Abstract
The recent advancement of dielectrophoresis (DEP)-enabled microfluidic platforms is opening new opportunities for potential use in cancer disease diagnostics. DEP is advantageous because of its specificity, low cost, small sample volume requirement, and tuneable property for microfluidic platforms. These intrinsic advantages have made it especially suitable for developing microfluidic cancer diagnostic platforms. This review focuses on a comprehensive analysis of the recent developments of DEP enabled microfluidic platforms sorted according to the target cancer cell. Each study is critically analyzed, and the features of each platform, the performance, added functionality for clinical use, and the types of samples, used are discussed. We address the novelty of the techniques, strategies, and design configuration used in improving on existing technologies or previous studies. A summary of comparing the developmental extent of each study is made, and we conclude with a treatment of future trends and a brief summary.
I. INTRODUCTION
Cancer is a disease resulting from irreversible modifications to the genetic material in cells, due to the continuous exposure of the cellular constituents to chemical carcinogens, ionization radiations, and reactive oxygen species.1 There are several types of cancers such as breast,2 prostate,3 oral,4 melanoma,5 bladder,6 and leukemia.7,8 The advancement of cancer cell research up to the cellular level has resulted in numerous diagnostic studies related to cancer cell detection. One promising research area in cellular-based cancer studies is the use of microfluidic platforms to manipulate cells.
Microfluidics has been used to promote the isolation, enrichment, and analyses of cancer cells. These cells can be isolated from a large population of other cell types based on one or several unique properties.9 The advantages of using microfluidic platforms stem from factors such as suitable scale, unique fluidic phenomena, the ability to integrate multiple functions, and their large surface-to-volume ratio.10,11 Additionally, the specificity of control of cell motion is another leverage of the microfluidic platforms.
The external forces applied to control and manipulate biological particles (bioparticles) in a microfluidic platform are known as bioparticle active manipulation forces.12 Dielectrophoresis (DEP) is one of the forces widely applied to cells in microfluidic platforms to control their motions. Dielectrophoresis (DEP) is an electro-kinetic phenomenon describing the motion of a neutral particle or cell in a non-uniform electric field due to polarization effects.13 The dielectrophoretic force is the result of non-uniform polarization generated by the interaction of induced dipoles with the externally applied alternating current (AC) or direct current (DC) electric field gradients. It enables the separation of particles and cells based on their different polarizabilities to their suspending medium manipulation. DEP is advantageous because it enables precise control of cells using small sample quantity.14 In this review, we categorize DEP platforms such as electrode-based DEP (eDEP),15 contactless-based DEP (cDEP),16–18 insulator-based DEP (iDEP),19 and optically induced dielectrophoresis (ODEP).20
While a number of excellent reviews on DEP-based manipulation of bioparticles are available, reviews specifically focusing on the application of DEP confined to cancer studies are limited. Çetin and Li21 reviewed the modeling of DEP-based manipulation and recent applications of DEP based studies on bioparticles and non-bioparticles. Qian et al.22 described the fundamentals of DEP while introducing five distinct DEP techniques on bioparticles, namely, capturing, focusing, characterizing, pairing, and separating. Another review by Demircan et al.23 categorized DEP applications according to the bioparticles used such as blood and plasma separation, fetal cell and blood separation, microorganisms, stem cells, and cancer cells. Furthermore, Chen et al.24 discussed the isolation of rare cells based on the dielectrophoretic signature as one of the four categories of microfluidic applications.
Briefly, circulating tumor cells (CTCs) are tumor cells which have been shed into blood from a primary tumor.25 Isolating CTCs is challenging because CTCs can be very rare within the bloodstream and maintaining CTC cell viability26 and purity27 is important. Changes in the viability of isolated CTCs will affect the morphology and molecular functionalities, which hinders further biochemical or cell-based assays after isolation.26 Several reviews focused on DEP applications in CTC studies. For example, Patil et al.28 reviewed recent work which exploits the physical characteristics of tumor cells to efficiently isolate them and discussed the intricate design perspective of experimental and commercialized DEP integrated microfluidic devices for efficient in vitro cancer diagnosis and prognosis. Yu et al.29 focused on nanotechnological miniaturization strategies of lab-on-a-chip (LOC) and the advancements in fabrication of LOC. They discussed three types of DEP devices, namely, eDEP, cDEP, and iDEP. In another review, Cima et al.30 reported on technologies for label-free separation of CTCs, with a brief description and discussion of a few dielectrophoretic based device studies. The review of Cima et al.30 covered the recent advances in label-free approaches to isolate and manipulate CTCs based on their biomechanical and electrical properties. These reviews briefly describe examples of the DEP method, highlighting the advantages and drawbacks of these technologies and the status of implementation in clinics. The coverage of these reviews is very limited and does not cover quantification and performance comparison of the devices.
Pethig31 covered the status of theory of dielectrophoresis and the developments of DEP devices for biomedical applications. The review of Jubery, Srivastava, and Dutta32 is comprehensive on existing models on DEP, consisting of the effective moment Stokes-drag (EMDS) method to more recently developed numerical methods accounting for the size, shapes, and types of bio-particles. Both reviews have limited coverage on cancer cells as the former did not review cancer enabled microfluidic platforms, while the latter did a review of some platforms but primarily focusing only on yeast, bacteria, and proteins. The review of Khamenehfar and Li33 presented a brief report on DEP-based separation of cancer cells focusing on breast and leukemia cells only. More recent reviews provide different angles of the development of microfluidic devices for cancer cells. Perez-Gonzalez et al.34 reviewed extensively microfluidic devices for cancer cells with emphasis on immunomagnetic-affinity-based devices and surface-plasmon-resonance microfluidic sensors. We found a lack of coverage on operating conditions of the DEP devices covered such as multi-parametric settings. Meanwhile, the review of Adekanmbi and Srivastava35 on microfluidic applications for diseases such as cancer, malaria, Human African trypanosomiasis (HAT), dengue, and Anthrax falls short in comparing the advantages and disadvantages of the DEP devices with respect to cancer diagnostics. This information would be useful for future developments of DEP-based microfluidic devices for cancer diagnostics.
Here, the limitations of current cancer diagnosis technologies, which indicate the importance of microfluidics based devices for cancer diagnostics, are reviewed. Several microfluidic techniques for cancer cell manipulation are introduced. The importance of DEP manipulation in cancer cell microfluidics is then highlighted, followed by a review of the recent literature regarding the use of dielectrophoretic forces to sort and manipulate cancer cells. The aim of this review is to make an in-depth comparison and critical review that provides an insight into the latest advancements of DEP based microfluidics for cancer cell manipulation and diagnostics. This knowledge would be useful for future research involving DEP-based microfluidic devices for cancer diagnostics.
II. CANCER, TUMOR, AND CANCER DIAGNOSTIC DEVICES
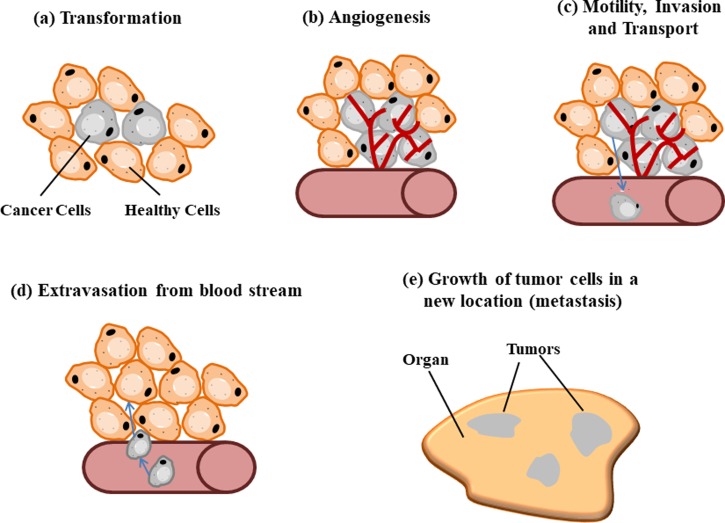
Cancer is a complex group of diseases characterized by the uncontrolled growth and spread of abnormal cells and has long been the leading cause of death in many countries. A tumor is formed by cells that multiply excessively and uncontrollably, and the resulting growth can be either malignant (cancerous) or benign (non-cancerous).36 Primary tumor cells that develop in epithelial cells or in the blood and lymphatic system (carcinomas)37 can spread via the vascular or blood vessel network as circulating tumor cells (CTCs). CTCs form secondary tumors called metastasis tumors which cause a majority of cancer-related fatalities.38,39 Figure 1 shows the metastasis process of tumor cells in the body. CTCs can be more easily accessed and repeatedly through blood,40 and CTC isolation from a blood sample is increasingly shown to be reliable in early detection and molecular characterization of cancer at diagnosis. Early and accurate detection of cancer is highly important for clinical diagnosis, effective toxicity monitoring, and, ultimately, the successful treatment of cancers, thus improving the chances of survival.41
FIG. 1.
Metastasis Process of Cancer: (a) Transformation of healthy cells to cancer cells. (b) Proliferation of tumor cells. (c) Tumor cells enter blood stream forming circulating tumor cells (CTCs). (d) CTCs adhere to endothelial cells at the wall of blood stream and migrate out into a new location. CTCs invade cells in other parts of the body. (e) CTCs adhere and proliferate/metastasize to form tumor(s).
