Abstract
While hexavalent chromium [Cr(VI)] is generally considered as a genotoxic environmental carcinogen, studies showed that Cr(VI) exposure also causes epigenetic changes. However, whether Cr(VI)-caused epigenetic dysregulations plays an important role in Cr(VI) carcinogenicity remain largely unknown. The aim of this study was to determine if chronic low dose Cr(VI) exposure causes epigenetic changes, the underlying mechanism and whether chronic low dose Cr(VI) exposure-caused epigenetic dysregulation contributes causally to Cr(VI)-induced cancer stem cell (CSC)-like property and cell transformation. Two immortalized human bronchial epithelial cell lines (BEAS-2B and 16HBE) were exposed to 0.25 μM of K2Cr2O7 for 20 and 40 weeks to induce cell transformation, respectively. Cr(VI)-induced epigenetic changes were examined in Cr(VI)-transformed cells and Cr(VI) exposure-caused human lung cancer tissues. Pharmacological inhibitors and gene knockdown experiments were used to determine the role of epigenetic dysregulation in Cr(VI) carcinogenicity. We found that chronic Cr(VI) exposure causes epigenetic dysregulation as evidenced by the increased levels of histone H3 repressive methylation marks (H3K9me2 and H3K27me3) and the related histone-lysing methyltransferases (HMTases). Pharmacological inhibition or knockdown of HMTases reduces H3 repressive methylation marks and malignant phenotypes of Cr(VI)-transformed cells. Moreover, knockdown of HMTases in parental cells significantly reduces chronic Cr(VI) exposure-induced CSC-like property and cell transformation. Further mechanistic study revealed that knockdown of HMTases decreases Cr(VI) exposure-caused DNA damage. Our findings indicate that chronic Cr(VI) exposure increases H3 repressive methylation marks by increasing the related HMTases expression; and that increased expression of HMTases plays a causal role in Cr(VI)-induced CSC-like property and cell transformation.
Keywords: hexavalent chromium, cell transformation, carcinogenesis, epigenetics, histone methyltransferase, cancer stem cell-like cell
1. Introduction
Hexavalent chromium [Cr(VI)], one of the most common environmental pollutants, is classified as a Group I human carcinogen causing lung and other cancers (IARC, 1990; Stout et al., 2009). Due to the widespread industrial use of chromium, a large amount of Cr(VI) has been released into the environment. For example, hundreds of large toxic waste sites in the United States known as Superfund sites contain Cr(VI) as a major pollutant (ATSDR, 2000). Thus, general population exposure to Cr(VI) through contaminated air, water, soil and food is common, representing a significant environmental health concern. Although Cr(VI) is a well-recognized human carcinogen, the mechanism of Cr(VI) carcinogenicity has not been well understood.
Once entering cells, Cr(VI) undergoes a series of metabolic reductions and generates reactive Cr metabolites and reactive oxygen species, which produce various genotoxic effects (Shi et al., 2004; Yao et al., 2008; Salnikow and Zhitkovich, 2008; Wise et al., 2008; Ovesen et al., 2014). As a result, Cr(VI) is generally considered as a genotoxic carcinogen. However, accumulating evidence shows that Cr(VI) exposure also causes various epigenetic changes (Brocato and Costa, 2013; Chervona et al., 2012; Martinez-Zamudio and Ha, 2011; Arita and Costa, 2009).
Epigenetics refers to heritable alterations in the pattern of gene expression that are not caused by changes in DNA sequences, but are mediated by DNA methylation, histone posttranslational modifications (acetylation, methylation, etc.), and non-coding RNAs (Waldmann and Schneider, 2013; Dawson and Kouzarides, 2012). In general, acetylation of histones H3 and H4, and methylation of H3 lysine 4 (H3K4) are usually associated with gene expression; but DNA methylation, and methylation of H3 lysine 9 (H3K9) and H3 lysine 27 (H3K27) cause the compaction of chromatin leading to gene silencing. It is now well established that epigenetics is often dysregulated in cancer and that epigenetics dysregulations play important roles in cancer initiation and progression (Waldmann and Schneider, 2013; Dawson and Kouzarides, 2012). Cancer stem cells (CSCs) are cancer cells possessing characteristics of normal stem cells. CSCs or CSC-like cells are considered as cancer initiating and maintaining cells (Nguyen et al., 2012). Studies showed that dysregulation of epigenetics plays crucial roles in producing CSCs or CSC-like cells (Shukla and Meeran, 2014).
