Abstract
Visual information processing in animals with large image forming eyes is carried out in highly structured retinotopically ordered neuropils. Visual neuropils in Drosophila form the optic lobe, which consists of four serially arranged major subdivisions; the lamina, medulla, lobula and lobula plate; the latter three of these are further subdivided into multiple layers. The visual neuropils are formed by more than 100 different cell types, distributed and interconnected in an invariant highly regular pattern. This pattern relies on a protracted sequence of developmental steps, whereby different cell types are born at specific time points and nerve connections are formed in a tightly controlled sequence that has to be coordinated among the different visual neuropils. The developing fly visual system has become a highly regarded and widely studied paradigm to investigate the genetic mechanisms that control the formation of neural circuits. However, these studies are often made difficult by the complex and shifting patterns in which different types of neurons and their connections are distributed throughout development. In the present paper we have reconstructed the three-dimensional architecture of the Drosophila optic lobe from the early larva to the adult. Based on specific markers, we were able to distinguish the populations of progenitors of the four optic neuropils and map the neurons and their connections. Our paper presents sets of annotated confocal z-projections and animated 3D digital models of these structures for representative stages. The data reveal the temporally coordinated growth of the optic neuropils, and clarify how the position and orientation of the neuropils and interconnecting tracts (inner and outer optic chiasm) changes over time. Finally, we have analyzed the emergence of the discrete layers of the medulla and lobula complex using the same markers (DN-cadherin, Brp) employed to systematically explore the structure and development of the central brain neuropil. Our work will facilitate experimental studies of the molecular mechanisms regulating neuronal fate and connectivity in the fly visual system, which bears many fundamental similarities with the retina of vertebrates.
Keywords: Drosophila, optic lobe, neuropil, connectivity, development, digital model
INTRODUCTION
Animals with complex visually guided behaviors possess image forming eyes whose neuronal projections to the brain form precise retinotopic maps. In vertebrates, image processing already takes place in the retina. Photoreceptors (rods and cones) of the outer plexiform layer project their short axons towards the inner layer, formed by first order visual interneurons- the bipolar cells. Bipolar cells target the basally located layer of the second order visual interneurons, the ganglion cells. Several types of local interneurons (amacrine cells, horizontal cells) laterally connect bipolar cells and ganglion cells (Baier 2013; Masland 2001; Masland and Raviola, 2000). Ganglion cell axons leave the eye through the optic stalk and project in a retinotopically ordered manner to the contralateral optic tectum and dorsal thalamus. In arthropods with large image forming eyes, including Drosophila, visual information is collected by the compound eye., which is formed by a large number of repetitive modules called ommatidia. Each ommatidium possesses six outer photoreceptors (R1-R6) and two inner photoreceptors (R7-R8). Photoreceptors project in a retinotopic order to the optic lobe, part of the brain that processes exclusively visual information, and that has been homologized with the inner layers of the vertebrate retina and the tectum (Joly et al., 2016; Sanes and Zipursky, 2010; Erclik et al., 2009; Cajal and Sanchez, 1915)
The Drosophila optic lobe has become a propitious model system for analyzing the structure, development and function of neural networks (Langen et al., 2015; Silies et al., 2014; Apitz and Salecker, 2014; Wernet et al., 2014; Brand and Livesey, 2011; Sanes and Zipursky, 2010; Fischbach and Hiesinger, 2008). The optic lobe of the adult fly has four main compartments (“optic ganglia”), the lamina, medulla, lobula and lobula plate (Fig.1), each of which is further subdivided into multiple layers. Photoreceptors involved in motion detection (R1-6) terminate in the lamina; R7 and R8, responsible for color vision, project to the outer medulla (Hadjieconomou et al., 2011; Meinertzhagen and Hanson, 1993; Braitenberg 1967; Trujillo-Cenóz 1965). This ordered projection subdivides the lamina and medulla into stereotyped, repetitive units, called cartridges in the lamina and columns in the medulla (Fig.1). Lamina interneurons (L1-L5), targeted by photoreceptors R1-R6 project to the medulla. Medulla intrinsic neurons targeted by R7/8 and by L1-L5, interconnect the distal and proximal layers of the medulla [medulla intrinsic neurons (Mi)] or different columns [multicolumnar distal medulla intrinsic neurons (Dm), multicolumnar proximal medulla intrinsic neurons (Pm)]. Transmedullary neurons (Tm/TmY) project to the lobula/lobula plate (Bausenwein et al., 1992; Fischbach and Dittrich, 1989; Fig.1). Medulla intrinsic neurons and transmedullary neurons comprise a large number of subtly different types. Neurons of the lobula plate (T2-T5; Scott et al., 2002; Fischbach and Dittrich, 1989) form complex, retinotopically-organized connections between medulla, lobula and lobula plate (Fig.1). Aside from these columnar (LC) neurons, the lobula/lobula plate, as well as the medulla, possess many different types of visual projection neurons whose axons transmit processed visual information to the central brain (Wernet et al., 2014; Aptekar et al., 2015).
Figure 1.
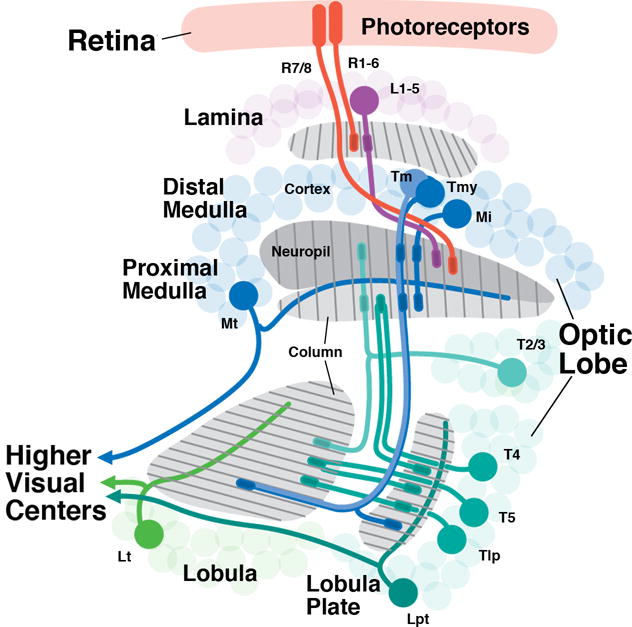
Overview of neuronal connectivity in the Drosophila visual system. Photoreceptors (red) are located in the retina and project in a precise retinotopic manner to the visual processing center of the brain, the optic lobe. The optic lobe has four synaptic compartments (from distal to proximal): lamina, medulla, lobula, and lobula plate. The medulla synaptic layer is subdivided to the distal and proximal region separated by the serpentine layer. R1-R6 photoreceptors terminate in the lamina while R7-R8 terminate in the medulla. Lamina interneurons (L1-5; purple), targeted by R1-R6 project to the distal medulla; medulla local interneurons (medulla intrinsic neurons; Mi) interconnect the distal and proximal medulla; projection neurons (Tm, Tmy) connect to the lobula and lobula plate. Columnar neurons also connect lobula and lobula plate (Tlp), and lobula/lobula plate to the medulla (T2-5). Large tangential neurons of the medulla (Mt), lobula (Lt) and lobula plate (Lpt) transmit processed visual information to the central nervous system. Several types of local neurons interconnecting neighboring medulla columns (Dm, Pm), projection neurons (T1, Y), as well as columnar projections neurons of the lobula are omitted.
All neurons of the four optic ganglia are produced by a small epithelial placode (“optic placode) that invaginates from the neurectoderm of the embryonic head (Green et al., 1993). In the late embryo, the optic placode splits into two layers called the outer (OOA) and inner (IOA) optic anlage. The OOA gives rise to the neurons of the lamina (L1-L5) and medulla (Mi, Dm, Pm, Tm, TmY, Mt), while the IOA produces neurons of the lobula and lobula plate (e.g., T2-5; Apitz and Salecker, 2015; 2014; Li et al., 2013a; 2013b; Suzuki et al., 2013; Hasegawa et al., 2011; 2013). Given the large number of optic lobe neurons (about 100,000, compared to approximately 20,000 in the central brain), proliferation of the optic anlagen continues throughout a long period of development, from embryo to early pupa. It is further boosted by a two-phase mechanism which is unique to the visual system. In a first phase, the optic anlagen grow by symmetric cell division to a size of several thousand epithelial progenitor cells (Ngo et al., 2010; Egger et al., 2007; Hofbauer and Campos-Ortega, 1990). During the second phase, which begins halfway through the larval period, the progenitors undergo an epithelial-mesenchymal transition (EMT), becoming neuroblasts that enter a phase of asymmetric cell division, each neuroblast producing a lineage comprising in the order of 100 neurons (K.N. and V.H., unpublished). The molecular pathways controlling the EMT of optic lobe progenitors has been elucidated by recent studies (Apitz and Salecker, 2015; Morante et al., 2013; Orihara-Ono et al., 2011; Egger et al., 2010; Ngo et al., 2010; Reddy et al., 2010; Yasugi et al., 2008, 2010). By contrast, little is known about the specification of fate and connectivity of the multitude of neurons making up the optic lobe. For the medial domain within the OOA, which gives rise to the medulla neurons,.cell fate appears to be mainly linked to the time of birth within the lineage produced by a progenitor (Bertet et al., 2014; Li et al., 2013a, 2013b; Hasegawa et al., 2011, 2013; Suzuki et al., 2013; Morante et al., 2011). On the other hand, the five different types of lamina neurons, derived from the lateral margin of the OOA, seem to be specified already at the level of the progenitor itself and are not time-dependent (Pinero et al., 2014; Selleck et al., 1992; Selleck and Steller, 1991). Nothing is known about the underlying mechanism of how the diverse cell types in the lobula and lobula plate are generated.
The analysis of optic lobe development is complicated by the fact that the optic anlagen and the neurons/nerve fibers they produce undergo complex morphogenetic movements before adopting their final position in the adult brain (Meinertzhagen and Hanson, 1993). For example, in the early larva the inner and outer optic anlagen form two C-shaped epithelia directly apposed to each other. Subsequently, while both anlagen grow in size, several morphogenetic processes happen at the same time. First, both anlagen give rise to neuroblasts; secondly, neuroblasts divide in a plane different from that of the cells that form part of the epithelial anlagen. Neuroblast divisions result in the generation of neurons which assemble into the cell body rinds (cortices) of the optic ganglia. Already during late the larval stage, these neurons project axons that form the rudimentary beginnings of the optic neuropils. As the neuropils and cellular cortices of the optic ganglia grow they dramatically change their position relative to each other. For example, the cortices of the lamina and lobula plate direct neighbors in the late larva, become far removed from each other in the pupa. The lamina, initially contiguous with and oriented at a right angle to the medulla, moves over the medulla. These and other large-scale movements are instrumental in shaping the architecture of the fiber masses interconnecting the optic ganglia, i.e., the outer and inner chiasm. In previous studies of optic lobe development, some of the morphogenetic movements have been inferred from comparisons between initial (i.e., larval) stages and the adult configuration. However, many of the morphogenetic events remain unclear.
In order to help filling this gap of knowledge, we have used a set of global markers for neurons and axon tracts, as well as specific markers for individual neural lineages, to reconstruct the three-dimensional architecture of the Drosophila optic lobe during sequential larval and pupal stages. We were able to separately recognize the primordia, cell body rinds, neuropils and axonal connections of the four optic ganglia and generate sets of annotated confocal z-projections and 3D digital models of these structures for representative stages. Our data illustrate the coordinated growth of the optic neuropils and their interconnecting fiber tracts, which follows a temporal gradient reflecting the posterior-to-anterior gradient of eye development. The data also document the movements that change the position and orientation of the optic neuropils and thereby shape the retinotopically ordered structure of the optic chiasms. Finally, we use the global markers N-cadherin and Bruchpilot (Brp) to define within the developing neuropil of the medulla and lobula complex discrete layers that can be correlated with the system of layers based on specific cell types (Fischbach and Dittrich, 1989). Our work, intended as a “user’s guide” for students of fly visual system development, will facilitate future genetic studies that rely on the interpretation of complex expression patterns and phenotypes.
MATERIALS AND METHODS
Fly Stocks
Flies were grown at 25°C using standard fly media unless otherwise noted. The following transgenic strains were used in this study (original source in parentheses): esg-Gal4 (B. Edgar); insc-Gal4; UAS-mCherry (Bloomington Stock Center, Stock #8751); Ln-Gal4 (L. Zipursky); forNP79-Gal4 (Kyoto Stock Center; Stock #103517); NP3233-Gal4 (Kyoto Stock Center; Stock #113173); bsh-Gal4 (M. Sato; Hasegawa et al., 2011); DrxGMR77F09-Gal4 (Janelia Research Campus FlyLight Gal4 Collection; Bloomington Stock Center, Stock #46986); acj6PG63-Gal4 (L. Luo; Potter et al., 2010); wg-Gal4 (Giraldez et al., 2002); Vsx-Gal4 (T. Erclik); UAS-mCD8GFP on X, II, III (Bloomington Stock Center; Stock #5130, #5136, #5137) was used to recombine the various Gal4 drivers. Genotypes used for labeling abbreviated cell types in the figures are listed below as a reference. Global markers are not listed. Refer to the main text for full name of cell types.
|
| |
| Genotype | Cell types |
|
| |
| esg-Gal4, UAS-myr::RFP | OPC/IPC ep |
|
| |
| forNP79-Gal4, UAS-mCD8::GFP | IPC nbs |
|
| |
| UAS-Flp, Act5C>y+>Gal4/+; forNP79-Gal4, UAS-mCD8::GFP/+; 10xUAS-IVS-mCD8::GFP/+ | Pupal T neurons Lt neurons |
|
| |
| UAS-mCD8::GFP; Ln-Gal4 | L3/L4 neurons |
|
| |
| Drx-Gal4/10xIVS-mCD8::GFP | T2-5 neurons |
|
| |
| Vsx-Gal4, UAS-mCD8::GFP | TM neurons |
|
| |
| acj6PG63-Gal4, UAS-mCD8GFP | T4/T5 neurons |
|
| |
| bsh-Gal4, UAS-mCD8::GFP | Mi1/L5 neurons |
|
| |
| insc-Gal4; UAS-mCherry | NBs; pan-neuronal |
|
| |
| JupiterG001147::GFP | neurons and axons |
|
| |
| c202-Gal4, UAS-mCD8::GFP | L1 neurons |
|
| |
| UAS-Flp, Act5C>y+>Gal4/+; wg-Gal4, UAS-mCD8::GFP/+; 10xUAS-IVS-mCD8::GFP/+ | MT neurons |
|
| |
| wg-Gal4, UAS-mCD8::GFP | MT neurons |
|
| |
| NP3233-Gal4, UAS-mCD8::GFP | ALG |
|
| |
| Nrv2-Gal4, UAS-mCD8::GFP | neuropil glia |
|
| |
Additional Markers
|
| |
| Markers | Cell types |
|
| |
| FasII | L4/L5 |
|
| |
| 24B10 | R7/R8 |
|
| |
| Dpn | neuroblasts |
|
| |
| Dac | LA, LO, LP neurons |
|
| |
Immunohistochemistry
The following antibodies were provided by the Developmental Studies Hybridoma Bank (Iowa City, IA): mouse anti-Neurotactin (BP106, 1:10), mouse anti-Neuroglian (BP104, 1:30), mouse anti-Fas2 (1:4), mouse anti-Chaoptin (24B10, 1:10) and rat anti-DN-Cadherin (DN-EX #8, 1:20); mouse anti-Dlg (4F3, 1:10). Additional primary antibodies used in this study were: rabbit anti-Synaptotagmin (1/500; gift from H. Bellen), rabbit anti-V5 (1/100; #R960-25, Invitrogen). Secondary antibodies, IgG (Jackson ImmunoResearch; Molecular Probes) were used at the following dilutions: Dynalight 649-conjugated anti-rat (1:400), Cy5 anti-rat (1:700); AlexaFluor 488-conjugated anti-mouse (1:500) and 546-conjugated anti-mouse (1:500). Larvae and pupae were staged as previously described (Bainbridge and Bownes, 1981). Fixation procedures for larval, pupal and adult optic lobes varied and are described below. For larval brains, dissected tissues were fixed in 3.7% formaldehyde in PBS (137 mM NaCl, 2.7 mM KCl, 5.63 mM Na2HPO4, 6.37 mM KH2PO4; pH = 7.4) for 25 min. Staged pupal tissues were dissected and fixed in 4% methanol-free paraformaldehyde (PFA) in PBT (PBS with 0.1% Triton X-100) for 40 min at 4°C. Tissues were permeabilized in PBT (PBS with 0.3% Triton X-100) and immunofluorescence was performed using standard procedures with the exception of BP106 (Neurotactin, Nrt) and BP104 (Neuroglian, Nrg) labeling. For BP106/BP104 labeling in the pupae, dissected tissues were fixed on ice in PBS in PFA for 35-90 min (depending on the stage). Tissues were dehydrated and stored in ethanol at −20°C overnight. Tissues were rehydrated on ice and standard immunolabeling was performed. Adult tissues were fixed in 4% PFA in PBT (PBS with 0.3% Triton X-100). Tissues were mounted in Vectashield mounting medium (Vector, Burlingame, CA, #H1000).
Lineage Tracing
Lineage tracing experiments were performed for for-Gal4 and wg-Gal4 by crossing to transgenic flies with the genotype: UAS-Flp, Act5C-FRT(stop, y+)FRT-Gal4;;10xUAS-mCD8GFP/TM3, Kr-Gal4, UAS-GFP. Staged pupae were grown at 25°C, dissected, and brain tissues were harvested at desired time points for downstream immunofluorescence.
EdU Labeling
5-ethynyl-2′-deoxyuridine (EdU) assays (Invitrogen) was used to label cells in S-phase. 2nd and early 3rd Instar larvae with the genotype insc-Gal4; UAS-mCherry (to globally label axon tracts and neuroblasts) at 72h ALH were fed media containing 16 μg/ml bromophenol blue and 130 μM EdU for four hours. Larvae with EdU incorporated were dissected either immediately (Fig. 5M-N) or chased to wandering third instar larvae stage (Fig. 5O-P) and fixed in 3.7% formaldehyde in PBS (pH = 7.4) and permeabilized with 0.3% PBT. Samples were incubated in an optimized EdU reaction (provided by the manufacturer) containing: 425 μl Click-iT Reaction Buffer, 20 μl CuSO4, 1.2 μl AlexaFluor 647 Azide, 1 μl anti-GFP (Fig. 5O-P; polyclonal antibody conjugated to AlexaFluor 488; Molecular Probes, Cat No #A-21311), 50 μl 10x Click-iT Buffer Additive for 90 min and whole-mounted in Vectashield.
Figure 5.
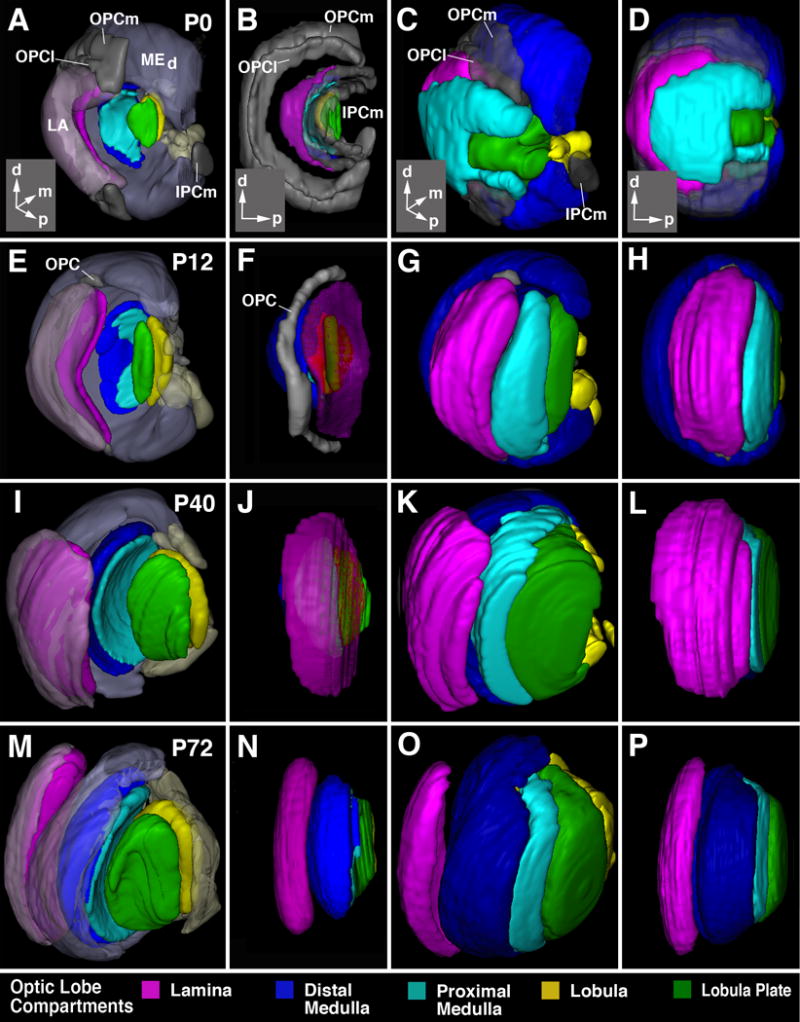
Morphogenesis of the optic lobe. All panels show digital 3D models at the pupariation (P0; first row, A-D), 12h after puparium formation (P12; second row, E-H), 40h after puparium formation (P40; thrird row, I-L), and 72h after puparium formation (P72; bottom row, M-P). Panels of first column (A, E, I, M) and third column (B, F, J, N) present view from dorso-lateral posterior; second column (B, F, J, N) and fourth column (D, H, L, P) show lateral view. Insets at lower right corner of panels of first row (A-D) delineate orientation of panels (a anterior; d dorsal; l lateral; p posterior). In panels of first and second column, developing neuropils are shown in saturated colors, using the color code indicated at bottom of figure. In these two columns, the epithelial part of the outer and inner proliferation centers (OPC, IPC) is in gray. In panels of first column, cortices of the lobula plate/proximal medulla are omitted to permit view of the neuropils. The cortex of the distal medulla (MEd) and lamina (LA) is rendered in semi-transparent, unsaturated colors. In second column, all cortices are omitted. In third and fourth column, cortices are shown in saturated colours.
