Abstract
Several 3D cell culture systems are currently available to create liver organoids. In general, these systems display better physiologic and metabolic aspects of intact liver tissue, compared with 2D culture systems. However, none of these reliably mimic human liver development, including parallel formation of hepatocyte and cholangiocyte anatomical structures. Here, we show that human fetal liver progenitor cells self-assemble inside acellular liver extracellular matrix (ECM) scaffolds to form 3D liver organoids that recapitulated several aspects of hepato-biliary organogenesis and resulted in concomitant formation of progressively more differentiated hepatocytes and bile duct structures. The duct morphogenesis process was interrupted by inhibiting Notch signaling, attempting to create a liver developmental disease model with a similar phenotype of Alagille syndrome. In the current study, we created an in vitro model of human liver development and disease, physiology and metabolism, supported by liver ECM substrata. We envision that it will be used in the future to study mechanisms of hepatic and biliary development, and for disease modeling and drug screening.
Keywords: Extracellular matrix, Liver development, 3D liver tissue, Organogenesis, Disease modeling
Until recently, most of the available liver cell culture models have been standard 2D (monolayer) systems. However, these models didn’t accurately mimic human liver tissue development and physiology. Improvements in cell culture techniques have enabled the creation of tissue models that better mimic human liver tissue and can advance our understanding of liver disease origin and progression, and aid in the development of novel and improved treatments (1). Currently, there are several models of hepatic micro-tissue available that display human specific metabolism and physiologic responses (2–8), but none are accurately able to mimic facets of liver organogenesis. Hence, the lack of key cell types, or the limitations observed in the replication of organ development are now central milestones being addressed by scientists to make these organoids more complex and mature. Alternatives such as humanized animal liver models (9–11), which can reproduce some human specific drug metabolism, pathogen-host interaction and disease in vivo also cannot replicate human liver development (12) and are restricted to phenomena occurring at a post-natal stage. Our prior research (13), using an intact lobe of an acellular liver extracellular matrix (ECM), perfused with human fetal liver progenitor cells (hFLPCs), showed organization of liver-like tissue with partially functional hepatocytes and biliary ductal structures. In the current study, we created human liver organoids that were self-assembled in vitro from hFLPCs seeded onto small, acellular liver-specific ECM discs. These 3D liver organoids demonstrated simultaneous human hepato-biliary organogenesis and partial metabolic and secretory functions of intact human liver tissue. Beyond the applications for human liver development research and modeling of liver congenital diseases, this system may be incorporated in high-throughput platforms for drug activity and toxicity screening.
Materials and Methods
Sources of Supplies and Reagents (all companies are in the USA unless stated otherwise). Abcam, Cambridge, United Kingdom; Advanced Bioscience Resources, Alameda, CA; American Bio, Natick, MA; Amresco, Solon, Ohio; BD Biosciences, Franklin Lakes, NJ; Bethyl Laboratories, Montgomery, TX; Bioassay Systems, Hayward, CA; Cell Sciences, Canton, MA; Cell Signaling Technology, Beverly, MA;Cole-Palmer, Vernon Hills, IL; Dako, Carpinteria, CA; Invitrogen, Carlsbad, CA; Johnson and Johnson, Arlington, TX; Leica Biosystems, Buffalo Grove, IL; Leica GmbH, Germany; Life Technologies, Carlsbad, CA; Marshall Bioresources, NY; Miltenyi Biotec Inc., Auburn, Ca; Olympus, Tokyo; Peprotech, Rocky Hill, NJ; PhoenixSongs Biologicals, Branford, CT; Phenomenex Inc., Torrance, CA; Qiagen, Maryland; Roche Life Sciences, Indianapolis, IN; Santa Cruz Biotechnology, Dallas, TX; J.L. Shepherd and Associates, Inc., San Fernando, CA; Sigma Aldrich, St. Louis, Mo; Sakura Finetek, Torrance, CA; Tree Star Inc., Ashland, OR; Worthington Biochemical Corporation, CLS-4, New Jersey;
Liver harvesting and decellularization
4–5 weeks old ferret (Marshall Bioresources) livers were utilized throughout all the experiments for decellularization and disc preparation. A detailed description of ferret liver harvesting and decellularization has been described previously (14). Briefly, livers were harvested with intact vessels, and the portal vein was cannulated with 16-guage cannulae (Cathlon Clear – Johnson & Johnson). The livers were then connected to a pump (Masterflex L/S peristaltic pump with Masterflex L/S easy load pump head and L/S 14-gauge tubing, Cole-Palmer) and perfused with 2 liters of distilled water at the rate of 6 ml/min. The livers were then perfused with 4 liters of detergent made up of 1% Triton-X 100 (Sigma-Aldrich Co) with 0.1% Ammonium hydroxide (Sigma-Aldrich). Finally, the livers were perfused with 8 liters of distilled water to wash out the decellularization detergent.
Acellular liver disc preparation
To obtain liver discs, decellularized livers were cut into small lobes and embedded in OCT in plastic molds (Sakura Finetek) and flash frozen with liquid nitrogen. These cryopreserved liver lobes were mounted onto a cryotome (Leica CM1950) to obtain liver ECM discs. The cryotome temperature was set around −8 to −10 C to maintain the liver lobes at warmer temperatures, facilitating thick and intact sectioning of liver lobes. The sections were cut at 300 μm thickness. To generate a disc from the liver sections, an 8mm diameter biopsy punch was used which was equipped with a plunger in order to place the discs in a 48 well plate. The 48 well plates were kept inside the cryotome until required numbers of discs were obtained. The discs were then airs dried for up to 4–6 hours or until they were almost dry. This is a critical step in disc preparation to preserve the intactness of the discs. Following the drying step, the discs were washed carefully with multiple washes of PBS and kept in PBS at 4C until ready for sterilization. The discs were sterilized by gamma irradiation at a dose of 1.5 Mrad (J.L. Shepherd and Associates).