Microfluidic platforms that can detect cancer cells and CTCs have been studied and developed, which make use of phenomena related to biomolecular interactions of cells42 and physical properties of cells.43 Microfluidic platforms based on biomolecular interactions of cancer cells use biomarkers44 to facilitate isolation, trapping, detection, and immobilization of target cells. A biomarker is designated as a substance or activity that can be objectively measured and evaluated as an indicator for a normal biological process, pathogenic process, or pharmacological responses to a therapeutic intervention.45 The physical properties of cells include the dielectric properties of cells and media, cell size, and cell shape. Forces known as bioparticle manipulation forces (BMFs) can be applied onto the cells and will cause the cells to respond based on their specific physical properties. These forces include electrophoresis,46 dielectrophoresis,47 magnetophoresis,48 optical tweezing,49 thermophoresis,50 and acoustophoresis,51 and they can be incorporated into microfluidic platforms for particle manipulation. The fundamentals and a brief description of each BMF were covered in a recent review by Ali et al.12
The selection of a suitable BMF for cancer or CTC diagnostic devices must take into account the physical properties of cancer cells. Some physical properties of cancer cells include an average cell diameter of 10.3–32.39 μm,52–55 they are electrically neutral,56 and they are able to preserve cell viability and cell purity for downstream analysis.57 A suitable BMF for developing a cancer diagnostic microfluidic platform must be selected based on these physical properties. Hydrodynamic based microfluidic systems are prone to be hindered due to clogging by particles in the flowing medium58 and may not be suitable cells which are larger than metal or synthetic particles. The electric field of electrophoresis influences charged particles,59 which poses a problem in manipulating cancer cells (bioparticles)53 which are neutrally charged.56 Magnetophoresis requires complex preparations of a conductive suspending medium, which complicates the purification procedure and may limit the availability of the target cells.12,60 Meanwhile, optical tweezing requires complicated setup procedures and instruments,61 while acoustophoresis faces the challenge of chaotic streaming flow from acoustic waves62 inside a complex microfluidic channel.63
In dielectrophoresis (DEP), particles are temporarily polarized, by a spatially non-uniform electric field,64 establishing dipoles which induce unequal columbic forces causing the particles to move.65 The incorporation of dielectrophoretic force into microfluidic platforms for cancer cell diagnosis is advantageous because it has high selectivity, is label-free, can manipulate neutral bioparticles, has ease of fabrication, can analyze with high sensitivity, and is compatible for microfluidic devices.66,67 The Sec. III covers in detail fundamentals of DEP theory and also briefly describes the different categories of DEP platforms that have been developed.
III. THEORY OF DIELECTROPHORESIS (DEP)
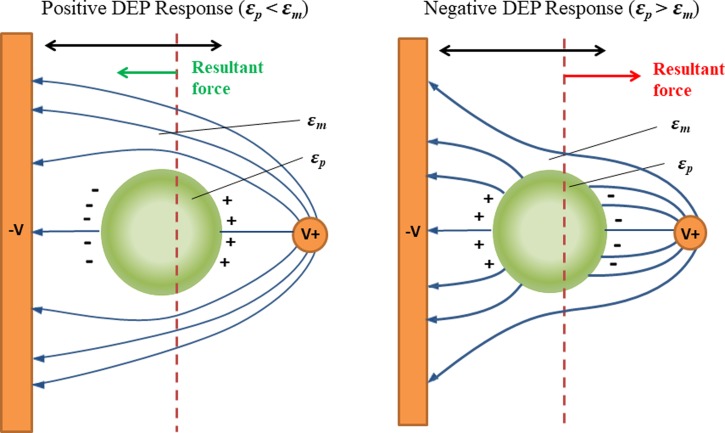
DEP is the motion of particles in a spatially non-uniform electric field. The spatially non-uniform electric field induces unequal columbic forces, temporarily polarizing the particles, and causes the particles to move in the direction of increasing or decreasing field intensity. The physical properties of the particles and the medium such as permittivity and conductivity determine how the particle polarizes.68 Figure 2 shows the DEP force of a polarized particle.
FIG. 2.
Polarization of particles causes dielectrophoretic response; negative DEP, the particle is attracted to the region with low electrical field density, and positive DEP, the particle is attracted to the opposite. Reproduced with permission from Doh and Cho, Sens. Actuators, A 121, 59 (2005). Copyright 2005 Elsevier.124
Pohl68 first established the approximate model of conventional DEP on the basis of the classical Maxwell electromagnetic field theory
| (1) |
where r is the radius of a spherical particle, is the absolute permittivity of the media, is the root mean square value of the electric field, is the real part of the Clausius–Mossotti (CM) factor, which refers to positive dielectrophoresis (pDEP) and negative dielectrophoresis (nDEP), and is given by
| (2) |
where is the absolute permittivity of a particle, and are the conductivity of the medium and the particle, and and are the complex permittivity of the medium and the particle, which are related to the conductivity, σ and the angular frequency, ω of the electric field
| (3) |
where , is the relative permittivity, is the electric conductivity, and is the angular frequency of the applied electric field. This equation reveals that the CM factor not only depends on the dielectric properties of the particles and medium but also on the frequency of the applied field.
Biological particles such as bacteria, viruses, spores, yeast, and other eukaryotic cell types as well as proteins, nucleic acids, and other biomolecules have a more complicated internal structure than a solid, homogeneous particle.21,69 These complications does not change fundamental physics but must be taken into account for expressions of the dipole moment and by modifying the DEP force.21
For biological particles, the common approach for theoretical modeling is the concentric multishell model70 in which the simplest case is the single, spherical shell model. In the single, spherical shell model,71,72 the original two-layer particle (Fig. 3) replaces a homogenous sphere with an effective complex permittivity of
| (4) |
where is defined as70
| (5) |
FIG. 3.
Schematic illustration of the single-shell model. Reproduced with permission from Çetin and Li, Electrophoresis 32, 2410 (2011). Copyright 2011 John Wiley and Sons.
The single-shell model can be extended to multi-shell models,72 which can adequately model the effective dipole moment of a typical mammalian cell73 which consists of a highly conducting cytoplasm surrounded by an insulating membrane, which is known as the protoplast model.74 The method of electrorotation (ROT), time domain dielectric spectroscopy (TDDS),75 or single-cell dielectric spectroscopy76 can be used to measure the dielectric properties of the cells. The commonly used method that is well developed is ROT where the applied rotating electric field is measured as a function of field frequency.21 The electric field induces torque that causes cells to rotate. Dielectric properties of cells are estimated by optimizing the parameters of the single-shell77 or multi-shell78 model to fit the experimental ROT spectrum data. The estimated properties are used to determine the DEP spectra of cells.
DEP equations are used in simulations to optimize platform design, and the experimental setup of DEP based microfluidic platforms incorporates studies of factors such as fluid flow velocity, microchannel dimensions, ion-diffusion, and effect of the electric field gradient on target cells.79–81 For example, Shim et al.81 conducted simulation of the parabolic Poiseuille flow velocity profile and ion diffusion zone of a continuous flow dielectrophoretic field flow fractionation (FFF) microfluidic device. This study optimizes the ratio of the microchannel dimensions (channel length and height), flow rate, and electric field gradient, which ensures that target cells are separated by pDEP force in a thin lamina (skim layer) that flows beneath the main eluate stream from peripheral blood mononuclear (PBMN) cells. In another simulation by Smith, Huang, and Kirby,79 the probability of contamination by PBMN was reduced, while the capture probability of target pancreatic cells was increased via Monte Carlo simulation using cell trajectories in a range of geometries and applied electric fields. A previous simulation on cell transport within a microfluidic obstacle array caused by fluid advection and DEP forcing was first carried out. Another simulation by Hwang et al.80 determined optimal outlet channel flow rates for deflection of target cells for establishing a high efficiency and high throughput device.
The understanding of the effect of electric fields on the movement of particles for precise control enables the development of DEP enabled microfluidic platforms. The Sec. IV covers the different categories of DEP enabled microfluidic platforms that have been developed.
IV. CATEGORIES OF DEP ENABLED MICROFLUIDIC PLATFORMS
The use of microelectrodes in microfluidic platforms is advantageous based on several reasons.35 Micron-sized electrodes increase the dielectrophoretic force experienced by particles near the electrode as demonstrated by direct dimensional analysis since dielectrophoretic force is dependent on the square of the electric field (V2 m−3). The micron-sized gap of micro-sized electrodes is able to generate sufficient dielectrophoretic force at smaller voltages to manipulate particles. Additionally, the viability of bioparticles is preserved as a result of miniaturization that reduces Joule heating effects and electrode decay.82
Microelectrode fabrication is possible with the advances in soft lithography techniques that are capable of creating the precise electrode shapes which generate the required specific field gradients for DEP manipulation.83 Some DEP techniques have been developed that do not require soft lithography to fabricate the electrodes but instead use light or ionic liquid to form the electrodes that induce dielectrophoretic force. Changes in the electrode shape or orientation and modifying the frequency and phase of electric signals applied provide varying degrees of dielectrophoretic control on the bioparticles such as trapping, sorting, electro-rotation, and traveling-wave DEP effects.84 Here, we categorize the different DEP systems that have been developed into different categories: (1) electrode-based DEP (eDEP); (2) insulator-based DEP (iDEP); (3) contactless DEP (cDEP); and (4) optical-based DEP (oDEP).
A. eDEP
eDEP designs have electrodes that are positioned inside the microchannel and directly in contact with the sample and the suspending medium.85 Microelectrodes in eDEP designs are commonly fabricated using metals such as gold86 and indium tin oxide (ITO)87 with an adhesion layer of titanium88 or chrome.89 Metal electrodes have disadvantages of fouling and electrolysis at high frequencies,90 resulting in gas generation or dissolution of the electrodes which will damage the biological sample. The use of ITO is advantageous due to its low capacitive current and its fabrication practicability as it allows for easy micropatterning.91
B. iDEP
This design uses external electrodes since the electrodes can be separately fabricated to reduce fabrication costs. The non-uniform electric field is induced by an insulating hurdle or obstacle in the microchannel and not the external electrodes which are placed at the outlet ports.92 A disadvantage of iDEP is Joule heating from the high electric field intensity in the microchannel. Biological fluids are highly conductive, and high temperatures in the microchannel can lead to cell death.93
C. cDEP
In cDEP, microfluidic channels are filled with a high-conductive liquid and used as electrodes. Thin insulating barriers that exhibit capacitive behavior separate the liquid electrodes from the sample channel. Electric gradients are created in the sample channel when electric potentials are applied across these side channels inducing DEP known as cDEP.34
D. oDEP
Another DEP technique developed is the oDEP which addresses the issue of delicate and complex fabrication processes with the use of optical images to form the electrodes.27,94 The non-uniform electric-field is produced when light is projected onto a photo-conductive layer, which excites the electron-hole pairs and significantly decreases the electrical impedance of the illuminated area. Operation of the oDEP device is less technically demanding as it uses computer-interfaced control.27
V. CANCER CELL ENABLED DEP PLATFORMS
In this section, DEP platforms incorporating cancer cells, also known as cancer cell enabled DEP platforms, are presented. The platforms are categorized according to the target cancer cells used. A summary of the operating conditions and performance of these platforms is presented in Table I with the available information reported in the respective studies.
TABLE I.
