Abstract
In Drosophila, long-term memory (LTM) requires the cAMP-dependent transcription factor CREBB, expressed in the mushroom bodies (MB) and phosphorylated by PKA. To identify other kinases required for memory formation, we integrated Trojan exons encoding T2A-GAL4 into genes encoding putative kinases and selected for genes expressed in MB. These lines were screened for learning/memory deficits using UAS-RNAi knockdown based on an olfactory aversive conditioning assay. We identified a novel, conserved kinase, Meng-Po (MP, CG11221, SBK1 in human), the loss of which severely affects 3 hr memory and 24 hr LTM, but not learning. Remarkably, memory is lost upon removal of the MP protein in adult MB but restored upon its reintroduction. Overexpression of MP in MB significantly increases LTM in wild-type flies showing that MP is a limiting factor for LTM. We show that PKA phosphorylates MP and that both proteins synergize in a feedforward loop to control CREBB levels and LTM. key words: Drosophila, Mushroom bodies, SBK1, deGradFP, T2A-GAL4, MiMIC
Research organism: D. melanogaster
Introduction
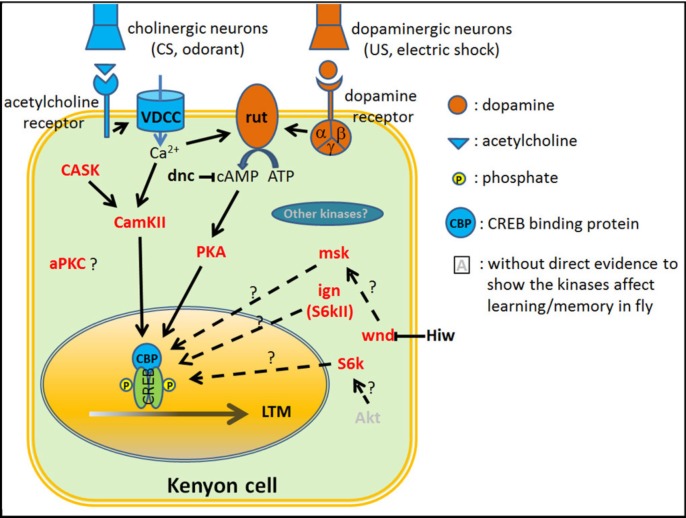
Forward genetic screens coupled with classical conditioning paradigms (Tully and Quinn, 1985) have been successful in identifying many molecular determinants of learning and memory in Drosophila (Guven-Ozkan and Davis, 2014). Among the genes identified are several components of the cAMP signaling pathway, including dunce (a cAMP specific phosphdiesterase), rutabaga (a cAMP specific adenylyl cyclase), Dc0 (Protein Kinase A, PKA) and CREBB (a transcription factor) (see Figure 1—figure supplement 1 for these and others) (McGuire et al., 2005). In adult flies, these genes act in cells of the mushroom bodies (MB), higher brain centers responsible for associating conditioned stimuli (CS) and unconditioned stimuli (US), and for storing these associations (Dubnau and Tully, 1998). Associations are stored by the MB Kenyon cells (KC), which are activated by CS—such as odors—via cholinergic transmission from olfactory projection neurons and US—such as electric shock—via dopamine signaling (McGuire et al., 2005). Acetylcholine receptors permit Ca2+ entry into KCs, while dopamine receptors activate a Ca2+/Calmodulin-responsive adenylyl cyclase (rutabaga), which acts as a coincidence detector (Busto et al., 2010). cAMP produced by Rutabaga activates PKA and triggers a downstream MAP kinase cascade, which leads to the phosphorylation and activation of the transcription factor CREBB (Impey et al., 1999). CREBB binds to cAMP response elements (CRE), activating the transcription of genes required for long term memory formation (DeZazzo and Tully, 1995).
CREBB is phosphorylated by several kinases, including PKA and CamKII, two kinases known to be involved in memory formation (Horiuchi et al., 2004; Mayr and Montminy, 2001) (Figure 1—figure supplement 1). Here we describe the identification of a novel gene Meng-Po (MP), that is required for memory formation but not for learning. The MP protein regulates the stability of CREBB together with PKA. In the absence of MP the protein stability of CREBB is affected, and removing a single copy of MP and PKA leads to a dramatic loss of CREBB and memory formation. In addition, overexpression of MP strongly promotes memory formation, indicating that MP is not only required but that it also plays an instructive role in memory formation.
Results
Genes encoding protein kinases expressed in MB in adult brain and behavioral consequences of RNA interference
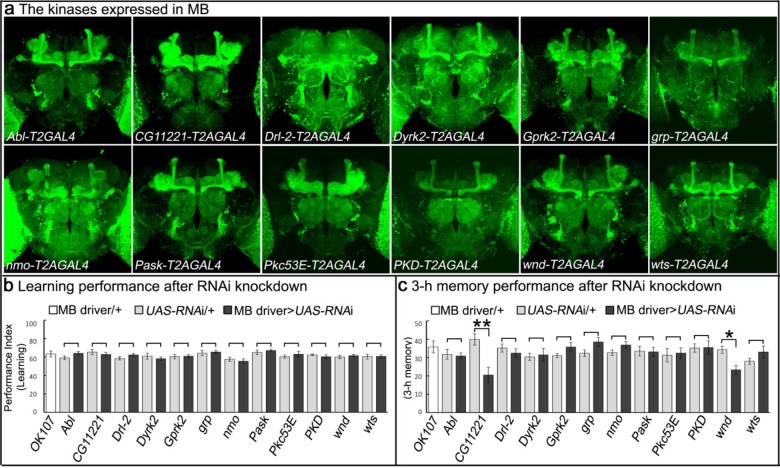
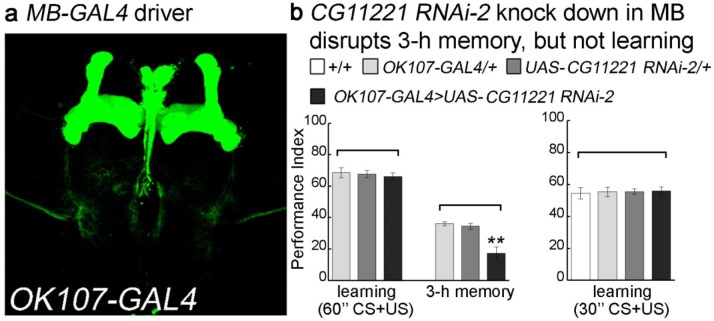
To identify novel protein kinases involved in learning/memory, we developed a strategy that allows us to determine which kinases are expressed in MBs. We selected 27 putative kinase-encoding genes for which fly lines were available that contained intronic insertions of the Minos Mediated Integration Casette (MiMIC) (Nagarkar-Jaiswal et al., 2015; Venken et al., 2011). These MiMICs contain a swappable cassette, allowing integration of any DNA using recombinase-mediated cassette exchange (RMCE). We replaced these cassettes with a Trojan exon encoding SA-T2A-GAL4 to permit the detection of cells expressing the kinase-encoding genes using UAS-mCD8::GFP (Diao et al., 2015). Of the 27 putative protein kinase genes screened, we found 12 that are expressed in MBs (Figure 1a; Figure 1—figure supplement 2). To determine if these 12 kinases expressed in MB play a role in learning and memory we knocked down their expression using UAS-RNAi expressed under the control of the MB-selective OK107-GAL4 driver (Figure 1—figure supplement 3a) and tested the performance of learning and 3 hr memory in knockdown flies upon an olfactory aversive conditioning assay (Figure 1; Figure 1—figure supplement 3b). In Drosophila, Pavlovian olfactory aversive learning requires coincidence detection of a conditioned stimulus (CS), an odor, and an unconditioned stimulus (US), an electric shock. Through a single training session using a T-maze assay in which flies are exposed to 12 CS-US pairings in one min, flies can associate CS with US and learn to avoid the odor paired with electric shock (Tully and Quinn, 1985). After training, flies can learn (tested immediately) and form an intermediate-term/3 hr memory (tested after 3 hr) (Margulies et al., 2005). We identified two putative kinases, CG11221 and wallenda (wnd), which when knocked down, cause significant reductions in performance index for 3 hr memory, but not for learning (Figure 1b–c). Overexpression of wnd has been documented to enhance memory in Drosophila (Huang et al., 2012), but CG11221 has not been previously characterized in flies. We therefore tested the effects of knocking down CG11221 expression using a second RNAi expressed specifically in MB neurons to confirm the memory loss phenotype (Figure 1—figure supplement 3b). To provide additional evidence that CG11221 is not required for learning, we used a short CS/US association protocol (30’ CS + US instead of 60’) to train flies. Knockdown of MP did not affect learning, further indicating that MP is not required for learning (Figure 1—figure supplement 3b).
Figure 1. Genes encoding kinases expressed in MB of adult brains and behavioral consequences of RNA interference.
(a) Expression patterns of genes encoding protein kinases in adult brains. The 12 protein kinases shown here are expressed in MB. (b–c) Screening results of behavioral assays by RNAi knockdown. The MB-GAL4 driver, OK107 was used to drive the UAS-RNAi. Flies were raised at 18°C until eclosion, transferred to 25°C for 3 days and behavioral assays were performed to test (b) learning and (c) 3 hr memory. The mean ±SEM is plotted for each genotype; n = 8 for each group. *p<0.05, **p<0.01.
Figure 1—figure supplement 1. Overview of the proteins required for olfactory aversive learning/memory formation in Drosophila.
Figure 1—figure supplement 2. The T2A-GAL4 expression patterns of protein kinases that are not expressed in MB.
