread as has also been described for R. prowazekii [40]. This was indicated by the irregularly infected cell layers. R. helvetica AS819 did not produce cytopathogenic effects in Vero E6 and L929 cells which is in agreement with the first description of this species [25]. In addition, actin polymerization due to R. helvetica AS819 was not detected in both cell lines which has also been described for R. helvetica elsewhere [25]. This adds to the previous suggestion that R. helvetica lacks intracellular motility which makes it unlikely to invade nuclei [25,41]. Moreover, plaque formation in Vero E6 and L929 cells was absent after 21 d of infection suggesting that R. helvetica does not spread from cell to cell. Interestingly, this is in contrast to Rolain et al. [42] who reported formation of small plaques after up to 8 d of incubation using the same cell lines. This difference might be due to strain-specific variation in growth characteristics and virulence as has been described for different strains of R. rickettsii [43–45]. Moreover, the number of serial passages of cell lines and rickettsia can influence growth characteristics [42–43,46].
RickA-mediated nucleation of actin plays a role in the intracellular spread of some species [16,47]. Although R. helvetica AS819 possesses a gene encoding for RickA, a bacterial actin nucleator most closely related to WASP/N-WASP-family proteins [19], no host actin polymerization (HAP) was observed. This has also been described for R. raoultii where rickA expression is not sufficient to promote actin-based motility [29]. Furthermore, R. felis and R. parkeri possess genes encoding full-length RickA but lack spread by HAP [48]. A disruption of the rickA coding sequence as has been shown in R. peacockii and REIS [49–50] was not confirmed in R. helvetica AS819 by our sequence analyses. In addition to RickA, Sca2 has been suggested to be necessary for the actin-based motility of rickettsiae [18]. Cardwell and Martinez [22] demonstrated that the first third of the Sca2 passenger domain is highly conserved among SFG rickettsia with the exception of a 39 amino acid deletion. They showed that the deletion of residues 309 to 347 resulted in complete abolishment of actin assembly in R. conorii [22]. Sca2 aa-sequence analysis from R. helvetica AS819 revealed a disrupted aa-sequence resulting in a non-functional protein. This is in concordance to the R. helvetica type strain C9P9. In this strain, sca2 has been deemed a pseudogene with one or more fragments that don’t span the complete protein [48]. Most likely, this might be responsible for the lack of actin tails observed in R. helvetica AS819-infected cell lines. Sca2 also seems to play an important role in the initial bacterial-host interaction and Sca2 of R. conorii mediates both adhesion and invasion of mammalian cells in vitro [22]. However, our R. helvetica isolate AS819 did not exhibit an appreciable defect in adherence or invasion of Vero and L929 cells in vitro. This might be attributed to other highly conserved proteins which may compensate for the lack of functional Sca2 [22]. Only recently, Sca4 was identified as another protein from SFG rickettsia that promotes spread [23]. Specifically, Sca4 of R. parkeri binds to the cell-adhesion protein vinculin and inhibits its activity thereby reducing intercellular tension forces [23,51]. For R. helvetica isolate AS819 a 639 aa-stretch was identified that revealed 95% similarity to Sca4 of R. helvetica C9P9. For the latter species Sca4 has been supposed to be a truncated protein [48] which most certainly might also be the case for the strain investigated in this study.
Cell-to-cell spread is one crucial step in the intracellular life cycle of several pathogens including rickettsiae. Actin-based motility contributes to cell-to-cell spread and dissemination within the host. In contrast to other rickettsiae, R. helvetica isolate AS819 did not spread directly from cell to cell by actin-based motility presumably due to a deletion in the predicted Sca2 protein. As Sca2 is needed for virulence [14] our results suggest less virulence and pathogenicity of R. helvetica isolated from ixodid ticks in Germany.