Confocal Microscopy
Staged Drosophila larval and pupal brains labeled with suitable markers were viewed as whole-mounts by confocal microscopy [LSM 700 Imager M2 using Zen 2009 (Carl Zeiss Inc.); lenses: 40× oil (numerical aperture 1.3)]. Complete series of optical sections were taken at 2-μm intervals. Captured images were processed by ImageJ (=Fiji; National Institutes of Health, http://rsbweb.nih.gov/ij/) and Adobe Photoshop. Measurement of distances were carried out with the “Analyze>Measure” tool of Image J.
Generation of three-dimensional models
Digitized images of confocal sections were imported using Trak-EM2 plug-in in FIJI software (Cardona et al., 2012; Schindelin et al., 2012). Since sections were taken from focal planes of one and the same preparation, there was no need for alignment of different sections. The optic lobe compartments (neuropils and cortices) were manually segmented using a global marker for neural tracts (BP106 from P0 to P32; BP104 from P32 onwards) and the synaptic marker, DN-Cadherin (DN-Cad) within a series of confocal images. All surface rendered digital atlas models were generated using 3-dimensional viewer as part of the FIJI software package.
Animations
Fiji Trak-EM2 plug-in was used to film the optic lobe compartments in 3D and rotating at 360°. They were saved in AVI format (audio video interleave) at no compression and 15fps (frames per second). After being saved, we used Movavi Video Suit (Copyright © 2017, Movavi), to edit the videos and add text. Once finished we saved in AVI format at 29fps. A video converter was used to convert the video to MPEG (moving pictures experts group). The videos are between 24 seconds and 1 minute 12 seconds.
RESULTS
Architecture of the optic lobe primordia in the late larva
The outer and inner optic anlagen of the early larva start out as small, crescent-shaped epithelial layers (OOAep, IOAep; Fig. 2A, B). The posterior, concave side of both optic anlagen is defined by the larval optic neuropil which consists of the afferent larval photoreceptors (Bolwig’s nerve) and their target neurons (Sprecher et al., 2011; Fig. 2C, D) (Animated model in supplementary file S3). As larval development progresses the OOA and IOA grow tangentially by symmetric cell division (described in Egger et al., 2007; Ngo et al., 2010; Yasugi et al., 2008). The curvature increases, such that the dorsal and ventral tips of the OOA and IOA come in close contact (Fig. 2F, G). When referring to the topography of the optic anlagen, we distinguish a medio-lateral axis, which corresponds roughly to the medio-lateral body axis and a dorso-ventral axis, which is curved, so that both the dorsal and ventral tips come to lie posteriorly (Fig. 2B, E). Around 48h ALH, the OOA and IOA start to convert into asymmetrically dividing neuroblasts (nb; Fig. 2H-J; animated models in supplementary file S4 and S5) which give rise to the distinct primordia of the optic ganglia. At this stage, the optic anlagen are more often termed outer proliferation center (OPC) and inner (IPC) proliferation centers of the optic lobe in the recent literature (Fig. 2K-M; Apitz and Salecker, 2015; Bertet et al., 2014; Li et al., 2013b; Brand and Livesey, 2011). We will adopt this terminology when referring to the late larva and pupa and will reserve “optic anlagen” for the neuroepithelia of the early stages.
Figure 2.
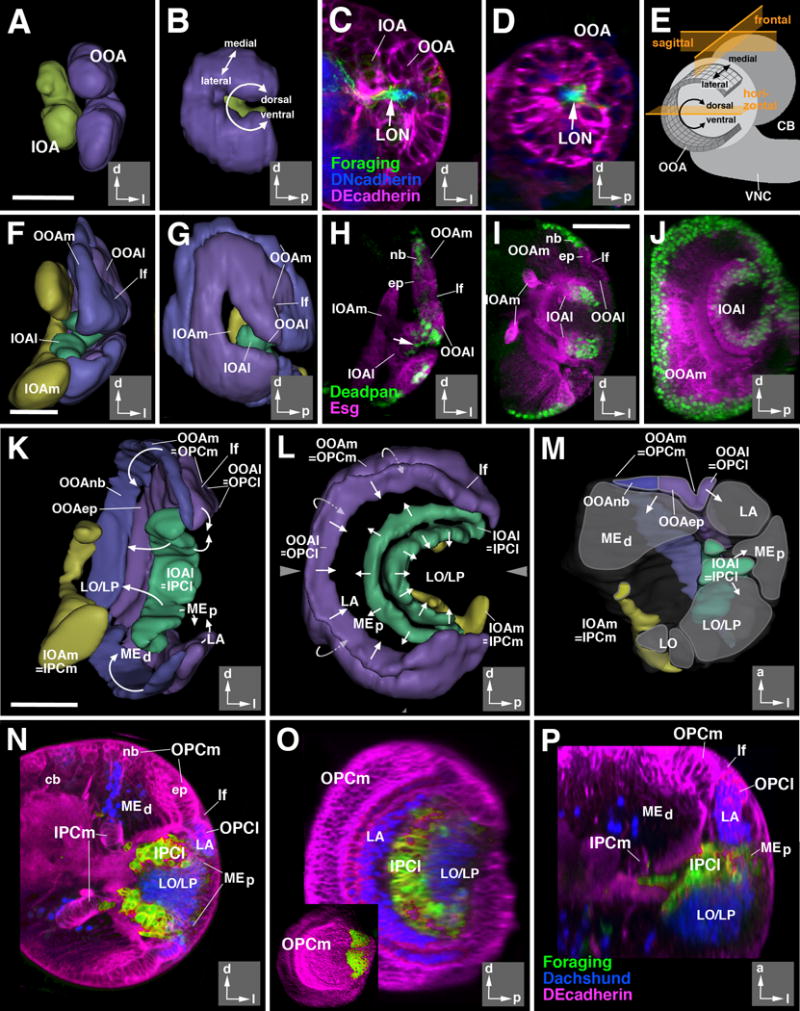
Architecture of the larval optic lobe primordium. 3D digital models and confocal sections of the developing larval optic lobe at the 2nd instar (32hr; A-D), early 3rd instar (72hr; F-H), and late 3rd instar (120hr; I-P) larval stages. Models present two different views for all three stages: posterior view (A, F, K), and lateral views (B, G, L). Insets at lower right corner of panels delineate orientation (a anterior; d dorsal; l lateral; p posterior). The outer optic anlage (OOA; for late stages called outer proliferation center, OPC) is shown in purple (epithelial; lateral part; OOAep) and blue (neuroblasts; medial part; OOAnb), respectively. The inner optic anlage (IOA; later stages: IPC) is shown in yellow (epithelial medial part; IPCm) and green (mesenchymal lateral part; IPCl). Panel (M) shows a cutaway model of the optic lobe of a late larva. In this model, the optic lobe is digitally cut along a horizontal plane [indicated by arrowheads in adjacent panel (L)], and a dorsal view of the cut surface of the bottom part is presented. Areas shaded in gray correspond to the primordia of the optic ganglia (LA lamina; LO lobula; LP lobula plate; MEd distal medulla; MEp proximal medulla) generated by the optic proliferation centers. Arrows in (K-M) indicate the direction in which optic proliferation centers convey postmitotic neurons. Drawing in panel (E) presents a schematic lateral view of larval brain (CB central brain; VNC ventral nerve cord) which illustrates the shape and location of the epithelial outer optic anlage. Note the curvature of the dorso-ventral axis which places the dorsal and ventral edge of the OOA close to each other. The three planes of sectioning used in this and the following figures (sagittal or parasagittal, frontal, horizontal) are indicated in orange. (C-D, H-J, N-P): z-projections of confocal sections (approximately 15-20 μm thickness) of the optic lobe; (C, F, H, I, N) are frontal sections, (D, J, O) are parasagittal sections, and (P) is a horizontal section. The epithelium of the optic anlagen is either marked by DE-Cadherin (in red; C-D, N-P) or mCD8-GFP driven by esg-Gal4 (in red; H-J). Neuroblasts are marked by Deadpan (in green, H-J). Larval optic neuropil (LON) is marked by foraging (forNP0079-Gal4>UAS-mCD8-GFP; green in C-D), and the central brain (CB) neuropil is marked by Discs-large (in blue, C-D). In panels (N-P) Dachshund (blue) labels neural precursors of the lamina (LA; Pineiro et al., 2014) and the T4/T5 neurons of the lobula/lobula plate (LO/LP). forNP0079-Gal4 >UAS-mCD8-GFP; green) exclusively labels the lateral domain of the inner proliferation center (IPCl). Inset in (O) shows expression of a wingless reporter (wg-Gal4>UAS-mCD8-GFP) in the dorsal and ventral tips of the outer proliferation center (OPC).
Abbreviations used in this and all following figures: cb central brain; cl medulla column; cor cortex; ep epithelium; IOA inner optic anlage; IOAl lateral part of inner optic anlage; IOAm medial part of inner optic anlage; IPCl lateral domain of inner proliferation center; IPCm medial domain of inner proliferation center; L columnar lamina neurons; LA lamina; lf lamina furrow; LO lobula; LON larval optic neuropil; LP lobula plate; Lt lobula tangential neurons; mc medulla cortex; ME medulla; MEd distal medulla; MEp proximal medulla; Mt medulla tangential neurons; nb neuroblast; np neuropil; OOA outer optic anlage; OOAep epithelial domain of the OOA; OOAl lateral part of outer optic anlage; OOAm medial part of outer optic anlage; OOAnb neuroblast domain of the OOA; OOC outer optic chiasm; OPCl lateral domain of outer proliferation center; OPCm medial domain of outer proliferation center; T2-5 classes of columnar T-shaped neurons interconnecting lobula complex and medulla; Tlp columnar trans lobula plate neurons; Tm columnar transmedulla neurons; Tmy columnar transmedulla y-shaped neurons; TMP transient medulla plexus; VLP ventrolateral protocerebrum; VNC ventral nerve cord.
Bars: 20 μm (A-D; F-H); 50μm (I-J; K-P)
Molecular markers expressed in the optic proliferation centers and at later stages allow one to identify precursors of different cell types at the larval stage and to follow the fates of the different primordia throughout metamorphosis. To globally label neuronal cell bodies and nerve fibers, we used antibodies against Neurotactin (BP106) or Neuroglian (BP104) (Hortsch et al., 1990a, 1990b). Specific markers for subpopulations of optic lobe neurons expressed from early to late stages of development included Ln-Gal4 (L3/L4 neurons of the lamina; Zhu et al., 2009), Dac (all lamina neurons, columnar neurons of the lobula and lobula plate T4/T5; Mardon et al., 1994), bsh-Gal4 (medulla intrinsic neuron Mi1; Hasegawa et al., 2011), Vsx1-Gal4 (columnar neurons connecting distal medulla with proximal medulla, lobula and lobula plate, Tm, TmY; Erclik et al., 2008), Acj6-Gal4 and forNP0079-Gal4 (columnar neurons connecting the lobula, lobula plate and proximal medulla, Tlp, T2-T5; Potter et al., 2010b) and wg-Gal4 (tangential neurons of the medulla and lobula, among others; Bertet et al., 2014; Fig. S1).
At the late larval stage, the optic proliferation centers have produced four main masses of immature neurons (“optic lobe primordia”) laid out in a way depicted in Fig. 2K-P (animated model in supplementary files S5, S7). The OPC is divided by the lamina furrow into a small lateral domain (OPCl) and large medial domain (OPCm) which, by that stage, consists mostly of asymmetrically dividing neuroblasts (Fig. 2N). The OPCl generates the primordium of the lamina; the OPCm gives rise to the large primordium of the medulla (Fig. 2M, P), containing all of the different types of columnar medulla neurons (Mi, Tm, TmY, Dm, Pm; see Fig.1). The OPCm/l (visible as epithelial, Crb-positive structures) persist into the early pupa (P12); by P18 Crb-labeling has become very weak (arrow in Fig.3I), and has disappeared by P24 (not shown). The IPC of the late larva also splits up into a large lateral component (IPCl), consisting of neuroblast-like (non-epithelial) progenitors and a medial, epithelial component (IPCm; Fig. 2K-P). As shown in a recent study, there continue to be cells delaminating from the IPCm and migrating towards the IPCl (Apitz and Salecker, 2015). The IPCl gives off neuronal precursors in two directions, producing two separate primordia (Fig. 2L; supplementary file S5): (1) cells given off posteriorly into the concavity of the crescent-shaped IPCl become the columnar elements of the lobula and lobula plate (T4/T5 and, possibly, Tlp; Fig.1) that form the lobula plate cortex; (2) cells moving antero-laterally will become the T-shaped neurons (T2/T2a/T3) connecting medulla and lobula complex (Fischbach and Dittrich, 1989; Fig.1) and C2/C3 neurons that connect medulla and lamina (Fischbach and Dittrich; not shown). We will call this part of the optic lobe cortex, which during pupal development is clearly set apart from other domains, the “posterior medulla cortex”, thereby distinguishing it from the “distal medulla cortex”, formed by the OPC.
Figure 3.
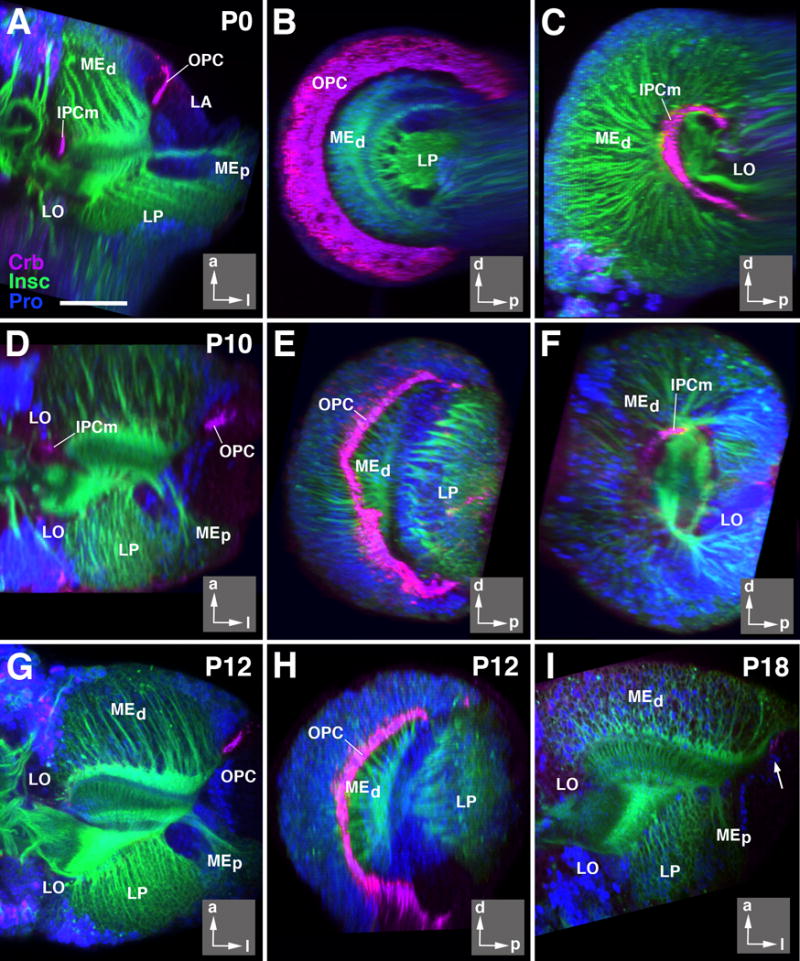
Neuroepithelial optic anlagen disappear during early metamorphosis. All panels show z-projections of confocal sections of optic lobe labeled with anti-Crumbs (Crb; magenta; apical surface of neuroepithelial cells), insc-Gal4 > UAS-mcd8GFP (green; neuronal cell bodies in cortex and axonal tracts) and nuclear marker (blue). Panels of left column (A, D, G) and panel (I) show horizontal sections; middle column (B, E, H) and (C, F) represent parasagittal sections (B, E, H: level of outer proliferation center; C, F: inner proliferation center).Insets at lower right corner of panels delineate orientation of panels (a anterior; d dorsal; l lateral; p posterior). Panels of upper row (A-C) depict white prepupa (P0); second row (D-F) 10h after puparium formation (P10); third row (G, H) 12h after puparium formation (P12), and 18h after puparium formation (I; P18). Note that Crb-positive epithelial cells are no longer visible for inner proliferation center after P10; for OPC, Crb-labeling has all but disappeared by P18 (arrow in I). For abbreviations, see legend of Figure 2. Bar: 50 μm
The IPCm, similar to the OPC described above, dissociates over the course of early metamorphosis. By P12, no Crb-positive cells remain (Fig.3G). Surrounding the larval and early pupal IPCm are several clusters of neuronal precursors which coalesce into a coherent lobula cortex, located medially of the lobula plate cortex (see below; Fig.5C, G, K, O). The development of these dispersed clusters of cells into different classes of lobula neurons can only be followed with specific markers and will not be considered here.
The dorsal and ventral domains of both OPC and IPC differ in their developmental potential from the central domain. Defined by the expression of the morphogen Wingless (Wg) (Bertet et al., 2014; shown in green inset in Fig. 2O), they give rise to many types of glial cells, and therefore were called “glial proliferation zone” (GPZ; Edwards and Meinertzhagen, 2010; Chotard and Salecker, 2007). In addition, the dorsal and ventral tips of the OPC produce several neuronal cell types, among them most, if not all, tangential elements of the medulla (Bertet et al., 2014).
Morphogenesis of the optic lobe during early metamorphosis
As a starting point for the following descriptions, Figure 4A-C schematically depicts the way in which neurons and their processes are arranged to compose the adult optic lobe. Neuronal cell bodies of columnar neurons (the large majority of cells of the optic lobe) form a layer (cortex or rind), several cell diameters deep (Fig. 4A). For the medulla and lamina, this layer is oriented in a parasagittal plane and is defined by a horizontal (antero-posterior) axis, and a vertical (dorso-ventral) axis. The lobula and lobula plate are oriented perpendicularly to the medulla (Fig. 4A-C); their horizontal axis points medio-laterally (Fig. 4A). Neuronal processes assemble in a second layer, the neuropil, which co-extends alongside the corresponding cortex (Fig. 4A). The large majority of neurons of the optic lobe are columnar neurons, which give off one main fiber directed perpendicularly to the plane of the cortex, thereby passing through the neuropil and (in case of projection neurons interconnecting different neuropils) exiting the neuropil at the side opposite to the cortex (Fig. 4A, blue neuron). The axis defined by the direction of these columnar axons will be referred to as “z-axis” (Fig. 4A). The main fiber of each columnar neuron forms branches at defined locations along the z-axis. The branch location of certain types of neurons were used to define discrete layers within the medulla and lobula/lobula plate (Fig. 4A, hatched lines; Fischbach and Dittrich, 1989).
Figure 4.