Isolation of hFLPCs
Human fetal livers were obtained from Advanced Bioscience Resources and were at developmental stages between 18 and 21 weeks of gestation. A detailed description for isolation of hFLPCs has been described previously (14). Briefly, non-hepatic tissue was removed by scalpels, and livers were enzymatically digested at 37°C by 6mg/ml collagenase type IV (Worthington Biochemical Corporation) and 2000 units of deoxyribonuclease (Roche Life Sciences). Following digestion, hematopoietic and nonparenchymal cells were separated from the parenchymal cell fraction by density gradient using Histopaque-1077 (Sigma-Aldrich, 10771). The lower fraction cell pellet was re-suspended in Kubota’s medium(15) and plated onto Collagen-IV (5 μg/cm2) (Sigma Aldrich, C5533) and Laminin (1 μg/cm2) (BD Biosciences, Cat# 354259) coated 15-cm culture plates and incubated at 37°C. The cells were washed next day to remove blood cells and were maintained in Kubota’s medium for up to 7 days.
hFLPCs seeding on acellular discs and Matrigel®
hFLPCs were harvested from culture plates using collagenase IV and counted. Sterilized discs were incubated with Kubota medium for 30–45 minutes prior to cell seeding and then air dried in biosafety cabinet. hFLPCs (3×105 – 5×105 cells) were suspended in 10 ul volume in seeding medium for each disc. The cell suspension was slowly pipetted on top of each disc and incubated for about an hour at 37°C for attachment before putting additional seeding medium. As a 2D control, same numbers of hFLPCs were seeded on a collagen IV and laminin coated 48 well tissue culture plates. Matrigel® (Corning, Cat# 3562354) control experiments used hFLPCs (1×105) suspended in 60 μl LDEV-free Matrigel® (5.5μg/ml) and placed in a well of a 96 well plate, allowed to become a gel at 37C for 30 minutes and were supplemented with seeding medium. Next day, the discs and cells in 2D and Matrigel® were incubated with liver differentiation medium made of advanced RPMI containing factors, Ascorbic Acid (10mg/L), Dexamethasone (10−7 M), cAMP (2.45mg/L), hProlactin (1μg/L), hGlucagon (1mg/L), hEGF (40μg/L, R&D Systems), Niacinamide (5mM), Tri-iodothyronine (0.67μg/L), Alpha-Lipoic Acid (0.105mg/L), (D-Ala2, D-Leu5)-Enkephalin Acetate (0.056μg/L), HGF (20ng/ml, Peprotech), Free Fatty Acid Mix (76μl/L), Human Growth Hormone (3.33μg/L), HDL (10mg/L, Cell Sciences), Oncostatin M (10μg/L, Peprotech). All factors were obtained from Sigma Aldrich unless stated otherwise. The culture medium was changed every 24 hours and the discs were cultured for up to 3 weeks before harvesting them after 1 week and 3 weeks for immunohistochemical and molecular analysis. In some experiments 10 nM DAPT was added to the media to inhibit Notch signaling.
Albumin and Urea assays
For hepatocyte functional analysis, culture medium was collected at 7, 14 and 21 days from cells growing on discs, matrigel and in 2D culture. For standard control, adult human hepatocytes were grown in collagen I/Matrigel sandwich culture and medium was collected after 48 hours. The media was stored at −80 until it was used for analysis. For analyzing albumin synthesis, ELISA assay was performed using human albumin ELISA kit (Bethyl Laboratories Inc., E101). At least 3 different media samples were used and analyzed in triplicates. The albumin concentrations were normalized per ng of DNA. Urea was measured by colorimetric assay using Quantichrom Urea Assay Kit (Bioassay systems, DIUR-500) and normalized per ng of DNA.
Drug metabolism assay
Liver organoids were incubated with Phenobarbital (1mM) for inducing CYP450 enzymes for up to 48 hours prior to adding Diazepam and 7-ethoxy coumarin for drug metabolism assay. The culture medium was collected at 3, 6, 12 and 24 hours after addition of Diazepam and 7-ethoxy coumarin. Tri-chloroacetic acid was added to media at 1:4 volume and the samples were incubated overnight at 4C. The samples were then centrifuged and the supernatant was collected and stored at −20C for LC-MS analysis. The metabolites were analyzed using HPLC/MS/MS. The auto sampler and HPLC was a refrigerated Reliance Stacker and dual pump HPLC from Spark Holland. The triple quadrupole mass spectrometer was a Quattro II with a Z-spray interface in the positive ion mode from MicroMass/Waters. The gradient used for column elution was based on the mix of the two following solvents:
HPLC solvent A = H2O: methanol 95:5 with 0.15% formic acid;
HPLC solvent B = methanol.
The gradient of A:B was as follow:
0 min – 95:5, 10min – 30:70, 20min – 30:70, 22min – 95:5, 30min – 95:5 with flowrate of 200uL/min. The source temperature was 80C and the desolvation temperature was 250C using nitrogen. 25uL of cell media was injected onto a Hypersil C18 BD 2.0mm × 150mm column (Phenomenex Inc. USA) at 50C.
The monitored MRM pairs were:
| 7-OH coumarin | 339m/z>163m/z ce=20eV |
| Temazepam | 301m/z>255m/z ce=22eV |
| Oxazepam | 287m/z>241m/z ce=22ev |
| Diazepam | 285m/z>193m/z ce=30ev |
| Nordiazepam | 271m/z>140m/z ce=30eV |
| 7-ethoxycoumarin | 191m/z>163m/z ce=20eV |
| 4-OH coumarin | 163m/z>121m/z ce=24eV |
| 7-OH coumarin | 163m/z>107m/z ce=24ev |
Quantitation of the metabolites was performed using response curves in cell media from 10pg/uL to 1000pg/uL of each metabolite vs 500pg/uL of internal standard 4-OH coumarin.
Immunofluorescence analysis
The discs were fixed in 10% neutral-buffered formalin, tissue processed and paraffin embedded for histological analysis. 5 μm sections were cut from the embedded paraffin blocks using a Leica microtome (Leica Biosystems) and stained with hematoxylin and eosin using an Autostainer XL (Leica Biosystem). For immunofluorescence analysis, the slides were deparaffinized and the antibody target retrieval was carried out using Target Retrieval Solution (Dako, S1700). Following target retrieval, the slides were treated with 1% Sodium Borohydride in PBS to reduce the tissue auto-fluorescence. The slides were then blocked for 30 minutes using serum free protein block (Dako, X0909). Primary antibodies were diluted in antibody diluent (Dako, S0809) prior to adding it on the slides. The slides were incubated over night at 4C with primary antibodies (Supplementary Table 1). The next day, slides were washed (3X) with 1X Tris Buffered Saline with Tween 20 (TBST) for 10 minutes. The slides were then incubated with appropriate Alexa Fluor secondary antibodies for 30 minutes and then washed 3X with 1X TBST. The slides were cover slipped with Prolong Gold Antifade Reagent with DAPI (Cell Signaling Technology, #8961). Imaging was done using either Leica DM4000B (Leica Biosystems) or Olympus Fluoview FV10i. (Olympus, Tokyo)
Gene expression studies
DNA and RNA from the liver organoids were extracted using Allprep DNA/RNA mini kit (Qiagen). cDNA was synthesized from 200–400 ng of total RNA using Superscript III first strand synthesis (Life Technologies). RT-PCR was performed using SuperScript III One-Step RT-PCR system with Taq DNA Polymerase (Life Technologies). The primers used are listed in Supplemental Table 2. Expression of genes analyzed was normalized using GAPDH as a housekeeping gene. The PCR mix was run on a 1% Agarose (American Bio) in a Tris Acetate (Amresco) buffer. The bands were quantified by densitometry analysis using Image J software (n= 3–5). For cytochrome enzymes gene expression analysis, the organoids were not incubated with Phenobarbital prior to the analysis. For quantitative real time PCR, PCR reactions were carried out with Power SYBR Green PCR Master Mix (Life Technologies, Cat. No. 4367659) with primer concentration of 300 nM, and 5–10 ng of cDNA. Each reaction was split into three 10 μl technical replicates and put into 384 well PCR plate. The conditions for the reactions were set at 95°C for 10min, 40 cycles of 95°C for 15s and 60°C for 1min. Ct values were determined by the ABI 7900HT Fast Real-Time PCR system and used for further data analysis. Unpaired t-test was performed to determine statistical significance of the samples.