The Cr(VI)-caused epigenetic change was first reported in a study showing that exposure to 20–50 μM of potassium chromate for 2 h silenced the G12 cell gpt transgene expression by increasing DNA methylation (Klein et al., 2002). A subsequent study found that exposure to 10–200 mg/l of potassium chromate for 3 days caused a genome-wide DNA hypermethylation in B. napus L. plants in a dose-dependent manner (Labra et al., 2004). Studies on human lung tumor tissues from workers exposed to chromate revealed increased DNA methylation levels in the promoter regions of several tumor suppressor genes (Ali et al., 2011; Kondo et al., 2006). In addition, human cell culture studies also showed that treatment with 5–50 μM of Cr(VI) for 1, 2, or 24 h causes various histone posttranslational modifications in liver and lung cancer cells (Schnekenburger et al., 2007; Sun et al., 2009; Zhou et al., 2009). While these studies clearly showed that Cr(VI) exposure is able to cause epigenetic changes, the mechanisms of Cr(VI) causing epigenetic changes remain largely unclear. Moreover, it is not clear whether the reported epigenetic changes also exist in cells transformed by chronic low dose Cr(VI) exposure (such as 0.125 or 0.25 μM for 5 to 6 moths). Furthermore, it is unknown whether Cr(VI)-caused epigenetic changes play a causal role in Cr(VI)-induced cell transformation and tumorigenesis. The objective of this study is to determine if chronic low dose Cr(VI) exposure causes epigenetics alterations, the underlying mechanism and whether Cr(VI)-caused epigenetic dysregulations contribute causally to chronic Cr(VI) exposure-induced cancer stem cell (CSC)-like property and cell transformation.
2. Materials and Methods
2.1 Cell culture
Immortalized human bronchial epithelial BEAS-2B and 16HBE cells were purchased from American Type Culture Collection (ATCC, Manassas, VA) and generously provided by Dr. Dieter C. Gruenert (University of California San Francisco, San Francisco, CA), respectively. BEAS-2B cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 5% fetal bovine serum (FBS) and 16HBE cells were cultured in Minimum Essential Media (MEM) supplemented with 10% FBS. The immortalized p53 intact human bronchial epithelial cell line (HBEC3-KT) was obtained from Dr. John D. Minna (University of Texas Southwestern Medical Center, Dallas, TX) and cultured in chemically defined serum-free medium (K-SFM) (Invitrogen, Carlsbad, CA) as described in detail in our recent publication (Wang et al., 2011).
2.2 Cell transformation by chronic low dose Cr(VI) (K2Cr2O7) exposure
BEAS-2B and 16HBE cells were first treated with different doses of K 2Cr2O7 (0.125, 0.25, 0.5 and 1 μM) for 72 h to determine the cytotoxic effect of Cr(VI). It was found that the maximal dose that had no obvious effect on the viability and proliferation of BEAS-2B and 16 HBE cells was 0.25 μM of K2Cr2O7. This Cr(VI) dose was then chosen for chronic cell transformation experiment following our published protocol (Wang et al., 2011). Briefly, BEAS-2B and 16HBE cells were continuously exposed to vehicle control (H2O) or 0.25 μM of Cr(VI) (K2Cr2O7). When reaching about 80–90% confluence after 72 h Cr(VI) exposure, cells were sub-cultured. Cr(VI) was then freshly added to cells each time after overnight cell attachment. Soft agar colony formation assay was performed after every 4-week Cr(VI) exposure to assess cell transformation. This process was repeated in BEAS-2B and 16HBE cells for 20 and 40 weeks, respectively.
2.3 Soft agar colony formation assay
The soft agar colony formation assay reflecting cell anchorage-independent growth was carried out in 60-mm cell culture dishes in triplicates for each group as previously described (Yang et al., 2005). Briefly, cultured cells were collected by trypsinization and suspended in DMEM (for BEAS-2B cells) or MEM (for 16HBE cells) containing 10% FBS at a concentration of 0.5 × 104 cells/ml. Normal melting point agar (5 ml of 0.6% agar in DMEM or MEM containing 10% FBS) was placed into each 60-mm cell culture dish as the bottom agar. After solidification of the bottom agar, 4 ml of cell mixture consisting of 2 ml of cell suspension (0.5 × 104 cells/ml) and 2 ml of 0.8% lower melting point agar in DMEM or MEM containing 10% FBS were poured over the bottom agar. After solidification of the upper agar, 3 ml of DMEM or MEM containing 10% FBS were added, and dishes were incubated at 37°C in a humidified 5% CO2 atmosphere. For soft agar colony formation assays with G9a inhibitor BX01294 and EZh2 inhibitor DZNeP, Cr(VI)-transformed cells were treated with a vehicle control or BIX01294 (2.5 μM) or DZNeP (0.25 μM) for 72h, then collected by trypsinization and neutralization with culture media and used for soft agar colony formation assays. The same concentration of vehicle control or inhibitors was included in the top agar. Colony formation in the agar was stained with 0.003% crystal violet, photographed and counted (if > 100 μm) after 4-week incubation.