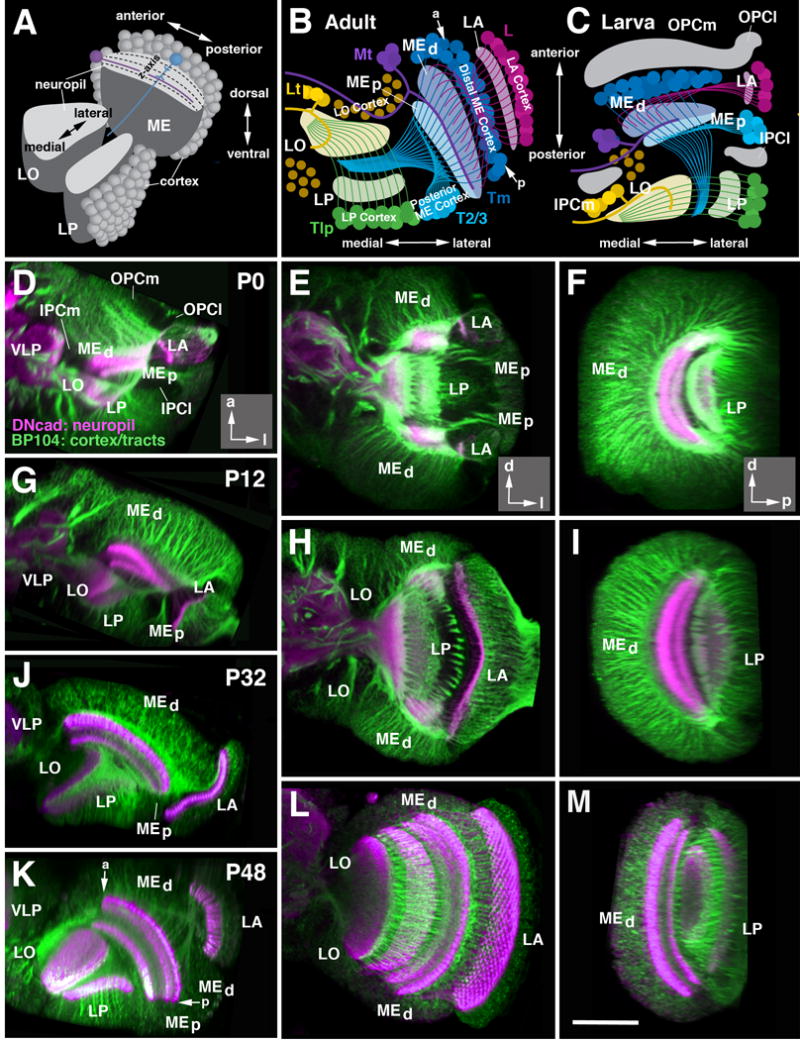
Morphogenesis of the optic lobe during metamorphosis. (A) Schematic cutaway model of the adult optic lobe, delineating orientation of medulla (ME), lobula (LO) and lobula plate (LO) relative to the body axis. For each of the optic ganglia, the cellular cortex and underlying neuropil is shown. Neuropils are subdivided into layers along the z-axis, as shown for the medulla (hatched lines). The large majority of neurons in all optic ganglia are columnar neurons (represented by the medulla neuron shown in blue) whose main fiber penetrates the neuropil along the z axis. Tangential neurons (purple) project parallel to horizontal plane of the neuropil. (B-C) are schematic horizontal sections of the optic lobe of the adult (B) and late larva (C). The location of optic lobe neuropils and orientation of major fiber systems is indicated by shaded areas and lines; the color code introduced here is consistently employed throughout the remainder of the figures (magenta: lamina; blue: distal medulla; cyan: proximal medulla; green: lobula plate; yellow: lobula). In (C), location of the optic anlagen (gray) is indicated. Major systems of columnar neurons include lamina neurons (L), transmedullary neurons (Tm), neurons interconnecting medulla and lobula/lobula plate (T2/T3), and lobula plate neurons (Tlp). Also shown are tangential neurons of medulla (Mt) and lobula (Lt). Note that primordia of optic ganglia and fiber systems are already formed in late larva, but undergo a rotation to reach their adult configuration. “a” and “p” in panel (B) indicate anterior and posterior edge of medulla, respectively. For further detail, see text.
(D-M) Z-projections of confocal stacks (15-20 μm thickness) of optic lobe labeled with anti-Neuroglian (BP104; green; neuronal cell bodies in cortex and axonal tracts) and anti-DNcadherin (DNcad; magenta; neuropil). Panels of left column (D, G, J, K) show horizontal sections; middle column (E, H, L) represents frontal sections; right column (F, I, M) parasagittal sections. Insets at lower right corner of panels of row (D, E, F) delineate orientation of panels (a anterior; d dorsal; l lateral; p posterior). Panels of upper row (D-F) depict white prepupa (P0); second row (G-I) 12h after puparium formation (P12); third row (K-M) 48h after puparium formation (P48). A horizontal section for P32 is shown in panel (J). For abbreviations, see legend of Figure 2. Bar: 50 μm (D-M)
Unlike most columnar neurons, tangential neurons (Fig. 4A, purple neuron) are confined to the edges of a compartment, and extend neurites oriented parallel to the plane of the corresponding neuropil (Fig. 4A). For example, the tangential neurons of the medulla (Mt) flank the anterior edge of this compartment and extend fibers (“Cucatti’s bundle) that penetrate the medulla neuropil anteriorly (Fig. 4B). Among columnar neurons, only few classes, notably the T2/T3 neurons (light blue neurons in Fig. 4B), are arranged in a way similar to tangential neurons. T2/T3 cell bodies fill a bar-shaped volume located along the junction between posterior medulla cortex and lateral lobula plate cortex (Fig. 4B).
In summary, we can define in the adult optic lobe an outer part (lamina and medulla) and an inner part (lobula and lobula plate). In the outer part, cortex and neuropil form alternating layers aligned roughly along the antero-posterior axis, with the lamina cortex being located most laterally, followed by the lamina neuropil, then the distal medulla cortex, and finally the medulla neuropil (Fig.4B). In the inner optic lobe, the neuropils of the lobula and lobula plate, aligned along the medio-lateral axis, are right next to each other. Most columnar neurons constituting these neuropils form a cortex located posterior of the lobula plate. In this posterior cortex, T2/T3 neurons, whose processes connect the LO/LP with the proximal medulla and make up for much of the volume of the proximal medulla, occupy a lateral territory; we will call this the “posterior medulla cortex” in the following (Fig.4B; note that in the supplementary animations, this cortex is annotated as “proximal medulla”). T4/T5 and Tlp are located medially (lobula plate cortex; Fig.4B). Tangential neurons of the medulla and lobula, as well as the populations of columnar projection neurons connecting the lobula/lobula plate to the central brain, form a cortex located anterior of the lobula (lobula cortex; Fig.4B).
The above described architecture of the optic lobe compartments can be already recognized in the late larva (Fig. 4C), and can be best appreciated if looking at a horizontal section of the optic lobe (Fig. 4D, G, J and K) or animated digital 3D models (supplementary file S6). Neuronal precursors form cortices flanked by underlying neuropil primordia. However, the orientation of the compartments, with the exception of the lamina, differs significantly from the adult configuration. The horizontal axis of the medulla is directed medio-laterally in the larva (Fig. 4C), and then rotates to antero-posteriorly in the adult (Fig. 4B). For the lobula/lobula complex, the larval postero-anterior axis reflects the adult medio-lateral axis (compare Fig. 4C to Fig. 4B). As stated earlier (see Fig. 2A-E), the vertical, dorso-ventral axis of all compartments is bent in the larva, so that both dorsal and ventral tip point posteriorly. During metamorphosis, the optic lobe compartments undergo extensive proliferation, fiber growth, and morphogenetic movements. These events are illustrated in Figs.4-7 and animated digital 3D models (supplementary files S6, S8-11). They will be discussed in the order: (1) reorientation of compartment axes; (2) directed growth; (3) formation of fiber systems (outer and inner optic chiasm); (4) rearrangement of neuronal cell bodies.
Figure 7.
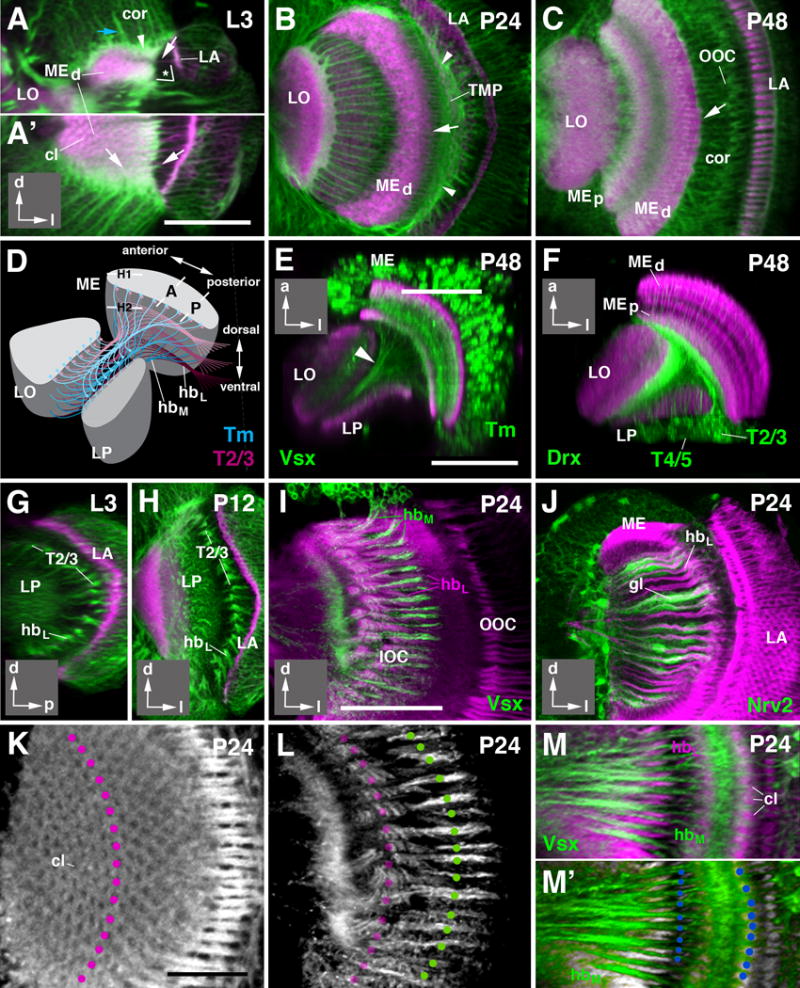
Development of fiber systems forming inner and outer chiasms. (A-C) Confocal sections of the optic lobe at third instar larva (L3, A-A′), 24h after puparium formation (P24; B) and 48h after puparium formation (P48; C). (A) represents a horizontal section; (A′, B, C) are frontal sections. Insets at lower right corner of these and following panels delineate orientation of panels (a anterior; d dorsal; l lateral; p posterior). Neuronal cell bodies and fibers (green) are labeled by anti-Neurotactin (A-B) or anti-Neuroglian (C); anti-DNcadherin (magenta) labels neuropil and some fiber systems, including axons of lamina neurons. At the larval stage up to P24, the thin neuropils of the medulla (MEd) and lamina (LA) are oriented roughly perpendicularly to each other (* in A). Axons of lamina neurons, as well as of R7/8 photoreceptors (white arrows in A and A′) penetrate the lamina neuropil and then pass tangentially over the surface of distal medulla. Terminals of retina/lamina fibers form regularly spaced, DNcadherin-positive clusters that foreshadow the medulla columns (cl in A′). Neurons forming the medulla cortex (cor in A) are located medially of the lamina. Axons of medulla neurons (blue arrow in A) intersect the plate of lamina axons at a right angle before entering the medulla neuropil. Outgrowing processes of several subpopulations of immature medulla neurons form a transient fibrous plexus located distal (outside) of the lamina axons (arrowhead in A, B). By P48, the lamina has shifted anteriorly, such that the planes of the lamina neuropil and medulla neuropil are roughly parallel to each other (see also Fig.5E, F). The masses of axons connecting lamina and medulla neuropil (arrow in C) are dragged into the medulla cortex (cor). The crossing of these axons, representing the outer optic chiasm (OOC), falls into the center of the medulla cortex. (D) Schematic illustrating the main fiber systems of the inner optic chiasm (IOC). Masses of axons formed by the transmedullary neurons (Tm and TmY; shown in blue) are split into horizontal slices (hbM), whereby each slice contains axons from one horizontal row of medulla columns. Similarly, axons of T neurons connecting medulla columns with lobula and lobula plate (only T2/3 shown, in magenta) form equal number of slices (hbL). Slices of transmedullary axons and T axons alternate, giving the inner optic chiasm, as seen in frontal sections (see panels G-M below) its characteristic striated appearance. (E-F) Horizontal sections of optic lobe 48h after puparium formation (P48), showing medulla Tm neurons (E; green, labeled by Vsx1-Gal4) and lobula plate T neurons (F; green, labeled by Drx-Gal4) fiber systems. Neuropil is labeled by anti-DNcadherin (magenta). (G-J) Frontal confocal sections of optic lobe of larva and early pupa, illustrating spatial arrangement of fiber systems of inner optic chiasm during early metamorphosis. In (G,H), plane of section is slightly posterior of the inner optic chiasm, as indicated by “P” in panel (D). Axon bundles of T2/3 neurons (hbL; green, labeled by anti-Neurotactin) are seen as they enter the chiasm from posteriorly. (I, J) Confocal sections representing plane “A” as shown in panel (D). General neuronal labeling by anti-Neuroglian (magenta). In (I), bundles of Tm neurons (hbM), labeled by Vsx1-Gal4 (green) alternate with bundles of T neurons (hbL). In (J), glia of inner optic chiasm (labeled by Nrv2-Gal4; green) is seen to surround individual hbL bundles. (K-M′) One-to-one numerical relationship between the fiber bundles formed by transmedullary neurons and T neurons. General neuronal labeling by anti-Neuroglian (white in (K, L, M′; magenta in M). (K) shows tangential section of medulla at level indicated by “H1” in panel (D). Note regular hexagonal arrangement of medulla columns (cl). One vertical row of columns is indicated by magenta circles. (L) presents tangential section of the same preparation at a deeper level, proximal to the medulla neuropil, indicated by “H2” in panel (D). Faint magenta circles indicate where the vertical row of columns visible in (K) project onto the plane shown in (L). Green circles are placed on all hbL bundles. Note identical number and spacing of columns and hbL bundles. (M, M′) represent a section at the plane indicated by “A” in panel (D). The medulla neuropil is cross-sectioned, and medulla columns (cl) are clearly visible. Vsx-Gal4 labels hbL bundles formed by Tm neurons (green). Note equal number and spacing of these bundles and columns, as indicated by blue circles in (M′). For abbreviations see legend of Figure 2. Bars: 50 μm (A-C; E, F; G-J); 25 μm (K-M′)
Changes in the axes of optic lobe compartments
The first change to occur in the orientation of optic lobe primordia is the straightening of the dorso-ventral axis (“optic lobe straightening”), which happens during the first 24 hours of metamorphosis, and can be best appreciated when looking at parasagittal sections (Fig. 4F, I, M) of the brain or lateral views of the 3D digital models (Fig. 5D, H, L, P; supplementary file S11). At 12h after pupariation (P12), the shape of the primordia of the lamina and medulla has changed from its original C-configuration to that of slightly curved “banana”. This change equally affects the shape of the cell body layers (Fig. 5B, F) and the underlying neuropils (Fig.4F, I and Fig.5B, F). The case of the inner ganglia, lobula and lobula plate is slightly different. At the larval stage, the IPC (gray; inner C-shaped structure, Fig. 5B) follows the same curvature as the OPC (gray; outer C-shaped structure, Fig. 5B). As a result, the clusters of immature neurons budded off from the IPC towards the convex side and concave side also fill a curved volume (green and yellow, Fig. 5C-D) which then straightens out during early metamorphosis (green and yellow, Fig. 5G-H). However, the primordia of the neuropils of the lobula plate and lobula (green and yellow, respectively; Fig. 5A-B) exhibit a relatively straight dorso-ventral (vertical) axis from the larval stage when they first appear (Figs.4F; 5A-B; supplementary file S6, S11).
The second large-scale movement that takes place during the first half of metamorphosis is a global rotation (“optic lobe rotation”) of the optic lobe around a vertical axis. This movement can be best visualized in horizontal sections (dorsal view) of the optic lobe (Fig. 4D, G, J, K), but is also apparent when looking at dorso-lateral views or lateral views of the corresponding digital 3D models (Fig.5C, D, G, H). The medulla cortex and neuropil is oriented medio-laterally in the larva and early pupa (Fig.4D, G). It then rotates by almost 90°, and from about 40 APF onward, is aligned roughly with the antero-posterior axis (Fig.4K). Due to the curvature of the medulla (and the overlying lamina and retina), the anterior edge of the medulla remains located more medially than the posterior edge (arrows “a” and “p” in Fig. 4B, K). The medulla cortex, located anteriorly of the medulla neuropil (if discounting for the curvature of the dorso-ventral axis) in the larva and early pupa (Fig. 4C-D, G), shifts to a position antero-lateral of the neuropil in the pupa and adult (Fig. 4J-K). The lobula (LO)/lobula plate (LP) performs a similar rotation as the medulla. In the larva, the horizontal axis of the LO/LP neuropils has a posterior-to-anterior orientation (yellow and green, Fig. 4C; LO and LP in Fig. 4D). The IPCl-derived immature neurons (green neurons; Fig. 4C) forming the LP cortex are located laterally of their neuropils (Fig.4C, D). Towards mid-pupal stages, the orientation of the LP rotates clockwise. As a result of this rotation, the cortex of cell bodies of the LP comes to lie posteriorly of the neuropils (Fig. 4B, K). Concomitantly, cell bodies of neurons produced by the IPCm (yellow in Fig. 4B-C) and initially located medially of the lobula neuropil (Fig. 4C), shift anteriorly to become the anterior cortex of the lobula (Fig.4B, K).
The third movement shaping the optic lobe affects the lamina, which pivots anteriorly around the vertical axis (“lamina rotation”). This movement occurs at a later stage, following the global optic lobe rotation described above. In the larva, immature lamina (LA) neurons are budded off posteriorly, away from the OPCl (Huang and Kunes, 1996; Fig. 4C-D). As a result, the lamina is “attached” to the lateral edge of the medulla, and the plane of the lamina stands orthogonally to that of the medulla. This relationship between lamina and medulla remains constant until approximately 32h APF (Fig. 4J). Subsequently, between 32h and 48h APF), the lamina rotates and “floats” anteriorly over the medulla cortex, so that the plane of the lamina now lies parallel to that of the medulla (Langen et al., 2015; Fig.4B, K; see also supplementary file S11, and snapshots of models in Fig.5). As a result of the lamina rotation, the outer optic chiasm adopts its characteristic X-shape (see section below).
Directed growth of the optic lobe compartments
During the first third of pupal development, all optic lobe compartments grow in size along the horizontal axis by a factor of approximately 4 (lamina), and 2-3 (medulla, lobula/lobula plate; Fig. 4). There is little or no growth along the vertical axis. To understand the growth of the optic lobe, discussed in more detail below, it is helpful to briefly recapitulate the development of the eye (Kumar et al., 2010; Ready et al., 1976), whose photoreceptors innervate the optic lobe. The eye and optic lobe compartments are divided into modular units, the ommatidia (retina), cartridges (lamina), and columns (medulla, lobula/lobula plate). Units form vertical and horizontal rows. Photoreceptors R1-6 of the posterior-most vertical row of ommatidia project to the posterior-most vertical row of lamina cartridges; these cartridges, as well as R7/R8 of the posterior ommatidia, are connected to the most anterior vertical row of medulla columns (Meinertzhagen and Hanson, 1993). In turn, the most anterior vertical row of medulla columns is connected to the most medial vertical row of columns in the lobula complex (Fig. 4B). These crossed connections create the outer and inner optic chiasm (Fig. 4B; see below). Along the dorso-ventral axis, connections are not crossed (Fig. 4E-F, H-I, L-M): dorsal ommatidia project to dorsal lamina cartridges and dorsal medulla columns, which in turn project to dorsal lobula columns; ventral units in the eye project to ventral units of the optic lobe.