Flow Cytometry
Following cell harvesting from culture dishes, a sample of cells were taken for fluorescence-activated cell-sorting analysis (FACS analysis). We performed this analysis on 5 different human fetal liver samples. Briefly, 200.000 cells were loaded per tube and fixed in 4% paraformaldehyde and permeabilized with 0.1% saponin for intracellular markers. All cell samples were then pelleted and extensively blocked with the use of Fc Blocking Reagent (Miltenyi Biotec Inc) for 15 minutes. Immunolabelling followed immediately with antibodies raised against EpCAM PE (EBA-1) (Santa Cruz Biotechnology), ICAM1 APC (HA58) (BD Biosciences), α fetoprotein (C-19) (Santa Cruz Biotechnology) plus donkey anti-goat AF594 (Invitrogen), albumin FITC (Bethyl Laboratories), cytokeratin 18 (NCL-CK18) (Leica Biosystems GmbH) plus goat anti-mouse AF633 (Invitrogen), α-smooth muscle actin (ab5694) (Abcam) plus donkey anti-rabbit AF488 (Invitrogen), CD105 PE (266) (BD Biosciences), CD31 APC (WM59) (BD Biosciences) for 30 minutes in the dark at 4C, followed by incubation with secondary antibody where needed for additional 30 minutes after 3 washes. Negative controls were also prepared with mouse monoclonal anti-KLH IgG1 isotype control antibodies FITC, PE and APC (340755, 340761, and 340754, BD Biosciences). In the case of the goat anti-α fetoprotein, mouse anti-CK18, and rabbit anti-α-smooth muscle actin antibodies, staining with secondary antibody alone was used as negative control. Cell fluorescence was measured immediately after staining with a Becton Dickinson FACSCalibur flow cytometer (BD Biosciences) and all data analyzed using FlowJo software v. 7.1.3 (Tree Star Inc).
Results
Lineage specification of human liver progenitors inside liver extracellular matrix
To create human liver organoids we used our previously described decellularization method to fabricate liver extracellular matrix (ECM) scaffolds (13, 14, 16, 17), sectioned the scaffold and seeded sections (8mm ECM discs) with hFLPCs cultures that contained liver stromal (25–40%) and endothelial (5–15%) cells (Suppl. Fig. 1). Within 2–3 weeks post-seeding and culture in liver differentiation medium (LDM), the cells and ECM self-assembled into 3D organoid structures with progressive cellular organization and differentiation (Supp. Fig. 2A, B and C). Large clusters of cells expressing hepatoblast markers (ALB+/CK19+/EpCAM+) were observed after 1 week, suggesting lineage restriction to the hepatoblast (Fig. 1A, Top). After 3 weeks, there were clear changes in cell phenotype including ALB+/CK19−/EpCAM− clusters and ALB−/CK19+/EpCAM+ ductal structures, suggesting parallel lineage specification into hepatocytes and polarized cholangiocytes, respectively (Fig. 1A, Bottom). In contrast, hFLPCs seeded in Matrigel® maintained their progenitor phenotype (ALB+/CK19+/EpCAM+) even after 3 weeks in culture, suggesting no parallel lineage specification into hepatocyte and cholangiocytes (Supp. Fig. 3A). We also identified asymmetrical primitive ductal structures with half of the circumference lined by CK19+ cells and the other by ALB+ cells, suggesting an intermediate phase of lineage specification(18) (Fig. 1B). Progressive maturation of hFLPCs into hepatocytes was assessed by immunostaining, showing clusters of cells expressing both alpha fetoprotein (AFP) and albumin after 1 week of culture, similar to fetal liver tissue (Fig. 1C, left panels). In contrast, after 3 weeks of culture the hFLPCs completely lost AFP expression, suggestive of committed progenitors or a stage of adult liver cells (Fig. 1C, right panels). However, there were several non-stained cells, which constitute a mixed population of stromal cells (data not shown). Gene expression analysis showed expression of HNF4α, a hepatocyte differentiation regulator, in organoids after 3 weeks of culture, to levels that were higher than hFLPCs, but lower than those detected in adult liver (Fig 1D). Cells in Matrigel® showed higher levels of HNF4α compared to hFLPCs but the levels were lower compared to the organoids suggesting less hepatocytic maturation in matrigel (Sup. Fig. 3B). The differentiation results were further supported by expression of SOX9, a biliary marker (Supp. Fig. 3B). Similarly, expression of HNF6, a cholangiocyte differentiation major regulator, progressively increased in organoids between 1 and 3 weeks of culture, and was higher than in hFLPCs and adult liver tissue (Fig 1D). However, the expression level of HNF1β, another regulator of cholangiocyte differentiation and bile duct formation, was higher in liver progenitors, and showed similar lower expression after 1 and 3 weeks of culture, comparable to adult liver (Fig. 1D). Together, these results suggest premature hepatocytic and biliary phenotypes.
Figure 1. Lineage specification of human fetal liver progenitor cells (hFLPCs) and formation of liver organoids.