2.4 Suspension culture spheroid formation assay
The spheroid formation assay reflecting the stem cell property was performed following the published protocol (Shaheen et al., 2016; Qiu et al., 2012; Dontu et al., 2003) with minor modifications. Briefly, single cells were plated in ultralow attachment 24-well culture plates (Corning, Corning, NY) at a density of 2.5 × 103 cells per well suspended in serum-free DMEM or MEM containing human recombinant basic fibroblast growth factor (bFGF, 20 ng/ml), human recombinant epidermal growth factor (EGF, 20 ng/ml) (R&D, Minneapolis, MN), B27 (50 times diluted from the original 50x stock solution, Invitrogen, Carlsbad, CA) and heparin (4 μg/ml, Sigma). Plates were incubated at 37°C in a humidified 5% CO2 atmosphere. For spheroid formation assays with G9a inhibitor BX01294 and EZh2 inhibitor DZNeP, Cr(VI)-transformed cells were treated with a vehicle control or BIX01294 (2.5 μM) or DZNeP (0.25 μM) for 72h, then collected by trypsinization and neutralization with culture media and used for spheroid formation assays. The same concentration of vehicle control or inhibitors was included in the sphere-forming culture media. Spheres were viewed, photographed and counted (if > 100 μm) under a phase-contrast microscope after 10-day culture.
2.5 Nude mouse xenograft tumorigenesis study
Passage-matched control and Cr(VI)-transformed BEAS-2B cells (1.5 × 106 cells in 0.1 ml of 1:1 growth factor-reduced matrigel and PBS) were injected subcutaneously into the right flank of female nude mice (Nu/Nu, Charles River laboratories, four mice in each group). Animals were maintained under specific pathogen-free conditions, and animal protocols were reviewed and approved by the University of Kentucky Institutional Animal Care and Use Committee. All mice were euthanized 14 weeks after injection, and the xenograft tissues were harvested and fixed with 10% formalin solution for histology analysis.
2.6 Western blot analysis
Cells were lysed using lysis buffer following our published protocol (Yang et al., 2006; Wang et al., 2014) and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) (10 to 30 μg of protein/lane). The following primary antibodies were used: anti-H3 acetylation, anti-H3K4me2, anti-H3K36me2, anti-H3K79me2, anti-H3K9me2, anti-H3K27me3, anti-total H3, anti-G9a, anti-SUV39H1 (suppressor of variegation 3–9 homolog 1), and anti-EZH2 (enhancer of zeste homolog 2) (Cell Signaling Technology, Beverly, MA); anti-GLP (G9a-like protein) (Abcam, Cambridge, MA); anti-phospho-histone H2A.X (Ser139) (γH2AX), and anti-β-actin (Millipore Sigma, St. Louis, MO).
2.7 Generation of G9a, SUV39H1 and EZH2 shRNA stable knockdown BEAS-2B cells and cell transformation experiment by chronic low dose Cr(VI) exposure
Vector control and G9a, SUV39H1 and EZH2 stable knockdown cells were generated by transducing cells with control (pLKO.1-puro) or G9a, SUV39H1 and EZH2 short hairpin RNA (shRNA) expressing lentiviral particles, respectively. The control and G9a, SUV39H1 and EZH2 shRNA lentiviral constructs were purchased from Sigma (St Louis, MO) and the lentiviral particles were packaged following our published protocol (Zhao et al., 2011). Cells were transduced with lentiviral particles and selected with puromycin. Specific knockdown of each gene was confirmed by Western blot. For Cr(VI) cell transformation experiment, vector control and each gene specific knockdown BEAS-2B cells were continuously exposed to 0.125 μM of K2Cr2O7 for 30 weeks as described above.
2.8 Measurement of intracellular chromium levels
Intracellular chromium level was determined using an inductively-coupled plasma–optical emission spectrometer (Varian 710-ES ICP8OES) following the published protocol (Ott et al., 2004; Messer et al., 2006). Briefly, parental BEAS-2B cells, shRNA vector control cells and each HMTase stable knockdown cells were cultured to about 80–90% confluent, then washed with PBS and incubated with warm serum-free DMEM, treated with 10 μM of of K2Cr2O7 for 3 h at 37°C. At the end of the treatment, media were removed and cells were washed with PBS twice. Cells were collected by trypsinzation and cell pellets were washed with PBS and used for the analysis of protein concentrations and intracellular chromium levels as previously described (Ott et al., 2004; Messer et al., 2006).
2.9 Immunofluorescence (IF) staining of cultured cells and human lung tissue sections
The IF staining of phosphor-histone H2A.X (Ser139) (γH2AX) in cultured cells was carried out as previously described (Peterson-Roth et al., 2005). The IF staining of HMTases SUV39H1 and EZH2 in human lung tissues was performed as described in our previous publications (Humphries et al., 2014; Zhao Y et al., 2010). The IF staining pictures taken under a Nikon fluorescent microscope are the overlaid images of γH2AX, SUV39H1 or EZH2 staining in red fluorescence with nuclear 4’6-diamidino-2-phenylindole (DAPI) staining in blue fluorescence. The images were overlaid using Nikon NIS-Elements software.
2.10 Statistical analysis
The statistical analyses for the significance of differences in presented numerical data (mean ± SD) were carried out by testing different treatment effects using two-tailed t-tests for comparison of two data sets or one-way analysis of variance (ANOVA) for multiple data sets. A p-value of <0.05 was considered statistically significant.