The ordered connectivity between eye and optic lobe compartments is reflected during development by the temporal sequence in which the units of the visual system are born and differentiate. In the eye imaginal disc, ommatidia located in a given vertical row differentiate at the same time. The vertical row formed by the first born ommatidia comes to lie at the posterior edge of the eye; more anterior rows are gradually added, at a pace of one row every 90 min (Meinertzhagen and Hanson, 1993; Selleck and Steller, 1991). In other words, the eye primordium grows and differentiates along the horizontal axis from posterior to anterior. The growth of optic lobe compartments along the horizontal axis also shows a temporal order, as depicted schematically in Fig. 6A-C. This was demonstrated in numerous studies for the lamina, where, during the larval stage, the first born (presumptive) posterior vertical row of cartridges differentiates concomitantly with the incoming axons of the posterior row of photoreceptor axons (Meinertzhagen and Hanson, 1993; Huang and Kunes, 1996; Selleck et al., 1992; Selleck and Steller, 1991). More and more anterior cartridges are added until the final size of the lamina is reached around 40h APF (Fig. 6D-F). A similar gradient in differentiation is detectable in the deeper compartments of the optic lobe when using markers for specific cell types. Labeling of a late larval or early pupal optic lobe with antibody against FasII or Ln-Gal4 reporter shows L3/4 neurons located in the posterior lamina, projecting towards the medial medulla (Fig. 6D-E). The size and staining intensity of axonal terminal in the medulla clearly follows a medio-lateral gradient, with the most medial ones (produced by the first born L3/4 neurons) being the largest and most intensely labeled (Fig. 6E). A similar gradient is visible when labeling medullary Tm/TmY neurons around 24hrs APF using Vsx1-Gal4 (Fig. 6H). Medially located neurons are strongly labeled and send long axons with terminal arbors towards the posterior part of the lobula. Vsx expression of neurons and axons becomes increasingly fainter when moving laterally in the medulla, or anteriorly in the lobula. Also neurons derived from the IPC, labeled by DrxR77F09-Gal4 (Fig. 6G) or acj6-Gal4 (Fig. 6I) exhibit this gradient. The expression of Acj6 in neurons generated by the IPC in the late larva already reveals a clear gradient (Fig. 6J-L). As described in a previous section, the IPCl is formed by a crescent-shaped array of neuroblasts which buds off immature neurons both posteriorly (i.e., into the cavity of the IPCl), as well as antero-laterally. The first expression of Acj6 occurs in the very center of the mass of immature neurons located in the IPCl cavity (Fig. 6J-K), furthest away from the IPCl neuroblasts. When adjusting for the curvature of the IPCl, this translates into the anterior-to-posterior gradient in neuronal differentiation described above (Fig. 6I, L).
Figure 6.
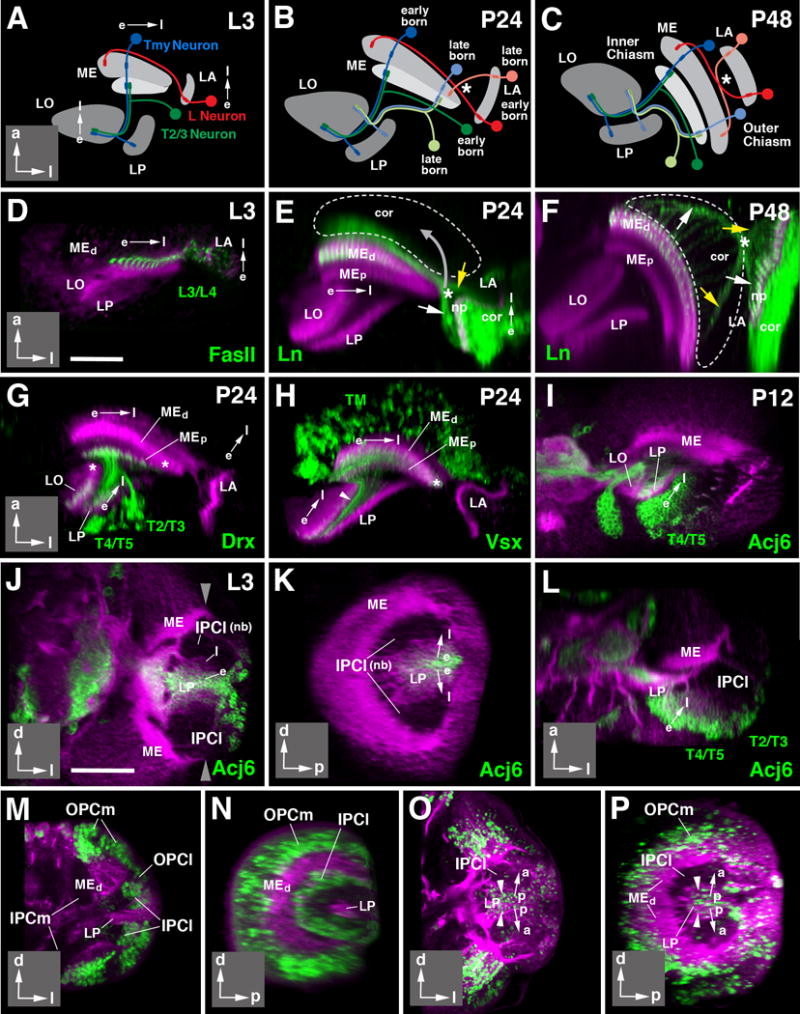
Directed growth and differentiation of the optic lobe. (A-C) Schematic horizontal sections of optic lobe of late larva (L3; A), 24h pupa (P24; B), and 48h pupa (P48; C). Neuropils are shaded in gray. Direction of growth in these and following panels is indicated by arrows (e early born; l late born). Insets at lower right corner of panels delineate orientation of panels (a anterior; d dorsal; l lateral; p posterior). Early born neurons are represented in saturated colors (lamina L neuron: red; medulla Tmy neuron: blue; medulla-lobula neuron (T2/T3): green); late born neurons of the same classes are shown in light colors in panels (B, C). (D-F): Directed differentiation of the lamina and distal medulla. Z-projections of horizontal confocal sections of the optic lobe at L3 (D), P24 (E), and P48 (F). Optic lobe neuropils in these and all following panels are labeled by anti-DNcadherin (magenta). Lamina projection neurons (L3/L4) are labeled by anti-Fasciclin II (FasII; green in D) or ln-Gal4>UAS-mCD8-GFP (green in E, F). Note that early born, more mature L3/4 axon terminals are thicker and show stronger GFP signal than later born immature terminals. Hatched lines in (E) and (F) outline boundaries of medulla cortex. White arrows in these panels point at early born L3/4 axons, yellow arrows at late born ones. Asterisk marks position where these axons cross. Note that position of crossing point moves as the lamina shifts forward and becomes oriented parallel to the medulla between P24 and P48 (movement indicated by gray arrow in E), resulting in formation of the outer optic chiasm (indicated by asterisk in F). (G-I) Directed differentiation of the proximal medulla and lobula/lobula plate. Panels show horizontal sections of optic lobes of 24h (P24) or 12h (P12) pupae. Discrete neuron populations are labeled by Drx-Gal4>UAS-mCD8-GFP (T2-T5 neurons, in G), Vsx-Gal4> UAS-mCD8-GFP (Tm neurons, in H), and acj6-Gal4> UAS-mCD8-GFP (T4/5 neurons in I). For all of these labeled neuron populations, terminal fibers are more pronounced in, or restricted to, the earlier born, more mature parts of the proximal medulla, lobula and lobula plate. Later formed, immature parts of these neuropils [asterisks in (G, H)] are devoid of fibers at P24. (I) and (J-L) show gradient in neuronal maturation in lobula plate cortex. T4/5 neurons, forming a major subpopulation of cells located in this cortex, show strong expression of the acj6 marker in posterior, more mature part of the cortex; expression decreases towards anteriorly. This gradient appears in the late larva, and reflects birth order of neurons, as shown in (J-L). The three panels represent frontal (J), sagittal (K) and horizontal (L) section of late larval (L3) optic lobe. Anti-DNcadherin (magenta) labels neuropil (strong label) and cell bodies/fibers (faint label). The C-shaped inner proliferation center [IPCl; unlabeled, consisting of dividing neuroblasts (nb)] buds off progeny towards the center. Earlier born cells are pushed centrally (away from the IPCl) by later born ones. Earlier born, more mature cells start expressing acj6; later born, less mature cells, closer to the IPCl (l) are acj6-negative. (M-P) demonstrate the directed growth in the IPCl using EdU incorporation. (M, N) represent frontal section (M) and sagittal section (N) of 76h larva pulsed with EdU over 4h prior to fixation. EdU label (green) is incorporated into the IPCl and the OPC. (O, P) present a frontal section (O) and sagittal section (P) of wandering third instar larva pulsed between 72and 76h, and chased to fixation at 100h. EdU-positive cells have vanished from the IPCl and are found in neural precursors, last divided between 72 and 76h, and now located in center of cell mass that becomes lobula plate cortex (LP). General label (magenta) of neurons (cortex, neuropils) by insc-Gal4; UAS-mCherry (M-N) or Jupiter::GFP (O-P). For other abbreviations, see legend of Fig.2. Bars: 25 μm (D-I); 50 μm (J-P)
The temporal gradient in differentiation exhibited by optic lobe neurons along the horizontal axis reflects a similar sequence in neuronal birthdates. This was shown in several previous studies for the OPC, which gives rise to the neurons of the lamina and medulla in a strict posterior-to-anterior and medial-to-lateral order, respectively (Piñeiro et al., 2014; Ngo et al., 2010; Yasugi et al., 2008; Egger et al., 2007; Meinertzhagen and Hanson, 1993; Hofbauer and Campos-Ortega, 1990). A similar order is observed in the IPCl (Fig. 6M-P). A pulse of EdU applied to a mid-third instar larva labels the crescent-shaped array of neuroblasts of the IPCl (Fig. 6M-N). When chasing the pulse to the late larval stage, labeling is seen in the center of the mass of immature neurons (Fig. 6O-P; see also Apitz and Salecker, 2015). This pattern indicates that each round of neurons born from the IPCl pushes the previously born neurons away from the IPCl, creating a peripheral-to-central (i.e., anterior-to-posterior) gradient in neuronal birth.
Development of the fiber systems of the optic lobe
The axons of optic lobe neurons form two major fiber systems, the outer and inner optic chiasm; which connect the lamina to the medulla, and medulla to the lobula/lobula plate, respectively. The outer chiasm is comprised of axons of L neurons and R7/8 photoreceptors, as well as a number of centripetal neurons projecting from the medulla to lamina (Strausfeld, 1976; Fischbach and Dittrich, 1989). These fibers form thin bundles extending parallel to the horizontal plane, each bundle comprising the elements connecting one cartridge of the lamina to one column of the medulla (Fig.7A, B). Fiber bundles are surrounded by processes of glia and form a thin layer directly adjacent to the distal surface of the medulla neuropil (Fig.7A-C).
The crossing of fibers of the lamina-to-medulla axon bundles in the horizontal plane can be seen as a direct result of the sequential timing of neuronal birth and differentiation, as described in the previous section, and shown in Fig. 6A-C (schematically) and Fig. 6D-F (labeling of L neurons). Axons of the first-born L neurons and R7/8 receptors extend all the way from their lateral origin in the lamina primordium (OPCl) to the medial edge of the OPC, where the first-born medulla neurons are located (Fig. 6A, D; Fig.6B, E, white arrow). Axons of later born neurons terminate further laterally in the medulla, thereby crossing their older siblings (Fig. 6B, E, yellow arrow). In the larva and early pupa, the characteristic shape of the chiasm (i.e., “cross”) is not yet visible, due to the fact that the lamina is oriented perpendicularly to the medulla. As the lamina rotates relative to the medulla, effectively aligning both neuropils along the antero-posterior axis (Fig. 6C, F) axons are pulled into the cross-shaped configuration characteristic for the outer optic chiasm (Fig.6C, F).
The spatial reorganization of lamina and medulla axon bundles during lamina rotation entails that axons are “dragged” through the mass of neurons forming the medulla cortex (Fig. 6E, F). Thus, prior to lamina rotation, the layer of bundles of L axons/photoreceptors enters the medulla from posteriorly (yellow arrow in Fig. 6E; see also Fig. 7B). After lamina rotation, this point has moved forward (yellow arrow in Fig. 6F), resulting in L fiber bundles radiating throughout the medulla cortex (Fig. 6F, 7C). This suggests that cell bodies of the medulla cortex do not tightly adhere to each other, but are able to let fibers glide past them over long distances.
The inner optic chiasm is more complex than the outer chiasm, since it is comprised of several different systems of fibers growing in different directions, as shown schematically in Fig. 7D. First there are the medullary systems, comprised of the axons of columnar Tm and TmY neurons that interconnect columns of the medulla with columns of the lobula/lobula plate in a retinotopic manner (blue in Fig. 7D; labeled by Vsx1-Gal4 in green in Fig. 7E). These fibers are directed orthogonally to the plane of the medulla. After exiting the medulla neuropil, fibers converge and enter into the cleft between lobula and lobula plate, extending parallel to the surface of these neuropils (Fig. 7D-E). Secondly, one observes the lobula/lobula plate systems (purple in Fig. 7D), including the different classes of T neurons with cell bodies located posterior to the lobula plate, which form bundles of fibers with a trajectory that is orthogonal to the medullary Tm/TmY system. There are two main subsystems, T2/T3 neurons interconnecting the medulla with the lobula, and T4 neurons connecting medulla and lobula plate (Fischbach and Dittrich, 1989; see Fig. 1). For clarity sake, only T2/T3 are shown in Fig.7D and will be considered in the following. T2/T3 axons enter into the cleft between medulla and lobula/lobula plate from posteriorly and extend forward, running parallel to the inner surface of the medulla (purple in Fig.7D; labeled by Drx-Gal4 in Fig. 7F). These fibers then make a sharp turn medially, entering the space between lobula and lobula plate.
Similar to what has been discussed for the outer chiasm above, the crossing of fibers of the Tm/TmY and T systems that generate the inner optic chiasm is also the result of the order in which these cells are born and differentiate (Fig. 6A-C). Early Tm/TmY neurons appear in the presumptive anterior medulla and project their axons to what will become the medial lobula complex (Fig. 6A). Likewise, early differentiating T neurons reach towards and innervate the presumptive anterior medulla, and the medial lobula complex (Fig. 6A). Later neurons innervate more posterior positions of the medulla; on their way towards more lateral domains of the lobula complex, they have to cross the earlier formed fibers, thus forming the inner optic chiasm (Fig. 6B).
A remarkable feature of the three-dimensional organization of the inner optic chiasm is the alternating arrangement of the two fiber systems originating in the medulla (Tm/TmY) and lobula complex (T2/T3/T4), respectively (Fig. 7D). When viewed in frontal sections, the axons of the T2/T3 system leave the posterior cell cortex as a series of approximately 30 regularly spaced bundles (“horizontal bundles”; “hbL” in Fig.7) arranged in a vertical row (Fig. 7G-I; section “P” indicated in Fig. 7D). Each bundle is surrounded by a layer of neuropil glia, called “outer chiasm giant glial cells” (Edwards and Meinertzhagen, 2010; Tix et al., 1994; “gl” in Fig. 7J). At more anterior levels (section “A” indicated in Fig. 7D), approaching the cleft between lobula and lobula plate, each horizontal bundle splays out into a fan-shaped array of fibers. The T2/3 bundles alternate with stacks of axons of medullary Tm/TmY neurons (“hbM” in Fig. 7D, I, M). Numerically, stacks of T2/3 axons and Tm/TmY axons correspond to the number of horizontal rows of medulla columns (Fig. 7K-M). This 1:1 relationship between medulla columns and fiber bundles of the inner chiasm can be directly seen in sections aligned with the cleft between lobula and lobula plate (section “A” in Fig. 7D; Fig. 7M, M′). It appears that each stack of Tm/TmY fibers is generated by the axons emanating from all medulla columns of one horizontal row. Similarly, a T2/3 stack contains all of the axons connecting one horizontal row of medulla columns with the corresponding row of lobula columns (data not shown).
Movement of neuronal cell bodies in the optic lobe cortices
The original position of immature neurons in the larval optic lobe primordium is determined by the position of its progenitor, as well as its date of birth (Piñeiro et al., 2014; Li et al., 2013a; Li et al., 2013b; Suzuki et al., 2013; Hasegawa et al., 2011). This relationship has been investigated in detail for the OPC, but, as shown above (see Fig. 6K, P), also applies for the neurons produced by the IPC. However, cell bodies of several classes of neurons change their position during the course of metamorphosis, as shown for several classes of medulla neurons, including Mi1 (labeled by the expression of bsh-Gal4; Fig. 8A-C), and a subset of Tm/TmY neurons expressing Vsx1-Gal4 (Erclik et al., 2017; Fig. 8C-F). Mi1 neurons are among the first born medullary neurons, and consecutively occupy a deep position in the primordium of the medulla cortex, furthest away from their neuroblasts of origin (Hasegawa et al., 2011; Fig. 8A). By 24h APF, this position has changed: Mi1 cell bodies have moved along the z-axis and are now located relatively superficially in the developing medulla cortex; they will maintain this position throughout later stages into the adult (white arrowhead in Fig. 8B; Hasegawa et al., 2011). In the case of neurons expressing Vsx, the movement occurs along the vertical plane. In the larva and early pupa, Vsx expression is confined to neurons located in a central domain of the medulla primordium (Erclik et al., 2008; 2017; Fig. 8D, E). By 48h APF, Vsx-positive neuronal cell bodies have spread out along the vertical plane to occupy all domains of the medulla cortex (Fig. 8F).
Figure 8.
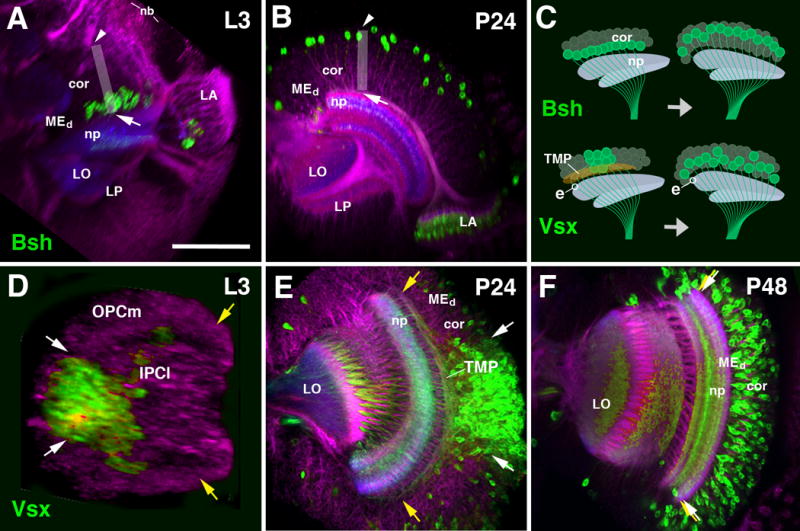
Cell body movements in the medulla cortex. (A, B) Horizontal confocal sections of optic lobe of late larva (L3, A) and early pupa (P24, B). Medulla intrinsic neurons (Mi1) are labeled by bsh-Gal4 (green); general labeling of neurons by anti-Neuroglian (red) and neuropil by anti-DNcadherin (blue). The vertical axis of the medulla cortex is shown by gray rectangle. Shortly after their birth, Mi1 cell bodies are close to medulla neuropil [arrow in (A)]; after 24h, they have shifted to distally towards the outer surface of the medulla cortex [arrowhead in (B)]. Top part of panel of (C), showing a schematic frontal section of medulla cortex and neuropil, schematically depict the cell body movement. (D-F) Confocal sections of optic lobe of late third instar (L3, D), early pupa (P24, E) and mid-stage pupa (P48, F). Labeling of Tm neurons by Vsx1-Gal4 (green). General labeling of neurons by anti-Neuroglian (red) and neuropil by anti-DNcadherin (blue). Vsx1-positive neurons originate from a central domain of the outer proliferation center [OPCm; in between white arrows in (D)], whereas the dorsal and ventral wings of the OPCm (yellow arrows) are devoid of this cell type. The same spatial restriction is still visible at P24 (E), where Vsx1-positive cell bodies are located in the central part of the cortex. Note that axons of the neurons at the dorsal rim of the Vsx1-positive cluster (white arrow) project dorsally, traverse the cortex and enter the neuropil at its very dorsal edge (yellow arrow); likewise, neurons at the ventral rim project ventrally. The mass of these fibers, which fill the deeper part of the medulla cortex, form the transient medullary fiber plexus (TMP; see Fig.6B and 9R). By P48 (F), cell bodies have spread out to fill the entire cortex (white arrows and yellow arrows coincide). All Tm axons extend parallel, and the TMP has disappeared. The movement of cell bodies is depicted schematically in bottom part of panel (C). The entry point of a representative Vsx1-positive axon, indicated by “e”, remains the same before and after the movement of cell bodies. For other abbreviations see legend of Fig.2. Bar: 50 μm
It is important to note that the above described movements only affect the position of neuronal cell bodies, not the (immature) terminal arborization formed in the medulla neuropil. Shortly after the birth of a neuron, it extends a process that enters the emerging medulla neuropil at a defined position (e.g., “e” in Fig.8C). For most neurons, this position corresponds to the position of the cell body within the cortex. By contrast, the Vsx-positive neurons, “delivered” only in the central part of the medulla, emit axons that fan out along the vertical axis (yellow arrows, Fig. 8E; see also Erclik et al., 2017). Axons of neurons located at a dorsal position within the Vsx-expressing cluster do not directly enter the neuropil, but project dorsally towards the dorsal edge of the medulla neuropil; likewise, neurons located ventrally in the Vsx domain project towards the ventral edge of the neuropil (yellow arrows, Fig. 8E). During the movement of the cell bodies that occurs between 24h and 48h APF, the position where axons enter the neuropil (and, presumably, form contacts with their targets) does not change; instead, the position of a cell body within the cortex approaches the position of its axon in the neuropil.