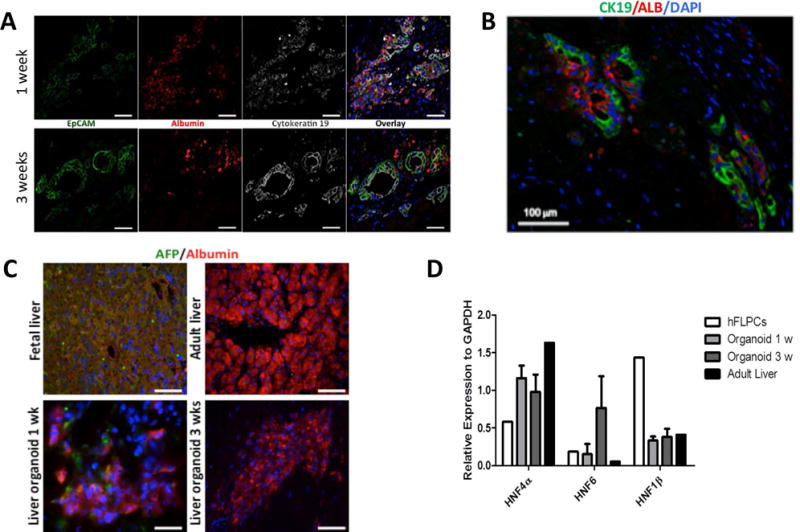
A) Distribution and phenotypic characteristics of hFLPCs during 1 (top panel) and 3 (bottom panel) weeks of differentiation in culture. Cells were stained for epithelial cell adhesion molecule (EpCAM), albumin (ALB), cytokeratin19 (CK19) and for cell nuclei (DAPI). Scale bar is 20 μm. B) Immunostaining of liver organoids after 3 weeks of differentiation shows ductal structures containing cells expressing both CK19 (green) and albumin (red). C) Expression of α-fetoprotein (AFP, green) and albumin (ALB, red) in liver organoids after 1 and 3 weeks differentiation and in fetal and adult liver tissues. Scale bar is 20 μm. D) RT-PCR analysis of the expression of hepatic transcription factors hepatocyte nuclear factor (HNF) 4α, HNF6 and HNF1β in freshly isolated hFLPCs, liver organoids after 1 and 3 weeks differentiation, and in adult liver tissue.
Liver tissue contains specific ECM molecules that surround the different liver zones and regulate specific cell differentiation, function and regeneration (19). We observed normal patterning of ECM molecules around the bile duct structures and the hepatocyte clusters in the liver organoids (Supplemental Fig. 2D and 12). Specifically, CK19+ bile ducts were surrounded by laminin and collagen IV, which are involved in the duct morphogenetic process, while Albumin+ hepatocytes were surrounded by collagen I and fibronectin.
Hepatocytic maturation of human liver progenitor cells
Immunohistological characterization of the hepatocyte clusters showed expression of several hepatocytic markers such as HNF4α, α1-antitrypsin (A1AT) and CYP3A4, after 3 weeks in culture (Fig. 2A). RT-PCR analysis confirmed the expression of mature hepatocyte markers including G6PC, AST and TAT (Fig. 2B) and the differentiated liver organoids also showed significantly higher albumin and urea secretion compared with hFLPCs differentiated in culture plates and Matrigel® (Fig. 2C and Supp. Fig. 4). Due to presence of mix population of cells within the discs including hepatocytes, the levels of albumin and urea are lower compared to 2D culture of adult hepatocytes (302 ng/ml/ng DNA and 1.25 mg/dl/ng DNA respectively) when normalized using DNA content. To further evaluate the metabolic maturation of hFLPCs, gene expression analysis demonstrated increased expression of cytochrome P450 3A4 (CYP3A4) at 3 weeks compared with 1 week of culture, whereas the expression of cytochrome P450 3A7 (CYP3A7) was slightly decreased after 3 weeks of culture (Fig. 2D). These isoforms represent post-natal and fetal isoforms of CYP450, respectively. In addition, different levels of expression of other CYP450 isoforms were confirmed by RT-PCR analysis, including CYP2B6, CYP2C9 and CYP2E1 (Fig. 2D). Finally, to demonstrate metabolic activity we incubated the liver organoids with diazepam and 7-ethoxy coumarin and detected phase I metabolites temazepam (CYP3A4) and nordiazepam (CYP2C19/3A4), as well as, 7-hydroxy coumarin (CYP2A6/2E1/1A2), respectively (Supplemental Fig. 6). However, we failed to detect phase II glucuronic acid conjugated metabolite. The liver organoids also expressed significantly higher levels of bile acid transporters expressed in hepatocytes such as Bile Salt Export Pump (BSEP) and Sodium taurocholate co-transporting polypeptide (NTCP) compared to hFLPCs (Supp. Fig. 5), thus, further confirming progressive hepatic differentiation and maturation within the liver organoids.
Figure 2. Hepatocytic maturation and differentiation of hFLPCs in liver organoids.
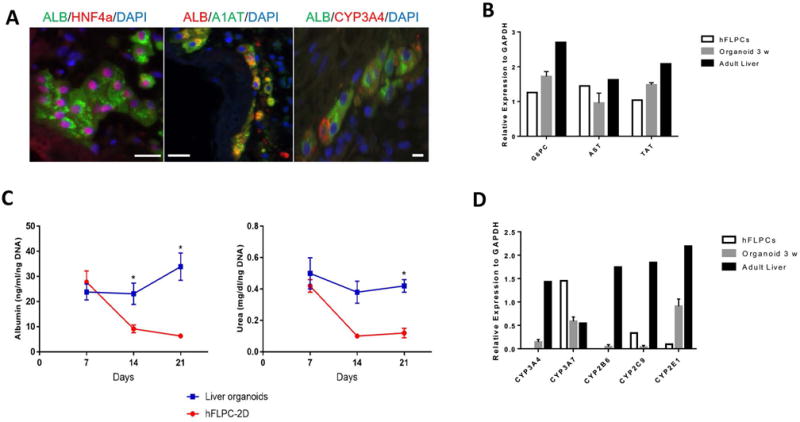
A) Immunostaining of liver organoids after 3 weeks differentiation showing clusters of cells positively stained for mature hepatocyte markers such as albumin (ALB), HNF4α, α-1 antitrypsin (A1AT) and CYP3A4. Scale bar is 20 μm. B) RT-PCR analysis of glucose-6-phosphatase (G6PC), aspartate aminotransferase (AST), and tyrosine aminotransferase (TAT) in liver organoids after 1 and 3 weeks differentiation and in adult liver tissue. C) Measurements of albumin and urea in conditioned media of liver organoids and hFPLCs in culture dishes during 3 weeks of differentiation (* = p<0.05). D) RT-PCR analysis of the fetal isoform cytochrome CYP3A4, adult isoform CYP3A7, CYP2B6, CYP2C9 and CYP2E1 in freshly isolated hFLPCs, liver organoids after 3 weeks differentiation and in adult liver tissue.