Figure 1—figure supplement 3. Reducing the levels of CG11221 in MB affects 3 hr memory formation.
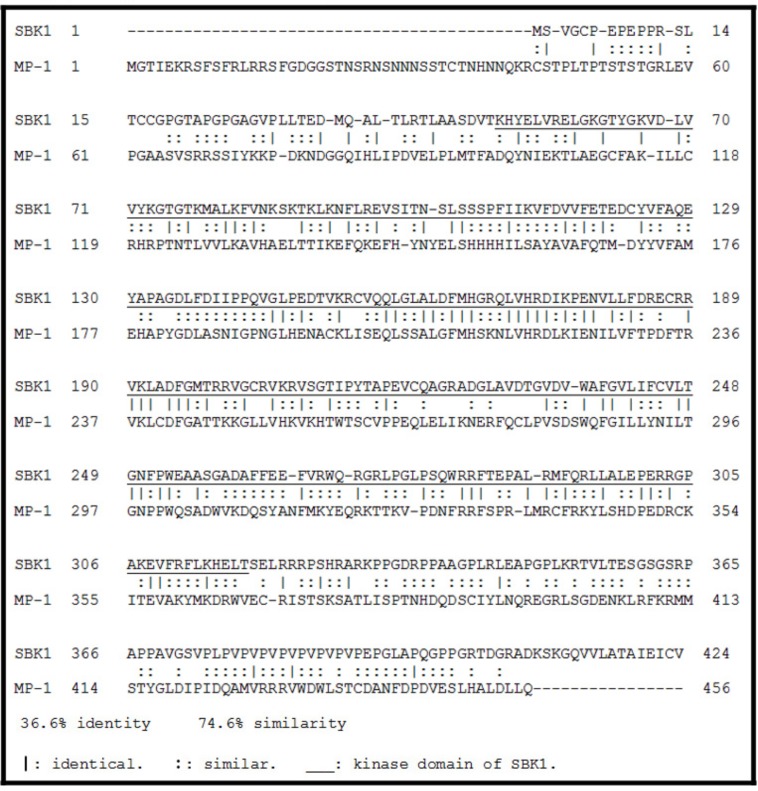
Figure 1—figure supplement 4. Comparison of protein sequence between human SBK1 and fly MP (CG11221).
Figure 1—figure supplement 5. MP is expressed ubiquitously.
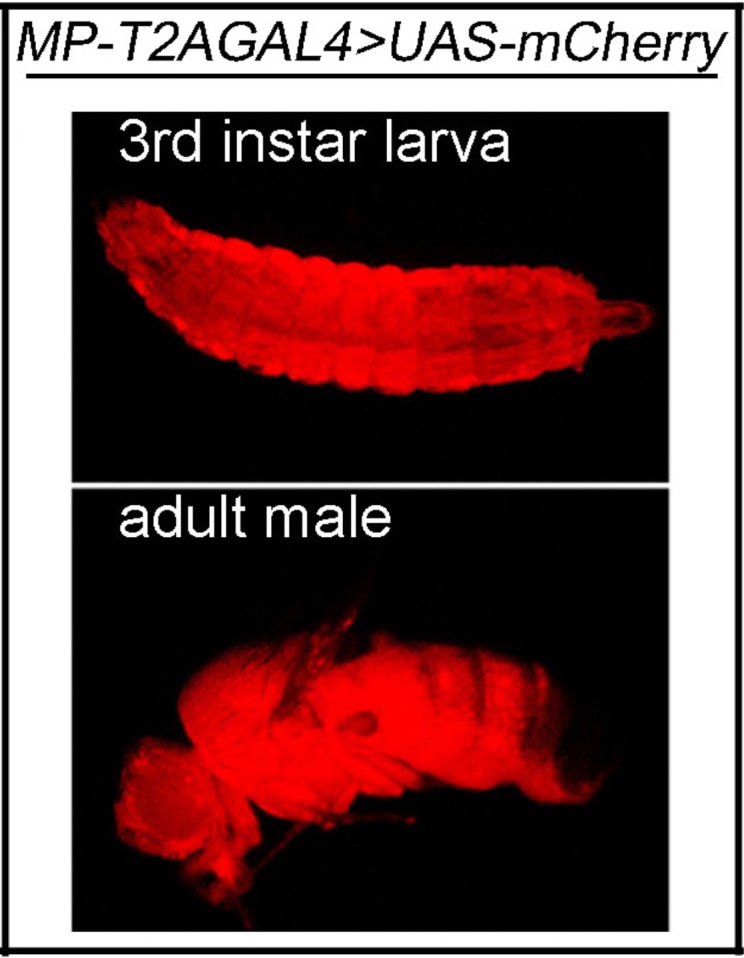
CG11221 is an evolutionarily conserved serine/threonine protein kinase (human SBK1: 37% identity and 75% similarity, Figure 1—figure supplement 4). SBK1 is expressed in the hippocampus, cortex, and cerebellum of adult rodents and loss of SBK1 in mice causes partial embryonic lethality (Nara et al., 2001; Skarnes et al., 2011). The CG11221MI03008-T2A-GAL4; UAS-mCherry flies exhibit broad expression of the gene in third instar larvae and adult flies (Figure 1—figure supplement 5). As shown in Figure 1a, the gene is also expressed in the adult brain and is prominent in the MB. In light of its importance in memory, we renamed CG11221, Meng-Po (MP), for the Lady of Forgetfulness, a character in Chinese mythology who ensures that people are ready for reincarnation by providing the ‘Tea of Forgetfulness’ so they lose the memory associated with their former life.
Loss of MP causes a loss of memory
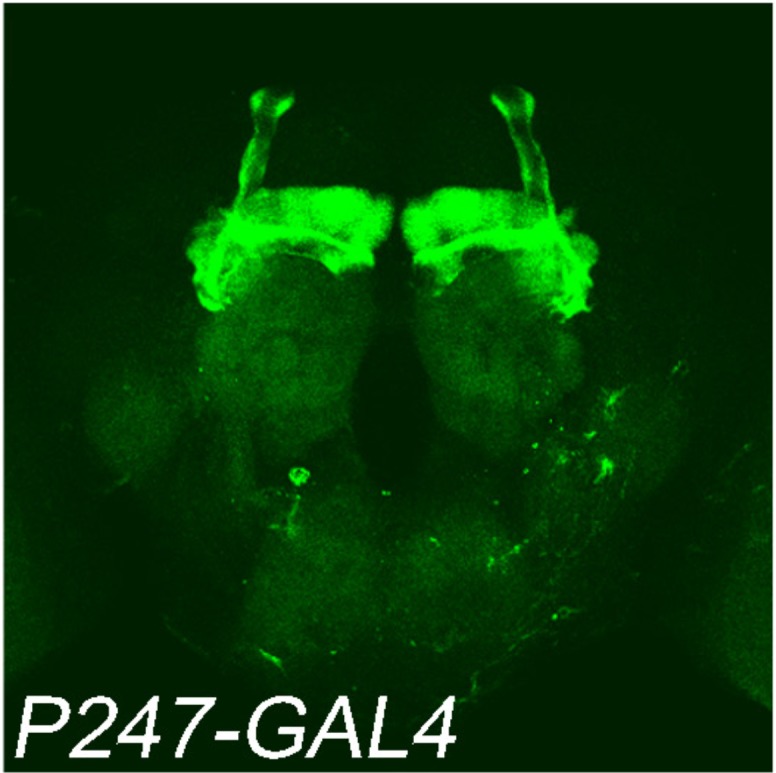
To determine whether the memory deficits resulting from loss of MP are due to its activity in adult MB neurons or are a result of its developmental expression, we used the deGradFP method (Caussinus et al., 2011; Nagarkar-Jaiswal et al., 2015) to selectively decrease levels of MP protein in MB of 5 day old flies. To do so, we replaced the MiMIC insertion of the CG1122103008 in the first coding intron with a SA-GFP-SD in-frame with the MP coding sequence using a previously described technique (Nagarkar-Jaiswal et al., 2015). This manipulation insures the expression of an internally-tagged MP-GFP-MP (MP-GFP) fusion protein. Flies homozygous for the modified MP-GFP gene were viable and anti-GFP staining of homozygous MP-GFP flies revealed expression in MB (Figure 2a). Furthermore, these flies did not exhibit obvious viability or learning/memory defects (Figure 2—figure supplement 1), indicating that the MP-GFP fusion protein retains its function. Crossing MP-GFP flies to flies that express UAS-deGradFP under the control of the MB-specific GAL4 driver P247 (Figure 2—figure supplement 2) resulted in progeny in which the MP-GFP fusion protein was substantially depleted from MB when flies were raised at 28°C, but not at 18°C. By shifting flies between these temperatures we can reversibly eliminate the tagged protein from MB neurons. Hence, the protein is present during development and early adulthood (Figure 2a,a’), avoiding developmental requirements. The temperature shift leads to a loss of MP in 5 day old flies (Figure 2a,b’). Upon a two day recovery period at 18°C, the protein levels are restored (Figure 2a,c’). In summary, the deGradFP technique with the MP-GFP flies provides a precise tool for manipulating and monitoring MP levels in specific tissues in adult flies in a reversible manner.