The development of layers within the optic lobe neuropils: medulla
The fact that branching of different populations of neurons occurs at specific locations along the z-axis of the neuropil was used to define layers in the medulla, lobula and lobula plate (Fischbach and Dittrich, 1989). For example, the upper strata of the medulla (layers m1-m5) are defined by the endings of lamina neuron classes L1-L5. Photoreceptors R7 have their terminal bulbs right underneath L5, thereby defining layer m6. Medulla intrinsic neurons (Mi), as well as Tm and TmY neurons, define the layers of the proximal medulla (m7-m10). In an analogous manner, 6 layers were defined for the lobula, and four for the lobula plate (Fischbach and Dittrich, 1989).
Layers can also be recognized by globally labeling the neuropil with markers for proteins enriched in synapses, like Synaptotagmin (Syt), Bruchpilot (Brp, nc82), Discs-large (Dlg) or DN-cadherin (DNcad), the markers used in this study (Fig.S2). Some layers are characterized by higher levels of DNcad or Brp labeling than others. Using DNcad in conjunction with specific layer markers, we reconstructed the development of neuropil layering in the optic lobe. The results, depicted in Figs. 9-12, will be presented in the following, starting with the development of the medulla, and ending with the lobula/lobula plate.
Figure 9.
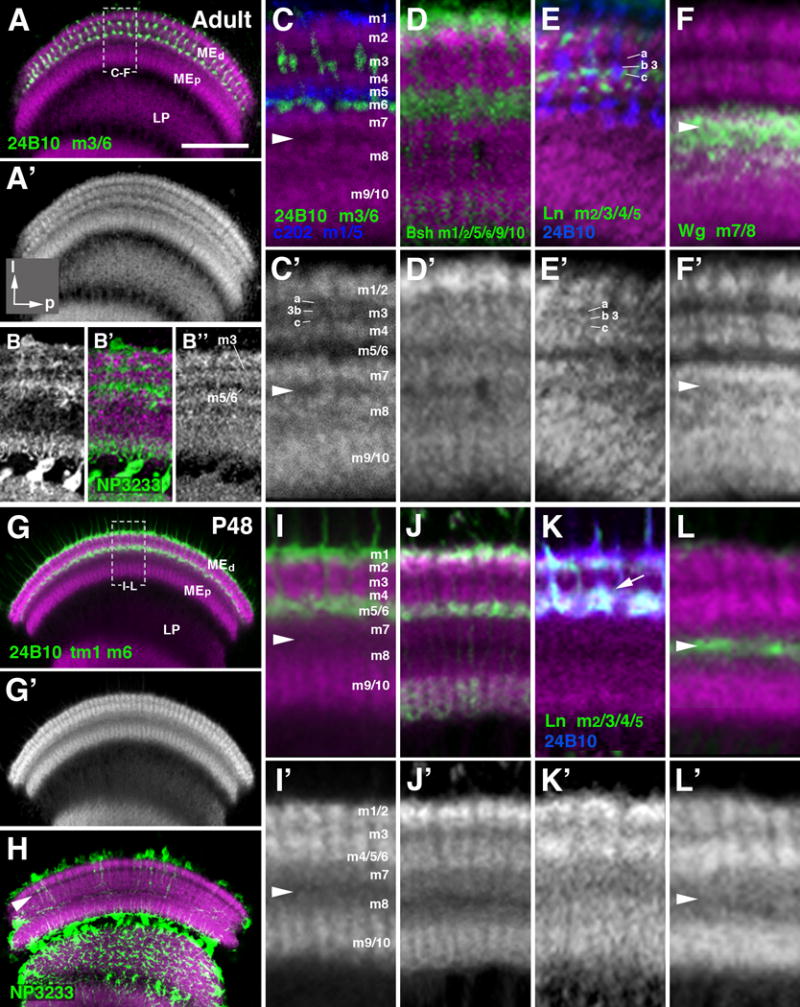
Layering of the medulla neuropil in the adult and mid-stage pupa. (A-L′) Frontal sections of medulla neuropile, labeled by anti-DNcadherin (magenta in A-L; white in A′, B″, C′-L′), which reveals distinct strata with different signal intensity. Double labeling with cell type-specific markers, traditionally used to define layers m1-m10, permits one to assign the DNcadherin pattern to these layers. The layer-specific markers used are: anti-Chaoptin (24B10; labeling of photoreceptors R7/8; green in A, C, I; blue in E, K); c202-Gal4 (labeling of lamina L1 neurons; blue in C); bsh-Gal4 (labels medulla Mi6 neurons; green in D, J); ln-Gal4 (labels lamina L3/4 neurons; green in E, K); wg-Gal4 (labels precursors of medulla tangential neurons; green in F, L). The medulla layers (m1-10) defined by the specifically neuronal populations are indicated next to the marker (lower left of panels (A-F) and (G, K). Note that anti-Chaoptin labels adult layers m3 and m6; at P8 and earlier (see Fig.9) it labels m6 and transient layer distal of m1 (“tm1”). The top two rows of panels (A-F′) show adult stage; bottom rows (G-L′) pupa at 48h after puparium formation (P48). For a given pair of panels (e.g., C, C′), the upper one shows anti-DNcadherin label (magenta) in conjunction with marker; the lower one is anti-DNcadherin only (in white). Panels of left column (A, A′, G, G′) present overview of medulla neuropil at low magnification; hatched lines outline domain of neuropil shown at high magnification in the four columns to the right. Panels (B-B″) and (H) show labeling of neuropil glia expressing the marker NP3233-Gal4 (green). White arrowhead in (C, C′, F, F′, I, I′, L, L′) indicates serpentine layer (boundary region between m7 and m8. White arrowhead in (H) marks the DNcadherinlow stratum (nascent layer m3) prior to the formation of significant glial processes. Note differences in anti-DNcadherin pattern between late pupal and adult stage illustrated in panels (C′) and (I′), respectively. Layer m3, in the adult, is represented by a thin band with moderate DNcadherin signal (“3b”) flanked by bands of low signal (“m3a”, “m3c”). In the 48h pupa, m3 is narrower and typically represented by a single band with low or moderate DNcadherin level. Most conspicuously, the DNcadherinlow band representing layers m5/6 in the adult (C, C′) is absent in the pupa (I, I′); here, terminals of m5/6 specific neurons (L1, R8), that coincide with a DNcadherinlow stratum in the adult, are found in the lower part of a DNcadherinhigh band (I). This band represents the “protolayer” for m4-m6. For other abbreviations, see legend of Fig.2. Bar: 50 μm (A, G)
Figure 12.
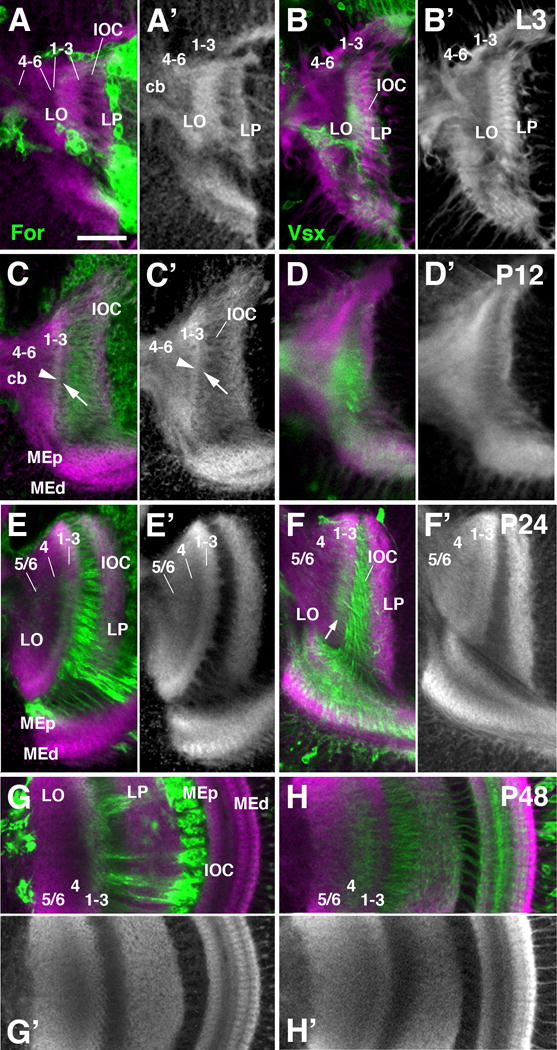
Layering of the lobula neuropil during metamorphosis. Frontal sections of the optic lobe labeled by for-Gal4 lineage tracing (green; A-A′, C-C′, E-E′, G-G′) and Vsx1-Gal4 (green; B-B′, D-D′, F-F′, H-H′) and counterstained with anti-DNcadherin (magenta in A-H; gray in A′-H′) at late larval stage (L3; first row; A-B′), 12h pupa (P12; second row; C-D′), 24h pupa (P24; third row; E-F′), and 48h pupa (P48; fourth and fifth row; G-H′). The tripartite subdivision of the lobula neuropil is visible from P24 onward (E, E′). Prior to this stage, a thin, DNcadherinrich band demarcates a protolayer for 1-3, in which for-Gal4-positive axon terminals are concentrated [arrowhead in (C)]; further proximally, the nascent lobula neuropil, which is in broad contact with the central brain neuropil (cb), has a moderate DNcadherin signal [protolayer 4-6 in (A-C′)]. Note that Vsx1-positive Tm axons, concentrated in proximal layers 4-6 in the adult lobula (see Fig.10D), are in the intermediate layer 4 at P48, and in distal layers 1-3 prior to that [arrow in (F)]. White arrows in (C, C′) point at superficial band of moderate DNcadherin signal that is continuous with the striated pattern of fiber bundles forming the inner optic chiasm (IOC). This band could correspond to layer of glial cells associated with the IOC. For abbreviations, see legend of figure 2. Bar: 25 μm
The superficial layers of the adult medulla, m1 and m2, show high DNcad signal, with m1 (blue in Fig. 9C; defined by c202-Gal4, which marks L1 neurons; Rister et al., 2007) exhibiting slightly less label than layer m2 (Fig. 9C, C′). Layers m3, m5 and m6 show the least amount of DNcad signal. M3, defined by the endings of the majority of R8 photoreceptors (Fig. 9A, C), is divided into a middle stratum (3b) with slightly higher DNcad intensity, flanked by two thin dark bands (3a, 3c; Fig. 9C′). The lower band receives terminals of L3, marked by the expression of Ln-Gal4 (Fig. 9E, E′). Layers m5 and m6, defined by the terminals of L5 and R7, respectively, are both very low in DNcad staining (Fig. 9C, C′). By contrast, the narrow m4 layer, receiving the terminals of L4 (marker: Ln-Gal4; FasII) shows a high DNcad signal (Fig. 9E, E′). M7 and m8 represent the domain where the lowest stratum of the distal medulla (m7) borders the upper stratum of the proximal medulla (m8). These two layers receive dense innervation by the large medulla tangential (Mt) neurons, which are derived from the GPZ of the OPC and can be labeled by lineage tracing cell populations derived from wg-Gal4 (see Materials and Methods; Bertet et al., 2014; Fig. 9F). Both layers exhibit moderate levels of DNcad and are separated by a thin band of low DNcad (arrowhead in Fig. 9C, C′, F, F′). The DNcad-poor band at the m7/8 boundary likely corresponds to the dense plexus of long fibers of medulla tangential neurons (Cucatti’s bundle) that are found at this position (Fischbach and Dittrich, 1989). The deep layers of the proximal medulla, m9 and m10, receive the terminal arborizations of Mi1 neurons, marked by bsh (Fig. 9D). They are indistinguishable in terms of DNcad intensity (Fig. 9D, D′).
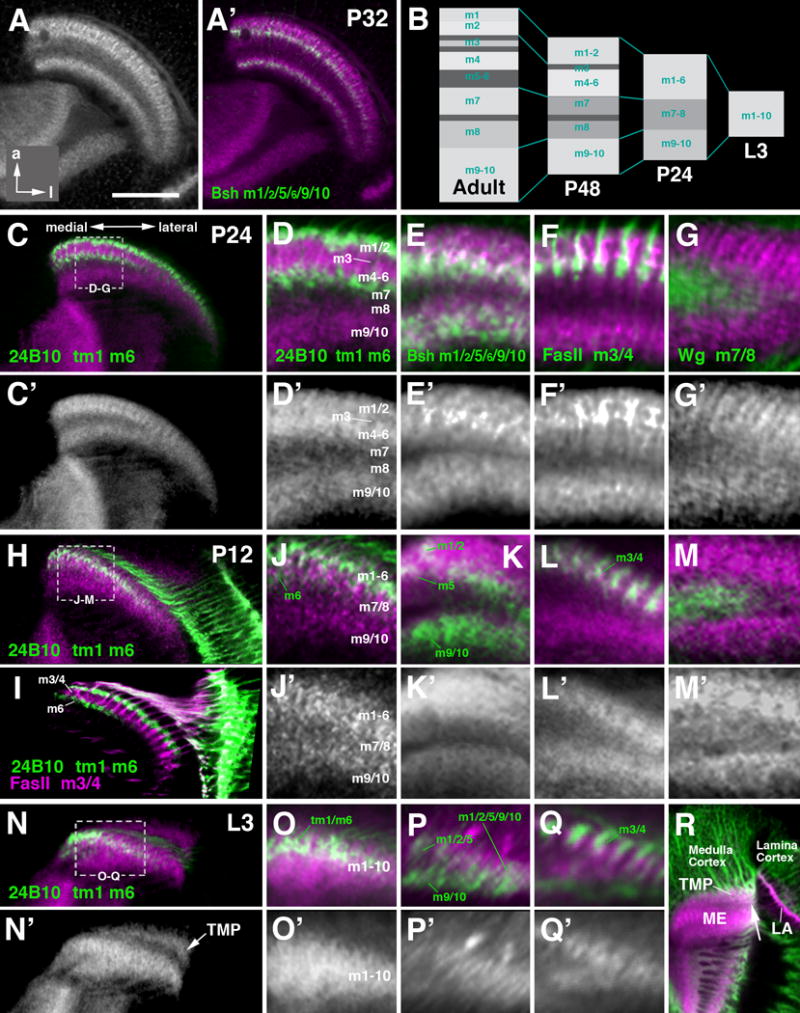
The layered organization of the medulla neuropil emerges during the second half of pupal development. At 72h APF, the pattern of DNcad labeling closely resembles the adult (not shown). Going backwards in time to 48h APF, the number of layers exhibiting different levels of DNcad intensity gets reduced, and the overall thickness of the medulla neuropil decreases, as schematically shown in Fig. 10B. At 48h APF, intermediate and deep layers (m7-m10) are thinner, but show a similar DNcad labeling as in adult, with m9/10, containing bsh-positive terminals of Mi1 (Fig. 9J, J′), at high intensity, and m7 and m8 at moderate density; whereas a band of very low DNcad signal marks the m7/m8 boundary (arrowhead in Fig. 9I, I′, L, L′). As in the adult, this band contains wg-positive tangential axons (Fig. 9L). By contrast, layers m1 to m6 of the distal medulla show a stratification of DNcad intensity that is simpler than the adult pattern (compare Fig. 9C′ to 9I′). Most notably, the wide layer that was lowest in DNcad signal in the adult, including m5 and m6, does not exist at 48h APF. The distal medulla consists of two layers of high DNcad signal, separated by a narrow band of low density. Based on the labeling of the superficial of the two DNcad-positive bands with bsh-Gal4 (Fig. 9J) and Ln-Gal4 (Fig. 9K), which are expressed in L1 (terminals in m1), L2 (terminals in m2), and L5 (terminals in m1 and m2); the more superficial DNCadhigh bands correspond to m1 and m2. The deep band harbors the terminals of L4 (labeled by Ln-Gal4), L5 (bsh-Gal4), and R7 (24B10), implying that this band represents the primordium, or “protolayer” (pm) for m4-6. Interestingly, the narrow stratum 3c, containing terminal arborizations of L3 (labeled by Ln-Gal4; arrow in Fig. 9K) is included in protolayer pm4-6. In other words, this protolayer encompasses the deep part of m3, and all of m4-m6 (schematically shown in Fig. 10B).
Figure 10.
Layering of the medulla neuropil in the late larva and early pupa. The design of this figure corresponds to that of the preceding Fig.8, presenting frontal sections of medulla neuropil labeled with anti-DNcadherin and layer-specific markers. Top row (A, A′) represents 32h pupa (P32); (B) schematically shows the developmental changes of the medulla neuropil layering as demonstrated with anti-DNcadherin from L3 to adult. Panels of the second and third row (C-G′) represent 24h pupa (P24); rows four and five (H-M′)12h pupa (P12); rows six and seven (N-R) late third instar larva (L3). (R) illustrates the transient medullary plexus (TMP), formed by subsets of medulla neurons, which does not form part of the medulla neuropil. The distal boundary of this neuropil is defined by the incoming lamina/retina axons, visible as a DNcadherinpoor band (arrow). For detail, see text. Bar: 50 μm (A, C, H, I, N)
Two events are temporally correlated with the transition in medulla layering that occurs between 48h APF and eclosion: the differentiation of neuropil glia, and the extension of photoreceptor R8 axon terminals, analyzed previously (Edwards et al., 2012; Ting et al., 2005). During early pupal stages, R8 terminals are held back at the surface of the medulla neuropil, where they form a “transient R8” layer superficial to m1 (Fig. 9I). Between 48h and 60h APF, terminals extend basally, to occupy their final position within m3. At around the same stage, astrocyte-like medulla neuropil glia (ALG), labeled by NP3233-Gal4 (Edwards and Meinertzhagen, 2010; Omoto et al., 2015), extend their processes into the neuropil. Processes become strongly concentrated around photoreceptor terminals in layers m3 and m6, as well as tangential axons at the m7/m8 boundary (Fig. 9B, H). In addition, the “chandelier glia” at the boundary between inner optic chiasm and medulla are strongly labeled (Fig.9B). It is possible that this layer-specific growth of glial processes, which do not form synapses, is causally related to the appearance of DNcad-negative bands in m3 and m5/6. However, the higher concentration of glia is not likely the only cause for decreased DNcad density, since the DNcadlow stratum demarcating layer m3 is present at P48, even though glial processes just start to form around that stage (Fig. 9H, arrowhead).
Going further backward in development to 32h APF (P32; Fig. 10A-A′), 24h APF (P24; Fig. 10C-G′) and earlier (P12; Fig. 10H-M′) towards late larval stages (Fig. 10N-R), the medulla neuropil is thinner and the DNcad banding pattern simpler. During these early stages, the medulla neuropil has a characteristic wedge-like shape. The pointed end of the wedge, which represents the earliest stage of development, is located laterally, where the still active OPC keeps adding neurons (Fig. 10C). Near the further developed medial edge of the medulla neuropil one can distinguish three layers: a superficial and a deep layer with strong and moderate DNcad signal, respectively, separated by a band of low signal (Fig. 10D, D′). Based on the expression of specific markers, the superficial DNcadhigh layer represents the protolayer for m1-m6 (pm1-6). From about P24 onward, a thin DNcadlow band demarcates the nascent m3 (Fig. 10B, D, D′). The deep DNcadmoderate layer corresponds to m9 and m10, and the intermediate DNcadlow layer contains m7 and m8 (Fig. 10B, D, D′). Detailed analysis of specific markers expressed at these early stages clearly demonstrates that, despite of the homogenous DNcad signal density, the protolayers are polarized: different cell types form nascent arborizations at different depth. For example, L3 and L4 (FasII) terminate in the center of protolayer pm1-6, where m3/4 will form later (Fig. 10F, I, L, Q); whereas terminals of L5 (bsh-Gal4) or R7 (24B10) terminate deeper (Fig.10D, E, J, K). On the other hand, it is also evident that different cell populations reach their ultimate level of termination at different time points. Markers expressed in L4 (Ln-Gal4; FasII), later forming spatially separate branches in m1 and m4 (Fig. 9E, K), have endings that span almost the entire thickness of protolayer m1-6 (Fig. 10F, L), suggesting that the filopodia of these neurons do not yet respond to cues that later lead to their separation into superficial (m1) and deeper (m4) branches.
Even at its earliest stage of development, when the medulla neuropil represents a single protolayer which includes all of m1-m10, a polarization is clearly noticeable. Axon tips later destined to occupy the distal layers of the medulla (e.g., L4) are concentrated in the upper strata of pm1-10 (Fig. 10Q); those neurons which will innervate the proximal medulla (e.g., Vsx; T4) cluster at the lower boundary of pm1-10 (not shown). Arborizations of neurons that will branch in both distal and proximal medulla (e.g. Mi1 neurons labeled by bsh-Gal4) initially fill the entire thickness of pm1-10 (Fig. 10P, right). As the transition from the single layer (pm1-10) to triple layer (pm1-6/pm7-8/pm9-10; right panel in Fig. 10B) occurs, bsh-positive terminals become segregated by the nascent pm7-8 into a deep layer in pm9-10, and a superficial layer in pm1-6 (left in Fig. 10P). The appearance of pm7-8 is correlated with and could be causally linked to the arrival of axons of tangential neurons (labeled by expression of wg-Gal4; Fig. 10G, G′, M, M′) which penetrate into the medulla protolayer from its lateral edge.