Bile duct morphogenesis inside the self-assembled liver organoids
The liver organoids exhibit different stages of bile duct morphogenesis including single or double ductal layers, transient asymmetrical primitive ductal structures, immature ducts without defined lumen and mature ductal structures, resembling duct developmental stages observed in human fetal liver (Fig. 3A). These stages were quantified as percentages of total biliary structures (CK19+/ALB−), showing 33% with single or double ductal layers, 47% immature ducts without defined lumen and 20% mature ductal structures (Fig. 3B). Proliferating cells were observed in some of the nascent immature ductal structures but were absent or detected only outside mature ducts, further indicating bile duct organogenesis in the liver organoids (Fig 3C and D).
Figure 3. Bile duct formation and cholangiocyte differentiation of hFLPCs in liver organoids.
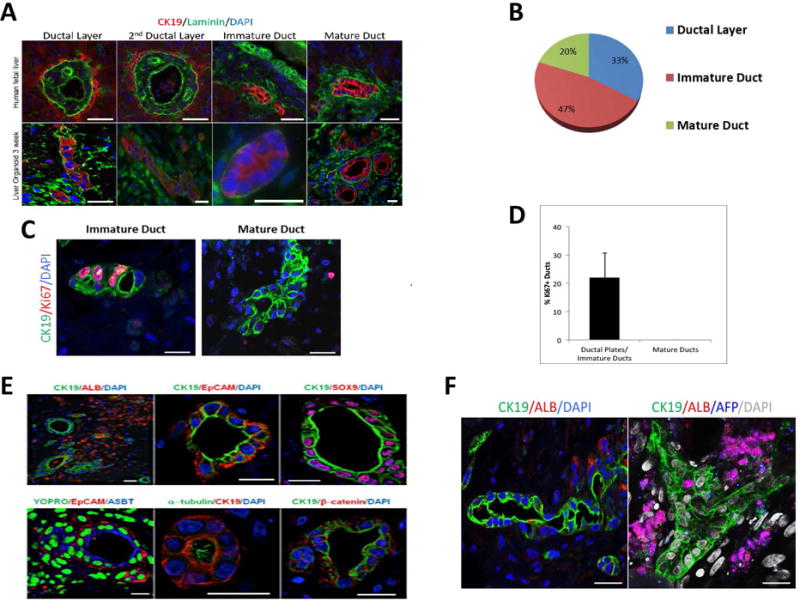
A) Different stages of ductal morphogenesis in human fetal liver tissue (top) and liver organoids after 3 weeks differentiation (bottom), stained with CK19 (red), laminin (green) (blue color denote DAPI-stained nuclei) staining. B) The relative proportions of the different stages of bile duct formations, as shown in A, were quantified and are presented as percent of total ductal structures. C) Proliferating cells are observed in immature (left panel), but not in more mature (middle panel), ductal structures and quantified (right panel). D) Graph representing percentage of Ki67 positive ducts in liver organoids. E) Characterization of ductal structures formed in liver organoids after 3 weeks of differentiation using antibodies against CK19, albumin, EpCAM, acetylated α-tubulin, apical sodium dependent bile transporter (ASBT), β-catenin and sex-determining region Y-box 9 (SOX9). F) A bile duct structure of more than 100μm surrounded by Albumin+ cells and branched biliary duct surrounded by AFP+/ALB+ hepatoblasts. Scale bar is 20 μm.
The ductal structures within the liver organoids were negative for albumin and positive for markers indicative of biliary tree progenitors and mature cholangiocytes including CK19, EpCAM and SOX9 (Fig 3E, top panels). Evidence that these ductal structures contain more mature cholangiocytes included 1) typical bile duct apical-basal polarity, 2) the presence of primary cilia (stained for α- acetylated tubulin) and a bile salt transporter (ASBT) in the apical membrane and 3) β -catenin on the baso-lateral membrane (Fig 3E, bottom panels). Moreover, we observed long ductular structures with lumen spanning for about several hundred micrometers (Fig. 3F) and a few branched ducts (Fig. 3F).
Signals from the portal mesenchyme, specifically Jagged1, activate the Notch2 receptor and guide bile duct morphogenesis (20, 21). The Notch ligand, Jagged1, is expressed in cells surrounding the ductal structures within the liver organoids (Fig. 4A). These cells were CK19- and albumin-negative, suggesting that they may represent the portal stellate cell population in the hFLPCs preparation (Supplemental figure 1). To further confirm the role of Notch signaling in bile duct development in the liver organoids, we supplemented the LDM medium with an inhibitor of Notch signaling, DAPT. This inhibition increased the number of biliary structures at the earliest stage of ductal formation (ductal layer) and showed a trend towards a decrease in the number of immature ducts, with a total absence of mature ductal structures (Fig. 4B). In parallel, RT-PCR analysis showed a significant reduction in the expression of transcription factors regulating cholangiocyte differentiation including HNF6 and Sox9 and mature markers of cholangiocytes such as AE2 and GGT1 (Fig 4C). To validate a reduction of Notch-2 signaling in our experimental setup, we performed a double immunofluorescence staining of Notch-2 NICD (notch intracellular domain) and CK19 in the DAPT inhibited and control liver organoids. The result was a visible decrease in Notch-2 NICD presence in the cells cultured in medium containing DAPT (Fig. 4D).
Figure 4. Role of Jagged 1 in bile duct development.