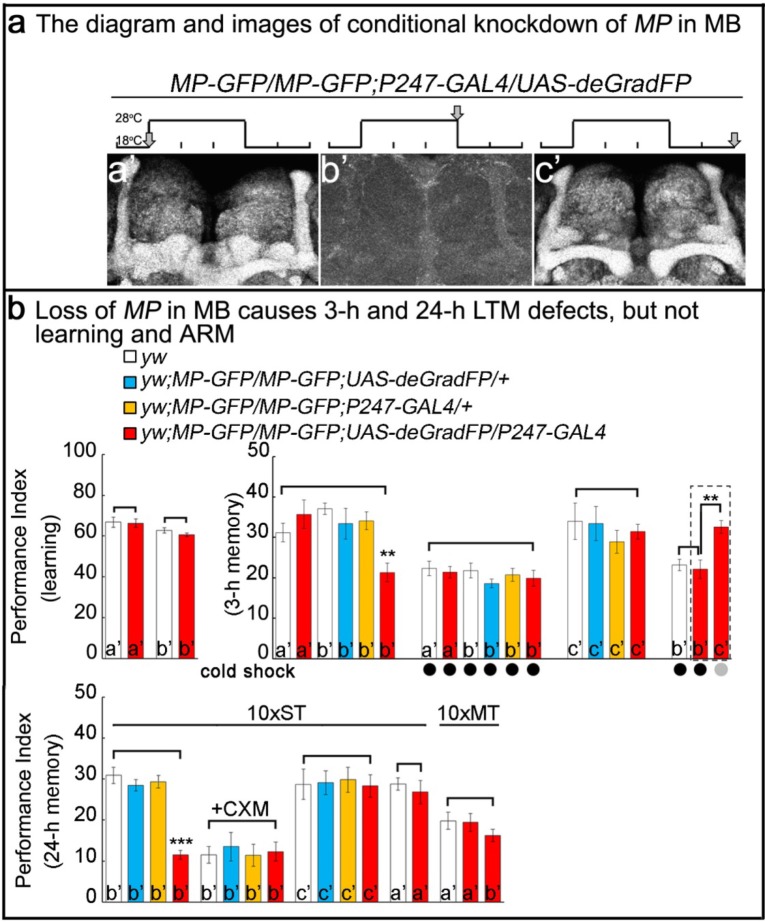
Figure 2. Loss of MP causes a loss of memory.
(a) UAS-deGradFP driven by MB driver, P247-GAL4. a'-c’: scheme for temporal control of deGradFP expression via temperature shift. (a’) 18°C; (b’) 28°C for three days; (c’) 28°C for three days, followed by a shift to 18°C for two days. MP-GFP-MP expression at the time points shown above by arrows. (b) Performance scores of learning, 3 hr memory and 24 hr memory. (a’), (b’) and (c’) are referred as the time points in (a). (b), learning) Learning is normal after knockdown of MP in MB by deGradFP at 28°C for 3 days (b’). Flies raised at 18°C are used as a control (a’). (b, 3 hr memory) 3 hr memory is impaired after knockdown of MP in MB (b’). However, the performance score of ARM is intact in MP knockdown flies which are treated with a cold shock and compared to flies raised at 18°C (3 hr memory a’ and b’+cold shock). In c’ condition, the flies show normal performance score of 3 hr memory. The 3 hr memory impairment (in b’+cold shock, right panel) can be fully rescued when the animals are shifted to 18°C for two days (the groups boxed in dashed line are the exactly same flies). (c), 24 hr memory) 24 hr LTM (10 x ST) is impaired upon knockdown of MP in MB (b’, red). After treatment with 35 mM cycloheximide (CXM), the MP knockdown flies (b’+CXM, red) don’t exhibit a performance that is worse than control flies. In the c’ conditions, the flies exhibit a normal performance score for the 24 hr memory assay. The performance of ARM (10 x MT) is intact. 10 x ST:10 times spaced training. 10 x MT:10 times massed training. The mean ±SEM is plotted for each genotype; n = 8 for each group. **p<0.01. ***p<0.001.
Figure 2—figure supplement 1. MP-GFP-MP animals have proper learning and memory.
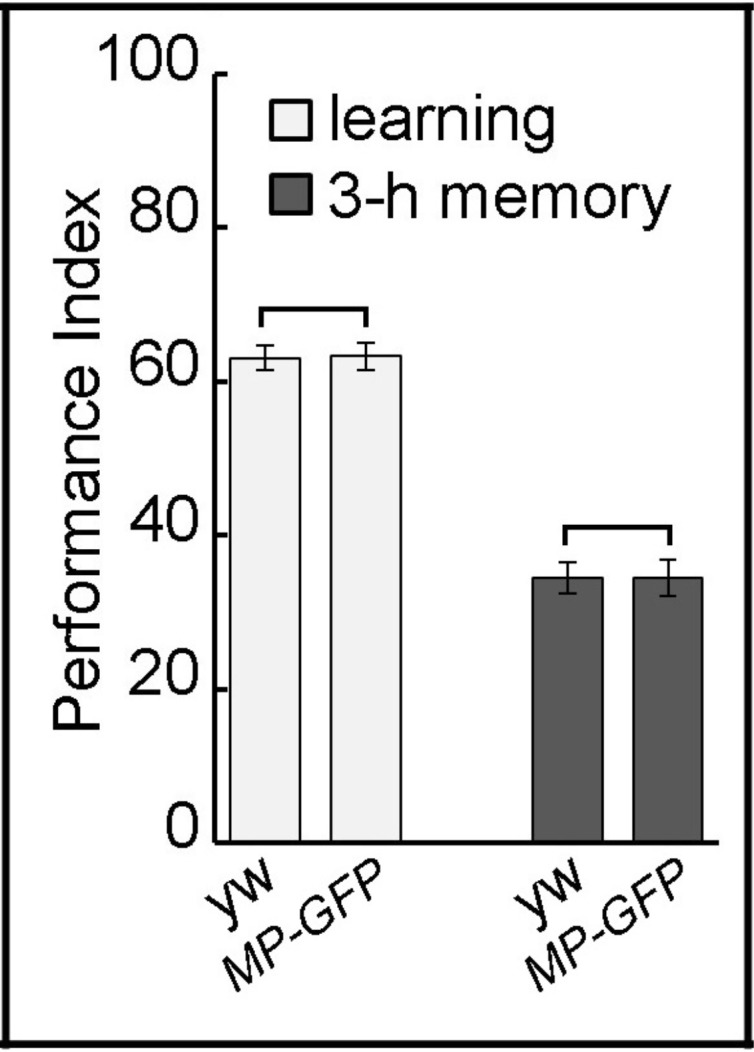
Figure 2—figure supplement 2. P247-GAL4 was used for protein knockdown.
Using this strategy, we reduced the level of MP-GFP in adult MB for three days (Figure 2a,b’) and assayed learning and memory performance in these flies. As shown in Figure 2b, flies with MP depleted in the MB (b’, red column) learn as well as y w control animals (b’, white). However, when tested 3 hr after training, these flies exhibited severe memory deficits (Figure 2b, 3-h memory, middle panel, b’, red column) when compared with either y w, other controls or animals of the same genotype tested prior to depletion of MP (Figure 2b, 3-h memory, middle panel). These results are consistent with those obtained by constitutive knockdown of MP expression using RNAi and demonstrate that MP is required in MB neurons specifically in adults (Figure 1c; Figure 1—figure supplement 3b).
It has been previously shown that the 3 hr memory has two distinct components: an anesthesia-resistant memory (ARM) and an anesthesia-sensitive memory (ASM), only the latter of which can be erased by cold-shock (Lee et al., 2011; McGuire et al., 2005). To determine whether MP functions in ARM, ASM or in both, we subjected trained animals in which MP-GFP had been depleted in the MB to cold-shock and found that the residual memory was unaffected (Figure 2b, 3-h memory, b’ with cold shock). Furthermore, the memory performance in these animals was statistically indistinguishable from that of control animals subjected to cold shock. These results suggest that MP is required for ASM, but not for ARM. Importantly, loss of 3 hr memory can be restored by placing the flies at 18°C for two days after knockdown (Figure 2b, 3-h memory, c’). To test whether the transient loss of MP in the MB has long-term consequences to the animal’s ability to form memories, we restored expression of MP-GFP after 3 days of depletion at 28°C by returning animals to 18°C. Their ability to learn and remember upon restoration of MP-GFP revealed complete recovery of memory function (Figure 2b, 3-h memory, right panel, column c’).
In parallel studies we also tested animals with reduced MP for deficits in 24 hr long-term memory, LTM, and radish dependent 24 hr ARM (Margulies et al., 2005). LTM requires CREBB as well as new protein synthesis (Fropf et al., 2013; Keene and Waddell, 2007) and is typically induced by repetitive (10x) spaced training (ST) at 15 min intervals. In contrast, 24 hr ARM does not require protein synthesis (Dubnau and Tully, 1998; Keene and Waddell, 2007) and is induced by repetitive mass training (MT) without the 15 min resting intervals. We found that MP is dispensable for 24 hr ARM (Figure 2b, 24-h memory, 10x MT), but is required for LTM (Figure 2b, 24-h memory, 10x ST, b’, red column). After treatment with 35 mM cycloheximide (CXM), an inhibitor for protein synthesis, the MP knockdown flies (Figure 2b, 24-h memory, 10x ST, b’+CXM, red column) do not perform worse than other control flies. This indicates that the loss of memory is protein synthesis-dependent. Loss of 24 hr LTM memory can be restored when returning the flies to 18°C for two days after knockdown. (Figure 2b, 24-h memory, 10x ST, c’). Hence, loss of MP affects neither learning nor ARM, but severely impairs 3 hr ASM and 24 hr LTM.