DEP-based cancer cell manipulation platforms and characteristics.
| Target cancer cell-line (References) | DEP platform characteristic | DEP platform type | Optimum operating conditions | Optimum performance of the platform for target cells | |
|---|---|---|---|---|---|
| Breast cancer cells | |||||
| MCF-7 (Kim et al.97) | 3D-asymmetric microelectrode dielectrophoretic activated cell sorter | eDEP | • Flow rate: 0.35 μl/min | ||
| • Frequency: 45 MHz | • 84% cell recovery | ||||
| • Voltage: 7 Vpp | |||||
| MCF-7 (Bhattacharya et al.98) | DC selective trapping iDEP platform | iDEP | • Electric field: 100 V/cm | • 68.5% cell viability after 2 min exposure to the electric field | |
| GX treated MCF-7-LCC9 (Soltanian-Zadeh et al.99) | Off-chip passivated-electrode insulator based Dielectrophoresis for separation | iDEP | • Flowrate: 20 μl/h | ||
| • Drug dosage: 500 nM • GX-treated cells | • 72% ± 1.1% trapping efficiency | ||||
| • Frequency: 1 MHz | |||||
| • Voltage: 80 Vpp | |||||
| GFP Labelled MDA-MB-231 (Alazzam et al.86) | Planar interdigitated transducer electrodes for separation | eDEP | • Flow rate: 6 μl/h | ||
| • Voltage: 15 Vpp | • Separation accuracy: 100% | ||||
| • Frequency: 40 kHz | • Purity: around 81% | ||||
| MDA-MB-231 (Li and Anand101) | Wireless electrode array | eDEP | • Flow rate: 0.1 ml/h | ||
| • Voltage: 248 Vpp | • N/A | ||||
| • Frequency: 40 kHz | |||||
| MDA-MB-231 (Henslee et al.16) | cDEP fluid electrode channel cell trapping device | cDEP | • Flow rate: 0.02 ml/h | ||
| • Voltage: 20 Vrms | • 90% or greater target cell trapping | ||||
| • Frequency: 180–210 kHz | |||||
| Leukemia | |||||
| HL-60 (Su et al.87) | DEP spring method using angled coplanar electrodes for electrical cell characterization | eDEP | • Min. - 1.47 S/m, max. - 0.35 S/m | • Characterization of approximately 10 cells/s | |
| • Algorithm accuracy of 96% | |||||
| K562 (Lee et al.106) | Gravitational and nDEP based electrode array cell sorter | eDEP | • Throughput: 17 000 cells/min | ||
| • Voltage: 7 Vpp | • Recovery rate: 49.42% | ||||
| • Frequency: 10 kHz | • Sorting efficiency: 94.74 ± 0.77% | ||||
| THP-1 (Yasukawa et al.107) | nDEP based electrode with the navigator and separator configuration for cell sorting by size | eDEP | • Flow velocity: 0.2 mm/s | ||
| • Voltage: Navigator - 15 Vpp • Separator – 14 Vpp | • 88% passing accuracy | ||||
| • Frequency: 100 kHz | |||||
| • 110 mS/m | |||||
| Jurkat T-lymphoblasts (Novickij et al.108) | High pulsed magnetic field inducing pDEP electric field coil | cDEP | • Frequency altered magnetic field: 1.3 MHz | ||
| • Average cell velocity: 73 μm/s | • N/A | ||||
| • Pulse repetitive frequency: 1–25 Hz | |||||
| Prostate cells | |||||
| ALDH expressing PC-3 cells (Salmanzadeh et al.122) | pDEP based selective trapping device | cDEP | • Flow rate: 0.2 ml/h | ||
| • Voltage: 293 Vrms | • Around 69% separation of PC-3 cells from non-PC-3 cells | ||||
| • Frequency: 200 kHz | |||||
| PC-3 live cells (Sun et al.114) | Ionic liquid electrode device for target cell separation | cDEP | • Flow rate: 50 μl/h | ||
| • Voltage: 53 Vrms | • 89.8% live cancer cells deflected to the designated outlet. | ||||
| Frequency: 50 kHz | |||||
| Live and dead PC-3 cells and blood (Chiu et al.20) | nDEP optically induced dielectrophoresis cell manipulation with laminar flow CTC cell isolation | ODEP | • Flow rate: 2.5 μL/min | ||
| • Voltage: 8 V | |||||
| • Frequency: 50 kHz | • Separation purity: 100% | ||||
| • Hollow circular light images: | • Cancer cell recovery rate: 41.5% | ||||
| • Fixed ID – 40 μm | |||||
| • Bandwidth – 40 μm | |||||
| • Moving velocity – 50 μm/s | |||||
| 22Rv1 cells (Khamenehfar et al.115) | Crossflow microfilter-DEP integrated single cancer cell isolation chip | eDEP | • Voltage: 11.5 V | ||
| • Frequency: 3 MHz | • Isolation time of cancer cells >60 s | ||||
| Ovarian cells | |||||
| MOSE cells (Čemažar et al.118) | Cell-scale micropillar (20 μm) pDEP cell isolation and trapping device | cDEP | • Flow rate: 20 μl/min | ||
| • Voltage: 300 Vrms | • 71%–90% cancer cell viability | ||||
| • Frequency: 50 kHz | • Sorting over 1 × 106 cancer cells/h | ||||
| Cervical cells | |||||
| HeLa cells from RBC (Chen et al.18) | pDEP stepping electric field cancer cell concentration and collection open-top chamber device | eDEP | • Voltage: 8 Vpp | ||
| • Frequency: 1 MHz | • 76%–80% cancer cell survival rate | ||||
| • Relay switching interval: 20 s | • 76%-80% cancer cell concentration | ||||
| • Cell concentration: 500 cells/ml | |||||
| HeLa cells (Das et al.119) | 3-inlet 3-outlet microfluidic platform with a tapered planar microelectrode | eDEP | • Voltage: 10 Vpp | ||
| • Frequency: 1 MHz | • N/A | ||||
| • Cell concentration: 106 cells/ml | |||||
| Colorectal cells | |||||
| HCT116 cells (Xing and Yobas120) | Continuous flow IDT-3D ring array microelectrode (40 μm) | eDEP | • Flow rate: 0.2 ml/h | ||
| • Voltage: 10 Vpp | • 94% cancer cell viability | ||||
| • Frequency: 100 kHz | • 82% cancer cell recovery | ||||
| • Cell concentration: 107 cells/ml | |||||
| Lung cells | |||||
| AS2-GFP from RBC and WBC cells (Cheng et al.88) | 3D LDEP nDEP lateral displacement CTC isolation system | eDEP | • Flow rate: 10 μl/min | ||
| • Voltage: 18 Vpp | • Recovery was greater than 85% and the isolation | ||||
| • Frequency: 10 kHz | • Purity was greater than 90% | ||||
| • Cell concentration: 600 cells/ml | • Enrichment factor of 105 | ||||
| • Channel length: 6 and 13 cm | |||||
A. Breast cancer cell
The use of breast cancer cells in microfluidic research is extensive due to their complexity and heterogeneity.95 Upon literature review and to the authors' best knowledge, MCF-7 and MDA-MB-231 are the most common epithelial breast cancer cell lines used in research on DEP enabled microfluidic platforms. This may be due to the popularity of these cell lines in breast cancer cell related research.96
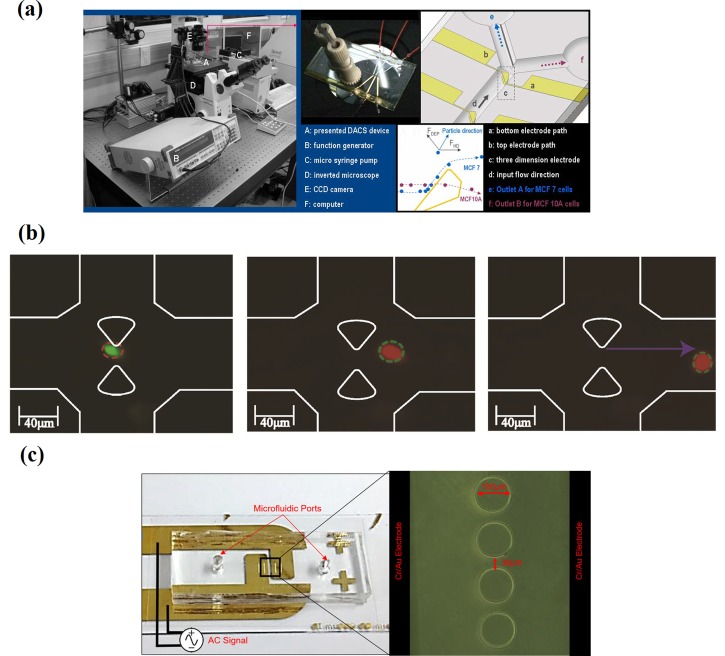
Kim et al.,97 Bhattacharya et al.,98 and Soltanian-Zadeh et al.99 conducted DEP-based separation experiments using the MCF-7 epithelial breast cancer cell line. The study by Kim et al.97 on a 3D-asymmetric microelectrode system device showed a 20% error in separation efficiency of MCF-7 cells from non-cancerous breast epithelial cells, MCF-10A cells [Fig. 4(a)]. They reported that a weak DEP force region (“blind spot”) in the DEP field generated by the pair of electrodes that caused small sized MCF-7 cells (within 20 μm) is found at the MCF-10A cell collecting outlet. Bhattacharya et al.98 studied selective trapping of MCF-7 cells on a DC iDEP in mixtures containing mammalian peripheral blood mononuclear cells (PBMCs) and MDA-MB-231. This device shows the selectivity of weakly metastatic MCF-7 versus highly invasive MDA-MB-231 from the cancer-cancer cell mixture, while in mixtures containing the cancer-PBMC mixture, PBMCs showed less favorable cell viability. Figure 4(b) shows the sequence of trapping of viable MCF-7 cells (red outline) between the tips of teardrop posts, while the smaller PBMCs (white contour lines) flow through the trapping region.
FIG. 4.
Dielectrophoretic platforms for MCF-7 cell manipulation. (a) Images and fundamental schematic of the microfluidic platform. Reproduced with permission from Kim et al., J. Mech. Sci. Technol. 23, 3132 (2010). Copyright 2010 Springer. (b) Snapshots depicting a selective trapping sequence for MCF-7 and MDA-MB-231 in an insulated-based dielectrophoretic (iDEP) device. Reproduced with permission Bhattacharya et al., Anal. Bioanal. Chem. 406, 1855 (2014). Copyright 2014 Springer. (c) Off-chip passivated electrode insulator-based dielectrophoretic (iDEP) platform. Reproduced with permission from Soltanian-Zadeh et al., Electrophoresis 70, 1 (2017). Copyright 2017 John Wiley and Sons.99
The single cell selectivity approach suggests that this device can be employed for the detection and enrichment of rare cancer cells in cancer diagnostics. Another study conducted by Soltanian-Zadeh et al.99 detected changes in the DEP profile of MCF-7 cells treated with anti-tumor drug GX using an off-chip passivated electrode iDEP platform [Fig. 4(c)]. This is advantageous compared to proliferation assays, a traditional drug assessment method, which is unable to detect the changes after drug treatment. However, iDEP requires large electric field magnitudes that result in limited frequency ranges due to equipment restrictions and large electronics due to a thick passivation layer (100 μm).