Supporting information
The sequences are trimmed at the end. Red: G-actin binding site; green: WH2 regions; blue: central region; orange: acidic domain. The proline-rich regions are highlighted in grey. A conserved motif can be seen at the end of the sequence, where Ф represents an aliphatic amino acid, X represents any residue, and R is a conserved arginine. Abbreviations: Rhel—R. helvetica, Rak—R. akari, Rmas—R. massiliae, Rrao—R. raoultii, Rfel—R. felis, Rcan—R. canadensis, Rsib—R. sibirica, Rafr—R. africae, Rcon—R. conorii, Rric—R. rickettsii, Rslo—R. slovaca, Rmon—R. monacensis.
(PDF)
Numbering above the alignment indicates amino acid numbering according to R. conorii Sca2 (accession number NP 359747). Dashes within the alignment indicate deletions. * indicate stop-codons. Amino acids 309 to 347, which have been described necessary for the stimulation of actin assembly by Sca2, are shaded in grey; WH2 domains are shaded in blue, and proline-rich domains are boxed in black. Abbreviations: Rcon—R. conorii, Rric—R. rickettsii, Rpar—R. parkeri, Rpea–R. peacockii, Rhel—R. helvetica.
(PDF)
Acknowledgments
The authors would like to thank Christina Speck for her valuable help in preparing figures. We thank Harald Weber for his valuable technical assistance.
Data Availability
All sequence files are available from the GenBank database (accession numbers MF163037, MF163038, MF163039, MF163040). The rest of relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Wölfel R, Terzioglu R, Kiessling J, Wilhelm S, Essbauer S, Pfeffer M, et al. Rickettsia spp. in Ixodes ricinus ticks in Bavaria, Germany. Ann N Y Acad Sci. 2006; 1078:509–511. doi: 10.1196/annals.1374.133 [DOI] [PubMed] [Google Scholar]
- 2.Silaghi C, Hamel D, Thiel C, Pfister K, Pfeffer M. Spotted fever group rickettsiae in ticks, Germany. Emerg Infect Dis. 2011; 17(5):890–892. doi: 10.3201/eid1705.101445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schorn S, Pfister K, Reulen H, Mahling M, Silaghi C. Occurrence of Babesia spp., Rickettsia spp. and Bartonella spp. in Ixodes ricinus in Bavarian public parks, Germany. Parasit Vectors. 2011; 15(4):135 doi: 10.1186/1756-3305-4-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schicht S, Schnieder T, Strube C. Rickettsia spp. and coinfections with other pathogenic microorganisms in hard ticks from northern Germany. J Med Entomol. 2012; 49(3):766–771. [DOI] [PubMed] [Google Scholar]
- 5.Fournier PE, Allombert C, Supputamongkol Y, Caruso G, Brouqui P, Raoult D. Aneruptive fever associated with antibodies to Rickettsia helvetica in Europe and Thailand. J Clin Microbiol. 2004; 42(2):816–818. doi: 10.1128/JCM.42.2.816-818.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nilsson K. Septicaemia with Rickettsia helvetica in a patient with acute febrile illness, rash and myasthenia. J Infect. 2009; 58(1):79–82. doi: 10.1016/j.jinf.2008.06.005 [DOI] [PubMed] [Google Scholar]
- 7.Nilsson K, Lindquist O, Påhlson C. Association of Rickettsia helvetica with chronic perimyocarditis in sudden cardiac death. Lancet. 1999; 354(9185):1169–1173. doi: 10.1016/S0140-6736(99)04093-3 [DOI] [PubMed] [Google Scholar]
- 8.Nilsson K, Elfving K, Pahlson C. Rickettsia helvetica in patient with meningitis, Sweden, 2006. Emerg Infect Dis. 2010; 16(3):490–492. doi: 10.3201/eid1603.090184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray GG, Weinert LA, Rhule EL, Welch JJ. The Phylogeny of Rickettsia Using Different Evolutionary Signatures: How Tree-Like is Bacterial Evolution? Syst Biol. 2016; 65(2):265–79. doi: 10.1093/sysbio/syv084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parola P, Paddock CD, Socolovschi C, Labruna MB, Mediannikov O, Kernif T, et al. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev. 2013; 26(4):657–702. Erratum in: Clin Microbiol Rev. 2014; 27(1):166. doi: 10.1128/CMR.00032-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silverman DJ, Wisseman CL Jr. In vitro studies of rickettsia-host cell interactions: ultrastructural changes induced by Rickettsia rickettsii infection of chicken embryo fibroblasts. Infect Immun. 1979; 26(2):714–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valbuena G, Walker DH. Changes in the adherens junctions of human endothelial cells infected with spotted fever group rickettsiae. Virchows Arch. 2005; 446(4):379–382. doi: 10.1007/s00428-004-1165-3 [DOI] [PubMed] [Google Scholar]
- 13.Walker DH, Ismail N. Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat Rev Microbiol. 2008; 6(5):375–386. doi: 10.1038/nrmicro1866 [DOI] [PubMed] [Google Scholar]
- 14.Ireton K. Molecular mechanisms of cell-cell spread of intracellular bacterial pathogens. Open Biol. 2013; 17;3(7):130079 doi: 10.1098/rsob.130079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teysseire N, Chiche-Portiche C, Raoult D. Intracellular movements of Rickettsia conorii and R. typhi based on actin polymerization. Res Microbiol. 1992; 143(9):821–9. [DOI] [PubMed] [Google Scholar]
- 16.Heinzen RA, Hayes SF, Peacock MG, Hackstadt T. Directional actin polymerization associated with spotted fever group Rickettsia infection of Vero cells. Infect Immun. 1993; 61(5):1926–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serio AW, Jeng RL, Haglund CM, Reed SC, Welch MD. Defining a core set of actin cytoskeletal proteins critical for actin-based motility of Rickettsia. Cell Host Microbe. 2010; 20;7(5):388–398. doi: 10.1016/j.chom.2010.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleba B, Clark TR, Lutter EI, Ellison DW, Hackstadt T. Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect Immun. 2010; 78(5):2240–2247. doi: 10.1128/IAI.00100-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gouin E, Egile C, Dehoux P, Villiers V, Adams J, Gertler F, et al. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature. 2004; 29;427(6973):457–461. doi: 10.1038/nature02318 [DOI] [PubMed] [Google Scholar]
- 20.Jeng RL, Goley ED, D'Alessio JA, Chaga OY, Svitkina TM, Borisy GG, et al. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell Microbiol. 2004; 6(8):761–769. doi: 10.1111/j.1462-5822.2004.00402.x [DOI] [PubMed] [Google Scholar]
- 21.Haglund CM, Choe JE, Skau CT, Kovar DR, Welch MD. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat Cell Biol. 2010; 12(11):1057–1063. doi: 10.1038/ncb2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cardwell MM, Martinez JJ. Identification and characterization of the mammalian association and actin nucleating domains in the Rickettsia conorii autotransporter protein, Sca2. Cell Microbiol. 2012; 14(9):1485–1495. doi: 10.1111/j.1462-5822.2012.01815.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamason RL, Bastounis E, Kafai NM, Serrano R, del Álamo JC, Theriot JA, et al. Rickettsia Sca4 reduces vinculin-mediated intercellular tension to promote spread. Cell. 2016; 167(3):670–683.e10. doi: 10.1016/j.cell.2016.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elfving K, Lukinius A, Nilsson K. Life cycle, growth characteristics and host cell response of Rickettsia helvetica in a Vero cell line. Exp Appl Acarol. 2012; 56(2):179–187. doi: 10.1007/s10493-011-9508-7 Erratum in: Exp Appl Acarol. 2012; 56(2):189–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beati L, Péter O, Burgdorfer W, Aeschlimann A, Raoult D. Confirmation that Rickettsia helvetica sp. nov. is a distinct species of the spotted fever group of rickettsiae. Int J Syst Bacteriol. 1993; 43(3):521–526. doi: 10.1099/00207713-43-3-521 [DOI] [PubMed] [Google Scholar]