3. Results
3.1 Cr(VI)-transformed human bronchial epithelial cells have higher levels of histone H3 repressive methylation marks (H3K9me2 and H3K27me3)
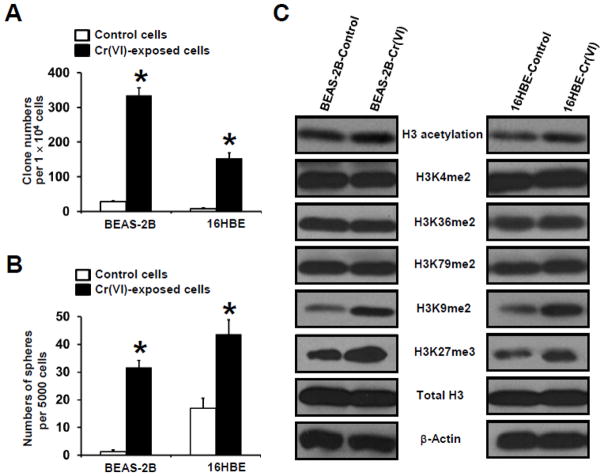
To study the mechanism of Cr(VI) carcinogenicity, we first established cell transformation models by chronically exposing two immortalized human bronchial epithelial cells (BEAS-2B and 16HBE) to a low dose of Cr(VI) (0.25 μM of K2Cr2O7) as lung cells are the major target of Cr(VI). The protocol of chronic Cr(VI) exposure and the determination of Cr(VI) exposure dose are described in Methods. It was found that BEAS-2B and 16HBE cells were malignantly transformed after exposure to 0.25 μM of K2Cr2O7 for 20 and 40 weeks, respectively, as demonstrated by (i) significantly increased colony formation in soft agar (Fig. 1A); (ii) significantly increased tumorigenic sphere formation under suspension culture condition (Fig. 1B); representative images of soft agar clone formation and suspension culture sphere formation are shown in Supplementary Fig. S1 and Fig. S2, respectively; and (iii) tumor formation upon inoculation of Cr(VI)-exposed cells into nude mice (Supplementary Fig. S3). The cancer stem cell (CSC)-like property of Cr(VI)-transformed cells was further evaluated by determining the spheroid forming capability of cells derived from Cr(VI)-transformed cells-produced spheres. It was found that cells dissociated from old spheres are able to form significantly more spheres (>2-fold), indicating that these sphere forming cells are capable of self-renewal, a distinct feature of stem cells.
Fig. 1. Chronic low dose Cr(VI) exposure induces cancer stem cell-like property and cell transformation and Cr(VI)-transformed cells have higher levels of histone H3 repressive methylation marks.
(A,B) Quantitation of chronic low dose Cr(VI) exposure-induced soft agar colony and suspension culture sphere formation by human bronchial epithelial cells (BEAS-2B and 16HBE). After exposure to 0.25 μM of Cr(VI) (K2Cr2O7) for 20 (BEAS-2B) and 40 (16HBE) weeks, passage-matched control and Cr(VI)-exposed BEAS-2B and 16 HBE cells were harvested for soft agar colony formation (A) and suspension culture spheroid formation (B) assay as described in Methods. The results are presented as means ± standard deviations (n=3). *p<0.05, compared to passage-matched vehicle (H2O) control-exposed cells. (C) Representative Western blot images for the analysis of histone H3 posttranslational modifications in passage- matched control and Cr(VI)-exposed BEAS-2B and 16 HBE cells. After exposure to 0.25 μM of Cr(VI) (K2Cr2O7) for 20 (BEAS-2B) and 40 (16HBE) weeks, passage-matched control and Cr(VI)-exposed BEAS-2B and 16 HBE cells were harvested for Western blot analysis as described in Methods. Similar results were obtained in two additional experiments.
To determine whether chronic low dose Cr(VI) exposure causes epigenetic changes, we analyzed histone H3 posttranscriptional modification levels in Cr(VI)-transformed cells by Western blot. Studies showed that differential H3 posttranslational modifications have distinct effects on chromatin structure and gene expression (Smolle and Workman, 2013; Greer and Shi, 2012). For example, H3 acetylation, methylation of H3 at lysine 4 (H3K4), 36 (H3K36), and 79 (H3K79) are correlated with euchromatin structure activating gene transcription; whereas methylation of H3 at lysine 9 (H3K9) and lysing 27 (H3K27) are associated with heterochromatin structure repressing gene transcription. We found that except a small increase of H3 acetylation levels, no obvious changes of levels of H3 active marks such as H3 lysine 4, 36, and 79 dimethylation (H3K4me2, H3K36me2 and H3K79me2) are observed in Cr(VI)-transformed both BEAS-2B and 16HBE cells (Fig. 1C). In contrast, drastic increases of H3 repressive methylation marks such as H3 lysine 9 dimethylation (H3K9me2) and H3 lysine 27 trimethylation (H3K27me3) are detected in Cr(VI)-transformed both BEAS-2B and 16HBE cells (Fig. 1C). Moreover, these increased levels of H3K9me2 and H3K27me3 are also detected in Cr(VI)-transformed cells cultured in the absence of Cr(VI) for 5 passages (data not shown), indicating that chronic low dose Cr(VI) exposure-caused epigenetic changes can be detected in the absence of further Cr(VI) exposure.