In the larva and first day of metamorphosis, the primordium of the medulla neuropil is capped distally by a layer of elevated DNcad intensity (arrow in Fig. 10N, N′). This transient layer, which decreases in thickness from laterally (earlier stages of medulla development) to medially (later stages), is formed by a plexus of immature fibers of medullary neurons which assemble in the deep layer of the cortex (transient medullary plexus, TMP; arrow Fig. 10N′; white arrowhead in Fig.7A-B). In other words, it does not become part of the medulla neuropil. Afferents from the lamina, which define the outer surface of the medulla neuropil (arrows in Fig. 7A-A′) penetrate in between the medulla neuropil and the TMP (arrowhead in Fig. 7A-A′, B).
The development of layers within the optic lobe neuropils: lobula complex
DNcad signal is distributed homogenously over the lobula plate (Fig. 11A′-D′), showing no correlation to the four layers defined on the basis of differential terminal arborization of T and Tlp neurons (Fischbach and Dittrich, 1989). Likewise, processes of neuropil glia are evenly distributed throughout the LP neuropil (Fig. 11A). In the lobula, DNcad labeling reveals three layers: a narrow distal layer of high DNcad signal, a wide proximal zone of moderate signal, and an intermediate layer of low signal (Fig. 11A′, C′). Glial density is correlated with this layering: processes of the lobula ALG are most concentrated in the intermediate and proximal stratum of the lobula, with the exception of a very thin surface layer in the distal lobula (Fig. 11A-B).
Figure 11.
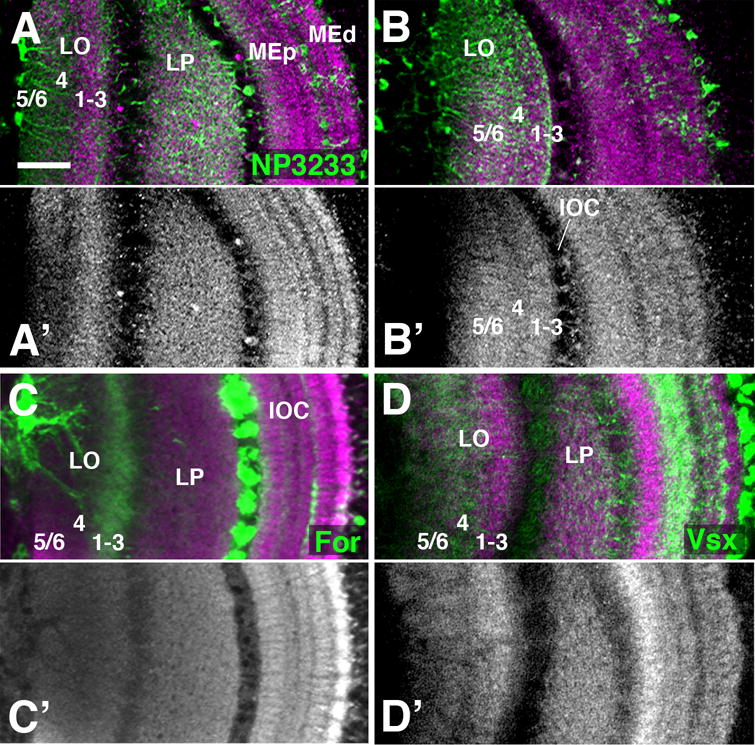
Layering of the late pupal lobula neuropil. (A-D′). Frontal sections of P72 optic lobe labeled with anti-DNcadherin (magenta in A-D; gray in A′-D′), and co-labeled with NP3233-Gal4 [expressed in glia; green in (A,B)]; for-Gal4 [lineage tracing; labels T neurons; green in (C)]; and Vsx1-Gal4 [labels Tm neurons; green in (D)]. (B, B′) represent a plane of section anterior to that in (A, A′), showing only the lobula (LO) and not the lobula plate (LP). The DNcadherin expression in the lobula neuropil reveals three layers, including a central layer with moderate signal intensity, flanked by a proximal and distal layer with higher intensity. Processes of neuropil glia occur at a slightly higher density in the intermediate layer than the flanking proximal and distal layers (B). The distal layer contains terminals of T neurons (C), identifying this wide band as combined lobula layer 1-3 (Fischbach and Dittrich, 1989). Vsx1-positive Tm neurons are confined to the proximal and intermediate layers, indicating that they comprise lobula layers 4-6, which receive the large majority of Tm/Tmy neurons (Dittrich and Fischbach, 1989). The lobula plate (LP) is uniformly labeled by DNcadherin and there appears to be no clear layer separation with the present markers. For other abbreviations, see legend of Fig.2. Bar: 25 μm
Labeling of columnar neurons innervating the lobula complex indicates that the DNcad-rich distal domain corresponds to layers 1-3, which are innervated by the T neurons labeled by acj6-Gal4 and for-Gal4LT (Fig.11C; Fig. S1). Acj6-positive and For-positive arborizations are distributed at a low level diffusely over the lobula plate (Fig. 11C; Fig. S1). Labeling with Vsx1-Gal4, expressed in a subset of Tm and TmY neurons (Li et al., 2013b; Erclik et al., 2008), also shows a diffuse innervation of the lobula plate, as well as a concentration of processes in the proximal and intermediate lobula (layers 4-6; Fig. 11D). This matches the previous description of Tm/TmY neurons (Fischbach and Dittrich, 1989), according to which most classes of these neurons (among them evidently the ones expressing Vsx) have terminals in the proximal lobula layers (Fig. S1). The proximal lobula is also strongly innervated by the classes of wide-field Lt neurons (Fischbach and Dittrich, 1989); a representative class of Lt neurons with arborizations in the proximal lobula, concentrated in layer 4, is marked by the expression of for-Gal4 (Fig. 11C).
Developmentally, a distinct lobula and lobula plate neuropil can be recognized from late larval stages onward. The lobula exhibits a subdivision into a distal, DNcad-rich domain and a proximal DNcad-poor domain already at the onset of metamorphosis (Fig. 12A′, B′). Immature, Acj6- and For-expressing T-neurons reach into the superficial layer (Fig. 12A), indicating that it constitutes the protolayer LO1-3. At P12, this protolayer is transiently subdivided into a more superficial stratum with moderate DNcad signal (white arrow in Fig. 12C-C′), and a deeper part with very high signal (white arrowhead in Fig. 12C-C′). By P24, the final pattern of DNcad expression is established, with evenly high signal in protolayer LO1-3, moderate signal in the deeper part of LO4-6, and low signal in the domain bordering LO1-3 (Fig. 12E-H′).
The projection of T neurons of the lobula complex is largely restricted to its proper neuropil protolayer (LO1-3) from early stages onward. This also applies to some other neuron populations, such as the Lt neurons labeled by for-Gal4, which from early pupal stages onward are concentrated in protolayer 4 (Fig. 12E, G). However, there are some groups of neurons that innervate the lobula at protolayers which do not ultimately correspond to the adult pattern. The Vsx-positive Tm/TmY neurons, whose final destination are the proximal strata of the lobula (Fig. 11D), initially terminate in the distal protolayer LO1-3 (Fig. 12D, D′). Between P24 and P48, Vsx-positive projections extend towards deeper layers to reach their final pattern at around P72.
Discussion
By reconstructing the global architecture and connectivity of the optic lobe at sequential stages of development, we provide a dynamic map that will help in future studies to understand the formation of specific neuronal circuits and to interpret experimental findings. Our analysis reveals three major structural/developmental hallmarks by which the optic lobe, compared to other regions of the fly brain, stands out: large scale neuronal movements, correlated temporal gradients in neuron production and differentiation, highly ordered retinotopic projections in between visual neuropils, and the formation of multiple layers within these neuropils.
Morphogenetic movements during optic lobe development
The position of cells in the optic lobe undergoes profound change between the late larval stages, when most cells are born, and the mid-pupal stage when the adult architecture and connectivity of the optic lobe is established. On the one hand, cell masses forming the cortices of the different optic lobe compartments move in toto; for example, the cortex of the medulla and lamina changes from its initial hemicylindrical shape to a rectangular shape. The lamina, initially perpendicular to the medulla, shifts forward and becomes oriented parallel to the medulla. On the other hand, neurons within a given cortex change position relative to each other; for example, medulla Tm neurons starting close to the center of the cortex shift in position all the way to the edges.
Long range cell migration occurs in numerous types of neural precursors in the developing vertebrate nervous system. It entails the protrusion and adhesion of the leading edge, followed by detachment of cell bodies and cytoskeletal contractions (Cooper, 2013). Cell migration has been extensively described in the cerebellum, neocortex, chick optic tectum and the mouse superior colliculus (SC; Watanabe and Yaginuma, 2015; Omi et al., 2014; Sugiyama and Nakamura, 2003; Tan et al., 2002). In these systems, neural precursors migrate at an early stage, prior to sending out axons and forming connections with other neurons. It stands to reason that the migration event plays an important role in establishing proper connectivity: neural precursors prevented from moving would not reach the domain where they are able to contact their proper synaptic targets.
The movements of neurons observed in the Drosophila optic lobe do not appear to follow the canonical mechanism of neuronal migration described in vertebrates. Thus, neuronal precursors emit axons that establish contact with the nascent neuropil already before their movement. This contact remains stable throughout development; only the cell bodies change their position, either along the z axis (e.g., from deep to superficial, as in case of the medulla Mi1 neurons), or along the vertical axis (as described here for the Vsx-positive Tm neurons). A similar type of movement was also described for several clusters of neurons in the central brain, where two hemilineages, produced by a common neuroblast, are located right next to each other in the larval brain, but move apart during the course of metamorphosis (Lovick et al., 2013). Likewise, several lineages located at the lateral surface of the central brain are displaced to dorsal or ventral positions by the growth of the optic lobe that occurs between early and late larval stages (Lovick et al., 2015). The fact that the displacement of neuronal cell bodies of some cell populations in the optic lobe and central brain occurs after the outgrowing fibers have reached and entered the neuropil also suggests that the movement is not essential for the connectivity of these neurons.
The mechanism by which cell bodies move in the Drosophila central brain or optic lobe has not been elucidated. The characteristic elongation and production of lamellipodia at the leading edge, described for “canonical” migration of neuronal precursors in vertebrates, is not apparent. This suggests that the movement is not active, but maybe caused by mechanical forces acting upon the cell bodies from the outside. In the central brain, such a passive movement seems to occur. Thus, if growth of the optic lobe was prevented by ablating optic lobe progenitors at an early stage (Lovick et al., 2015), central brain lineages did not move, but retained their position in the lateral brain cortex. At the same time, their axonal projection in the neuropile appeared unchanged, indicating that cell body movement is not essential for establishing connectivity.
Temporal gradients in neuronal birth and differentiation
A second hallmark of Drosophila optic lobe development is the presence of correlated temporal gradients underlying the birth and differentiation of neurons (reviewed in Campos-Ortega and Hartenstein, 1984; Meinertzhagen and Hanson, 1993). The first-born photoreceptors form a vertical row at the (presumptive) posterior edge of the retina. Their outgrowing axons encounter the row of first born lamina and medulla neurons, to which they establish contacts. Successively later born photoreceptors connect to later targets. We observed similar gradients in the deeper compartments of the optic lobe along either the mediolateral or anteroposterior axis: L3/4 and Tm/TmY neurons in the medulla; and IPCl derived proximal medulla and lobula plate neurons. The gradients in neuronal differentiation are reflective of the neuronal birthdates in both the OPC and IPC (Fig. 6M-P; Pineiro et al., 2014; Ngo et al., 2010; Yasugi et al., 2008; Egger et al., 2007; Hofbauer and Campos-Ortega, 1993; Meinertzhagen and Hanson, 1993). Additional studies are needed to address the question how precise the correlation between birthdate and connectivity in the deeper layers of the optic lobe really is; more importantly, the relationship between cell type, birth order, and connectivity needs to be established. Thus, incoming axons contact multiple target cell types, including different types of next-order projection neurons and local interneurons (Takemura et al., 2013). Given that, at least in the medulla, different types of neurons are born at different time points (Morante et al., 2013), it would be important to find out which of these is the “primary target” of the incoming axon, and whether this target (among all the cells that eventually are contacted by the axon) is always the first to differentiate.
Correlated temporal gradients underlying the formation of neurons and their targets have also been observed in other arthropod systems, notably crustaceans (Elofsson and Dahl, 1970; Harzsch et al., 1999). Here, three major growth zones, called P1, P2 and P3, were described for several species. P1 and P2 are neighboring each other in the head ectoderm, and produce, following the same posterior-to-anterior gradient as in insects, the retina (P1), lamina (P2), and medulla (P2). P3, located further medially in the ectoderm, is associated with the formation of the deep layers of the optic lobe. The minute visual system of the anostracan Daphnia magna, which possesses only 12 ommatidia projecting in a retinotopic manner on their target neuropil (lamina), has served as a classical model system to investigate the role of the temporal gradient in neuron production in controlling connectivity (Macagno, 1978; see below).
The temporal dynamics of neuronal birth and retinal axon extension has been documented in great detail in vertebrates. In Xenopus development, the retina grows by concentric accretion in which cells are added to the periphery where cell division is maintained at the dorsal and ventral ciliary margin (Hollyfield 1971; Straznicky and Gaze, 1971). This growth gradient results in a position-dependent birth order of retinal cells, whereby the oldest cells are located in the central retina and younger cells are located peripherally. Similar central-to-peripheral birth order of retinal cell types have also been reported in chicks, goldfish, several frog species, cat, and rat (Mednik and Spring, 1988; Drager 1985; Reh and Constantine-Paton, 1983; Rager 1980; Hollyfield 1968, 1971, 1972; Fujita and Horii, 1963; Sidman 1961). Unlike the retina, which shows a concentric mode of growth, the tectum grows directionally in many vertebrates including chick, fish and frogs (Crossland 1979; Crossland et al., 1975; Gaze et al., 1974; Meyer 1978; Straznicky and Gaze, 1972; LaVail, 1971a, b; LaVail and Cowan, 1971). Anterior neurons are the first to differentiate, and neurons are continuously added by a growth zone extending in a crescent-shaped domain along the lateral and posterior margin of the tectum.
The chronotopic organization of axon pathways in the visual system
Axonal connections within the visual system follow a strict retino-topic order, which makes it possible that an image of the visual field is generated in the brain. In flies, retinal photoreceptors project onto the lamina, whereby both dorso-ventral axis are maintained. At the next two stages, the projection of the lamina onto the medulla, and the medulla onto lobula/lobula plate, the dorso-ventral axis is maintained, but the antero-posterior axis is reversed in the two optic chiasms. The location of axons within the chiasms faithfully reflects the location of the cell body in the compartment of origin; for example, axons of neighboring neurons in the lamina are also adjacent to each other in the outer optic chiasm. This implies that, given the posterior-to-anterior gradient of neuronal birth in eye and lamina, the position of axons in the chiasms also reflects birthdate of the neuron of origin (chronotopy).
In vertebrates, the relationship between birthdates of neurons and their targets and neuronal connectivity is more complex. Projections between the photoreceptors and their targets in the retina (bipolar cells, retinal ganglion cells) are short, and maintain the axes of the visual field. The long projection of the retinal ganglion cells to the contralateral tectum, or thalamus, reverts both axes, with medial (=anterior) retinal ganglion cells connecting to the posterior tectum, and dorsal cells to the lateral (=ventral) tectum (Goodhill and Xu, 2005). This implies that birth order of retinal ganglion cells (central to peripheral) and their targets in the tectum (anterior to posterior) does not match. Even more, the divergent birth orders necessitate a constant reorganization (“shifting connections) of retinotectal projections (Reh and Constatine-Paton, 1984). Thus, as new neurons are constantly added at the posterior margin of the tectum, the early-born RGCs have to break their initially formed connections in order to maintain the retinotopic projection. This amazing shift in connectivity (which goes on while the visual system performs its function!) has been demonstrated in frogs and fish, as well as chick (McLoon 1985; Reh and Constatine-Raton, 1984; Easter and Struemer, 1984; Gaze et al., 1979; Schmidt 1977).
Whereas the birth order of retinal ganglion cells and their tectal targets is not correlated, a chronotopic order does exist in the axonal projection between retina and tectum, the optic nerve (retina to chiasm) and optic tract (chiasm to tectum. In goldfish and chick, newly grown fibers from the peripheral retina are added at the ventral surface of the nerve head (Finlay and Sengalaub, 1989; Easter et al., 1984; Easter et al., 1981; Rager 1980). In Xenopus, one also observes a chronotopic order of fibers which changes between optic nerve and tract (Taylor, 1987). In the nerve, axons are ordered concentrically, with younger axons originating in the retinal periphery, located at nerve perimeter, and older axons located at the nerve center. As the optic nerve passes through the chiasm, a reorganization of fibers occurs, such that young axons occupy a ventral position, and older fibers a dorsal position (Taylor, 1987). In mammalian vertebrates such as rodents, although there is a central-to-peripheral gradient of RGC formation (Drager 1985; Sidman 1961), there is no clear chronotopic ordering of the retino-collicular projection (Simon and O’Leary 1990, 1992). This is due to the fact that in mammals RGCs initially project to many incorrect targets. Secondary axonal pruning plays a crucial role in establishing the mammalian vertebrate retinotopic map.
It stands to reason that the correlated temporal gradients underlying the formation of visual neurons and their targets, and or the chronotopic ordering of the tracts by which visual neurons project on their targets, play a role in the process that controls the formation of the retinotopic projections. This has been directly confirmed in classical ablation experiments in the small crustacean Daphnia (Macagno, 1978). It was found that retinal axons form connections with target neurons in the lamina in the order in which they arrive; if retinal axons 1, 2, 3 normally contact targets A, B, C, and axon 2 is ablated, then the remaining axon 3 will contact target B. Thus, in the experimental situation, axon 3 is next in line after axon 1; it arrives in the lamina primordium adjacent to 1, and occupies the next available target, which is B. Thus, no specific “matching” of a retinal axon and its target is needed.
The mechanism controlling connectivity has to be different in the vertebrate visual system. First, as explained above, birth orders of RGCs and their tectal targets are not correlated. Accordingly, experiments where either retina or tectum was rotated, still allowed for an orderly formation of a retinotopic map (Sharma and Hollyfield, 1980). An abundance of studies has made it clear that the graded distribution of the repulsive signals of the ephrin family and their receptors in the RGCs and tectum, respectively, play a crucial role in the patterning of the retinotectal projection (Lemke and Reber, 2005). This does not exclude additional roles of local cell-cell interactions between the chronotopically ordered retinal axons, as shown by recent findings of Pittman et al. (2008) in zebrafish.
The case of Drosophila, despite its wide use as a favorable model system in developmental neurobiology, is unclear. Local interactions between retinal axons, mediated by signals such as Robo 2, are important for the projection from eye to lamina and medulla (Pappu et al., 2011). Global secreted signals, such as the ephrins, are present in the eye and optic lobe and could play a role similar to that described for vertebrates (Dearborn et al., 2012). Earlier experiments of Kunes et al. (1993) had also indicated that, along the dorso-ventral axis, chemical labels are important for the connections between retina and lamina. However, the importance of the correlated temporal gradients in neuronal birth that exist along the anterior-posterior axis has not been tested thus far.
Development of layering in the visual neuropils
Our data show that the global neuronal marker DNcad highlights the layers of the medulla and, to a lesser extent, the lobula. In previous studies, DNcad has been used to define discrete compartments in the neuropile of the central brain. In particular, the so called “structured neuropil”, including the central complex, mushroom body, or antennal lobe exhibits modular subdivisions that are revealed by DNcad, or other markers that primarily highlight synaptic density (Pereanu et al., 2010; Ito et al., 2014; Omoto et al., 2017). The simultaneous labeling of all medulla layers by global marking techniques, rather than specific genetic markers that can only visualize one type of neuron/layer at a time, will be useful for future studies relying on the anatomical mapping of individual neurons, or classes of neurons.