Overexpressing MP enhances 24 hr LTM and increases CREBB activity
To determine whether MP is not only necessary for LTM, but also acts as a limiting factor for LTM, we overexpressed MP in MB and analyzed 24 hr LTM. To avoid saturating LTM, as occurs when the spaced training is repeated 10x, we adopted a paradigm in which flies were subjected to only a 3X training paradigm (Lee et al., 2011) (3x ST). Conditional overexpression of MP in MB under the control of OK107-GAL4 was accomplished using a temperature-sensitive GAL4 inhibitor (Tub-GAL80ts), which blocks UAS-MP expression at 18°C (Figure 3a,a’–b’), but not 29°C (McGuire et al., 2004). Flies grown at 18°C and placed at 29°C for two days robustly expressed MP as detected by Western blot (Figure 3a,c’).
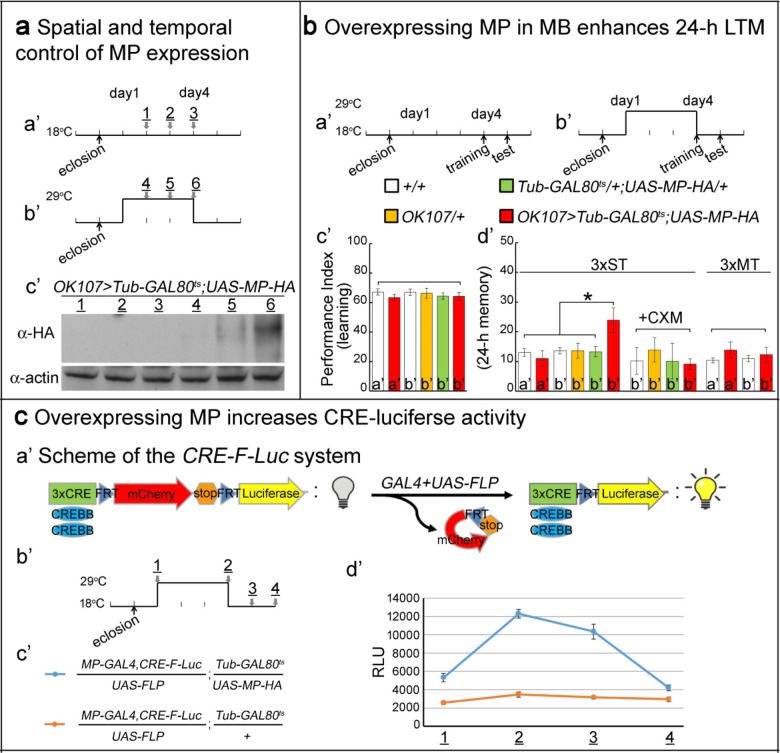
Figure 3. Overexpressing MP enhances 24 hr LTM and increases CREBB activity.
(a) Spatial and temporal control of MP expression was achieved with a MB-GAL4 driver and Tub-GAL80ts. Schemes are shown in (a’) and (b’). MP expression was assessed by Western blot with α-HA antibody in (c’). (b) Temporal overexpression of MP in MB enhances 24 hr LTM. Diagrams for MP overexpression, training and testing of behavior are shown in (a’) and (b’). The learning scores are normal after MP overexpression in (c’) (red column, b’). 24 hr LTM is significantly enhanced after 3x ST training in (d’) (red column, (b’). However, memory enhancement can be erased by feeding flies with cycloheximide. The 24 hr ARM is unaltered after MP overexpression in (d’) (red column, (b’), a’=no overexpression, b’=overexpression, and 3x ST (spaced training) or 3x MT (massed training). The mean ±SEM is plotted for each genotype; n = 8–12 for each group with 200 animals per group. *p<0.05. (c) Overexpressing MP increases CRE-luciferase activity. The CRE-F-Luc system is shown in (a’). The diagram is shown in (b’). The fly genotypes are shown in (c’). blue curve: MP-GAL4,CRE-F-Luc/UAS-FLP;Tub-GAL80ts/UAS-MP-HA. orange curve: MP-GAL4,CRE-F-Luc/UAS-FLP;Tub-GAL80ts/+. In (d’), the luciferase activity of the two genotypes of flies at the following time points: 1, 2, 3 and 4 which are shown in (b’). (1) at 18°C for one day after eclosion. (2) at 29°C for three days. (3) at 29°C for three days, then shift to 18°C for one day. (4) at 29°C for three days then shift to 18°C for two days. 10 fly heads (five males/five females) were collected for a single assay. RLU: relative luminescence unit. The mean ±SEM is plotted for each point; n = 6.
Flies were subjected to temperature protocols that either did (Figure 3b,b’) or did not (Figure 3b,a’) induce MP overexpression and we then tested their performance in both learning and LTM using the T-maze assay. The learning scores of Tub-GAL80ts/+;UAS-MP-HA/+;OK107-GAL4/+ flies temperature-shifted to 29°C, and therefore overexpressing MP, were the same as those of unshifted flies of the same genotype and those of other control groups (Figure 3b,c’). In contrast, the performance scores of these flies for 24 hr LTM were significantly enhanced after MP overexpression in MB (3x ST; Figure 3b,d’). The observed memory enhancement can be erased by feeding flies 35 mM CXM, indicating that enhanced memory is protein synthesis-dependent (3x ST +CXM; Figure 3b,d’). Moreover, this enhancement is specific to 24 hr LTM, and not 24 hr ARM induced by 3-times mass training (3x MT; Figure 3b,d’). Thus, MP is not only necessary for LTM, but also is also promoting LTM formation.
Given that CREBB is a central player in LTM formation, we tested its activity in MB during MP overexpression using a previously described luciferase assay. This assay relies on CREBB-mediated transcription of luciferase from the CRE-F-Luc construct, which contains three copies of the CREBB-binding cAMP Response Element (CRE), upstream of an FRT-flanked mCherry-encoding stop cassette and the firefly luciferase gene (Figure 3c,a’) (Tanenhaus et al., 2012). Spatial control of luciferase expression can be achieved by using the GAL4/UAS system to drive the cell-type specific expression of a UAS-FLP recombinase (Figure 3c,a’–b’). We created flies of the following genotype: MP-T2A-GAL4,CRE-F-Luc/UAS-FLP;Tub-GAL80ts/UAS-MP-HA (Figure 3c,c’). In these flies, GAL4 drives expression of UAS-MP-HA and UAS-FLP when the flies are shifted to 29°C. FLP removes the stop cassette in the CRE-F-Luc construct, allowing translation of luciferase in the MP-expressing neurons (Figure 3c,a’–b’). When assayed for luciferase activity, flies placed at 29°C for 3 days display a very significant increase—measured in relative luminescence units (RLU)—when compared to the flies kept at 18°C. The RLU gradually decrease to base line upon return to 18°C (Figure 3c,d’, blue). In contrast, the luciferase activity of control flies that do not carry the UAS-MP (genotype: MP-T2A-GAL4,CRE-F-Luc/UAS-FLP;Tub-GAL80ts/+) display no change in RLU when subjected to the same temperature shifts (Figure 3c,c’–d’, orange). In summary, overexpression of MP in MB significantly enhances 24 hr LTM and increases CRE-luciferase activity. Our results are consistent with a model in which MP facilitates LTM by positively regulating CREBB activity (Dubnau and Tully, 1998).
MP is a kinase, and loss of MP affects CREBB protein levels in MP gene trap animals
Given that MP encodes a putative kinase, we tested its ability to phosphorylate a known substrate of two other kinases, ERK and PKA. We expressed MP-HA in S2 cells, affinity purified the protein, and performed kinase assays on Myelin Basic Protein (MBP) (Martenson et al., 1983). As shown in Figure 4a, ERK, PKA and MP phosphorylate MBP. Given the CRE-luciferase assay results and the established regulation of CREBB by phosphorylation (Horiuchi et al., 2004; Tully et al., 2003), we hypothesized that CREBB is a substrate of MP. To determine if MP can phosphorylate CREBB we purified the CREBB protein from S2 cells and performed kinase assays. Unlike PKA, which is able to phosphorylate CREBB, MP was not able to phosphorylate CREBB (Figure 4b). Hence, although CREBB activity is dependent on MP levels, CREBB may not be a direct substrate of MP.
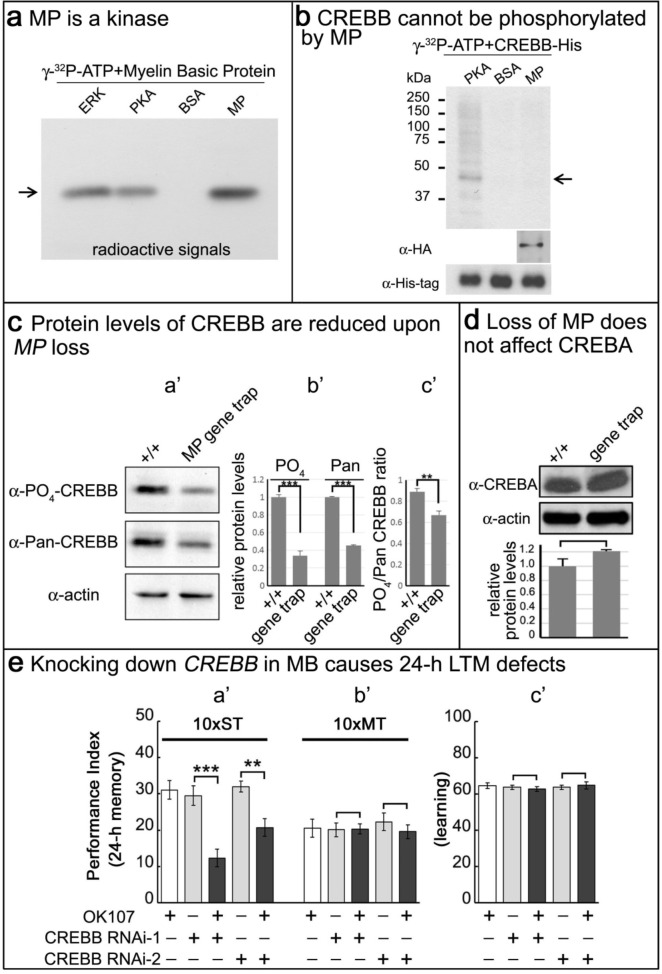
Figure 4. MP is a kinase, and loss of MP affects CREBB protein levels in MP gene trap animals.