3.2 The expression levels of several histone-lysine N-methyltransferases (HMTases) are increased in Cr(VI)-transformed cells and Cr(VI) exposure-caused human lung cancer tissues
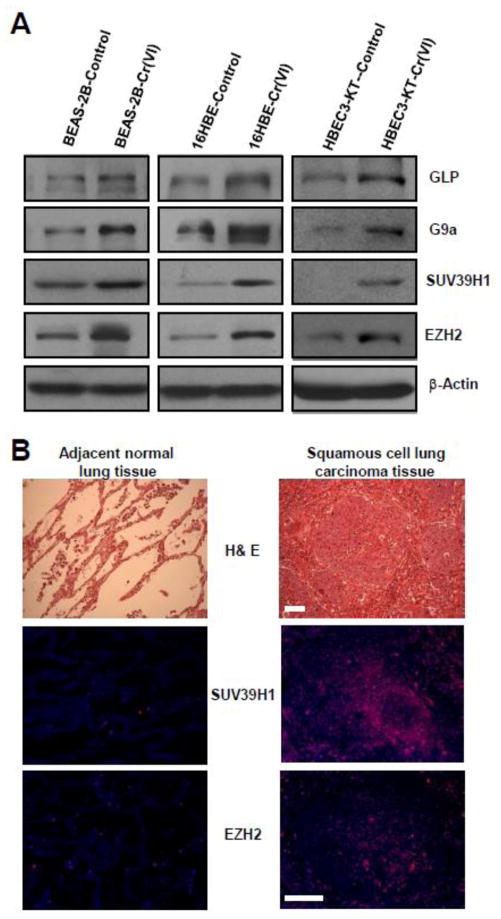
Next, we wanted to determine the mechanism by which chronic Cr(VI) exposure causes epigenetic alterations. Increases of H3 repressive methylation marks (H3K9me2 and H3K27me3) could be caused by either increased expression or activity of histone lysine methyltransferases (HMTases) or decreased expression or activity of histone lysine demethylases. Studies showed that several HMTases such as GLP (G9a-like protein), G9a, and SUV39H1 (suppressor of variegation 3–9 homolog 1) can cause H3K9 di- and trimethylation (Smolle and Workman, 2013; Greer and Shi, 2012). EZH2 (enhancer of zeste homolog 2), a core component of polycomb repressive complex (PRC2) capable of promoting CSC-like property, is a H3K27 methyltransferase (MTase) catalyzing trimethylation of H3K27 (Kondo, 2014; Chang et al., 2011). Our Western blot analysis revealed that in consistent with significantly higher levels of H3K9me2 and H3K27me3 shown in Fig. 1C, the levels of H3K9 MTases including GLP, G9a, SUV39H1, and the level of H3K27 MTase EZH2 are greatly increased in Cr(VI)-transformed BEAS-2B and 16HBE cells (Fig. 2A). Moreover, elevated levels of HMTases are also similarly observed in an immortalized p53 intact human bronchial epithelial cells (HBEC3-KT) exposed to Cr(VI) (0.25 μM) for 30 weeks (Fig. 2A). The HBEC3-KT cells were immortalized by expressing CDK4 and hTERT and have an intact p53 (Wang et al., 2011; Ramirez et al., 2004). These results indicate that chronic low dose Cr(VI) exposure is capable of increasing HMTases levels in immortalized human bronchial epithelial cells independent of their p53 status.
Fig. 2. The expression levels of several histone-lysine N-methyltransferases (HMTases) are increased in Cr(VI)-transformed cells and Cr(VI) exposure-caused human lung cancer tissues.
(A) Representative Western blot images for the analysis of several HMTases levels in passage-matched control and Cr(VI)-exposed human bronchial epithelial. After exposure to 0.25 μM of Cr(VI) (K2Cr2O7) for 20 (BEAS-2B), 40 (16HBE) and 30 (HBEC3-KT) weeks, passage- matched control and Cr(VI)-exposed BEAS-2B, 16 HBE and HBEC3-KT cells were harvested for Western blot analysis as described in Methods. Similar results were obtained in two additional experiments. (B) Representative images of H&E staining and immunofluorescence (IF) staining of SUV39H1 and EZH2 in the squamous cell lung carcinoma tissue and the adjacent normal lung tissue from a non-smoker worker exposed to chromate for 19 years. Similar staining results were obtained in lung cancer tissue from another non-smoker worker exposed chromate for 38 years. Scale bar: 100 μm.