Anti-DNcad-labeling also sheds light on the gradual emergence of medulla layers during pupal development; our description supports and extends previous detailed studies of Lee et al. (2001) and Nern et al. (2008) that addressed the function of DNcadherin in layer formation. The prominent layering of the insect medulla and deeper optic lobe compartments presents an interesting phenomenon that primarily relies on the subdivision of the cell membrane of individual neurons into small subcompartments that are able to interact only with specific synaptic partners. Thus, even though the fiber of medulla neurons TM1or TM3 extend throughout the entire thickness of the medulla neuropil, contact with their main presynaptic partners, L2 and L1, is made only at a specific depth which defines layer m2 and m1/m5, respectively (Fischbach and Dittrich, 1989; Takemura et al., 2008; 2013). This principle applies for all of the connections made between neurons in the medulla neuropil, as well as in the lobula and lobula plate. It should be noted that the layers as discussed here for the fly optic lobe neuropils are very different from the entities called “layers” in many regions of the vertebrate brain, such as the cerebral or cerebellar cortex, which are defined by entire cells, including cell bodies, and are generated by the migration of neuronal precursors. The layers of the optic tectum of vertebrates, on the other hand, bears much similarity to the insect optic neuropils: cell bodies are all located apically, near the ventricular layer, and send long dendritic fibers towards the basal (outer) surface; layers are generated by specific inputs that contact tectal dendrites only at specific depths (Meek, 1983; Baier, 2013).
There are different hypothetical mechanisms by which the layering of the medulla (and other visual neuropils) could evolve developmentally (Fig.13), depending on the time course in which input to the medulla arrives, and medulla interneurons emit branches at different positions. Layers could be established sequentially (Fig.13A), with one type of input and its corresponding postsynaptic branches appearing first (e.g., elements “b” and “c” in Fig.13A), followed gradually by others. The opposite extreme scenario (Fig.13B) would be if all layers develop simultaneously at an early stage and merely grow in thickness by increasing branch number and synapse formation. Both scenarios can be clearly dismissed by the observations reported here and in the previous literature. Thus, the definitive layers of the medulla visible from approximately 60h APF onward, are preceded by protolayers, in which terminal branches of different afferents and their targets, which are later separated in different layers, overlap (Fig.13C). This implies that many types of interneurons and afferents appear around the same stage (elements “a”-“f” in top panel of Fig.13C). Undifferentiated terminal branches largely overlap in a protolayer (“1/2/3/4” in Fig.13C). Protolayers may already be polarized, with elements later restricted to deep layers (e.g., “f”) occupying a deeper position within the protolayer, and vice versa. In subsequent steps, branches of different elements sort out, resulting in the gradual appearance of layers (Fig.13C, middle, bottom). Some elements (e.g., “c”, representing photoreceptor R8) may not initially form part of protolayers, but remain at the surface, and only later insert into one of the emergent layers.
Figure 13.
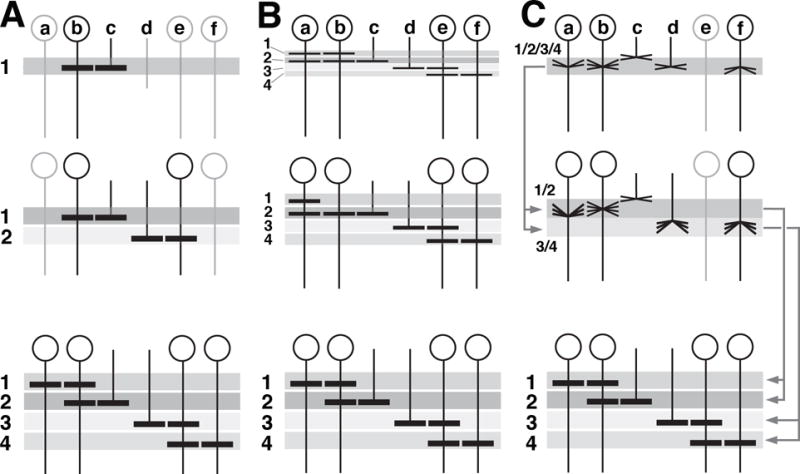
Hypothetical mechanisms of neuropil layer formation. (A-C) show time series (top: early stage; bottom: late stage) of schematic cross sections of layered neuropil, such as the medulla. Shaded horizontal bars (1-4) represent layers. Vertical elements (a-f) represent interneurons (a, b, e, f) and photoreceptor afferents (c, d) whose terminal branches (horizontal lines within layers) generate the layers. (A) Layers are formed sequentially. Elements b and c generate the top layer (1). Only these elements differentiate early (top). As a result, the only layer present at early stage is (1). Other elements and their layers follow at subsequent stages. (B) Layers form simultaneously; development merely consists in increase in branching/layer thickness. (C) Layers develop from protolayers. Most interneurons and afferents appear around the same stage (top) and form undifferentiated terminal branches that largely overlap in a protolayer (1/2/3/4). Protolayers may already be polarized, with elements later restricted to deep layers [e.g., (f)] occupying a deeper position within protolayer, and vice versa. Some elements (c, e) may not initially form part of protolayers.
The underlying molecular basis for restricting contacts to specific subdomains of the neuronal membrane could include highly localized expression of specific “recognition molecules” along the length axis of the columnar neuronal fibers, as well as diffusible attractive or repulsive signals acting on neurons and restricting their sites of contact to specific positions. In Drosophila, recent studies have provided some insight into the mechanisms guiding lamina neurons (L1-5) and photoreceptors (R7/8) to their proper layer in the medulla neuropil. R7 and R8 establish their final projections in the medullary m6 and m3 layers in a two-step process: R7/R8 first extend their growth cones to the respective temporary layers; by the mid pupal stage, they reach their final target destination (this work; Ting et al., 2005). The molecular network controlling the proper layer targeting of R7 includes Drosophila N-cadherin, along with the phospho-tyrosine phosphatases PTP69D and Lar; while R8 growth cone extension requires Capricious (Caps), Flamingo (fmi, a transmembrane Cadherin) and the putative receptor, Golden Goal (ggo), as well as Robo-3 and Slit (Kulkarni et al., 2016; Hofmeyer and Treisman, 2009; Nern et al., 2008; Tomasi et al., 2008; Chen and Clandinin, 2008; Ting et al., 2005; Lee et al., 2001). Netrins and its receptor, Frazzled (Fra) have been recently identified as additional players in controlling R8 layer specificity (Timofeev et al., 2012).
Supplementary Material
Figure S1 Representative markers and their expression in the Drosophila optic lobe. Panels of left column show horizontal z-projection of optic lobe of 24h pupa labeled with anti-DNcadherin (red) and various markers (green) indicated at lower left of panels. Cell types expressing markers are annotated in green lettering. Right column are semi-schematic line drawings of horizontal sections of optic lobe, illustrating specific cell types (camera lucida drawings of Golgi-impreganted neurons), including those labeled by markers shown to the left (after Fischbach and Dittrich, 1989; with permission).
Figure S2 Labeling of medulla neuropil layers by synaptic markers Bruchpilot (Nc82), Discs large (Dlg) and DNcadherin (DNcad) in late pupa (P64) and adult. Note that layers with low DNcad intensity are also low in Nc82 (arrows in C′, C″).
Movie S3 Animation of model of second instar larval optic lobe (L32)
The model includes the left brain hemisphere, rendered gray. The movie starts with an anterior view, and then rotates along the dorso-ventral axis. During the first full 360deg rotation, the inner optic anlage (IOA), rendered in chartreuse yellow, is shown. During the second rotation the outer optic anlage (OOA), rendered in light magenta, is added. At this stage both anlagen are small and crescent shaped, and consist only of a single, epithelial layer.
Movie S4 Animation of model of early third instar larval optic lobe (L72)
At this stage, the OOA and IOA each have divided into two distinct regions. In case of the IOA, only the medial part, rendered in light yellow, is still epithelial; the lateral part, in light green, has transformed into neuroblasts. For the OOA, the lateral part represents what is left of the epithelium (light magenta); the medial part (purple) is transformed into neuroblasts. The neuroblasts divide with a mitotic spindle oriented perpendicularly to the surface, thereby “budding” their daughter cells interiorly. These daughter cells (ganglion mother cells, first neurons, glial cells of the medulla. The are are rendered in dark blue.
The movie starts from an anterior view. During the first rotation, the medial and lateral domain of the IOA are visible; during the second rotation, the domains of the OOA are added. During the third turn, only the OOA is visible. During the fourth turn, the neurons/ganglion mother cells descending from the OOAepithelium are added to the OOA.
Movie S5 Animation of model of optic lobe at the transition between larva and pupa (P0). 1.Optic proliferation centers and neuronal cortices.
The two optic anlagen, both with an epithelial and a neuroblast domain rendered in the same colors as in the previous model, are still in existence. The difference to the L72 model is the increase in volume of the optic lobe neurons descended from the optic anlagen. These “squeeze” in between the IOA and OOA and push them aside.
The movie starts from an anterior view. During the first rotation, the IOA (light yellow: epithelial; light green: neuroblasts) is visible. During the second rotation, the OOA is added; the third rotation shows only OOA. During the fourth rotation, the neurons descending from the OOA are added. These comprise two groups: the neurons forming the medulla cortex (including all Mi neurons, Tm neurons, Tmy neurons) are located inward of the medial OOA. They are rendered in dark blue. The second group are the neurons of the lamina, budded off from the lateral OOA; they are rendered dark magenta. The fifth rotation adds the IOA (neuroblasts light green; epithelium light yellow); the sixth rotation then shows the neurons budded off from the lateral IOA. The lateral IOA gives off neurons to too sides. Neurons given off posteriorly, into the concavity of the C-shaped IOA, are the neurons forming the cortex of the lobula plate, including T4 and T5. They are rendered in dark green. Neurons given off towards laterally, and anteriorly (towards the convex side of the IOA) are the neurons that will form the posterior medulla cortex (T2/T3). They are rendered in turquois. Clusters of lobula neurons, possibly derived from the medial part of the IOA, are rendered in dark yellow.
Movie S6 Animation of model of optic lobe at the transition between larva and pupa (P0). 2.Neuronal cortices and neuropils.
The first rotation shows the cortices of the lamina (dark magenta) and medulla (dark blue). During the second rotation, the neuropils of the distal medulla and lamina are added (cyan and pink, respectively). These neuropils form thin sheaths underlying the cell body cortices, following their C-shaped curvature. Note that lamina cortex/neuropil is oriented perpendicular to the medulla. The third rotation shows the developing cortices of the lobula plate, proximal medulla (i.e., posterior medulla cortex) and lobula. To these, the corresponding neuropils, rendered in bright colors, are added during the fourth rotation. In the fifth rotation, only the inner neuropils (MEp, LP, LO) are visible; the sixth rotation shows all neuropils.
Movie S7 Combination of Models presented in S3, S4 and S6.
Movie S8-10 Animations of models of optic lobe at three pupal stages (S8: 12h after puparium formation; S9: 40h after puparium formation; S9: 72h after puparium formation). The sequence in which cortices and neuropils appear is the same described for the P0 model in S6: The first rotation shows the cortices of the lamina (dark magenta) and distal medulla (dark blue). During the second rotation, the neuropils of the distal medulla and lamina are added (cyan and pink, respectively). These neuropils form thin sheaths underlying the cell body cortices, following their C-shaped curvature. Note that lamina cortex/neuropil is oriented perpendicular to the medulla. The third rotation shows the developing cortices of the lobula plate, lobula, and the posterior medulla cortex. To these, the corresponding neuropils, rendered in bright colors, are added during the fourth rotation. In the fifth rotation, only the inner neuropils (MEp, LP, LO) are visible; the sixth rotation shows all neuropils.
Movie S11 Combination of Models presented in S6, S8-10.
Acknowledgments
This work was supported by NIH Grant R01 NS054814 to V.H., and a NSF Graduate Research Fellowship (No. DGE-0707424) to K.N.
References
- Apitz H, Salecker I. A region-specific neurogenesis mode requires migratory progenitors in the Drosophila visual system. Nat Neurosci. 2015;18:46–55. doi: 10.1038/nn.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apitz H, Salecker I. A challenge of numbers and diversity: neurogenesis in the Drosophila optic lobe. J Neurogenet. 2014;28:233–49. doi: 10.3109/01677063.2014.922558. [DOI] [PubMed] [Google Scholar]
- Aptekar JW, Keleş MF, Lu PM, Zolotova NM, Frye MA. Neurons forming optic glomeruli compute figure-ground discriminations in Drosophila. J Neurosci. 2015;35:7587–7599. doi: 10.1523/JNEUROSCI.0652-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier H. Synaptic laminae in the visual system: molecular mechanisms forming layers of perception. Annu Rev Cell Dev Biol. 2013;29:385–416. doi: 10.1146/annurev-cellbio-101011-155748. [DOI] [PubMed] [Google Scholar]
- Bainbridge SP, Bownes M. Staging the metamorphosis of Drosophila melanogaster. J Embryol Exp Morph. 1981;66:57–80. [PubMed] [Google Scholar]
- Bausenwein B, Dittrich APM, Fischbach KF. The optic lobe of Drosophila melanogaster II. Sorting of retinotopic pathways in the medulla. Cell Tissue Res. 1992;267:17–28. doi: 10.1007/BF00318687. [DOI] [PubMed] [Google Scholar]
- Bertet C, Li X, Erclik T, Cavey M, Wells B, Desplan C. Temporal patterning of neuroblasts controls Notch-mediated cell survival through regulation of Hid or Reaper. Cell. 2014;158:1173–1186. doi: 10.1016/j.cell.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braitenberg V. Patterns of projection in the visual system of the fly. I. Retina-lamina projections. Exp Brain Res. 1967;3:271–298. doi: 10.1007/BF00235589. [DOI] [PubMed] [Google Scholar]
- Brand AH, Livesey FJ. Neural stem cell biology in vertebrates and invertebrates: more alike than different? Neuron. 2011;70:719–729. doi: 10.1016/j.neuron.2011.05.016. [DOI] [PubMed] [Google Scholar]
- Cajal SR, Sanchez D. Contribucion al conocimiento de los centros nerviosos del los insectos. Trab Lab Invest Biol. 1915:1–167. [Google Scholar]
- Cardona A, Saalfeld S, Schindelin J, Arganda-Carreras I, Preibisch S, Longair M, Tomancak P, Hartenstein V, Douglas RJ. TrakEM2 software for neural circuit reconstruction. PLoS One. 2012;7:e38011. doi: 10.1371/journal.pone.0038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PL, Clandinin TR. The cadherin Flamingo mediates level-dependent interactions that guide photoreceptor target choice in Drosophila. Neuron. 2008;58:26–33. doi: 10.1016/j.neuron.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotard C, Salecker I. Glial cell development and function in the Drosophila visual system. Neuron Glia Biol. 2007;3:17–25. doi: 10.1017/S1740925X07000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantine-Paton M, Pitts EC, Reh TA. The relationship between retinal axon ingrowth, terminal morphology, and terminal patterning in the optic tectum of the frog. J Comp Neurol. 1983;218:297–313. doi: 10.1002/cne.902180306. [DOI] [PubMed] [Google Scholar]
- Cooper JA. Cell biology in neuroscience: mechanisms of cell migration in the nervous system. J Cell Biol. 2013;202:725–734. doi: 10.1083/jcb.201305021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossland WJ, Cowan WM, Rogers LA. Studies on the development of the chick optic tectum. IV. An autoradiographic study of the development of retino-tectal connections. Brain Res. 1975;91:1–23. doi: 10.1016/0006-8993(75)90463-1. [DOI] [PubMed] [Google Scholar]
- Dearborn RE, Jr, Dai Y, Reed B, Karian T, Gray J, Kunes S. Reph, a regulator of Eph receptor expression in the Drosophila melanogaster optic lobe. PLoS One. 2012;7(5):e37303. doi: 10.1371/journal.pone.0037303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dräger UC. Birth dates of retinal ganglion cells giving rise to the crossed and uncrossed optic projections in the mouse. Proc R Soc Lond B Biol Sci. 1985;224:57–77. doi: 10.1098/rspb.1985.0021. [DOI] [PubMed] [Google Scholar]
- Easter SS, Jr, Stuermer CA. An evaluation of the hypothesis of shifting terminals in goldfish optic tectum. J Neurosci. 1984;4:1052–63. doi: 10.1523/JNEUROSCI.04-04-01052.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easter SS, Jr, Rusoff AC, Kish PE. The growth and organization of the optic nerve and tract in juvenile and adult goldfish. J Neurosci. 1981;1:793–811. doi: 10.1523/JNEUROSCI.01-08-00793.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easter SS, Jr, Bratton B, Scherer SS. Growth-related order of the retinal fiber layer in goldfish. J Neurosci. 1984;4:2173–90. doi: 10.1523/JNEUROSCI.04-08-02173.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TN, Nuschke AC, Nern A, Meinertzhagen IA. Organization and metamorphosis of glia in the Drosophila visual system. J Comp Neurol. 2012;520:2067–2085. doi: 10.1002/cne.23071. [DOI] [PubMed] [Google Scholar]
- Edwards TN, Meinertzhagen IA. The functional organisation of glia in the adult brain of Drosophila and other insects. Prog Neurobiol. 2010;90:471–497. doi: 10.1016/j.pneurobio.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Boone JQ, Stevens NR, Brand AH, Doe CQ. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev. 2007;2:1. doi: 10.1186/1749-8104-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Gold KS, Brand AH. Notch regulates the switch from symmetric to asymmetric neural stem cell division in the Drosophila optic lobe. Development. 2010;137:2981–2987. doi: 10.1242/dev.051250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elofsson R, Dahl E. The optic neuropiles and chiasmata of Crustacea. Z Zellforsch Mikrosk Anat. 1970;107:343–60. doi: 10.1007/BF00336672. [DOI] [PubMed] [Google Scholar]
- Erclik T, Hartenstein V, McInnes RR, Lipshitz HD. Eye evolution at high resolution: the neuron as a unit of homology. Dev Biol. 2009;332:70–79. doi: 10.1016/j.ydbio.2009.05.565. [DOI] [PubMed] [Google Scholar]
- Erclik T, Hartenstein V, Lipshitz HD, McInnes RR. Conserved role of the Vsx genes supports a monophyletic origin for bilaterian visual systems. Curr Biol. 2008;18:1278–1287. doi: 10.1016/j.cub.2008.07.076. [DOI] [PubMed] [Google Scholar]
- Erclik T, Li X, Courgeon M, Bertet C, Chen Z, Baumert R, Ng J, Koo C, Arain U, Behnia R, del Valle Rodriguez A, Senderowicz L, Negre N, White KP, Desplan C. Integration of temporal and spatial patterning generates neural diversity. Nature. 2017;541:365–370. doi: 10.1038/nature20794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach KF, Dittrich APM. The optic lobe of Drosophila Melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 1989;258:441–447. [Google Scholar]
- Fischbach KF, Hiesinger PR. Optic lobe development. Adv Exp Med Biol. 2008;628:115–136. doi: 10.1007/978-0-387-78261-4_8. [DOI] [PubMed] [Google Scholar]
- Fujita S, Horii M. Analysis of cytogenesis in chick retina by tritiated thymidine autoradiography. Arch Histol Jpn. 1963;23:359–66. doi: 10.1679/aohc1950.23.359. [DOI] [PubMed] [Google Scholar]
- Gaze RM, Keating MJ, Chung SH. The evolution of the retinotectal map during development in Xenopus. Proc R Soc Lond B Biol Sci. 1974;185:301–30. doi: 10.1098/rspb.1974.0021. [DOI] [PubMed] [Google Scholar]
- Gaze RM, Keating MJ, Ostberg A, Chung SH. The relationship between retinal and tectal growth in larval Xenopus: implications for the development of the retino-tectal projection. J Embryol Exp Morphol. 1979;53:103–43. [PubMed] [Google Scholar]
- Goodhill GJ, Xu J. The development of retinotectal maps: a review of models based on molecular gradients. Network. 2005;16:5–34. doi: 10.1080/09548980500254654. [DOI] [PubMed] [Google Scholar]
- Green P, Hartenstein AY, Hartenstein V. The embryonic development of the Drosophila visual system. Cell Tissue Res. 1993;273:583–589. doi: 10.1007/BF00333712. [DOI] [PubMed] [Google Scholar]
- Hadjieconomou D, Timofeev K, Salecker I. A step-by-step guide to visual circuit assembly in Drosophila. Curr Opin Neurobiol. 2011;2:76–84. doi: 10.1016/j.conb.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Harzsch S, Benton J, Dawirs RR, Beltz B. A new look at embryonic development of the visual system in decapod crustaceans: neuropil formation, neurogenesis, and apoptotic cell death. J Neurobiol. 1999;39:294–306. [PubMed] [Google Scholar]
- Hasegawa E, Kaido M, Takayama R, Sato M. Brain-specific-homeobox is required for the specification of neuronal types in the Drosophila optic lobe. Dev Biol. 2013;377:90–99. doi: 10.1016/j.ydbio.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Hasegawa E, Kitada Y, Kaido M, Takayama R, Awasaki T, Tabata T, Sato M. Concentric zones, cell migration and neuronal circuits in the Drosophila visual center. Development. 2011;138:983–993. doi: 10.1242/dev.058370. [DOI] [PubMed] [Google Scholar]