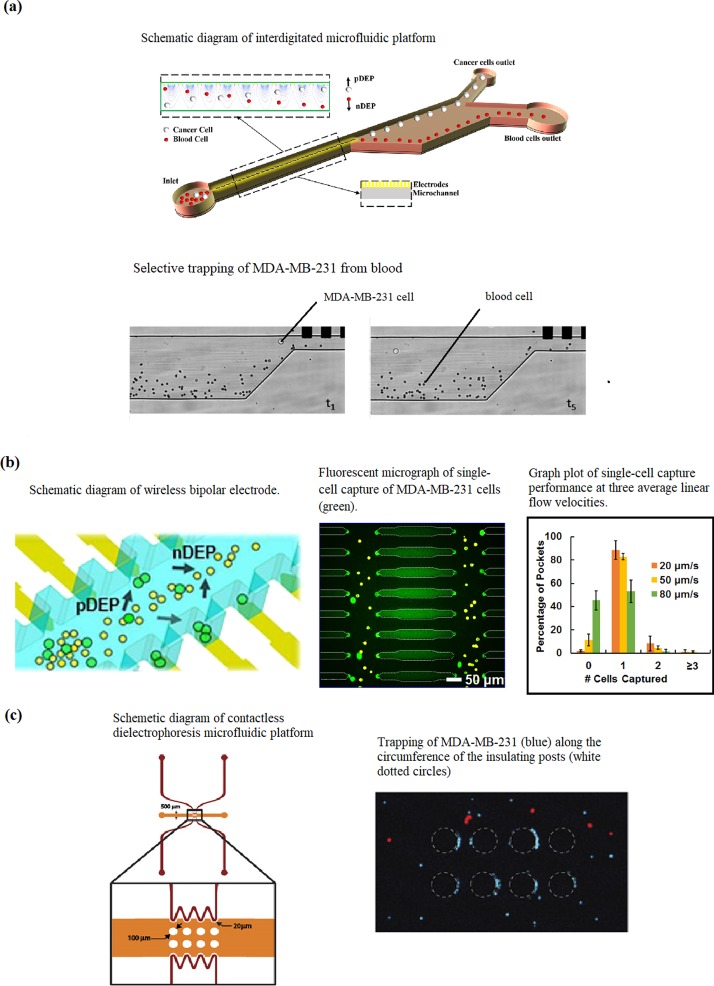
Alazzam et al.100 demonstrated continuous dielectrophoresis separation of green fluorescent protein (GFP)-labelled MDA-MB-231 cells from normal blood cells on a planar interdigitated electrode (IDT) transducer electrode platform [Fig. 5(a)]. The lateral design makes separation independent of gravity, which improves throughput and eases placement of the outlets for ease of collection of separated cells. The current design has a maximum flow rate for optimum separation at 6 μl/hand achieving 100% separation accuracy and a purity of around 81%. Further studies are required to support theoretical calculations that the flow rate can be increased by increasing the electrode pairs and distribution length.
FIG. 5.
(a) Interdigitated electrode (IDT) microfluidic device. Reproduced with permission from Alazzam et al., Electrophoresis 32, 1327 (2011).Copyright 2011 John Wiley and Sons. (b) Wireless bipolar electrode (BPE) microfluidic platform. Reproduced with permission from Li and Anand, J. Am. Chem. Soc. 139, 8950 (2017). Copyright 2017 ACS Publications. (c) Contactless dielectrophoretic (cDEP) microfluidic platform. Reproduced with permission from Henslee et al., Electrophoresis 32, 2523 (2011). Copyright 2011 John Wiley and Sons.
In another study, Li and Anand101 developed a high throughput, scalable wireless bipolar electrode (BPE) array and tested it on MDA-MB-231 cells [Fig. 5(b)]. The wireless feature of the device enables branching of the microchannel, which further increases throughput. They successfully demonstrated that the device was able to perform selective separation and single cell capture of MDA-MB-231 cells. Currently, further evaluation of the capture efficiency and the performance of the technique in clinical samples is in progress. Henslee et al.16 used a heterogeneous mixture containing MDA-MB-231, MCF-10A, and MCF-7 cells to represent early, intermediate, and late-staged breast cancer and achieved selective concentration of MDA-MB-231 cells using cDEP [Fig. 5(c)]. Future studies for this device are targeted on improving sensitivity and efficiency with the use of clinically relevant sample mixtures.
B. Leukemia cells
Cancer of the blood, commonly known as leukemia, is caused by uncontrolled proliferation and accumulation of leukocytes, one type of white blood cell (WBC).102 There are four main categories of leukemia: acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), chronic lymphocytic leukemia (CLL), and chronic myeloid leukemia (CML).103 There are many existing leukemia cell lines used in various research studies on leukemia,104,105 and we came across four common leukemia cell lines in our studies of DEP enabled platforms: HL-60, K-562, THP-1, and Jurkat T-lymphoblasts.
HL-60 cells were tested using a newly developed method, named as DEP spring by Su, Prieto, and Voldman.87 This device has an angled coplanar electrode configuration and incorporates an automated system to measure the electrical properties of cells at different frequencies and different medium conductivities under continuous flow conditions by determining the Re[fCM] data from the DEP and hydrodynamic drag force balance [Fig. 6(a)]. This provides insight into the heterogeneity of the electrical properties of the cells and overcomes the limitations reported in other studies that were only able to measure cell electrical properties in low-conductivity media.
FIG. 6.
(a) Image of a microfluidic device with the angled coplanar electrode configuration. Reproduced with permission from Su et al., Lab Chip 13, 4109 (2013). Copyright 2013 Royal Society of Chemistry Publishing (b) Motion of K-562 cells in the dielectrophoretic activated cell sorter. Reproduced with permission from Lee et al., Micro Nano Syst. Lett. 4, 2 (2016).106 Copyright 2016 Springer. (c) Timed sequence of the movement of THP-1 in the microelectrode array configuration. Reprinted with permission from Yasukawa et al., Sens. Actuators, B 186, 9 (2013). Copyright 2013 Elsevier.
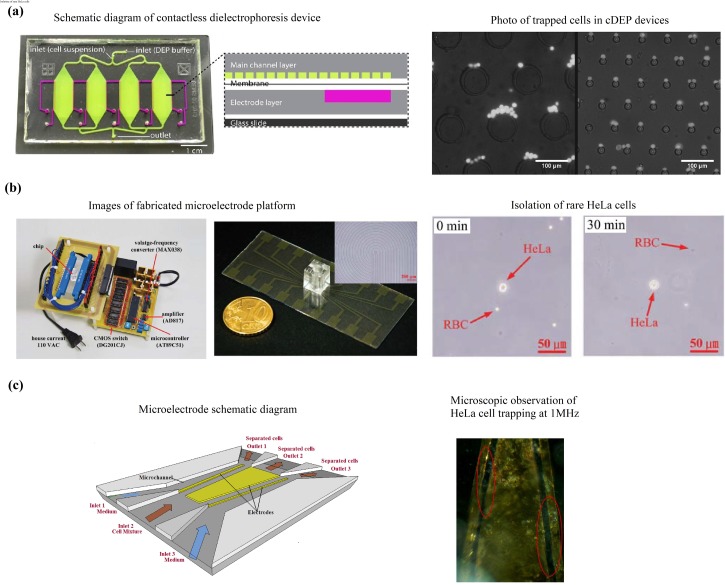
In another study, Lee et al.106 proposed a platform that integrates nDEP force and gravitational forces to sort K-562 cells [Fig. 6(b)]. The repeated nDEP barrier configuration was shown to be an effective method of enhancing separation efficiency, while the minimization of components reduced assembly difficulties, leaking between components, and cell adhesion in component gaps. The device achieved a maximum separation efficiency of 94.74 ± 0.77%, a throughput of 17 000 cells min−1, and a recovery rate of 62.5%. Some studies show potential but required further investigation into the methodology and performance optimization. Yasukawa et al.107 developed a microelectrode array configuration using repulsive nDEP forces induced by navigator and separator electrodes to guide and separate red blood cells (RBCs) and THP-1 cells [Fig. 6(c)]. Novickij, Grainys, and Novickij108 conducted experiments with Jurkat T-lymphoblasts using a pulsed magnetic field cDEP device.
C. Prostate cancer cell
Prostate cancer affects one in six men, with a 3.4% chance of death, and it is the second-leading cause of death in men.109,110 Prostate tumors grow slowly and have a long latent period that makes early diagnosis possible. The most common screening methods currently used for prostate cancer are digital rectal examination111 and serum prostate specific antigen (PSA) testing.112 Digital rectal examination is invasive,111 while serum PSA testing tends to cause over diagnosis and over treatment113 while lacking sensitivity for men who have elevated serum PSA but do not develop lethal prostate cancer. Here, DEP-enabled platforms using prostate cancer cell lines, PC-3 and 22Rv1, to develop simple, fast, and accurate early diagnostics will be discussed.
PC-3 has been used in DEP-enabled microfluidic platforms by Salmanzadeh et al.,17 Sun et al.,114 and Chiu et al.20 Salmanzadeh et al.17 demonstrated the separation of PC-3 cells and non-prostate cancer cells using cDEP [Fig. 7(a)]. The advantage of cDEP is the absence of contact between the electrodes and the sample inside the fluidic channels, which avoids any contaminating effects the electrodes may have on the sample. This method separates the cells based on the difference of membrane surface proteins and their electrical charge. The experiments showed that PC-3 exhibited stronger DEP force than other cells at a specific frequency and voltage. Sun et al.114 conducted continuous separation of PC-3 cells from polystyrene microbeads and between live and dead PC-3 cells using a metal-electrode-free device. The technique used is conductivity-induced dielectrophoresis with 3D self-assembled ionic liquid electrodes. The schematic diagram in Fig. 7(b) shows that particles with different electrical properties and sizes are deflected by the DEP force differently and separate from each other in the electrode region. The dashed circles represent the intermediate position of the particles. They reported that features of this device enhance cell viability such as low-voltage and low-frequency operating electric fields, there is no direct contact between cells and ionic liquid, and cells are collected in culture medium. Additionally, the absence of metal electrodes makes fabrication simpler and cheaper.