To establish the potential relevance of these findings in our Cr(VI)-transformed cells to Cr(VI) exposure-induced lung cancer in humans, we examined the levels of HMTases in Cr(VI) exposure-caused human lung cancer tissue sections and the matched-adjacent normal lung tissue sections from chromate workers. The details of workers’ chromate exposure were described in previous publications (Ali et al., 2011; Kondo et al., 2006). By immunofluorescence (IF) staining we found that the levels of HMTases SUV39H1 and EZH2 are significantly higher in Cr(VI) exposure-caused human lung cancer tissues than that in the adjacent normal lung tissues. The representative H&E histology and IF staining images of SUV39H1 and EZH2 from a non-smoker worker exposed to chromate for 19 years are shown in Fig. 2B. The antibodies for GLP and G9a IF staining did not work well (data not shown). These findings from Cr(VI)-exposed human lung tissue samples are consistent with that from our cell culture studies.
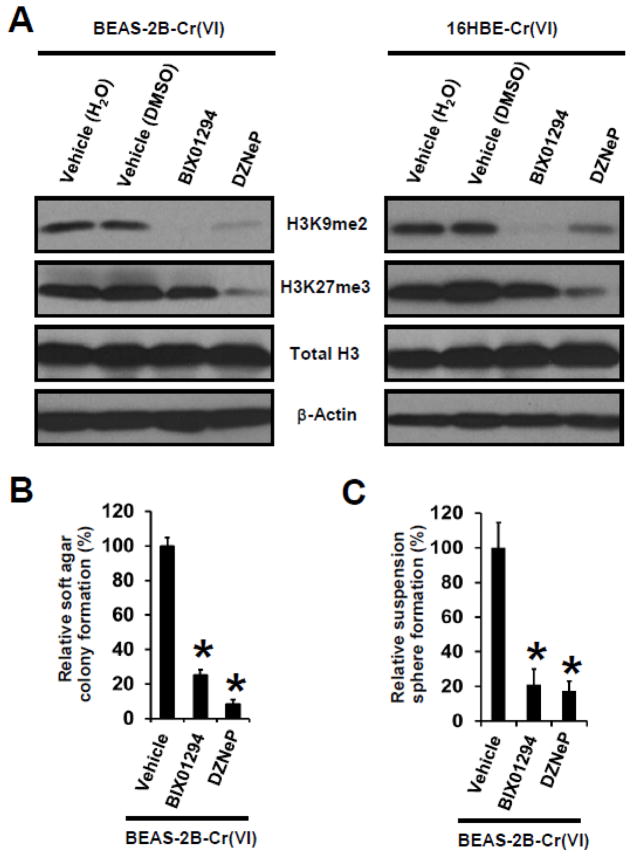
3.3 Inhibition or knockdown of HMTases in Cr(VI)-transformed cells abolishes H3 repressive methylation marks and reduces their transformed phenotypes
We next wanted to determine whether up-regulated HMTases play a role in the increases of H3 repressive methylation marks and investigate whether they have a role in maintaining the malignant phenotypes of Cr(VI)-transformed cells. As shown in Fig. 3A, treatment with a G9a inhibitor BIX01294 (Cayman Chemical, Ann Arbor, MI) (dissolved in H2O) efficiently reduced H3K9me2 levels but had no obvious effect on H3K27me3 levels in Cr(VI)-transformed cells. Treatment with an EZH2 inhibitor DZNeP (Selleckchem, Houston, TX) (dissolved in DMSO) reduced the levels of both H3K27me3 and H3K9me2 in Cr(VI)-transformed cells. These results suggest that either the presence of H3K27me3 is required for the presence of H3K9me2 or the EZH2 inhibitor DZNeP may also inhibit the activity of G9a or other H3K9 MTases. In either case, these findings indicate that the increases of H3 repressive methylation marks in Cr(VI)-transformed cells are caused by the up-regulated HMTases.
Fig. 3. Pharmacological inhibition of HMTases in Cr(VI)-transformed cells abolishes their H3 repressive methylation marks and reduces their transformed phenotypes.
(A) Representative Western blot images for the analysis of histone H3 repressive methylation marks in Cr(VI)-transformed cells. Cells were treated with vehicle control, a G9a inhibitor BIX01294 (2.5 μM) or an EZH2 inhibitor DZNeP (0.25 μM) for 72 h, and harvested for Western blot analysis. (B) Quantitation of effects of inhibition of G9a or EZH2 on soft agar colony and suspension culture sphere formation by Cr(VI)-transformed cells. Cells were treated with vehicle control, a G9a inhibitor BIX01294 (2.5 μM) or an EZH2 inhibitor DZNeP (0.25 μM) for 72 h, and harvested for soft agar colony and suspension culture sphere formation assay. Results are expressed as relative colony or sphere formation compared to vehicle-treated BEAS-2B-Cr(VI) cells (100%). Data are presented as mean ± SD (n=3). *p< 0.05, compared to vehicle-treated group. Similar results were obtained in two additional experiments.