- Hofbauer A, Campos-Ortega JA. Proliferation pattern and early differentiation of the optic lobes in Drosophila melanogaster. Roux’s Arch Dev Biol. 1990;198:264–274. doi: 10.1007/BF00377393. [DOI] [PubMed] [Google Scholar]
- Hofmeyer K, Treisman JE. The receptor protein tyrosine phosphatase LAR promotes R7 photoreceptor axon targeting by a phosphatase-independent signaling mechanism. Proc Natl Acad Sci U S A. 2009;106:19399–404. doi: 10.1073/pnas.0903961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollyfield JG. Differential addition of cells to the retina in Rana pipiens tadpoles. Dev Biol. 1968;18:163–79. doi: 10.1016/0012-1606(68)90041-9. [DOI] [PubMed] [Google Scholar]
- Hollyfield JG. Differential growth of the neural retina in Xenopus laevis larvae. Dev Biol. 1971;24:264–86. doi: 10.1016/0012-1606(71)90098-4. [DOI] [PubMed] [Google Scholar]
- Hortsch M, Bieber AJ, Patel NH, Goodman CS. Differential splicing generates a nervous system-specific form of Drosophila Neuroglian. Neuron. 1990a;4:697–709. doi: 10.1016/0896-6273(90)90196-m. [DOI] [PubMed] [Google Scholar]
- Hortsch M, Patel NH, Bieber AJ, Traquina ZR, Goodman CS. Drosophila neurotactin, a surface glycoprotein with homology to serine esterases, is dynamically expressed during embryogenesis. Development. 1990b;110:1327–1340. doi: 10.1242/dev.110.4.1327. [DOI] [PubMed] [Google Scholar]
- Huang Z, Kunes S. Signals transmitted along retinal axons in Drosophila: Hedgehog signal reception and the cell circuitry of lamina assembly. Development. 1998;125:3753–3764. doi: 10.1242/dev.125.19.3753. [DOI] [PubMed] [Google Scholar]
- Joly JS, Recher G, Brombin A, Ngo K, Hartenstein V. A Conserved Developmental Mechanism Builds Complex Visual Systems in Insects and Vertebrates. Curr Biol. 2016;26:R1001–R1009. doi: 10.1016/j.cub.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni A, Ertekin D, Lee CH, Hummel T. Birth order dependent growth cone segregation determines synaptic layer identity in the Drosophila visual system. Elife. 2016 Mar 17;5:e13715. doi: 10.7554/eLife.13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP. Retinal determination the beginning of eye development. Curr Top Dev Biol. 2010;93:1–28. doi: 10.1016/B978-0-12-385044-7.00001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunes S, Wilson C, Steller H. Independent guidance of retinal axons in the developing visual system of Drosophila. J Neurosci. 1993;13:752–767. doi: 10.1523/JNEUROSCI.13-02-00752.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langen M, Agi E, Altschuler DJ, Wu LF, Altschuler SJ, Hiesinger PR. The developmental rules of neural superposition in Drosophila. Cell. 2015;162:120–33. doi: 10.1016/j.cell.2015.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVail JH, Cowan WM. The development of the chick optic tectum. I. Normal morphology and cytoarchitectonic development. Brain Res. 1971a;28:391–419. doi: 10.1016/0006-8993(71)90053-9. [DOI] [PubMed] [Google Scholar]
- LaVail JH, Cowan WM. The development of the chick optic tectum. II. Autoradiographic studies. Brain Res. 1971b;28:421–41. [PubMed] [Google Scholar]
- Lee CH, Herman T, Clandinin TR, Lee R, Zipursky SL. N-cadherin regulates target specificity in the Drosophila visual system. Neuron. 2001;30:437–50. doi: 10.1016/s0896-6273(01)00291-4. [DOI] [PubMed] [Google Scholar]
- Lemke G, Reber M. Retinotectal mapping: new insights from molecular genetics. Annu Rev Cell Dev Biol. 2005;21:551–80. doi: 10.1146/annurev.cellbio.20.022403.093702. [DOI] [PubMed] [Google Scholar]
- Li X, Chen Z, Desplan C. Temporal patterning of neural progenitors in Drosophila. Curr Top Dev Biol. 2013a;105:69–96. doi: 10.1016/B978-0-12-396968-2.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Erclik T, Bertet C, Chen Z, Voutev R, Venkatesh S, Morante J, Celik A, Desplan C. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature. 2013b;498:456–462. doi: 10.1038/nature12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovick JK, Ngo KT, Omoto JJ, Wong DC, Nguyen JD, Hartenstein V. Postembryonic lineages of the Drosophila brain: I. Development of the lineage-associated fiber tracts. Dev Biol. 2013;384:228–57. doi: 10.1016/j.ydbio.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macagno ER. Mechanism for the formation of synaptic projections in the arthropod visual system. Nature. 1978;275:318–20. doi: 10.1038/275318a0. [DOI] [PubMed] [Google Scholar]
- Mardon G, Solomon NM, Rubin GM. Dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development. 1994;120:3473–3486. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- Masland RH. The fundamental plan of the retina. Nat Neurosci. 2001;4:877–886. doi: 10.1038/nn0901-877. [DOI] [PubMed] [Google Scholar]
- Masland RH, Raviola E. Confronting complexity: strategies for understanding the microcircuitry of the retina. Ann Rev Neurosci. 2000;23:249–284. doi: 10.1146/annurev.neuro.23.1.249. [DOI] [PubMed] [Google Scholar]
- McLoon SC. Evidence for shifting connections during development of the chick retinotectal projection. J Neurosci. 1985;5:2570–80. doi: 10.1523/JNEUROSCI.05-10-02570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek J. Functional anatomy of the tectum mesencephali of the goldfish. An explorative analysis of the functional implications of the laminar structural organization of the tectum. Brain Res. 1983;287:247–97. doi: 10.1016/0165-0173(83)90008-5. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Hanson TE. The development of the optic lobe. In: Bate M, Martinez-Arias A, editors. The development of Drosophila melanogaster. Cold Spring Habor Laboratory Press; Plainview: 1993. pp. 1363–1492. [Google Scholar]
- Meyer RL. Evidence from thymidine labeling for continuing growth of retina and tectum in juvenile goldfish. Exp Neurol. 1978;59:99–111. doi: 10.1016/0014-4886(78)90204-2. [DOI] [PubMed] [Google Scholar]
- Morante J, Vallejo DM, Desplan C, Dominguez M. Conserved miR-8/miR-200 defines a glial niche that controls neuroepithelial expansion and neuroblast transition. Dev Cell. 2013;27:174–187. doi: 10.1016/j.devcel.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morante J, Erclik T, Desplan C. Cell migration in Drosophila optic lobe neurons is controlled by eyeless/Pax6. Development. 2011;138:687–693. doi: 10.1242/dev.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Zhu Y, Zipursky SL. Local N-cadherin interactions mediate distinct steps in the targeting of lamina neurons. Neuron. 2008;58:34–41. doi: 10.1016/j.neuron.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo KT, Wang J, Junker M, Kriz S, Vo G, Asem B, Olson JM, Banerjee U, Hartenstein V. Concomitant requirement for Notch and Jak/Stat signaling during neuro-epithelial differentiation in the Drosophila optic lobe. Dev Biol. 2010;346:284–295. doi: 10.1016/j.ydbio.2010.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva C, Choi CM, Nicolai LJ, Mora N, De Geest N, Hassan BA. Proper connectivity of Drosophila motion detector neurons requires Atonal function in progenitor cells. Neural Dev. 2014;9:4. doi: 10.1186/1749-8104-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omi M, Harada H, Watanabe Y, Funahashi J, Nakamura H. Role of En2 in the tectal laminar formation of chick embryos. Development. 2014;141:2131–8. doi: 10.1242/dev.102905. [DOI] [PubMed] [Google Scholar]
- Omoto JJ, Keleş K, Lovick J, Nguyen B, Bolanos C, Frye MA, Hartenstein V. Developmentally- and functionally- distinct parallel neuronal populations relay visual information to the Drosophila central complex in the anterior visual pathway. Curr Biol. 2017;27:1098–1110. doi: 10.1016/j.cub.2017.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orihara-Ono M, Toriya M, Nakao K, Okano H. Downregulation of Notch mediates the seamless transition of individual Drosophila neuroepithelial progenitors into optic medullar neuroblasts during prolonged G1. Dev Biol. 2011;351:163–175. doi: 10.1016/j.ydbio.2010.12.044. [DOI] [PubMed] [Google Scholar]
- Pappu KS, Morey M, Nern A, Spitzweck B, Dickson BJ, Zipursky SL. Robo-3–mediated repulsive interactions guide R8 axons during Drosophila visual system development. Proc Natl Acad Sci U S A. 2011;108:7571–7576. doi: 10.1073/pnas.1103419108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeiro C, Lopes CS, Casares F. A conserved transcriptional network regulates lamina development in the Drosophila visual system. Development. 2014;14:2838–2847. doi: 10.1242/dev.108670. [DOI] [PubMed] [Google Scholar]
- Pittman AJ, Law MY, Chien CB. Pathfinding in a large vertebrate axon tract: isotypic interactions guide retinotectal axons at multiple choice points. Development. 2008;135:2865–71. doi: 10.1242/dev.025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rager GH. Development of the retinotectal projection in the chicken. Adv Anat Embryol Cell Biol. 1980;63:1–90. [PubMed] [Google Scholar]
- Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Reddy BV, Rauskolb C, Irvine KD. Influence of fat-hippo and notch signaling on the proliferation and differentiation of Drosophila optic neuroepithelia. Development. 2010;137:2397–2408. doi: 10.1242/dev.050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reh TA, Constantine-Paton M. Retinal ganglion cell terminals change their projection sites during larval development of Rana pipiens. J Neurosci. 1984;4:442–57. doi: 10.1523/JNEUROSCI.04-02-00442.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rister J, Pauls D, Schnell B, Ting CY, Lee CH, Sinakevitch I, Morante J, Strausfeld NJ, Ito K, Heisenberg M. Dissection of the peripheral motion channel in the visual system of Drosophila melanogaster. Neuron. 2007;56:155–170. doi: 10.1016/j.neuron.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Zipursky SL. Design principles of insect and vertebrate visual systems. Neuron. 2010;66:15–36. doi: 10.1016/j.neuron.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EK, Raabe T, Luo L. Structure of the vertical and horizontal system neurons of the lobula plate in Drosophila. J Comp Neurol. 2002;454:470–481. doi: 10.1002/cne.10467. [DOI] [PubMed] [Google Scholar]
- Selleck SB, Gonzalez C, Glover DM. Regulation of the G1-S transition in postembryonic neuronal precursors by axon ingrowth. Nature. 1992;355:253–255. doi: 10.1038/355253a0. [DOI] [PubMed] [Google Scholar]
- Selleck SB, Steller H. The influence of retinal innervation on neurogenesis in the first optic ganglion of Drosophila. Neuron. 1991;6:83–99. doi: 10.1016/0896-6273(91)90124-i. [DOI] [PubMed] [Google Scholar]
- Sharma SC, Hollyfield JG. Specification of retinotectal connexions during development of the toad Xenopus laevis. J Embryol Exp Morphol. 1980;55:77–92. [PubMed] [Google Scholar]
- Silies M, Gohl DM, Clandinin TR. Motion-detecting circuits in flies: coming into view. Annu Rev Neurosci. 2014;37:307–27. doi: 10.1146/annurev-neuro-071013-013931. [DOI] [PubMed] [Google Scholar]
- Simon DK, O’Leary DD. Relationship of retinotopic ordering of axons in the optic pathway to the formation of visual maps in central targets. J Comp Neurol. 1991;307:393–404. doi: 10.1002/cne.903070305. [DOI] [PubMed] [Google Scholar]
- Simon DK, O’Leary DD. Development of topographic order in the mammalian retinocollicular projection. J Neurosci. 1992;12:1212–32. doi: 10.1523/JNEUROSCI.12-04-01212.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprecher SG, Cardona A, Hartenstein V. The Drosophila larval visual system: high-resolution analysis of a simple visual neuropil. Dev Biol. 2011;358:33–43. doi: 10.1016/j.ydbio.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld NJ. The organization of the insect visual system (light microscopy). I. Projections and arrangements of neurons in the lamina ganglionaris of Diptera. Z Zellforsch. 1971;121:377–441. [Google Scholar]
- Straznicky K, Gaze RM. The growth of the retina in Xenopus laevis: an autoradiographic study. J Embryol Exp Morphol. 1971;26:67–79. [PubMed] [Google Scholar]
- Straznicky K, Gaze RM. The development of the tectum in Xenopus laevis: an autoradiographic study. J Embryol Exp Morphol. 1972;28:87–115. [PubMed] [Google Scholar]
- Sugiyama S, Nakamura H. The role of Grg4 in tectal laminar formation. Development. 2003;130:451–62. doi: 10.1242/dev.00232. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Kaido M, Takayama R, Sato M. A temporal mechanism that produces neuronal diversity in the Drosophila visual center. Dev Biol. 2013;380:12–24. doi: 10.1016/j.ydbio.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Takemura SY, Lu Z, Meinertzhagen IA. Synaptic circuits of the Drosophila optic lobe: the input terminals to the medulla. J Comp Neurol. 2008;509:493–513. doi: 10.1002/cne.21757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS. Fibre organization and reorganization in the retinotectal projection of Xenopus. Development. 1987;99:393–410. doi: 10.1242/dev.99.3.393. [DOI] [PubMed] [Google Scholar]
- Timofeev K, Joly W, Hadjieconomou D, Salecker I. Localized netrins act as positional cues to control layer-specific targeting of photoreceptor axons in Drosophila. Neuron. 2012;75:80–93. doi: 10.1016/j.neuron.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CY, Yonekura S, Chung P, Hsu SN, Robertson HM, Chiba A, Lee CH. Drosophila N-cadherin functions in the first stage of the two-stage layer-selection process of R7 photoreceptor afferents. Development. 2005;132:953–963. doi: 10.1242/dev.01661. [DOI] [PubMed] [Google Scholar]
- Tomasi T, Hakeda-Suzuki S, Ohler S, Schleiffer A, Suzuki T. The transmembrane protein Golden goal regulates R8 photoreceptor axon-axon and axon-target interactions. Neuron. 2008;57:691–704. doi: 10.1016/j.neuron.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Trujillo-Cenóz O. Some aspects of the structural organization of the arthropod eye. Cold Spring Harb Symp Quant Biol. 1965;30:371–382. doi: 10.1101/sqb.1965.030.01.037. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Yaginuma H. Tangential cell migration during layer formation of chick optic tectum. Dev Growth Differ. 2015;57:539–43. doi: 10.1111/dgd.12238. [DOI] [PubMed] [Google Scholar]
- Wernet MF, Huberman AD, Desplan C. So many pieces, one puzzle: cell type specification and visual circuitry in flies and mice. Genes Dev. 2014;28:2565–2584. doi: 10.1101/gad.248245.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasugi T, Sugie A, Umetsu D, Tabata T. Coordinated sequential action of EGFR and Notch signaling pathways regulates proneural wave progression in the Drosophila optic lobe. Development. 2010;137:3193–203. doi: 10.1242/dev.048058. [DOI] [PubMed] [Google Scholar]
- Yasugi T, Umetsu D, Murakami S, Sato M, Tabata T. Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development. 2008;135:1471–80. doi: 10.1242/dev.019117. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Nern A, Zipursky SL, Frye MA. Peripheral Visual Circuits Functionally Segregate Motion and Phototaxis Behaviors in the Fly. Curr, Biol. 2009;19:613–619. doi: 10.1016/j.cub.2009.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Representative markers and their expression in the Drosophila optic lobe. Panels of left column show horizontal z-projection of optic lobe of 24h pupa labeled with anti-DNcadherin (red) and various markers (green) indicated at lower left of panels. Cell types expressing markers are annotated in green lettering. Right column are semi-schematic line drawings of horizontal sections of optic lobe, illustrating specific cell types (camera lucida drawings of Golgi-impreganted neurons), including those labeled by markers shown to the left (after Fischbach and Dittrich, 1989; with permission).
Figure S2 Labeling of medulla neuropil layers by synaptic markers Bruchpilot (Nc82), Discs large (Dlg) and DNcadherin (DNcad) in late pupa (P64) and adult. Note that layers with low DNcad intensity are also low in Nc82 (arrows in C′, C″).
Movie S3 Animation of model of second instar larval optic lobe (L32)
The model includes the left brain hemisphere, rendered gray. The movie starts with an anterior view, and then rotates along the dorso-ventral axis. During the first full 360deg rotation, the inner optic anlage (IOA), rendered in chartreuse yellow, is shown. During the second rotation the outer optic anlage (OOA), rendered in light magenta, is added. At this stage both anlagen are small and crescent shaped, and consist only of a single, epithelial layer.
Movie S4 Animation of model of early third instar larval optic lobe (L72)
At this stage, the OOA and IOA each have divided into two distinct regions. In case of the IOA, only the medial part, rendered in light yellow, is still epithelial; the lateral part, in light green, has transformed into neuroblasts. For the OOA, the lateral part represents what is left of the epithelium (light magenta); the medial part (purple) is transformed into neuroblasts. The neuroblasts divide with a mitotic spindle oriented perpendicularly to the surface, thereby “budding” their daughter cells interiorly. These daughter cells (ganglion mother cells, first neurons, glial cells of the medulla. The are are rendered in dark blue.
The movie starts from an anterior view. During the first rotation, the medial and lateral domain of the IOA are visible; during the second rotation, the domains of the OOA are added. During the third turn, only the OOA is visible. During the fourth turn, the neurons/ganglion mother cells descending from the OOAepithelium are added to the OOA.
Movie S5 Animation of model of optic lobe at the transition between larva and pupa (P0). 1.Optic proliferation centers and neuronal cortices.
The two optic anlagen, both with an epithelial and a neuroblast domain rendered in the same colors as in the previous model, are still in existence. The difference to the L72 model is the increase in volume of the optic lobe neurons descended from the optic anlagen. These “squeeze” in between the IOA and OOA and push them aside.
The movie starts from an anterior view. During the first rotation, the IOA (light yellow: epithelial; light green: neuroblasts) is visible. During the second rotation, the OOA is added; the third rotation shows only OOA. During the fourth rotation, the neurons descending from the OOA are added. These comprise two groups: the neurons forming the medulla cortex (including all Mi neurons, Tm neurons, Tmy neurons) are located inward of the medial OOA. They are rendered in dark blue. The second group are the neurons of the lamina, budded off from the lateral OOA; they are rendered dark magenta. The fifth rotation adds the IOA (neuroblasts light green; epithelium light yellow); the sixth rotation then shows the neurons budded off from the lateral IOA. The lateral IOA gives off neurons to too sides. Neurons given off posteriorly, into the concavity of the C-shaped IOA, are the neurons forming the cortex of the lobula plate, including T4 and T5. They are rendered in dark green. Neurons given off towards laterally, and anteriorly (towards the convex side of the IOA) are the neurons that will form the posterior medulla cortex (T2/T3). They are rendered in turquois. Clusters of lobula neurons, possibly derived from the medial part of the IOA, are rendered in dark yellow.
Movie S6 Animation of model of optic lobe at the transition between larva and pupa (P0). 2.Neuronal cortices and neuropils.
The first rotation shows the cortices of the lamina (dark magenta) and medulla (dark blue). During the second rotation, the neuropils of the distal medulla and lamina are added (cyan and pink, respectively). These neuropils form thin sheaths underlying the cell body cortices, following their C-shaped curvature. Note that lamina cortex/neuropil is oriented perpendicular to the medulla. The third rotation shows the developing cortices of the lobula plate, proximal medulla (i.e., posterior medulla cortex) and lobula. To these, the corresponding neuropils, rendered in bright colors, are added during the fourth rotation. In the fifth rotation, only the inner neuropils (MEp, LP, LO) are visible; the sixth rotation shows all neuropils.
Movie S7 Combination of Models presented in S3, S4 and S6.
Movie S8-10 Animations of models of optic lobe at three pupal stages (S8: 12h after puparium formation; S9: 40h after puparium formation; S9: 72h after puparium formation). The sequence in which cortices and neuropils appear is the same described for the P0 model in S6: The first rotation shows the cortices of the lamina (dark magenta) and distal medulla (dark blue). During the second rotation, the neuropils of the distal medulla and lamina are added (cyan and pink, respectively). These neuropils form thin sheaths underlying the cell body cortices, following their C-shaped curvature. Note that lamina cortex/neuropil is oriented perpendicular to the medulla. The third rotation shows the developing cortices of the lobula plate, lobula, and the posterior medulla cortex. To these, the corresponding neuropils, rendered in bright colors, are added during the fourth rotation. In the fifth rotation, only the inner neuropils (MEp, LP, LO) are visible; the sixth rotation shows all neuropils.
Movie S11 Combination of Models presented in S6, S8-10.