FIG. 7.
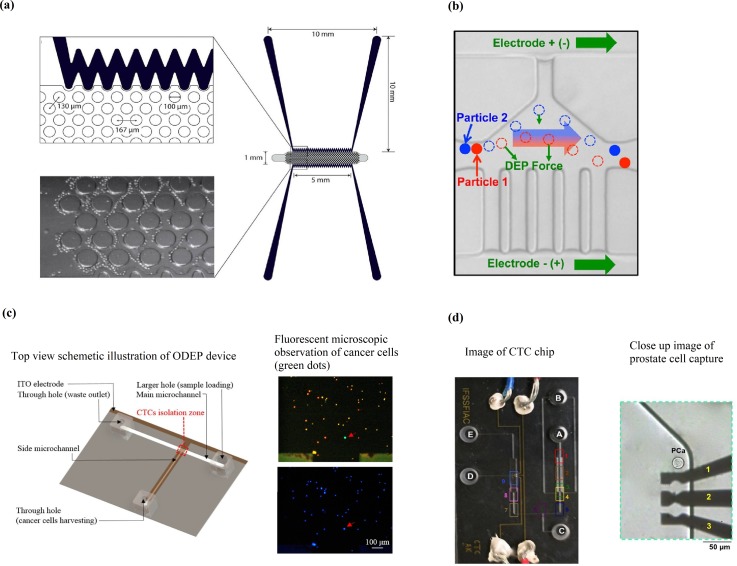
(a) Contactless DEP (cDEP) microdevice (bottom). Reproduced with permission from Salmanzadeh et al., Lab Chip 12, 182 (2012). Copyright 2012 Royal Society of Chemistry. (b) Schematic showing the mechanism of the particle and cell separation of the DEP device with ionic liquid electrodes. Reproduced with permission from Sun et al., Anal. Chem. 88, 8264 (2016). Copyright 2016 ACS Publications. (c) Optical induced dielectrophoretic (ODEP) device. Reproduced with permission from Chiu et al., Sci. Rep. 6, 32851 (2016). Copyright 2016 Nature. (d) Single-cell capture microfluidic platform. Reproduced from permission from Biomicrofluidics 9, 064104 (2015). Copyright 2015 AIP Publishing LLC.
Chiu et al.20 reported on the separation of PC-3 cells from leukocytes using an oDEP-based microfluidic device [Fig. 7(c)]. oDEP is advantageous due to the flexibility and user-friendly manner of creating or modifying the electrode layout. The electrode layout is designed and controlled with a computer interface and projected onto the microchannel via a digital projector. They characterized the light configurations and parameters for effective manipulation of leukocytes and PC-3 cells and reported an isolation purity of PC-3 cells of 100% and a cell recovery rate of 41.5%.
Khamenehfar et al.115 demonstrated single cell capturing of 22Rv1 cells using a cell retention chamber with a 3 DEP electrode configuration [Fig. 7(d)]. The microfluidic platform incorporates cross-flow microfilters to first remove red blood cells (RBCs), followed by selective trapping of 22Rv1 cells from white blood cells (WBCs) in the cell retention chamber. They were also able to demonstrate good cell viability and the ability for drug accumulation measurements from single cell trapping of 22Rv1 cells. While this study offers a better understanding of incorporating filtration and DEP techniques in label-free single cell trapping for multiple rounds of single cell measurements, it lacks in reporting on optimal operating parameters that are crucial for developing a clinically viable device.
D. Cervical and ovarian cancer cells
Cervical and ovarian cancers are malignancies of female reproductive organs.116 Cervical cancer is unstoppable growth of abnormal tissue within the cervix of the uterus. HeLa cells, derived from an aggressive cervical adenocarcinoma, are a commonly used cervical cancer cell line.117 Ovarian cancer research commonly uses mouse ovarian surface epithelial (MOSE) cells that are derived from mouse ovary cancer.
Čemažar et al.118 reported trapping of MOSE cells in a new high-throughput cDEP device [Fig. 8(a)]. The device achieved optimum cell viabilities of 71% and 81% in untrapped and trapped MOSE cells, respectively, and is capable of sorting over 106 cells/h. The improvements in the design include small trapping areas for even distribution of cells along the channel to increase throughput. The device also incorporates a multilayer design that enhances robustness and enables more electric field penetration through the insulating membrane to improve separation between similar cell types at lower frequency electric fields. These improvements alleviate the issues of cell-to-cell interactions of typical iDEP and cDEP devices. Additional efforts are needed to assess and quantify the enhanced selectivity of the device over the more conventional platform.
FIG. 8.
Contactless dielectrophoresis (cDEP) device. (a) Reproduced with permission from Biomicrofluidics 10, 014109 (2016). Copyright 2016 AIP Publishing LLC. (b) Stepping field dielectrophoretic device. Reproduced with permission from Chen et al., Biochip J. 8, 67 (2014). Copyright 2014 Springer. (c) 3-inlet 3-outlet tapered planar microelectrode. Reproduced with permission from Das et al., Med. Eng. Phys. 36, 726 (2014). Copyright 2014 Elsevier.
HeLa cells were also used in another study by Chen et al.18 in a polydimethylsiloxane (PDMS) chamber that concentrates the cancer cells with DEP force generated by stepping fields [Fig. 8(b)]. Target cells experienced pDEP and were collected in the center of the chamber. They achieved 76%–80% separation efficiency of HeLa cells from blood in the central electrode. They noted that further studies were needed to identify and optimize separation of cancer cells from WBCs, which are of similar size and shape. Das et al.119 characterized the DEP response of HeLa cells in phosphate buffer solution (PBS) on a 3-inlet 3-outlet microfluidic platform with a tapered planar microelectrode configuration [Fig. 8(c)]. It was determined that the crossover frequency at 10Vpp was between 700 kHz–1 MHz. Cell viability was not affected as the electric field from numerical simulations was lower than the electroporation threshold value.
E. Bone, colorectal, and lung cancer cells
This section summarizes the studies of the DEP-enabled microfluidic platform on cancer cells which are less reported, i.e., bone, colorectal, and lung cancer cells.
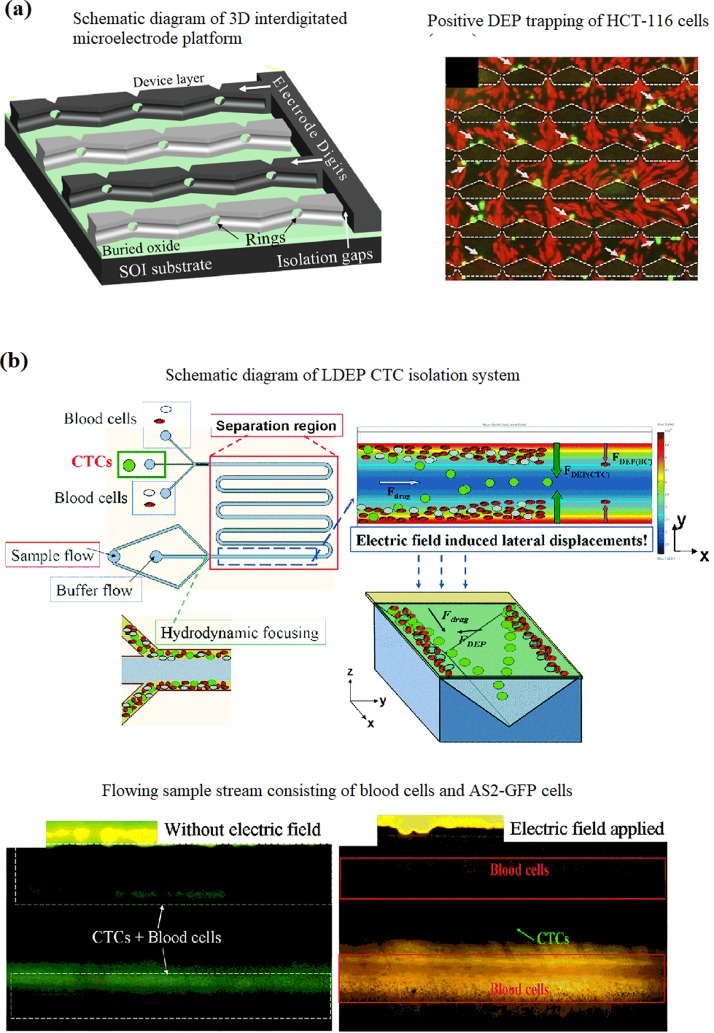
Xing et al.120 reported on a DEP-enabled IDT ring-array microelectrode for label-free enumeration of colorectal cancer cells, HCT-116, from blood lymphocytes. A 3D rendering of the overall design is shown in [Fig. 9(a)], which shows the electrode structure in a partially drawn PDMS cap. Two adjacent inlet ports that feature separate syringe pumps supply the chamber with cell suspension and elution buffer. A single outlet port collects the eluent. Selective trapping of HCT-116 cells (green) by pDEP when AC voltage is applied is indicated with arrows. Although the lymphocytes were under a weak pDEP, they appear to be unaffected by the field because of the overwhelming drag force of the flowing cell suspension.
FIG. 9.
(a) 3D interdigitated microelectrode platform. Reproduced with permission from Xing et al., Biosens. Bioelectron. 61, 434 (2014). Copyright 2014 Elsevier. (b) The lateral dielectrophoretic (LDEP) CTC isolation system. Reproduced with permission from Cheng et al., Lab Chip 15, 2950 (2015). Copyright 2015 Royal Chemistry of Society.
The device achieved a high loading density of 107 cells ml−1 with an 82% recovery rate of cancer cells and a removal of 99% of blood lymphocytes with 94% cell viability. The data show that the device has a highly effective/influential force field across the entire flow chamber. This design can be incorporated into electrical detection and enumeration of the cancer cells isolated from lymphocytes by measuring impedance or conductivity changes across the IDT microelectrodes before and after capturing cancer cells.
Ling et al.121 presented a periodic array of discrete but locally asymmetric triangular bottom microelectrodes and a continuous top electrode, and they achieved a separation efficiency of 82.8% of bone cancer cells, MG-63, from erythrocytes. This was a modified and an improved design from a previous study that had eliminated unwanted trapping of cells at the microelectrodes and a failure to promote effective separation of cells under pDEP. The periodic array design configuration was to replicate the non-uniform electric field cycles generated by the power on/off duty cycles to achieve cell separation with low cell adhesion. This study, however, did not present results on cell viability and design configuration of outlet channels for collecting the separated cells.