(a) MP can phosphorylate myelin basic protein similar to ERK and PKA. Arrow points to MBP. (b) CREBB is not phosphorylated by MP kinase. As a positive control, we used PKA to phosphorylate the CREBB-His tag protein. MP failed to phosphorylate CREBB under these conditions. BSA was used as a negative control. Arrow points to CREBB. (c) In (a’–b’), protein levels of phospho-CREBB and total CREBB in the absence of MP are decreased, and also the phosphorylation ratio of phospho-CREBB/total CREBB in (c’). Total CREBB was detected by anti-Pan-CREBB, phosphorylated CREBB was detected by anti-PO4-CREBB (**p<0.01. ***p<0.001). (d) Protein levels of CREBA are not affected in MP gene-trap flies. CREBA protein was detected by anti-CREBA. The protein levels were normalized to actin. (Student’s t test, n = 3). (e) Knocking down CREBB in MB with CREBB RNAi causes 24 hr LTM defects (10x ST, a’), but not ARM (10x MT, b’) and learning in (c’). 10x ST = 10 times spaced training. 10x MT = 10 times massed training. The mean ±SEM is plotted for each genotype; n = 8 for each group. **p<0.01. ***p<0.001.
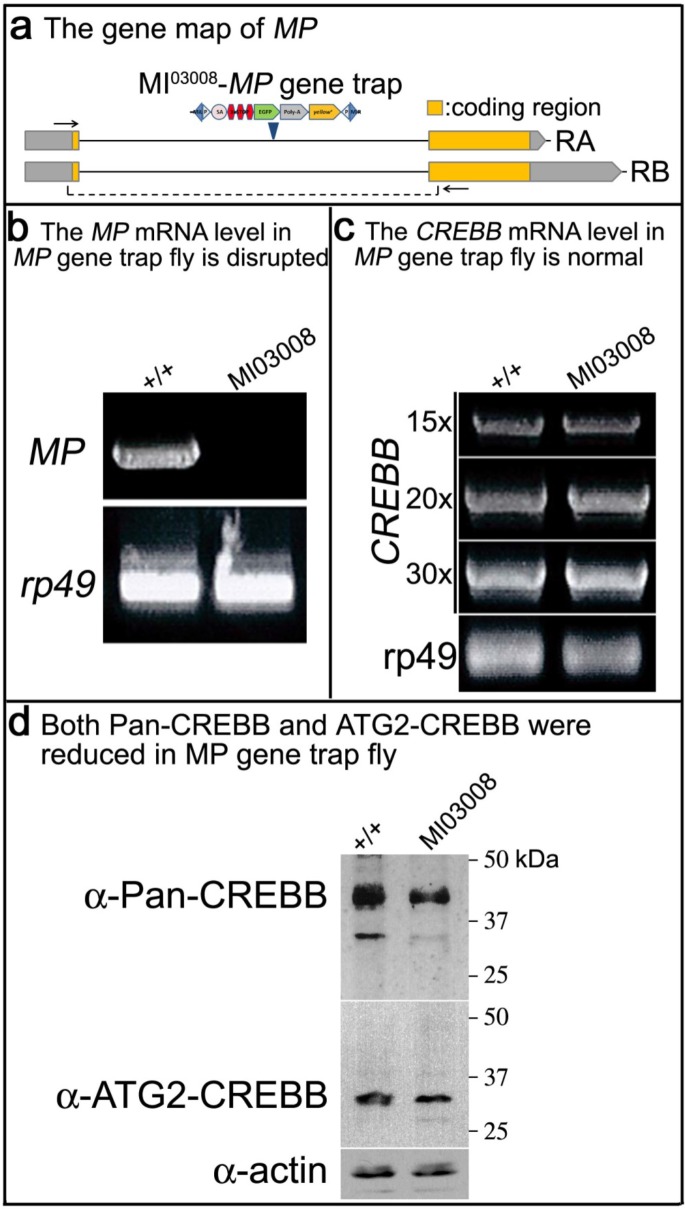
Figure 4—figure supplement 1. CREBB mRNA level is not affected in MP gene trap fly, but CREBB proteins are reduced.
To more generally assess how MP upregulates CREBB, we sought to determine how CREBB activity is affected by loss of MP function. The MI03008 MiMIC insertion in the MP gene functions as a gene trap (Figure 4—figure supplement 1a) and RT-PCR fails to amplify a product between these exons (Figure 4—figure supplement 1b). This and other evidence suggest that MI03008 is a severe loss of function allele of MP. To estimate the levels of CREBB in MI03008 mutants, we probed Western blots of protein isolated from the heads of adult MI03008/MI03008 animals using an anti-phospho-CREBB antibody (Fropf et al., 2013) and compared the signal to that observed in Western blots from heads of control animals. In parallel, we analyzed the signals obtained using a second antibody (Pan-CREBB) that assesses total CREBB protein levels. Both antibodies revealed an approximately 60% reduction in immunoreactivity in samples from MP mutant brains (Figure 4c,a’–b’). Note, that there is a minor reduction in the ratio of phospho-CREBB/total CREBB (Figure 4c,c’). In contrast, CREBA protein levels are not altered (Figure 4d), consistent with the observation that CREBA has no role in memory formation (Abrams and Andrew, 2005). Finally, CREBB mRNA levels are unchanged in MP mutants (Figure 4—figure supplement 1c), indicating that the observed reduction in CREBB protein is a post-transcriptional effect.
We also tested if ATG2-CREBB is affected. This CREBB isoform is encoded by a downstream, in-frame initiation codon of one of the transcripts of the CREBB gene, and corresponds to a transcriptional activator. It has been shown to be required for memory enhancement (Tubon et al., 2013). As shown in Figure 4—figure supplement 1d, we find that both the anti-Pan-CREBB antibody and the antibody recognizing ATG2-CREBB (Fropf et al., 2013) identify a ~30 kDa band that is reduced in MI03008 mutant brains. Hence, these data suggest that MP promotes the translation or stabilization of CREBB protein.
If CREBB mediates the effects of MP on LTM and loss of MP function reduces CREBB protein levels, then reducing CREBB in MB should phenocopy the effects of eliminating MP in MBs. To determine whether this is the case, we knocked down CREBB mRNA in adult MB using OK107-GAL4 and two independent UAS-RNAi lines by shifting 3–5 day old flies kept at 18°C to 25°C three days prior to testing. We find that decreasing CREBB in MB causes a reduction of 24 hr LTM with both RNAi knockdowns (10 x ST; Figure 4e,a’), but it does not affect learning (Figure 4e,c’) or ARM (10 x MT; Figure 4e,b’). Comparison of these results with the knockdown of MP in MBs (Figure 2) indicates that loss of MP or CREBB function results in very similar memory defects. The most parsimonious explanation of these data is that loss of the MP protein kinase leads to reduced levels of CREBB protein, which in turn leads to a severe LTM impairment.
MP is regulated by PKA
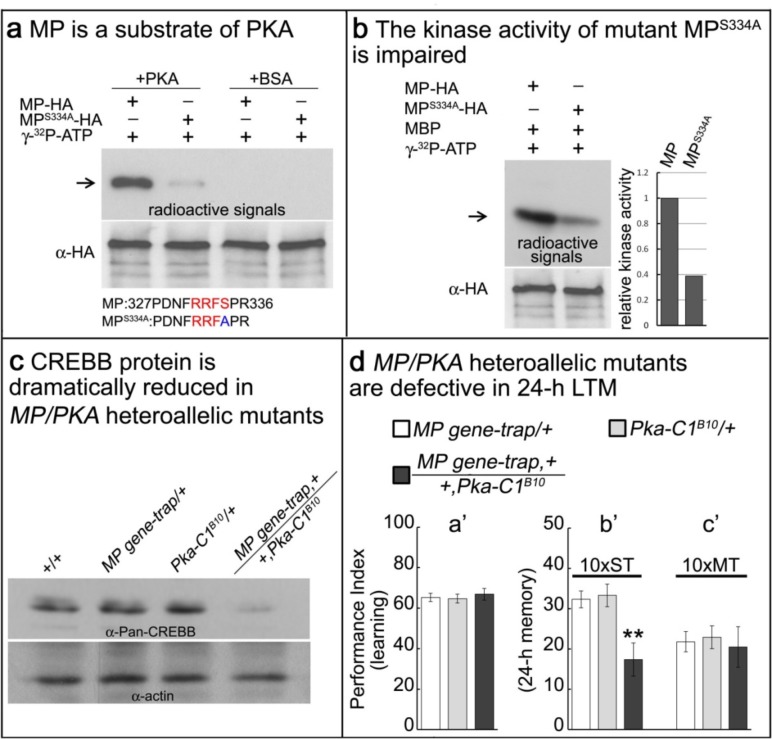
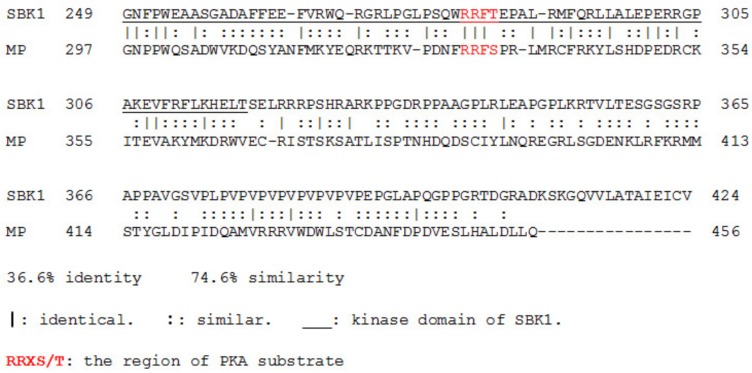
The MP protein contains the canonical PKA phosphorylation motif RRFS (Huang et al., 2005) at residues 331–334 (Figure 5—figure supplement 1). To determine whether serine residue 334 is a substrate of PKA we introduced a S334A mutation into MP (MPS334A) and examined phosphorylation of both the wild-type and mutant proteins by PKA. As shown in Figure 5a, MP is phosphorylated by PKA, but MPS334A is barely phosphorylated. We conclude that S334 is required for proper MP phosphorylation by PKA. To determine whether phosphorylation alters the MP kinase activity, we next assayed the kinase activities of both wild-type MP and MPS334A. As shown in Figure 5b, the kinase activity of MPS334A is significantly lower than that of wild-type MP.