While treatment with BIX01294 (2.5 μM) or DZNeP (0.25 μM) for 72 h showed no significant effect on Cr(VI)-transformed cell viability and proliferation as determined by Trypan blue staining and cell counting (data not shown), further functional analysis revealed that both inhibitors significantly reduced soft agar colony formation (Fig. 3B) and suspension culture sphere formation (Fig. 3C) by Cr(VI)-transformed BEAS-2B cells. We could not find a specific inhibitor for SUV39H1, so we stably and specifically knocked down SUV39H1 expression in Cr(VI)-transformed BEAS-2B cells and found that shRNA knockdown of SUV39H1 significantly reduces their level of H3K9me2, reduces their soft agar colony number by 70% and decreases their suspension culture sphere formation by 80% (data not shown). Since soft agar colony formation reflects the anchorage-independent growth of tumor cells; and suspension culture sphere formation by tumor cells reflects cancer stem cell (CSC)-like property, these results suggest that upregulation of HMTases play important roles in maintaining the malignant phenotypes of Cr(VI)-transformed cells.
3.4 Stable knockdown of HMTases in parental BEAS-2B cells significantly reduces chronic low dose Cr(VI) exposure-induced CSC-like property and cell transformation
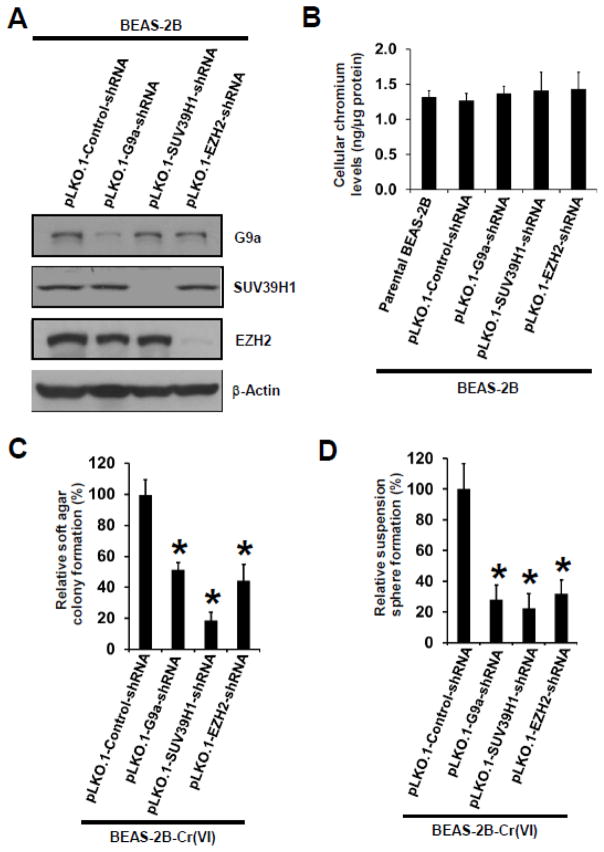
To further determine whether upregulation of HMTases plays a causal role in chronic low dose Cr(VI) exposure-induced CSC-like property and cell transformation, we generated shRNA vector control (pLKO.1-Control shRNA), G9a stable knockdown (pLKO.1-G9a shRNA), SUV39H1 stable knockdown (pLKO.1-SUV39H1 shRNA), and EZH2 stable knockdown (pLKO.1-EZH2 shRNA) BEAS-2B cells. Since it has been shown that G9a and GLP mainly exist and function as a G9a-GLP heteromeric H3K9 MTase complex; and that depleting one of these two HMTases causes the inactivation of the other (Shinkai and Tachibana, 2011), we did not generate GLP stable knockdown cells. Western blot analysis showed that G9a, SUV39H1 and EZH2 shRNA vectors efficiently and specifically knocked down (more than 80%) the level of G9a, SUV39H1 or EZH2, respectively (Fig. 4A).
Fig. 4. Stable knockdown of HMTases in parental BEAS-2B cells significantly reduces chronic low dose Cr(VI) exposure-induced CSC-like property and cell transformation.
(A) Representative Western blot images for the analysis of levels of HMTases in G9a, SUV391 and EZH2 shRNA stable knockdown BEAS-2B cells. Generation of shRNA vector control and each HMTase shRNA stable knockdown cells are described in Methods. (B) Measurement of intracellular chromium levels. Cells were treated with 10 μM of K2Cr2O7 for 3 h and collected for intracellular chromium level analysis as described in Methods. Data are presented as mean ± SD (n=3). (C,D) Quantitation of effects of stable knockdown of G9a, SUV39H1 or EZH2 on chronic Cr(VI) exposure-induced soft agar colony (C) and suspension culture sphere formation (D). Vector control and each HMTase stable knockdown BEAS-2B cells were exposed to 0.125 μM of Cr(VI) (K2Cr2O7) for 25 weeks as described in Methods. At the end of Cr(VI) exposure, cells were collected for soft agar colony and suspension culture sphere formation assays. Results are expressed as relative colony or sphere formation compared to Cr(VI)-exposed shRNA vector control cells (100%). Data are presented as mean ± SD (n=3). *p< 0.05, compared to Cr(VI)-exposed shRNA vector control cells. Similar results were obtained in two additional experiments.