Lung cancer cells, AS2-GFP, were separated from blood in a study by Cheng et al.88 using a 3D lateral dielectrophoretic (LDEP) configuration, which provides a long-range field gradient for rare cell isolation [Fig. 9(b)]. The induced LDEP forces on the blood cells and CTCs produce different LDEP velocities normal to the through-flow and balance the fluid viscosity at different equilibrium positions, resulting in the separation of CTCs and blood cells which are collected at a distance of 150–200 μm from the channel side walls by hydrodynamic focusing. When a determined electric field was applied, AS2-GFP cells experienced a higher LDEP force that induced a longer lateral displacement, which manipulated them into the middle region of the channel. Blood cells experienced a lower LDEP force that induced a shorter displacement, and they were only transported to a distance of ∼200 μm from the channel side walls. The device achieved an enrichment factor of 105 and a recovery rate of 85% from a 0.001% cancer cell sample. An optimal flow rate of 20 μl min−1 passing through a 6-cm long LDEP channel with an appropriate voltage at a frequency of 10 kHz was used. The throughput of this platform can be increased by increasing the channel length, which prolongs the particle residence time in the LDEP field. We emphasize that the fabrication of this platform is also simplified as the steps for electrode patterning and precise alignment in the top and bottom layers are eliminated.
VI. FUTURE TRENDS
Important developments are being made to further improve the capabilities of established techniques of DEP-enabled cancer cell manipulation microfluidic devices. These developments give insights into the requirements of various aspects of a clinically suitable cancer diagnostics device that is commercially viable. They address the needs by improving various design aspects of the microfluidic platforms, which we will briefly summarize here.
A focus in developing DEP platforms that are compatible with the existing and emerging cancer treatment approaches and modalities including chemo-, radiation, plasma, and hybrid onco-therapies is needed to address the deficiencies of methods currently available. This is demonstrated by Soltanian-Zadeh et al.99 where changes in drug-treated MCF-7 cells from untreated MCF-7 cells were detected with their iDEP platform which traditional methods have failed to detect. This shows that DEP devices offer the possibility for better post-treatment analysis and diagnosis than conventional and traditional methods.
Design flexibility is important in improving throughput and efficiency of the DEP device for fast and accurate cancer diagnostics. One strategy that enhances design flexibility is the use of a wireless bipolar electrode (BPE) array as demonstrated by Li and Anand.101 This electrode design removes the need for ohmic contact to individual array elements, which enables more flexible device configurations such as allowing non-target cells to pass through the array in the open-design and achieving high-fidelity single-cell capture for interconnected parallel microchannel design when pocket dimensions were matched to those of the cells. This is an important feature in developing single-cell sorting platforms which could be used to enumerate or identify the cells after the desired isolation/sorting has been achieved. An additional flexibility feature of the device is the scalability in the x- and y- directions to increase throughput of the device. Studies with other clinically relevant samples such as heterogeneous cell mixtures, samples at different stages of cancer, and samples with cancer cell concentration that resemble CTCs would further emphasize this application for cancer diagnostics.
Most DEP platforms available are demonstrated with regular laboratory cancer cell suspension mediums which have high conductivity. Low conductivity suspension mediums have a lower cross-over frequency,123 and hence, the ability for DEP platforms to operate at low frequencies is vital for low conductivity suspension mediums. Small electrodes experience high electrode interfacial impedance at low frequencies due to double layer capacitance.76 Ionic liquid electrodes have demonstrated the ability to perform sorting and impedance measurements of cancer cells at low frequencies.76,114 Additionally, ionic electrodes are advantageous for dielectric spectroscopy since the field in the measurement region is horizontal and perpendicular to the flow. Hydrodynamic focusing ensures passage of the cells in the center of the channel only, eliminating the influence of the cell trajectory with respect to the electrodes. The signal amplitude is determined by the cell size and cell dielectric properties, while the exact cell trajectory does not interfere. This increases the accuracy of dielectric characterization of cells. Another method that has demonstrated the ability to operate in a wide range of medium conductivities is the DEP spring method developed by Su et al.87 The ease of use of this device was enhanced with the incorporation of an automated system that measures the electrical properties of cells at different frequencies and different medium conductivities under continuous flow conditions by determining the Re[fCM] data from the DEP and hydrodynamic drag force balance.
VII. SUMMARY
In this review, recent technological advances in DEP-enabled cancer cell microfluidic platforms are discussed and critically reviewed. Emerging methods have been highlighted for their simplified and cost-efficient fabrication methods, improvement in cell sorting performances, ability to perform continuous sorting with heterogeneous cell samples, and single-cell trapping capabilities. Studies have also been conducted to further develop the portability and clinical viability functions of microfluidic devices. Many studies on microfluidics have demonstrated their biomedical applications in the CTC detection single-cell analysis and dielectric spectroscopy. These microfluidic technologies have shown promising opportunities in cancer cell studies through precise control of cells. Although there are still many challenges in further applications of DEP-enabled microfluidic platforms for cancer cell analysis, significant advances have been achieved in recent years. With continuous advancements and development, this technology could potentially and commercially compete or complement other approaches for companion diagnostics of cancer.
ACKNOWLEDGMENTS
The authors acknowledge support from the Ministry of Higher Education of Malaysia (MOHE) for support under Fundamental Research Grant Scheme (FRGS), FRGS/1/2015/TK04/MMU/02/9. The authors also thank the Director-General of Health Malaysia for his permission to publish this paper and the Director of the Institute for Medical Research for her support.
References
- 1. Panikkanvalappil S. R., MacKey M. A., and El-Sayed M. A., J. Am. Chem. Soc. 135, 4815 (2013). 10.1021/ja400187b [DOI] [PubMed] [Google Scholar]
- 2. Qiao G., Duan W., Chatwin C., Sinclair A., and Wang W., J. Phys.: Conf. Ser. 224, 12081 (2010). 10.1088/1742-6596/224/1/012081 [DOI] [Google Scholar]
- 3. Antfolk M., Antfolk C., Lilja H., Laurell T., and Augustsson P. E. R., Lab Chip 15, 2102 (2015). 10.1039/C5LC00078E [DOI] [PubMed] [Google Scholar]
- 4. Leroy J., Dalmay C., Landoulsi A., Hjeij F., Mélin C., Bessette B., Bounaix Morand Du Puch C., Giraud S., Lautrette C., Battu S., Lalloué F., Jauberteau M. O., Bessaudou A., Blondy P., and Pothier A., Sens. Actuators, A 229, 172 (2015). 10.1016/j.sna.2015.04.002 [DOI] [Google Scholar]
- 5. Zhang H., Tu E., Hagen N. D., Schnabel C. A., Paliotti M. J., Hoo W. S., Nguyen P. M., Kohrumel J. R., Butler W. F., Chachisvillis M., and Marchand P. J., Biomed. Microdevices 6, 11 (2004). 10.1023/B:BMMD.0000013361.03291.6c [DOI] [PubMed] [Google Scholar]
- 6. Babjuk M., Burger M., Zigeuner R., Shariat S. F., Van Rhijn B. W. G., Compérat E., Sylvester R. J., Kaasinen E., Böhle A., Palou Redorta J., and Rouprêt M., Eur. Urol. 64, 639 (2013). 10.1016/j.eururo.2013.06.003 [DOI] [PubMed] [Google Scholar]
- 7. Shields C. W., Johnson L. M., Gao L., and López G. P., Langmuir 30, 3923 (2014). 10.1021/la404677w [DOI] [PubMed] [Google Scholar]
- 8. Guo F., Li P., French J. B., Mao Z., Zhao H., Li S., Nama N., Fick J. R., Benkovic S. J., and Huang T. J., Proc. Natl. Acad. Sci. U.S.A. 112, 43 (2015). 10.1073/pnas.1422068112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tadimety A., Syed A., Nie Y., Long C. R., Kready K. M., and Zhang J. X. J., Inegr. Biol. 9, 22 (2017). 10.1039/C6IB00202A [DOI] [PubMed] [Google Scholar]
- 10. Cheng I. F., Chang H. C., Hou D., and Chang H. C., Biomicrofluidics 1, 021503 (2007). 10.1063/1.2723669 [DOI] [Google Scholar]
- 11. Mu X., Zheng W., Sun J., Zhang W., and Jiang X., Small 9, 9 (2013). 10.1002/smll.201200996 [DOI] [PubMed] [Google Scholar]
- 12. Ali M. A. M., (Ken) Ostrikov K., Khalid F. A., Majlis B. Y., and Kayani A. A., RSC Adv. 6, 113066 (2016). 10.1039/C6RA20080J [DOI] [Google Scholar]
- 13. Pohl H. A., J. Appl. Phys. 22, 869 (1951). 10.1063/1.1700065 [DOI] [Google Scholar]
- 14. Auerswald J., Linder V., and Knapp H. F., Microelectron. Eng. 73–74, 822–829 (2004). 10.1016/S0167-9317(04)00228-X [DOI] [Google Scholar]
- 15. Green N. G. and Morgan H., “AC electrokinetics,” in Encyclopedia of Microfluidics and Nanofluidics (Springer, 2002), Vol. 2, p. 8.