Figure 5. MP kinase is regulated by PKA.
(a) The potential PKA phosphorylation site of MP was altered to S334A. MP can be phosphorylated by PKA, but MPS334A is severely impaired. Arrow points to wild-type and mutant MP. (b) MPS334A has severely reduced kinase activity compared to wild-type MP. Arrow points to MBP. (c) CREBB protein levels are dramatically reduced in MP gene-trap +/+Pka-C1B10 heteroallelic flies. CREBB protein levels are probed with anti-Pan-CREBB antibody. (d) MP gene-trap +/+Pka-C1B10 heteroallelic flies have intact learning in (a’) and ARM in (c’), but show 24 hr LTM impairment in (b’). The mean ±SEM is plotted for each genotype; n = 8 for each group. **p<0.01.
Figure 5—figure supplement 1. Human SBK1 and fly MP (CG11221) both contain a PKA phosphorylation site.
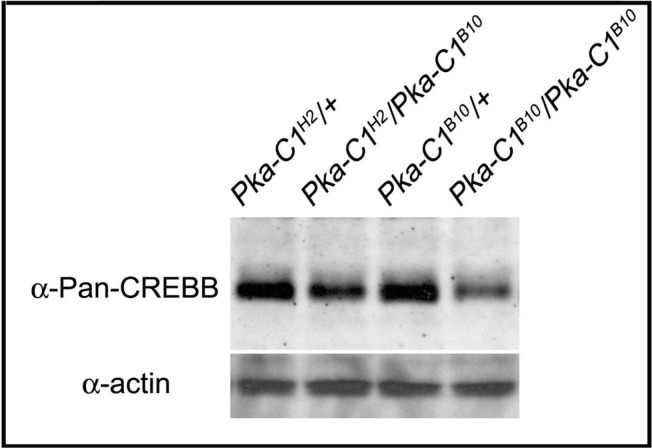
Figure 5—figure supplement 2. Protein levels of CREBB are reduced in Pka-C1 mutants.
The above data indicate that PKA activates MP in addition to activating CREBB. Loss of function mutations in PKA cause a subtle but significant reduction in CREBB (Figure 5—figure supplement 2). Hence, PKA and MP may work together in a coherent feedforward loop (Mangan et al., 2003) to upregulate CREBB activity and support LTM formation. To assess possible synergistic interactions between PKA and MP, we simultaneously reduced the protein levels of both MP and the catalytic subunit of PKA by creating MI03008,Pka+/MP+,Pka-C1B10 heteroallelic flies, and assessed the levels of CREBB protein by Western blot (Fropf et al., 2013). CREBB immunoreactivity in the heads of these animals is very severely reduced compared to that observed in the heads of either wild-type contols or animals with reduced gene dosage of only MP or Pka-C1 (Figure 5c). Simultaneous reduction of PKA and MP thus substantially potentiates the effects of reducing either protein alone, consistent with a model in which the two kinases act within the same signaling pathway to regulate CREBB activity and LTM. To determine if the MI03008,Pka+/MP+,Pka-C1B10 animals exhibit memory defects, we assayed their ability to both learn and form 24 hr LTM. We find that these flies exhibit normal learning (Figure 5d,a’) and ARM (10 x MT; Figure 5d,c’), but lack 24 hr LTM (10 x ST; Figure 5d,b’) consistent with the loss of CREBB function. Interestingly, flies heterozygous for either MP or Pka-C1 display neither learning nor LTM defect, suggesting a potent synergistic interaction and feedforward loop between PKA and MP.
Discussion
Using MiMIC technology, we converted 27 genes encoding putative protein kinases with the Trojan T2A-GAL4 exon and performed an image screen for genes expressed in MBs (Diao et al., 2015). This tagging approach is especially useful for genes that are expressed at low levels in the CNS. By tagging the proteins with GFP, a conditional and reversible knockdown can be achieved in almost any tissue or cell (Nagarkar-Jaiswal et al., 2015). This allowed us to identify a novel serine/threonine protein kinase, Meng-Po (MP), that is a critical player in LTM formation in Drosophila. MP is a homologue of SBK1 in mammals (Figure 1—figure supplement 4), a gene that is expressed in the hippocampus and the cortex (Nara et al., 2001; Skarnes et al., 2011). Loss of this gene in mice is associated with embryonic lethality (Skarnes et al., 2011), whereas in flies, loss of MP leads to a reduction in viability as well as sterility.
Our data show that CREBB stability is highly susceptible to loss of MP. CREBB activity is modulated by phosphorylation via PKA and CamKII in Drosophila (Horiuchi et al., 2004). Although our findings indicate that MP kinase activity is critical for maintaining CREBB levels and that MP kinase activity acts in synergy with PKA (Figure 6), we have not been able to demonstrate that CREBB is a direct target of MP. However, some kinases require a previously phosphorylated residue as part of their recognition sequence and we have not mixed various kinases with MP in our in vitro assays (Horiuchi et al., 2004). Hence, it remains to be established how CREBB is degraded in the absence of MP.
Figure 6. Model.
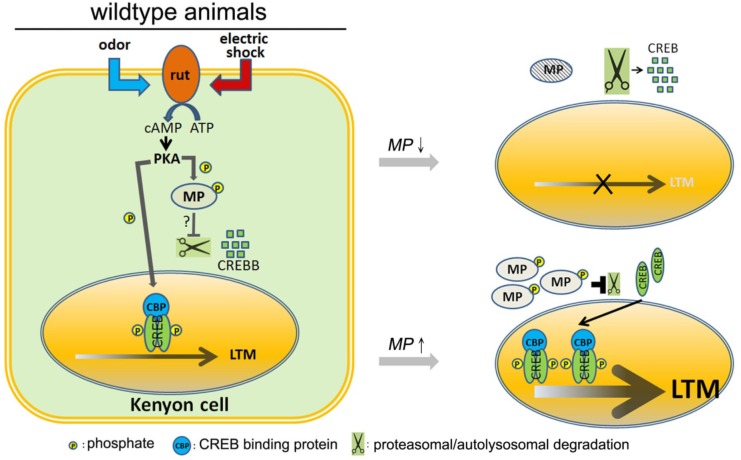
In wild-type animals, rutabaga (rut) as a coincidence detector can receive the odor and electric shock signals then activate cAMP-PKA signaling. MP is phosphorylated by PKA which maintains the CREBB levels, possibly by inhibiting proteasomal or autolysosomal degradation. It permits CREB-dependent LTM to form. Knocking down MP will unlock this inhibition and facilitate CREBB degradation thereby disrupting LTM. In contrast, overexpression of MP promotes LTM formation.
A reduction in CREB levels has been shown to be associated with an age-dependent memory loss in rodents. Interestingly, delivery of CREB protein in the hippocampus using somatic cell transfer attenuated LTM impairement (Mouravlev et al., 2006). However, no gene has so far been shown to affect CREBB stability in vivo and our findings that MP, together with PKA, synergize to dramatically affect CREBB levels via a feedforward loop (Mangan et al., 2003), reveal another mechanism to control CREBB levels during memory formation (Figure 6). This model is supported by the observation that overexpression of MP increases CREBB activity and promotes memory formation, suggesting that it is a central player in LTM.
Materials and methods
Fly strains
Fly strains were maintained on standard cornmeal-yeast-agar medium at 25°C, at 60–70% relative humidity and on a 12/12 hr light/dark cycle. The MiMICs were created in the Bellen lab (Diao et al., 2015; Nagarkar-Jaiswal et al., 2015; Venken et al., 2011) and P247-GAL4 (Zars, 2000), OK107-GAL4 (Pascual and Préat, 2001), Tub-GAL80ts, Pka-C1H2, PKa-C1B10, UAS-FLP, 20xUAS-6xmCherry and UAS-mCD8::GFP (Pfeiffer et al., 2010) were obtained from the Bloomington Drosophila Stock Center (USA). The Canton-S w1118 (iso1CJ) wild-type fly (Lee et al., 2011) was from Josh Dubnau. The CRE-F-Luc fly was from Jerry Yin (Tanenhaus et al., 2012). UAS-RNAi flies were from BDSC or VDRC. The stock information is listed on Supplementary file 1.