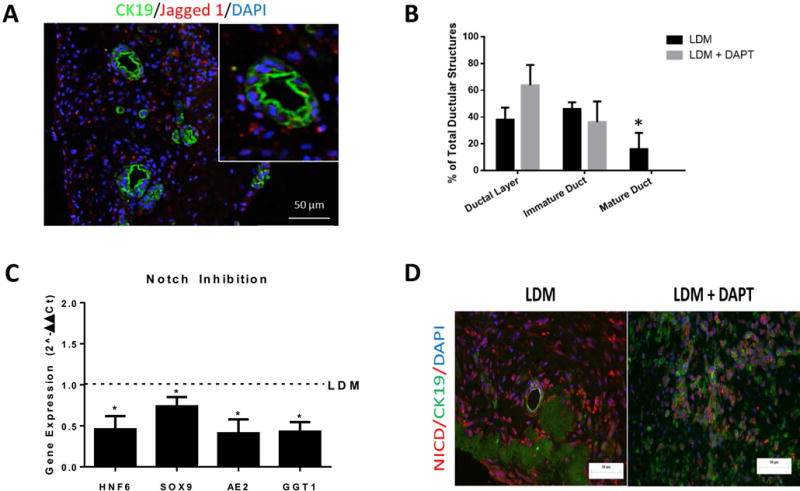
A) Jagged 1 expression in cells surrounding CK19-positive bile duct structures inside the liver organoids after 3 weeks differentiation (inset, a higher magnification of a ductal structure). B) Human FLPCs, seeded in liver ECM discs, were differentiated for 3 weeks in liver differentiation media (LDM) and LDM containing DAPT. The relative proportions of the different stages of bile duct formation were quantified and are presented as percent of total ductal structures. C) RT-PCR analysis of the expression of transcription factors regulating bile duct morphogenesis hepatic nuclear factor 6 (HNF6) and SOX9 and of mature markers of cholangiocytes AE2 and GGT1. D) NICD and CK19 expression in organoids treated with DAPT and control. * = p<0.05
Discussion
In vitro models of human tissue and organ development mostly utilize human embryonic and induced pluripotent stem cells (22, 23). However, these models do not fully recapitulate the simultaneous differentiation of liver progenitors to the hepatocytic and biliary fates, and the formation of liver with these two tissues present. The generation of bona fide mature bile ducts is especially difficult in vitro (21, 24), and requires the presence of a 3D environment for efficient cellular polarization and organization (25, 26). Scaffolds made from liver ECM were shown to possess the 3D environment that can guide differentiation and maturation of human liver progenitors (27). Here, we demonstrate a model of self-assembled human liver organogenesis. Liver ECM discs were used as scaffolds for hFLPCs, resulting in 3D organoids that recapitulated to some extent the hepato-biliary differentiation of the fetal liver, including liver metabolic and secretory functions and formation of biliary tubular structures. We propose that the unique structure and composition of the liver ECM provide an environment for specific cell-ECM interactions that lead to the concomitant differentiation of hepatoblasts into hepatocytes and cholangiocytes. An important aspect of the current study was the use of a single culture media combination to achieve both hepatocytic and biliary differentiation of the hFLPCs. Others have used defined serum-free media, as well as specialized biomaterials, to differentiate human embryonic and induced pluripotent stem cells into functionally mature hepatocytes (28, 29). Similarly, we compared Matrigel® with our scaffold and showed that hFLPCs exposed to Matrigel® in the same culture conditions did not completely mature into cholangiocytes and hepatocytes. Hence, these results clearly indicate that the liver ECM is superior in inducing maturation of hFLPCs into hepatocytes and biliary epithelium. Furthermore, we have documented the presence of laminin and collagen IV around the developing bile duct structures and collagen I and fibronectin in close proximity to the hepatocytes. Although we have not determined whether these molecules were already present in the acellular liver ECM preparations or secreted by the fetal cells, their presence at exact locations further strengthen our model. We have previously shown that hFLPCs cultured inside a ferret liver ECM developed into a native liver tissue including hepatocytic and biliary structures(13), suggesting a conservation of cell differentiation signals from the ECM among different species. Additionally, the double immunofluorescence staining of CK19/Laminin, CK19/Collagen IV, ALB/Fibronectin and ALB/Collagen I suggested that the CK19+ or ALB+ cells were not the cells expressing those ECM molecules (Supp. Fig 2D and 9). Hence, these provides a reliable indication that the fetal cells detected in these images synthesizing ECM molecules (collagen I+, collagen IV+, laminin+ or fibronectin+ cells) are most likely the stromal cell population present in the hFLPCs preparations.
Models of tissue development have important applications in the discovery and treatment of human diseases. Especially in the liver, the Gunn rat model of inherited bilirubin-UGT deficiency such as the Crigler-Najjar syndrome (30) and the inv mouse (partial deletion of the inversin gene) model of biliary atresia (BA) (31) have been particularly helpful in the study of hepatic and biliary diseases, respectively. However, these models are not optimal for the study of human-specific congenital diseases and corresponding new therapeutic targets, due to differences in liver fetal development between species. Hepatocyte maturation is a dynamic process highlighted by changes in levels of various cytokines and transcription factors associated with differentiation and maturation of hepatoblast into hepatocytes. Transcriptional switch from α-fetoprotein (AFP) to albumin is one of the hallmarks of hepatocyte maturation that was demonstrated in our liver organoid model. Unlike previously described bioengineered human liver tissue (7), our model also depicts a stepwise maturation process including inducible cytochrome P450 isoforms. However, we were not able to document complete hepatocytic maturation since the organoids were not able to complete the metabolism of diazepam and 7-ethoxy coumarin beyond phase I metabolites. Additional immunofluorescence staining of hepatocytes and cholangiocytes present in adult and fetal liver sections with mature markers previously used in our liver organoid analysis (Fig. 2 and 3), presented a phenotype for these cells more comparable to fetal/neonatal bile ducts and hepatocytes rather than adult. Some adult marker expression was similar to the adult liver (HNF4a, ALB, CYP3A4, CK19, etc). However, some were equally absent in fetal liver sections and in our organoids (AQ4, AE2, etc) (Supplemental Fig. 8, 9 and 10). These suggest that the differentiated cells within the organoids present an immature phenotype, which we believe could be potentially further matured in culture.
Models depicting development of the bile ductal structures are also important to study hereditary BA diseases. The liver organoids showed a progressive trend of developmental generation of bile duct structures by histology, but molecular analysis (RT-PCR) didn’t show progressive increase in mature cholangiocyte markers. Like mentioned above, we believe that this was probably due to a cellular phenotype more similar to a fetal/neonatal bile duct rather than an adult. In Alagille Syndrome, for example, mutations in Jagged1 lead to inactive Notch pathway, causing bile duct paucity or BA (32, 33). During fetal development, signals from adjacent cells, such as endothelial and stellate, regulate bile duct organization around the portal mesenchyme (20, 34–36). Flow cytometry (FACS) analysis showed that hFLPCs preparations contain a population of stromal and endothelial cells (Suppl. Fig. 1) that can be found surrounding hepatocyte clusters and biliary structures in liver organoids cultured for 3 weeks (Suppl. Fig. 11). Furthermore, this stromal population (αSMA+) found in hFLPCs preparations most probably contains the cells expressing the Notch ligand, Jagged 1, that supported bile duct development and maturation in the liver organoids(20). To validate the role of Notch signaling in bile duct development in the liver organoids, a Notch inhibitor, DAPT, which inhibits γ-secretase, was added to the liver differentiation culture media. We observed attenuated maturation of the bile duct structures along with significant reduction in the expression of transcription factors regulating bile duct development.