- 16. Henslee E. A., Sano M. B., Rojas A. D., Schmelz E. M., and Davalos R. V., Electrophoresis 32, 2523 (2011). 10.1002/elps.201100081 [DOI] [PubMed] [Google Scholar]
- 17. Salmanzadeh A., Romero L., Shafiee H., Gallo-Villanueva R. C., Stremler M. A., Cramer S. D., and Davalos R. V., Lab Chip 12, 182 (2012). 10.1039/C1LC20701F [DOI] [PubMed] [Google Scholar]
- 18. Chen G.-H., Te Huang C.-T., Wu H.-H., Zamay T. N., Zamay A. S., and Jen C.-P., Biochip J. 8, 67 (2014). 10.1007/s13206-014-8201-4 [DOI] [Google Scholar]
- 19. LaLonde A., Romero-Creel M. F., Saucedo-Espinosa M. A., and Lapizco-Encinas B. H., Biomicrofluidics 9, 064113 (2015). 10.1063/1.4936371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chiu T.-K., Chou W.-P., Huang S.-B., Wang H.-M., Lin Y.-C., Hsieh C.-H., and Wu M.-H., Sci. Rep. 6, 32851 (2016). 10.1038/srep32851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Çetin B. and Li D., Electrophoresis 32, 2410 (2011). 10.1002/elps.201100167 [DOI] [PubMed] [Google Scholar]
- 22. Qian C., Huang H., Chen L., Li X., Ge Z., Chen T., Yang Z., and Sun L., Int. J. Mol. Sci. 15, 18281 (2014). 10.3390/ijms151018281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Demircan Y., Özgür E., and Külah H., Electrophoresis 34, 1008 (2013). 10.1002/elps.201200446 [DOI] [PubMed] [Google Scholar]
- 24. Chen Y., Li P., Huang P., Xie Y., Mai J. D., Wang L., Nguyen N., and Huang T. J., Lab Chip 14, 626 (2014). 10.1039/c3lc90136j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Plaks V., Koopman C. D., and Werb Z., Science 341, 1186 (2013). 10.1126/science.1235226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. den Toonder J., Lab Chip 11, 375 (2011). 10.1039/c0lc90100h [DOI] [PubMed] [Google Scholar]
- 27. Huang S.-B., Wu M.-H., Lin Y.-H., Hsieh C.-H., Yang C.-L., Lin H.-C., Tseng C.-P., and Lee G.-B., Lab Chip 13, 1371 (2013). 10.1039/c3lc41256c [DOI] [PubMed] [Google Scholar]
- 28. Patil P., Madhuprasad M., Kumeria T., Losic D., and Kurkuri M., RSC Adv. 5, 89745 (2015). 10.1039/C5RA16489C [DOI] [Google Scholar]
- 29. Yu L., Ng S. R., Xu Y., Dong H., Wang Y. J., and Li C. M., Lab Chip 13, 3163 (2013). 10.1039/c3lc00052d [DOI] [PubMed] [Google Scholar]
- 30. Cima I., Wen Yee C., Iliescu F. S., Min Phyo W., Hon Lim K., Iliescu C., and Han Tan M., Biomicrofluidics 7, 11810 (2013). 10.1063/1.4780062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pethig R. and Pethig R., Biomicrofluidics 4, 022811 (2013). 10.1063/1.3456626 [DOI] [Google Scholar]
- 32. Jubery T. Z., Srivastava S. K., and Dutta P., Electrophoresis 35, 691 (2014). 10.1002/elps.201300424 [DOI] [PubMed] [Google Scholar]
- 33. Khamenehfar A. and Li P. C. H., Curr. Pharm. Biotechnol. 17, 810 (2016). 10.2174/1389201017666160301103509 [DOI] [PubMed] [Google Scholar]
- 34. Perez-Gonzalez V. H., Gallo-Villanueva R. C., Camacho-Leon S., Gomez-Quiñones J. I., Rodriguez-Delgado J. M., and Martinez-Chapa S. O., IET Nanobiotechnol. 10, 263 (2016). 10.1049/iet-nbt.2015.0060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adekanmbi E. O. and Srivastava S. K., Lab Chip 16, 2148 (2016). 10.1039/C6LC00355A [DOI] [PubMed] [Google Scholar]
- 36. Cooper G. M. and Hausman R. E., The Cell : A Molecular Approach ( Sinauer Associates, 2015). [Google Scholar]
- 37. Lodish H., Berk A., Kaiser C. A., Krieger M., and Bretscher A., Molecular Cell Biology ( W. H. Freeman, 1999). [Google Scholar]
- 38. Liberko M., Kolostova K., and Bobek V., Crit. Rev. Oncol. 88, 338 (2013). 10.1016/j.critrevonc.2013.05.002 [DOI] [PubMed] [Google Scholar]
- 39. Azevedo S. A. S., Follain G., Patthabhiraman S., Harlepp S., and Goetz J. G., Cell Adhesion and Migration 9, 345–356 (2015). 10.1080/19336918.2015.1059563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Toss A., Mu Z., Fernandez S., and Cristofanilli M., Ann. Transl. Med. 2, 108 (2014) 10.3978/j.issn.2305-5839.2014.09.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wolf A. M. D., Wender R. C., Etzioni R. B., Thompson I. M., D'Amico A. V., Volk R. J., Brooks D. D., Dash C., Guessous I., Andrews K., DeSantis C., and Smith R. A., CA-Cancer J. Clin. 60, 70 (2010). 10.3322/caac.20066 [DOI] [PubMed] [Google Scholar]
- 42. Chuaqui R. F., Herold K. E., and Rasooly A., Biosensors and Molecular Technologies for Cancer Diagnostics ( Taylor & Francis, 2012), pp. 3–40. [Google Scholar]
- 43. Harouaka R. A., Nisic M., and Zheng S.-Y., J. Lab. Autom. 18, 455 (2013). 10.1177/2211068213494391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li J., Li S., and Yang C. F., Electroanalysis 24, 2213 (2012). 10.1002/elan.201200447 [DOI] [Google Scholar]
- 45. Golubnitschaja O. and Flammer J., Surv. Ophthalmol. 52, S155 (2007). 10.1016/j.survophthal.2007.08.011 [DOI] [PubMed] [Google Scholar]
- 46. Deng Y., Yi L., Lin X., Lin L., Li H., and Lin J.-M., Talanta 144, 136 (2015). 10.1016/j.talanta.2015.05.065 [DOI] [PubMed] [Google Scholar]
- 47. Sano M. B., Caldwell J. L., and Davalos R. V., Biosens. Bioelectron. 30, 13 (2011). 10.1016/j.bios.2011.07.048 [DOI] [PubMed] [Google Scholar]
- 48. Schneider T., Moore L. R., Jing Y., Haam S., Williams P. S., Fleischman A. J., Roy S., Chalmers J. J., and Zborowski M., J. Biochem. Biophys. Methods 68, 1 (2006). 10.1016/j.jbbm.2006.02.011 [DOI] [PubMed] [Google Scholar]
- 49. Kayani A. A., Khoshmanesh K., Ward S. A., Mitchell A., and Kalantar-zadeh K., Biomicrofluidics 6, 31501 (2012). 10.1063/1.4736796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reichl M. R. and Braun D., J. Am. Chem. Soc. 136, 15955 (2014). 10.1021/ja506169b [DOI] [PubMed] [Google Scholar]
- 51. Burguillos M. A., Magnusson C., Nordin M., Lenshof A., Augustsson P., Hansson M. J., Elmér E., Lilja H., Brundin P., Laurell T., and Deierborg T., PLoS One 8, e64233 (2013). 10.1371/journal.pone.0064233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kang G., Kim Y.-J., Moon H.-S., Lee J.-W., Yoo T.-K., Park K., and Lee J.-H., Biomicrofluidics 7, 044126 (2013). 10.1063/1.4818838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang F., Yang X., Jiang H., Bulkhaults P., Wood P., Hrushesky W., and Wang G., Biomicrofluidics 4, 013204 (2010). 10.1063/1.3279786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kamyabi N. and Vanapalli S. A., Biomicrofluidics 10, 21102 (2016). 10.1063/1.4944057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lin B. K., McFaul S. M., Jin C., Black P. C., and Ma H., Biomicrofluidics 7, 17 (2013) 10.1063/1.4812688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jung J., Seo S.-K., Joo Y.-D., and Han K.-H., Sensors Actuators B Chem. 157, 314–320 (2011) 10.1016/j.snb.2011.03.042. [DOI] [Google Scholar]
- 57. Simon M. G., Li Y., Arulmoli J., McDonnell L. P., Akil A., Nourse J. L., Lee A. P., and Flanagan L. A., Biomicrofluidics 8, 64106 (2014). 10.1063/1.4902371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Destgeer G., Jung J. H., Park J., Ahmed H., and Sung H. J., Anal. Chem. 89, 736–744 (2017). 10.1021/acs.analchem.6b03314 [DOI] [PubMed] [Google Scholar]
- 59. Mark D., Haeberle S., Roth G., Stetten F. Von, and Zengerle R., NATO Sci. Peace Secur. Ser. A Chem. Biol. 2010, 305–376. 10.1007/978-90-481-9029-4-17 [DOI] [Google Scholar]
- 60. Zeng J., Deng Y., Vedantam P., Tzeng T.-R., and Xuan X., J. Magn. Magn. Mater. 346, 118 (2013). 10.1016/j.jmmm.2013.07.021 [DOI] [Google Scholar]
- 61. Huang N.-T., Zhang H.-L., Chung M.-T., Seo J. H., and Kurabayashi K., Lab Chip 14, 1230 (2014). 10.1039/C3LC51211H [DOI] [PubMed] [Google Scholar]
- 62. Destgeer G., Ha B. H., Park J., Jung J. H., Alazzam A., and Sung H. J., Phys. Procedia 70, 34 (2015). 10.1016/j.phpro.2015.08.028 [DOI] [Google Scholar]
- 63. Avesar J., Ben Arye T., and Levenberg S., Lab Chip 14, 2161 (2014). 10.1039/C4LC00013G [DOI] [PubMed] [Google Scholar]
- 64. Ohlin M., Iranmanesh I., Christakou A. E., and Wiklund M., Lab Chip 15, 3341 (2015). 10.1039/C5LC00490J [DOI] [PubMed] [Google Scholar]
- 65. Lenshof A., Magnusson C., and Laurell T., Lab Chip 12, 1210 (2012). 10.1039/c2lc21256k [DOI] [PubMed] [Google Scholar]
- 66. Gascoyne P. R. C. and Vykoukal J., Electrophoresis 23, 1973 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li M., Li W., Zhang J., Alici G., Wen W., Online R., Li M., Li W., Zhang J., Alici G., and Wen W., J. Phys. D: Appl. Phys. 47, 1 (2014) 10.1088/0022-3727/47/6/063001. [DOI] [Google Scholar]
- 68. Pohl H. A. and Crane J. S., J. Theor. Biol. 37, 1 (1972). 10.1016/0022-5193(72)90112-9 [DOI] [PubMed] [Google Scholar]