Plasmid constructs
The MP-HA and CREBB-6xHis cDNA were synthesized by GenScript. The cDNA fragments were double digested with EcoRI and XhoI and cloned into pUAST-attB. For site-directed mutagenesis of MP, the primer sets below were used:
MPS334A-F: 5’-CGCCGCTTCGCCCCCCGCCTGAT-3’
MP-R: 5’-CTTCAACAGCCCGTGGAACCACC-3’
The PCR reaction was performed with the Q5 Site-Directed Mutagenesis kit (NEB). The PCR product was ligated and transformed into E.coli competent cells, and the colonies were selected on ampicillin/LB agar plates. For transgenic animals, the UAS-MP-HA plasmid was extracted with HiPure Plasmid Midiprep kit (Invitrogen) for microinjection.
RT-PCR
Total RNA of flies was extracted with the RNAspin Mini kit (GE Healthcare). RT-PCR was performed with OneStep RT-PCR kit (QIAGEN). For RT-PCR, the primer sets below were used:
MP-RT-F: 5’-GAAAACAAGTCTTCAGAAATGGGCACTATCG-3’
MP-RT-R: 5’-AAAGTCCGGCGTGAAGACCAGGATAT-3’
CREBB-RT-F: 5’-ACAACAGCATCGTCGAGGAGAACG-3’
CREBB-RT-R: 5’-CGTGTTCGGTTCGGGCTTGATCTT-3’
rp49-RT-F: 5’-CCAAGGACTTCATCCGCCACC-3’
rp49-RT-R: 5’-GCGGGTGCGCTTGTTCGATCC-3’
Confocal imaging
Image processing was performed as described previously (Lee et al., 2011). Briefly, dissected brains were fixed in PBS with 4% paraformaldehyde at 4°C overnight, transferred to PBS with 2% Triton X-100 at room temperature, vacuumed for 1 hr and left overnight in the same solution at 4°C. For immunostaining of GFP, the samples were incubated with anti-GFP antibody conjugated with FITC (1:500) (Abcam) in PBS with 0.5% Triton X-100 overnight. Brains were cleared and mounted in RapiClear (SunJin Lab Co.) and imaged with a Zeiss LSM 880 Confocal Microscope under a 20 x or 40 x C-Apochromat water immersion objective lens.
Overexpression and pulldown of MP, MPS334A and CREBB in S2 cells
The Effectene transfection reagent (QIAGEN) was used to deliver DNA to S2 cells. For protein overexpression, 2 × 106 S2 cells were transferred into 5 ml fresh media overnight (Schneider’s Drosophila medium, Gibco). The cells were collected by centrifuging them for 2 min. UAS-MP-HA, UAS-MPS334A-HA or UAS-CREBB-6xHis were co-transfected with Act-GAL4 into S2 cells. Transfected cells were placed at room temperature for two days in medium, collected, and lysed with sample lysis buffer (50 mM Tris-Cl pH 7.5, 125 mM NaCl, 5% glycerol, 1% NP40, 1.5 mM MgCl2, 0.2 mM DTT) containing a protease inhibitor mix (cOmplete, Roche). The cell lysate was collected and α-HA agarose (EZview Red Anti-HA Affinity Gel, Sigma) or α-His resin (HisPur Ni-NTA Resin, ThermoFisher) was added for protein pulldown.
Western blot
Fly heads were collected and mashed in a sample lysis buffer. The samples with SDS sample buffer were boiled and run on a 10% SDS-PAGE gel and transferred to nitrocellulose membranes. Primary antibodies for PO4-CREBB (1:1000), pan-CREBB (1:5000) (Fropf et al., 2013), CREBA (1:5000) (Andrew et al., 1997) (Developmental Studies Hybridoma Bank), HA-tag (1:2000) (BioLegend), His-tag (1:2000) (Clontech) and actin (1:10,000) (Abcam) were used and detected by HRP conjugated secondary antibody (1:10,000) (Jackson ImmunoResearch). For detecting the ATG2-CREBB protein (Fropf et al., 2013), 50 adult brains were dissected and collected in 20 μL of the sample lysis buffer and placed at −80°C for overnight. After vortexing for one minute, samples were mixed with SDS sample buffer and boiled before loading into an SDS-PAGE gel and transferred to nitrocellulose. Primary antibodies for ATG2-CREBB (1:1,000) (Fropf et al., 2013) was used and detected by HRP conjugated secondary antibody (1:10,000) (Jackson ImmunoResearch). An ECL reagent kit (ThermoFisher) was used for Western blot detection.
Kinase assay
ERK and PKA were purchased from NEB. MP and MPS334A were overexpressed and purified from S2 cells. For kinase assay, the kinases (ERK: 0.01U (NEB), PKA: 0.1U (NEB), MP and mutant MP: 13.5 μl of α-HA-agarose pulldown) were added into a cocktail containing the kinase buffer (0.5 mg/ml BSA, 15 μM Tris-Cl pH7.5 in final) MgCl2/ATP (0.2 mM in final), γ-32P-ATP (10 μCi in final), and myelin basic protein (MBP) (1.2 mM in final). For testing if MP or CREBB are substrates of PKA, MP and CREBB were pulled down, and incubated at 30°C for 30 min. The reactions were terminated by adding 2x SDS sample buffer, boiling for 5 min, and run on SDS gels. After transferring to nitrocellulose membranes, radioactive signals were detected by CL-X posure film (ThermoFisher).
Luciferase activity assay
The luciferase assay system was from Promega. All groups of flies were raised in the same incubator in 12 hr light/12 hr dark conditions, and the CRE-luciferase assay was performed at the same time for all genotypes tested to avoid a circadian effect (Fropf et al., 2014). Briefly, fly heads were collected and homogenized in a reporter lysis buffer (10 heads/20 μl) and placed at −80°C overnight. The supernatant of lysate was collected and mixed with luciferase assay reagents (20 μl lysate/100 μl luciferase assay reagent) at RT for 1 min in 96-well plate. The luciferase activity was analyzed using an Optima luminescence reader (BMG LABTECH).
Behavioral assay
Behavioral assays were performed with balanced comparative groups which were trained and tested in parallel without blinding. For behavior assays, MB-GAL4 flies were outcrossed with Canton-S w1118 (iso1CJ) or y w flies for at least five generations, and Tub-Gal80ts, UAS-RNAi and UAS-MP-HA flies were outcrossed with iso1CJ. For RNAi knockdown, flies were raised at 18°C until eclosion then transferred to 25°C for 3 days. Aversive olfactory learning was performed using the T-maze apparatus (Tully and Quinn, 1985). In brief, one training session consists of approximately 100 flies that were electrically shocked while exposed to one of two odors (3-octanol and 4-methylcyclohexanol, Sigma). The shock was alternated between 3-octanol and 4-methylcyclohexanol. Flies trained by one training session and tested immediately are tested for learning, whereas those tested 3 hr later are assessed for 3 hr memory. Flies undergoing ten training sessions and tested 24 hr later are assessed for 24 hr memory or LTM. For cycloheximide (CXM, Sigma) feeding (DeZazzo and Tully, 1995), 35 mM CXM in 5% glucose was added on Whatman 3 MM filter paper in a bottle with fly food to feed flies overnight prior to performing training sessions. After training, the flies are placed in a vial containing CXM filter paper with fly food for another 24 hr prior testing.
Statistics
Statistical analyses were performed by KaleidaGraph 4.1 (Synergy software). Behavioral data were evaluated via one-way ANOVA followed with Tukey’s test for multiple comparisons (Lee et al., 2011). Data from two groups are analyzed by t-test. All data are presented as ‘mean ±SEM’. *p<0.05, **p<0.01, ***p<0.001.
Acknowledgements
We thank Amir Fayyazuddin for providing very insightful suggestions. We thank Josh Dubnau, Jerry Yin, and the Bloomington Drosophila Stock Center (NIH P40OD018537) and Vienna Drosophila Resource Center for flies. We thank Jerry Yin for sharing the α-ATG2-, α-Pan- and α-PO4-CREBB antibodies and the Developmental Studies Hybridoma Bank for α-CREBA antibodies. Confocal microscopy was supported by the BCM IDDRC Neurovisualization Core, which is supported by the NICHD (U54HD083092). We thank Jiangxing Lv, Yuchun He, Hongling Pan, Ying Fang, Qiaohong Gao, Zhihua Wang and Lily Wang for generating MiMIC GFP-tag and T2A-GAL4 fly stocks. This research was supported by NIH/NIGMS R01GM067858, the Robert A and Renee E Belfer Family Foundation and the Huffington Foundation. Hugo J Bellen is a Howard Hughes Medical Institute Investigator.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Hugo J Bellen, Email: hbellen@bcm.edu.
K VijayRaghavan, National Centre for Biological Sciences, Tata Institute of Fundamental Research, India.
Funding Information
This paper was supported by the following grants:
National Institute of General Medical Sciences R01GM067858 to Pei-Tseng Lee.
Eunice Kennedy Shriver National Institute of Child Health and Human Development U54HD083092 to Hugo J Bellen.
Robert A. and Renee E. Belfer Family Foundation to Hugo J Bellen.
Huffington Foundation to Hugo J Bellen.
Howard Hughes Medical Institute to Hugo J. Bellen.
Additional information
Competing interests
Reviewing editor, eLife.
No competing interests declared.
Author contributions
Conceptualization, Resources, Data curation, Validation, Methodology, Writing—original draft, Writing—review and editing.
Conceptualization, Data curation, Formal analysis, Validation, Methodology.
Resources.
Resources.
Resources, Writing—original draft, Writing—review and editing.
Conceptualization, Resources, Data curation, Supervision, Funding acquisition, Validation, Investigation, Writing—original draft, Project administration, Writing—review and editing.