- 26.Wölfel R, Essbauer S, Dobler G. Diagnostics of tick-borne rickettsioses in Germany: a modern concept for a neglected disease. Int J Med Microbiol 2008; 298 S1:368–374. [Google Scholar]
- 27.Schex S, Dobler G, Riehm J, Müller J, Essbauer S (2010). Rickettsia spp. in wild small mammals in Lower Bavaria, South-Eastern Germany. Vector Borne Zoonotic Dis. 2011; 11(5):493–502. doi: 10.1089/vbz.2010.0060 [DOI] [PubMed] [Google Scholar]
- 28.Daims H, Lücker S, Wagner M. Daime, a novel image analysis program for microbial ecology and biofilm research. Environ Microbiol. 2006; 8:200–213. doi: 10.1111/j.1462-2920.2005.00880.x [DOI] [PubMed] [Google Scholar]
- 29.Balraj P, El Karkouri K, Vestris G, Espinosa L, Raoult D, Renesto P. RickA expression is not sufficient to promote actin-based motility of Rickettsia raoultii. PLoS ONE 2008; 3(7):e2582 doi: 10.1371/journal.pone.0002582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis programme for Windows 95/98/NT. Nucl Acids Symp Ser. 1999; 41:95–98. [Google Scholar]
- 31.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panchal SC, Kaiser DA, Torres E, Pollard TD, Rosen MK. A conserved amphipathic helix in WASP/Scar proteins is essential for activation of Arp2/3 complex. Nat Struct Biol. 2003;10(8):591–598. doi: 10.1038/nsb952 [DOI] [PubMed] [Google Scholar]
- 33.Petney TN, Pfäffle M, Skuballa J. An annotated checklist of the ticks of Germany. Syst. Appl. Acarol. 2012; 17:115–170. [Google Scholar]
- 34.Fournier PE, Grunnenberger F, Jaulhac B, Gastinger G, Raoult D. Evidence of Rickettsia helvetica infection in humans, eastern France. Emerg Infect Dis. 2000; 6(4):389–392. doi: 10.3201/eid0604.000412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen H, Fournier PE, Pedersen IS, Krarup H, Ejlertsen T, Raoult D. Serological and molecular evidence of Rickettsia helvetica in Denmark. Scand J Infect Dis. 2004; 36(8):559–563. doi: 10.1080/00365540410020776 [DOI] [PubMed] [Google Scholar]
- 36.Cinco M, Luzzati R, Mascioli M, Floris R, Brouqui P. Serological evidence of Rickettsia infections in forestry rangers in north-eastern Italy. Clin Microbiol Infect. 2006; 12(5):493–495. doi: 10.1111/j.1469-0691.2006.01385.x [DOI] [PubMed] [Google Scholar]
- 37.Wölfel S, Speck S, Essbauer S, Thoma BR, Mertens M, Werdermann S, et al. High seroprevalence for indigenous spotted fever group rickettsiae in forestry workers from the federal state of Brandenburg, Eastern Germany. Ticks Tick Borne Dis. 2017; 8(1):132–138. doi: 10.1016/j.ttbdis.2016.10.009 [DOI] [PubMed] [Google Scholar]
- 38.Wisseman CL Jr, Waddell AD, Silverman DJ. In vitro studies on Rickettsia-host cell interactions: Lag phase in intracellular growth cycle as a function of stage of growth of infecting Rickettsia prowazeki, with preliminary observations on inhibition of rickettsial uptake by host cell fragments. Infect Immun. 1976; 13(6):1749–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marrero M, Raoult D. Centrifugation-shell vial technique for rapid detection of Mediterranean spotted fever rickettsia in blood culture. Am J Trop Med Hyg. 1989; 40(2):197–199. [DOI] [PubMed] [Google Scholar]
- 40.Wisseman CL Jr, Waddell AD. In vitro studies on rickettsia-host cell interactions: intracellular growth cycle of virulent and attenuated Rickettsia prowazeki in chicken embryo cells in slide chamber cultures. Infect Immun. 1975; 11(6):1391–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teysseire N, Chiche-Portiche C, Raoult D. Intracellular movements of Rickettsia conorii and R. typhi based on actin polymerization. Res Microbiol. 1992; 143:821–829. [DOI] [PubMed] [Google Scholar]