- 69. Vykoukal J. and Vykoukal D. M., Micro and Nano Manipulations for Biomedical Applications, in Dielectrophoretic Methods for Biomedical Applications ( Artech House, 2008), Ed. 1, pp. 179, Chap. 7. [Google Scholar]
- 70. Chikkaveeraiah B. V., Bhirde A. A., Morgan N. Y., Eden H. S., and Chen X., ACS Nano 6, 6546 (2012). 10.1021/nn3023969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Turcu I. and Lucaciu C. M., J. Phys. A: Gen. Phys. 22, 985 (1989). 10.1088/0305-4470/22/8/014 [DOI] [Google Scholar]
- 72. Jones T. B., Electromechanics of Particles ( Cambridge University Press, Cambridge, 1995), p. 265. [Google Scholar]
- 73. Gascoyne P. R. C. and Vykoukal J. V., Proc. IEEE 92, 22–42 (2004). 10.1109/JPROC.2003.820535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jones T. B., IEEE Eng. Med. Biol. Mag. 22, 33 (2003). 10.1109/MEMB.2003.1304999 [DOI] [PubMed] [Google Scholar]
- 75. Polevaya Y., Ermolina I., Schlesinger M., Ginzburg B.-Z., and Feldman Y., Biochim. Biophys. Acta, Biomembr. 1419, 257 (1999). 10.1016/S0005-2736(99)00072-3 [DOI] [PubMed] [Google Scholar]
- 76. Valero A., Braschler T., and Renaud P., Lab Chip 10, 2216 (2010). 10.1039/c003982a [DOI] [PubMed] [Google Scholar]
- 77. Yang J., Huang Y., Wang X., Wang X., Becker F. F., and Gascoyne P. R. C., Biophys. J. 76, 3307–3314 (1999). 10.1016/S0006-3495(99)77483-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chan K. L., Gascoyne P. R., Becker F. F., and Pethig R., Biochim. Biophys. Acta, Lipids Lipid Metab. 1349, 182 (1997). 10.1016/S0005-2760(97)00092-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Smith J. P., Huang C., and Kirby B. J., Biomicrofluidics 9, 14116 (2015). 10.1063/1.4908049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hwang B., Lee D., Kim B., and Lee J., J. Mech. Sci. Technol. 30, 3749 (2016). 10.1007/s12206-016-0737-x [DOI] [Google Scholar]
- 81. Shim S., Stemke-Hale K., Tsimberidou A. M., Noshari J., Anderson T. E., and Gascoyne P. R. C., Biomicrofluidics 7, 11807 (2013). 10.1063/1.4774304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Pethig R. and Markx G. H., Trends Biotechnol. 15, 426 (1997). 10.1016/S0167-7799(97)01096-2 [DOI] [PubMed] [Google Scholar]
- 83. Hughes M. P., Electrophoresis 23, 2569 (2002). [DOI] [PubMed] [Google Scholar]
- 84. Kirby B. J., Micro- and Nanoscale Fluid Mechanics : Transport in Microfluidic Devices ( Cambridge University Press, 2010). [Google Scholar]
- 85. Voldman J., Annu. Rev. Biomed. Eng. 8, 425 (2006). 10.1146/annurev.bioeng.8.061505.095739 [DOI] [PubMed] [Google Scholar]
- 86. Alazzam A., Mathew B., and Alhammadi F., J. Sep. Sci. 40, 1193 (2017). 10.1002/jssc.201601061 [DOI] [PubMed] [Google Scholar]
- 87. Su H. W., Prieto J. L., and Voldman J., Lab Chip 13, 4109 (2013). 10.1039/c3lc50392e [DOI] [PubMed] [Google Scholar]
- 88. Cheng I.-F., Huang W.-L., Chen T.-Y., Liu C.-W., Lin Y.-D., and Su W.-C., Lab Chip 15, 2950 (2015). 10.1039/C5LC00120J [DOI] [PubMed] [Google Scholar]
- 89. Kayani A. A., Khoshmanesh K., Nguyen T. G., Kostovski G., Chrimes A. F., Nasabi M., Heller D. A., Mitchell A., and Kalantar-zadeh K., Electrophoresis 33, 2075 (2012). 10.1002/elps.201200083 [DOI] [PubMed] [Google Scholar]
- 90. Gencoglu A. and Minerick A., Lab Chip 9, 1866 (2009). 10.1039/b820126a [DOI] [PubMed] [Google Scholar]
- 91. Fiedler S., Shirley S. G., Schnelle T., and Fuhr G., Anal. Chem. 70, 1909 (1998). 10.1021/ac971063b [DOI] [PubMed] [Google Scholar]
- 92. Srivastava S. K., Gencoglu A., and Minerick A. R., Anal. Bioanal. Chem. 399, 301 (2011). 10.1007/s00216-010-4222-6 [DOI] [PubMed] [Google Scholar]
- 93. Shafiee H., Caldwell J. L., Sano M. B., and Davalos R. V., Biomed. Microdevices 11, 997 (2009). 10.1007/s10544-009-9317-5 [DOI] [PubMed] [Google Scholar]
- 94. Hwang H., Lee D.-H., Choi W., and Park J.-K., Biomicrofluidics 3, 14103 (2009). 10.1063/1.3086600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Holliday D. L. and Speirs V., Breast Cancer Res. 13, 215 (2011). 10.1186/bcr2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Cailleau R., Olivé M., and Cruciger Q. V., In Vitro 14, 911 (1978) 10.1007/BF02616120. [DOI] [PubMed] [Google Scholar]
- 97. Kim Y., Lee J., An J., Lee S. H., and Kim B., J. Mech. Sci. Technol. 23, 3132 (2010). 10.1007/s12206-009-0910-6 [DOI] [Google Scholar]
- 98. Bhattacharya S., Chao T.-C., Ariyasinghe N., Ruiz Y., Lake D., Ros R., and Ros A., Anal. Bioanal. Chem. 406, 1855 (2014). 10.1007/s00216-013-7598-2 [DOI] [PubMed] [Google Scholar]
- 99. Soltanian-Zadeh S., Kikkeri K., Shajahan-Haq A. N., Strobl J., Clarke R., and Agah M., Electrophoresis 38, 1988 (2017). 10.1002/elps.201600447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Alazzam A., Stiharu I., Bhat R., and Meguerditchian A. N., Electrophoresis 32, 1327 (2011). 10.1002/elps.201000625 [DOI] [PubMed] [Google Scholar]
- 101. Li M. and Anand R. K., J. Am. Chem. Soc. 139, 8950 (2017). 10.1021/jacs.7b03288 [DOI] [PubMed] [Google Scholar]
- 102. Terwilliger T. and Abdul-Hay M., Blood Cancer J. 7, e577 (2017). 10.1038/bcj.2017.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Hernández J. A., Land K. J., and McKenna R. W., Cancer 75, 381 (1995). [DOI] [PubMed] [Google Scholar]
- 104. Drexler H. G., Dirks W. G., Matsuo Y., and MacLeod R. A. F., Leukemia 17, 416 (2003). 10.1038/sj.leu.2402799 [DOI] [PubMed] [Google Scholar]
- 105. MacLeod R. A. F., Nagel S., Scherr M., Schneider B., Dirks W. G., Uphoff C. C., Quentmeier H., and Drexler H. G., Curr. Med. Chem. 15, 339 (2008). 10.2174/092986708783497319 [DOI] [PubMed] [Google Scholar]
- 106. Lee D., Hwang B., Choi Y., and Kim B., Int. J. Precis. Eng. Manuf. 17, 247 (2016). 10.1007/s12541-016-0032-x [DOI] [Google Scholar]
- 107. Yasukawa T., Yamada J., Shiku H., Mizutani F., and Matsue T., Sens. Actuators, B 186, 9 (2013). 10.1016/j.snb.2013.05.048 [DOI] [Google Scholar]
- 108. Novickij V., Grainys A., and Novickij J., IET Nanobiotechnol. 8, 118 (2014). 10.1049/iet-nbt.2012.0039 [DOI] [PubMed] [Google Scholar]
- 109. Jemal A., Siegel R., Ward E., Hao Y., Xu J., and Thun M. J., CA-Cancer J. Clin. 59, 225 (2009). 10.3322/caac.20006 [DOI] [PubMed] [Google Scholar]
- 110. Sardana G., Dowell B., and Diamandis E. P., Clin. Chem. 54, 1951 (2008). 10.1373/clinchem.2008.110668 [DOI] [PubMed] [Google Scholar]
- 111. Schro F. H., Van Der P., Beemsterboer P., Kruger B., and Rietbergen J., J. Natl. Cancer Inst. 90, 1817 (1998). 10.1093/jnci/90.23.1817 [DOI] [PubMed] [Google Scholar]
- 112. Legler J. M., Feuer E. J., Potosky A. L., Merrill R. M., and Kramer B. S., Cancer Causes Control 9, 519 (1998). 10.1023/A:1008805718310 [DOI] [PubMed] [Google Scholar]
- 113. Etzioni R., Penson D. F., Legler J. M., di Tommaso D., Boer R., Gann P. H., and Feuer E. J., J. Natl. Cancer Inst. 94, 981 (2002). 10.1093/jnci/94.13.981 [DOI] [PubMed] [Google Scholar]
- 114. Sun M., Agarwal P., Zhao S., Zhao Y., Lu X., and He X., Anal. Chem. 88, 8264 (2016). 10.1021/acs.analchem.6b02104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Khamenehfar A., Beischlag T. V., Russell P. J., Ling M. T. P., Nelson C., and Li P. C. H., Biomicrofluidics 9, 064104 (2015). 10.1063/1.4934715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Markovic N. and Markovic O., What Every Woman Should Know about Cervical Cancer ( Springer Netherlands, Imprint, Springer, Dordrecht, 2008). [Google Scholar]
- 117. Kniss D. A. and Summerfield T. L., Reprod. Sci. 21, 1015 (2014). 10.1177/1933719114522518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Čemažar J., Douglas T. A., Schmelz E. M., and Davalos R. V., Biomicrofluidics 10, 014109 (2016). 10.1063/1.4939947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Das D., Biswas K., and Das S., Med. Eng. Phys. 36, 726 (2014). 10.1016/j.medengphy.2013.12.010 [DOI] [PubMed] [Google Scholar]
- 120. Xing X., Poon R. Y. C., Wong C. S. C., and Yobas L., Biosens. Bioelectron. 61, 434 (2014). 10.1016/j.bios.2014.05.054 [DOI] [PubMed] [Google Scholar]
- 121. Ling S. H., Lam Y. C., and Chian K. S., Anal. Chem. 84, 6463 (2012). 10.1021/ac300079q [DOI] [PubMed] [Google Scholar]
- 122. Salmanzadeh A., Sano M. B., Gallo-Villanueva R. C., Roberts P. C., Schmelz E. M., and Davalos R. V., Biomicrofluidics 7, 11809 (2013). 10.1063/1.4788921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Di Martino R., Camarda M., Cascio M., Gallo M., Magliano A., Baldo S., Romano A., Minafra L., Forte G. I., Russo G., Gilardi M. C., Di Raimondo F., Scalese S., and La Magna A., Sens. Bio-Sens. Res. 7, 162 (2016). 10.1016/j.sbsr.2015.12.001 [DOI] [Google Scholar]
- 124. Doh I. and Cho Y. H., Sens. Actuators, A 121, 59 (2005). 10.1016/j.sna.2005.01.030 [DOI] [Google Scholar]