Additional files
References
- Abrams EW, Andrew DJ. CrebA regulates secretory activity in the Drosophila salivary gland and epidermis. Development. 2005;132:2743–2758. doi: 10.1242/dev.01863. [DOI] [PubMed] [Google Scholar]
- Akalal DB, Yu D, Davis RL. A late-phase, long-term memory trace forms in the γ neurons of Drosophila mushroom bodies after olfactory classical conditioning. Journal of Neuroscience. 2010;30:16699–16708. doi: 10.1523/JNEUROSCI.1882-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew DJ, Baig A, Bhanot P, Smolik SM, Henderson KD. The Drosophila dCREB-A gene is required for dorsal/ventral patterning of the larval cuticle. Development. 1997;124:181–193. doi: 10.1242/dev.124.1.181. [DOI] [PubMed] [Google Scholar]
- Busto GU, Cervantes-Sandoval I, Davis RL. Olfactory learning in Drosophila. Physiology. 2010;25:338–346. doi: 10.1152/physiol.00026.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers D, Davis RL, Kiger JA. Defect in cyclic AMP phosphodiesterase due to the dunce mutation of learning in Drosophila melanogaster. Nature. 1981;289:79–81. doi: 10.1038/289079a0. [DOI] [PubMed] [Google Scholar]
- Caussinus E, Kanca O, Affolter M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nature Structural & Molecular Biology. 2011;19:117–121. doi: 10.1038/nsmb.2180. [DOI] [PubMed] [Google Scholar]
- DeZazzo J, Tully T. Dissection of memory formation: from behavioral pharmacology to molecular genetics. Trends in Neurosciences. 1995;18:212–218. doi: 10.1016/0166-2236(95)93905-D. [DOI] [PubMed] [Google Scholar]
- Diao F, Ironfield H, Luan H, Diao F, Shropshire WC, Ewer J, Marr E, Potter CJ, Landgraf M, White BH. Plug-and-play genetic access to drosophila cell types using exchangeable exon cassettes. Cell Reports. 2015;10:1410–1421. doi: 10.1016/j.celrep.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drier EA, Tello MK, Cowan M, Wu P, Blace N, Sacktor TC, Yin JC. Memory enhancement and formation by atypical PKM activity in Drosophila melanogaster. Nature Neuroscience. 2002;5:316–324. doi: 10.1038/nn820. [DOI] [PubMed] [Google Scholar]
- Dubnau J, Tully T. Gene discovery in Drosophila: new insights for learning and memory. Annual Review of Neuroscience. 1998;21:407–444. doi: 10.1146/annurev.neuro.21.1.407. [DOI] [PubMed] [Google Scholar]
- Fropf R, Tubon TC, Yin JC. Nuclear gating of a Drosophila dCREB2 activator is involved in memory formation. Neurobiology of Learning and Memory. 2013;106:258–267. doi: 10.1016/j.nlm.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fropf R, Zhang J, Tanenhaus AK, Fropf WJ, Siefkes E, Yin JC. Time of day influences memory formation and dCREB2 proteins in Drosophila. Frontiers in Systems Neuroscience. 2014;8:43. doi: 10.3389/fnsys.2014.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JM, Hodge JJ. CASK regulates CaMKII autophosphorylation in neuronal growth, calcium signaling, and learning. Frontiers in Molecular Neuroscience. 2013;6:27. doi: 10.3389/fnmol.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven-Ozkan T, Davis RL. Functional neuroanatomy of Drosophila olfactory memory formation. Learning & Memory. 2014;21:519–526. doi: 10.1101/lm.034363.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi J, Jiang W, Zhou H, Wu P, Yin JC. Phosphorylation of conserved casein kinase sites regulates cAMP-response element-binding protein DNA binding in Drosophila. Journal of Biological Chemistry. 2004;279:12117–12125. doi: 10.1074/jbc.M212839200. [DOI] [PubMed] [Google Scholar]
- Huang HD, Lee TY, Tzeng SW, Horng JT. KinasePhos: a web tool for identifying protein kinase-specific phosphorylation sites. Nucleic Acids Research. 2005;33:W226–W229. doi: 10.1093/nar/gki471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Zheng X, Zhao H, Li M, Wang P, Xie Z, Wang L, Zhong Y. A permissive role of mushroom body α/β core neurons in long-term memory consolidation in Drosophila. Current Biology. 2012;22:1981–1989. doi: 10.1016/j.cub.2012.08.048. [DOI] [PubMed] [Google Scholar]
- Impey S, Obrietan K, Storm DR. Making new connections: role of ERK/MAP kinase signaling in neuronal plasticity. Neuron. 1999;23:11–14. doi: 10.1016/S0896-6273(00)80747-3. [DOI] [PubMed] [Google Scholar]
- Keene AC, Waddell S. Drosophila olfactory memory: single genes to complex neural circuits. Nature Reviews Neuroscience. 2007;8:341–354. doi: 10.1038/nrn2098. [DOI] [PubMed] [Google Scholar]
- Lee PT, Lin HW, Chang YH, Fu TF, Dubnau J, Hirsh J, Lee T, Chiang AS. Serotonin-mushroom body circuit modulating the formation of anesthesia-resistant memory in Drosophila. PNAS. 2011;108:13794–13799. doi: 10.1073/pnas.1019483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhang X, Hu W, Liang X, Zhang F, Wang L, Liu ZJ, Zhong Y. Importin-7 mediates memory consolidation through regulation of nuclear translocation of training-activated MAPK in Drosophila. PNAS. 2016;113:3072–3077. doi: 10.1073/pnas.1520401113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Zaslaver A, Alon U. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. Journal of Molecular Biology. 2003;334:197–204. doi: 10.1016/j.jmb.2003.09.049. [DOI] [PubMed] [Google Scholar]
- Margulies C, Tully T, Dubnau J. Deconstructing memory in Drosophila. Current Biology. 2005;15:R700–R713. doi: 10.1016/j.cub.2005.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martenson RE, Law MJ, Deibler GE. Identification of multiple in vivo phosphorylation sites in rabbit myelin basic protein. The Journal of Biological Chemistry. 1983;258:930–937. [PubMed] [Google Scholar]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nature Reviews Molecular Cell Biology. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Science Signaling. 2004;2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Deshazer M, Davis RL. Thirty years of olfactory learning and memory research in Drosophila melanogaster. Progress in Neurobiology. 2005;76:328–347. doi: 10.1016/j.pneurobio.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Mouravlev A, Dunning J, Young D, During MJ. Somatic gene transfer of cAMP response element-binding protein attenuates memory impairment in aging rats. PNAS. 2006;103:4705–4710. doi: 10.1073/pnas.0506137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkar-Jaiswal S, Lee PT, Campbell ME, Chen K, Anguiano-Zarate S, Gutierrez MC, Busby T, Lin WW, He Y, Schulze KL, Booth BW, Evans-Holm M, Venken KJ, Levis RW, Spradling AC, Hoskins RA, Bellen HJ. A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. eLife. 2015;4:e05338. doi: 10.7554/eLife.05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nara K, Akasako Y, Matsuda Y, Fukazawa Y, Iwashita S, Kataoka M, Nagai Y. Cloning and characterization of a novel serine/threonine protein kinase gene expressed predominantly in developing brain. European Journal of Biochemistry. 2001;268:2642–2651. doi: 10.1046/j.1432-1327.2001.02157.x. [DOI] [PubMed] [Google Scholar]
- Pascual A, Préat T. Localization of long-term memory within the Drosophila mushroom body. Science. 2001;294:1115–1117. doi: 10.1126/science.1064200. [DOI] [PubMed] [Google Scholar]
- Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip N, Acevedo SF, Skoulakis EM. Conditional rescue of olfactory learning and memory defects in mutants of the 14-3-3zeta gene leonardo. Journal of Neuroscience. 2001;21:8417–8425. doi: 10.1523/JNEUROSCI.21-21-08417.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putz G, Bertolucci F, Raabe T, Zars T, Heisenberg M. The S6KII (rsk) gene of Drosophila melanogaster differentially affects an operant and a classical learning task. Journal of Neuroscience. 2004;24:9745–9751. doi: 10.1523/JNEUROSCI.3211-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoulakis EM, Davis RL. Olfactory learning deficits in mutants for leonardo, a Drosophila gene encoding a 14-3-3 protein. Neuron. 1996;17:931–944. doi: 10.1016/S0896-6273(00)80224-X. [DOI] [PubMed] [Google Scholar]
- Tanenhaus AK, Zhang J, Yin JC. In vivo circadian oscillation of dCREB2 and NF-κB activity in the Drosophila nervous system. PLoS One. 2012;7:e45130. doi: 10.1371/journal.pone.0045130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubon TC, Zhang J, Friedman EL, Jin H, Gonzales ED, Zhou H, Drier D, Gerstner JR, Paulson EA, Fropf R, Yin JC. dCREB2-mediated enhancement of memory formation. Journal of Neuroscience. 2013;33:7475–7487. doi: 10.1523/JNEUROSCI.4387-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. Journal of Comparative Physiology A. 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- Tully T, Bourtchouladze R, Scott R, Tallman J. Targeting the CREB pathway for memory enhancers. Nature Reviews Drug Discovery. 2003;2:267–277. doi: 10.1038/nrd1061. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, Bellen HJ. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nature Methods. 2011;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zars T. Behavioral functions of the insect mushroom bodies. Current Opinion in Neurobiology. 2000;10:790–795. doi: 10.1016/S0959-4388(00)00147-1. [DOI] [PubMed] [Google Scholar]