- 42.Rolain JM, Maurin M, Vestris G, Raoult D. In vitro susceptibilities of 27 rickettsiae to 13 antimicrobials. Antimicrob Agents Chemother. 1998; 42(7):1537–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson JW, Pedersen CE Jr. Plaque formation by strains of spotted fever group rickettsiae in monolayer cultures of various cell types. J Clin Microbiol. 1978; 7(4):389–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hackstadt T, Messer R, Cieplak W, Peacock MG. Evidence for proteolytic cleavage of the 120-kilodalton outer membrane protein of rickettsiae: identification of an avirulent mutant deficient in processing. Infect Immun. 1992; 60(1):159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eremeeva ME, Dasch GA, Silverman DJ. Quantitative analyses of variations in the injury of endothelial cells elicited by 11 isolates of Rickettsia rickettsii. Clin Diagn Lab Immunol. 2001; 8(4):788–796. doi: 10.1128/CDLI.8.4.788-796.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox HR. Cultivation of rickettsiae of the Rocky Mountain spotted fever, typhus and Q fever groups in the embryonic tissues of developing chicks. Science. 1941; 94(2444):399–403. doi: 10.1126/science.94.2444.399 [DOI] [PubMed] [Google Scholar]
- 47.Gouin E, Gantelet H, Egile C, Lasa I, Ohayon H, Villiers V, et al. A comparative study of the actin-based motilities of the pathogenic bacteria Listeria monocytogenes, Shigella flexneri and Rickettsia conorii. J Cell Scie. 1999; 112(Pt 11):1697–1708. [DOI] [PubMed] [Google Scholar]
- 48.Gillespie JJ, Simran JK, Sayeedur Rahman M, Rennoll-Bankert K, Sears KT, Beier-Sexton, et al. Secretome of obligate intracellular Rickettsia. FEMS Microbiol Rev. 2015; 39:47–80. doi: 10.1111/1574-6976.12084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simser JA, Rahman MS, Dreher-Lesnick SM, Azad AF. A novel and naturally occurring transposon, ISRpe1 in the Rickettsia peacockii genome disrupting the rickA gene involved in actin-based motility. Mol Microbiol. 2005; 58(1):71–79. doi: 10.1111/j.1365-2958.2005.04806.x [DOI] [PubMed] [Google Scholar]
- 50.Gillespie JJ, Joardar V, Williams KP, Driscoll T, Hostetler JB, Nordberg E, et al. A Rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J Bacteriol. 2012; 194(2):376–394. doi: 10.1128/JB.06244-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park H, Lee JH, Gouin E, Cossart P, Izard T. The rickettsia surface cell antigen 4 applies mimicry to bind to and activate vinculin. J Biol Chem. 2011; 286(40):35096–103. doi: 10.1074/jbc.M111.263855 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sequences are trimmed at the end. Red: G-actin binding site; green: WH2 regions; blue: central region; orange: acidic domain. The proline-rich regions are highlighted in grey. A conserved motif can be seen at the end of the sequence, where Ф represents an aliphatic amino acid, X represents any residue, and R is a conserved arginine. Abbreviations: Rhel—R. helvetica, Rak—R. akari, Rmas—R. massiliae, Rrao—R. raoultii, Rfel—R. felis, Rcan—R. canadensis, Rsib—R. sibirica, Rafr—R. africae, Rcon—R. conorii, Rric—R. rickettsii, Rslo—R. slovaca, Rmon—R. monacensis.
(PDF)
Numbering above the alignment indicates amino acid numbering according to R. conorii Sca2 (accession number NP 359747). Dashes within the alignment indicate deletions. * indicate stop-codons. Amino acids 309 to 347, which have been described necessary for the stimulation of actin assembly by Sca2, are shaded in grey; WH2 domains are shaded in blue, and proline-rich domains are boxed in black. Abbreviations: Rcon—R. conorii, Rric—R. rickettsii, Rpar—R. parkeri, Rpea–R. peacockii, Rhel—R. helvetica.
(PDF)
Data Availability Statement
All sequence files are available from the GenBank database (accession numbers MF163037, MF163038, MF163039, MF163040). The rest of relevant data are within the paper and its Supporting Information files.