Besides providing a better model for human liver development, the liver organoids may be used for drug development and toxicity screening applications. New drugs are usually tested in rodent models for predicted toxicity; however, rodent metabolism is different than human. Cultures of human hepatocytes have been developed to address this limitation. The self-assembled organoids present significantly more complex liver structures compared with two-dimensional (2D) in vitro hepatocyte culture and simple 3D gel-based hepatocyte culture systems and thus, have a higher potential to accurately predict drug metabolism and toxicity. The current study presents self-assembled liver organoids consisting of hepatocytes and biliary structures. The liver organoids replicate some of the key processes of fetal liver development, which can be manipulated in vitro to hinder bile duct development creating a model exhibiting a phenotype similar to Alagille Syndrome. These results are similar in many aspects to previous data from other in vitro models using differentiated cholangiocytes from pluripotent stem cells(37–39). However, none of these previous studies presented long or branched biliary ducts or the step-by-step developmental stages of bile duct organogenesis. Furthermore, our model has the potential to become an in vitro system to test drug teratogenesis in the liver, including a novel class of drugs targeting the Notch pathway (γ-secretase inhibitors) that are under clinical trials for multiple diseases (40). Nonetheless, future efforts should address the lack of fully developed non-parenchymal components, such as the vasculature and mesenchyme, as well as the presence of cells from the innate immune system (Kupffer cells), with important roles in the inflammation cascade. Collectively, the liver organoids developed here can serve as a tool to study congenital diseases, develop novel therapeutic strategies, and provide important information to guide research towards creating functional tissues for transplantation in patients suffering from end-stage diseases.
Supplementary Material
Acknowledgments
PMB was funded by Fundación ARAID, Zaragoza, Spain; H2020-MSCA-IF-2014-660554 (Marie Curie Actions – European Commission); and PI15/00563 grant from Instituto de Salud Carlos III, Spain. Dr. Lola Reid, PhD made suggestions for improvements in the experiments and in the manuscript and Dr. Eliane Wauthier assisted in preparation of media and of cells. The Microscopy Services Laboratory in Pathology and Laboratory Medicine is funded by a core facility grant (NIH P30DK34987, core director Victoria Madden PhD) and by the Lineberger Cancer Center grant (NCI grant CA016086).
Abbreviations
- ECM
Extracellular matrix
- hFLPCs
human fetal liver progenitor cells
- LDM
Liver differentiation medium
- ALB
Albumin
- AFP
Alpha feto protein
- EpCAM
Epithelial Cell Adhesion Molecule
- CK19
Cytokeratin 19
- HNF1β
Hepatocyte Nuclear factor 1 beta
- HNF4α
Hepatocyte Nuclear Factor 4, alpha
- HNF6
Hepatocyte Nuclear Factor 6
- A1AT
α1-antitrypsin
- G6PC
Glucose 6 phosphatase
- AST
Aspartate transaminase
- TAT
Tyrosine Aminotransferase
- CYP3A4
cytochrome P450 3A4
- CYP3A7
cytochrome P450 3A7
- SOX9
(SRY (sex determining region Y)-box 9)
- ASBT
Apical sodium dependent bile salt transporter
- DAPT
N-[(3,5-Difluorophenyl)acetyl]-L-alanyl-2-phenyl]glycine-1,1-dimethylethyl ester
- AE2
Anion Exchange Protein 2
- GGT1
Gamma-glutamyl transferase 1
Contributor Information
Dipen Vyas, Email: dipen2707@gmail.com.
Pedro M. Baptista, Email: pbaptista.iacs@aragon.es.
Matthew Brovold, Email: mbrovold@wakehealth.edu.
Emma Moran, Email: emoran3@gmail.com.
Matthew Brovold, Email: mbrovold@wakehealth.edu.
Brandon Gaston, Email: brandon.gaston@ucsf.edu.
Chris Booth, Email: cbooth6@jhmi.edu.
Michael Samuel, Email: michael.samuel@thermofisher.com.
Anthony Atala, Email: aatala@wakehealth.edu.
Shay Soker, Email: ssoker@wakehealth.edu.
References and Notes
- 1.Godoy P, Hewitt NJ, Albrecht U, Andersen ME, Ansari N, Bhattacharya S, Bode JG, et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol. 2013;87:1315–1530. doi: 10.1007/s00204-013-1078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sampaziotis F, Cardoso de Brito M, Madrigal P, Bertero A, Saeb-Parsy K, Soares FA, Schrumpf E, et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat Biotechnol. 2015 doi: 10.1038/nbt.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogawa M, Ogawa S, Bear CE, Ahmadi S, Chin S, Li B, Grompe M, et al. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat Biotechnol. 2015 doi: 10.1038/nbt.3294. [DOI] [PubMed] [Google Scholar]
- 4.Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotechnol. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 5.Drewitz M, Helbling M, Fried N, Bieri M, Moritz W, Lichtenberg J, Kelm JM. Towards automated production and drug sensitivity testing using scaffold-free spherical tumor microtissues. Biotechnol J. 2011;6:1488–1496. doi: 10.1002/biot.201100290. [DOI] [PubMed] [Google Scholar]
- 6.Katsuda T, Kojima N, Ochiya T, Sakai Y. Biliary Epithelial Cells Play an Essential Role in the Reconstruction of Hepatic Tissue with a Functional Bile Ductular Network. Tissue Eng Part A. 2013 doi: 10.1089/ten.TEA.2013.0021. [DOI] [PubMed] [Google Scholar]
- 7.Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 8.Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, Ellis E, et al. Long-Term Culture of Genome-Stable Bipotent Stem Cells from Adult Human Liver. Cell. 2014 doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tateno C, Yoshizane Y, Saito N, Kataoka M, Utoh R, Yamasaki C, Tachibana A, et al. Near completely humanized liver in mice shows human-type metabolic responses to drugs. Am J Pathol. 2004;165:901–912. doi: 10.1016/S0002-9440(10)63352-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azuma H, Paulk N, Ranade A, Dorrell C, Al-Dhalimy M, Ellis E, Strom S, et al. Robust expansion of human hepatocytes in Fah-/-/Rag2-/-/Il2rg-/- mice. Nat Biotechnol. 2007;25:903–910. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen AA, Thomas DK, Ong LL, Schwartz RE, Golub TR, Bhatia SN. Humanized mice with ectopic artificial liver tissues. Proc Natl Acad Sci U S A. 2011;108:11842–11847. doi: 10.1073/pnas.1101791108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baptista PM, Siddiqui MM, Lozier G, Rodriguez SR, Atala A, Soker S. The use of whole organ decellularization for the generation of a vascularized liver organoid. Hepatology. 2011;53:604–617. doi: 10.1002/hep.24067. [DOI] [PubMed] [Google Scholar]
- 14.Baptista PM, Vyas D, Moran E, Wang Z, Soker S. Human liver bioengineering using a whole liver decellularized bioscaffold. Methods Mol Biol. 2013;1001:289–298. doi: 10.1007/978-1-62703-363-3_24. [DOI] [PubMed] [Google Scholar]
- 15.Kubota H, Reid LM. Clonogenic hepatoblasts, common precursors for hepatocytic and biliary lineages, are lacking classical major histocompatibility complex class I antigen. Proc Natl Acad Sci U S A. 2000;97:12132–12137. doi: 10.1073/pnas.97.22.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baptista PM, Orlando G, Mirmalek-Sani SH, Siddiqui M, Atala A, Soker S. Whole organ decellularization – a tool for bioscaffold fabrication and organ bioengineering. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:6526–6529. doi: 10.1109/IEMBS.2009.5333145. [DOI] [PubMed] [Google Scholar]
- 17.Baptista PM, Moran EC, Vyas D, Ribeiro MH, Atala A, Sparks JL, Soker S. Fluid Flow Regulation of Revascularization and Cellular Organization in a Bioengineered Liver Platform. Tissue Eng Part C Methods. 2016;22:199–207. doi: 10.1089/ten.tec.2015.0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antoniou A, Raynaud P, Cordi S, Zong Y, Tronche F, Stanger BZ, Jacquemin P, et al. Intrahepatic bile ducts develop according to a new mode of tubulogenesis regulated by the transcription factor SOX9. Gastroenterology. 2009;136:2325–2333. doi: 10.1053/j.gastro.2009.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maher JJ, Bissell DM. Cell-matrix interactions in liver. Semin Cell Biol. 1993;4:189–201. doi: 10.1006/scel.1993.1023. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann JJ, Zovein AC, Koh H, Radtke F, Weinmaster G, Iruela-Arispe ML. Jagged1 in the portal vein mesenchyme regulates intrahepatic bile duct development: insights into Alagille syndrome. Development. 2010;137:4061–4072. doi: 10.1242/dev.052118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev Cell. 2010;18:175–189. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 22.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 23.Medvinsky A, Livesey FJ. On human development: lessons from stem cell systems. Development. 2015;142:17–20. doi: 10.1242/dev.114868. [DOI] [PubMed] [Google Scholar]
- 24.Navarro-Alvarez N, Soto-Gutierrez A, Kobayashi N. Hepatic stem cells and liver development. Methods Mol Biol. 2010;640:181–236. doi: 10.1007/978-1-60761-688-7_10. [DOI] [PubMed] [Google Scholar]
- 25.Ader M, Tanaka EM. Modeling human development in 3D culture. Curr Opin Cell Biol. 2014;31:23–28. doi: 10.1016/j.ceb.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 26.Chistiakov DA. Liver regenerative medicine: advances and challenges. Cells Tissues Organs. 2012;196:291–312. doi: 10.1159/000335697. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Cui CB, Yamauchi M, Miguez P, Roach M, Malavarca R, Costello MJ, et al. Lineage restriction of human hepatic stem cells to mature fates is made efficient by tissue-specific biomatrix scaffolds. Hepatology. 2011;53:293–305. doi: 10.1002/hep.24012. [DOI] [PubMed] [Google Scholar]
- 28.Medine CN, Lucendo-Villarin B, Storck C, Wang F, Szkolnicka D, Khan F, Pernagallo S, et al. Developing high-fidelity hepatotoxicity models from pluripotent stem cells. Stem Cells Transl Med. 2013;2:505–509. doi: 10.5966/sctm.2012-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villarin BL, Cameron K, Szkolnicka D, Rashidi H, Bates N, Kimber SJ, Flint O, et al. Polymer Supported Directed Differentiation Reveals a Unique Gene Signature Predicting Stable Hepatocyte Performance. Adv Healthc Mater. 2015;4:1820–1825. doi: 10.1002/adhm.201500391. [DOI] [PubMed] [Google Scholar]
- 30.Gunn CK. Hereditary Acholuric Jaundice in the Rat. Can Med Assoc J. 1944;50:230–237. [PMC free article] [PubMed] [Google Scholar]
- 31.Mazziotti MV, Willis LK, Heuckeroth RO, LaRegina MC, Swanson PE, Overbeek PA, Perlmutter DH. Anomalous development of the hepatobiliary system in the Inv mouse. Hepatology. 1999;30:372–378. doi: 10.1002/hep.510300223. [DOI] [PubMed] [Google Scholar]
- 32.McCright B, Lozier J, Gridley T. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development. 2002;129:1075–1082. doi: 10.1242/dev.129.4.1075. [DOI] [PubMed] [Google Scholar]
- 33.Geisler F, Nagl F, Mazur PK, Lee M, Zimber-Strobl U, Strobl LJ, Radtke F, et al. Liver-specific inactivation of Notch2, but not Notch1, compromises intrahepatic bile duct development in mice. Hepatology. 2008;48:607–616. doi: 10.1002/hep.22381. [DOI] [PubMed] [Google Scholar]
- 34.Raynaud P, Carpentier R, Antoniou A, Lemaigre FP. Biliary differentiation and bile duct morphogenesis in development and disease. Int J Biochem Cell Biol. 2011;43:245–256. doi: 10.1016/j.biocel.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 35.Zong Y, Stanger BZ. Molecular mechanisms of bile duct development. Int J Biochem Cell Biol. 2011;43:257–264. doi: 10.1016/j.biocel.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fabris L, Cadamuro M, Libbrecht L, Raynaud P, Spirli C, Fiorotto R, Okolicsanyi L, et al. Epithelial expression of angiogenic growth factors modulate arterial vasculogenesis in human liver development. Hepatology. 2008;47:719–728. doi: 10.1002/hep.22015. [DOI] [PubMed] [Google Scholar]
- 37.Sampaziotis F, Cardoso de Brito M, Madrigal P, Bertero A, Saeb-Parsy K, Soares FA, Schrumpf E, et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat Biotechnol. 2015;33:845–852. doi: 10.1038/nbt.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogawa M, Ogawa S, Bear CE, Ahmadi S, Chin S, Li B, Grompe M, et al. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat Biotechnol. 2015;33:853–861. doi: 10.1038/nbt.3294. [DOI] [PubMed] [Google Scholar]
- 39.Kamiya A, Chikada H. Human pluripotent stem cell-derived cholangiocytes: current status and future applications. Curr Opin Gastroenterol. 2015;31:233–238. doi: 10.1097/MOG.0000000000000180. [DOI] [PubMed] [Google Scholar]
- 40.Andersson ER, Lendahl U. Therapeutic modulation of Notch signalling–are we there yet? Nat Rev Drug Discov. 2014;13:357–378. doi: 10.1038/nrd4252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