As downregulation of HMTases G9a, SUV39H1 and EZH2 will reduce the levels of histone H3 repressive methylation marks thus impacting gene expression, we first determined the effect of knocking down these HMTases on cellular Cr(VI) uptake and responses of cells to acute Cr(VI) exposure. It was found that knockdown of each HMTase has no significant effect on cellular Cr(VI) uptake, as no significant differences of intracellular chromium levels were observed after exposure to 10 μM of Cr(VI) (K2Cr2O7) for 3 h (Fig. 4B). Interestingly, it was found while G9a or SUV39H1 stable knockdown itself had no obvious effect on BEAS-2B cell viability and growth, it reduced the maximal Cr(VI) dose that is not cytotoxic to BEAS-2B cells from 0.25 μM to 0.125 μM after 72 h Cr(VI) exposure (data not shown). This finding suggests that G9a and SUV39H1 have important roles for cell survival under Cr(VI) exposure. Stable knockdown of EZH2 showed no significant effect on BEAS-2B cell viability, proliferation and responses to acute Cr(VI) exposure (data not shown). Based on these findings, we chronically exposed shRNA vector control cells, G9a, SUV39H1 and EZH2 stable knockdown cells to 0.125 μM of Cr(VI) (K2Cr2O7) for 25 weeks for cell transformation experiment as described in Methods. It was found that compared to shRNA vector control cells, G9a, SUV39H1 or EZH2 stable knockdown significantly reduced chronic Cr(VI) exposure-induced soft agar colony formation and suspension sphere formation (Fig. 4C and D). These findings indicate that upregulation of HMTases plays a causal role in chronic Cr(VI) exposure-induced CSC-like property and cell transformation.
3.5 Stable knockdown of HMTases significantly reduces Cr(VI) exposure -caused DNA damage
Next, we wanted to further explore the potential mechanism by which knocking down HMTases reduces Cr(VI)-induced CSC-like property and cell transformation. It is believed that both genetic and epigenetic changes play critical roles in cancer initiation and progression; and that the crosstalk between epigenetic and genetic effects exists (You and Jones, 2012; Avgustinova and Benitah, 2016; Baylin and Jones, 2016). While studies showed that Cr(VI) exposure causes both genetic and epigenetic changes, it is not known whether Cr(VI)-caused epigenetic changes contribute significantly to its genotoxic effects. Since epigenetic silencing of genes with critical functions in DNA repair could increase DNA damage and gene mutations causing cell transformation and tumorigenesis, we wanted to determine whether Cr(VI) exposure-caused histone H3 repressive methylation (epigenetic effect) via upregulation of HMTases plays a role in Cr(VI)-caused DNA damage (genetic effect).
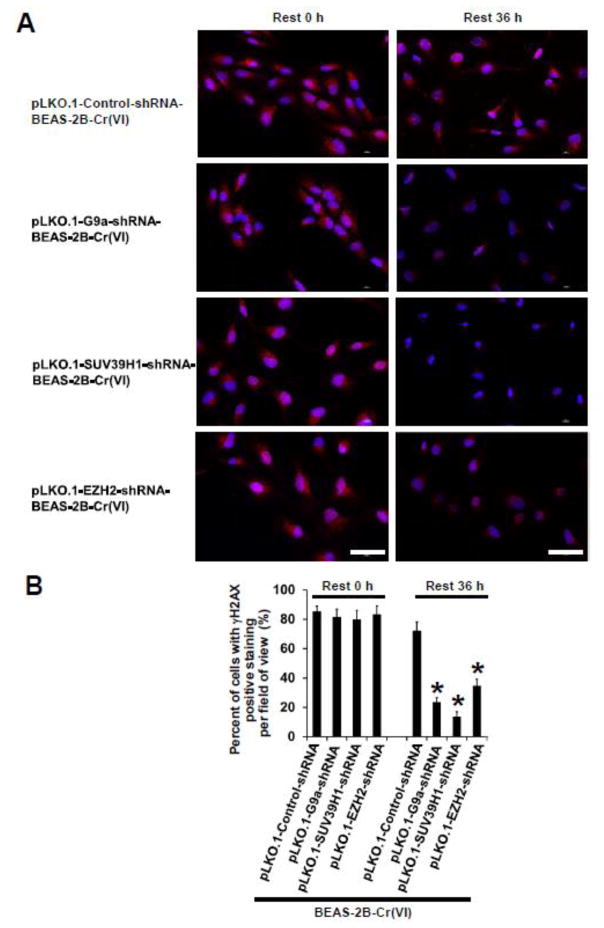
We challenged chronic low dose Cr(VI)-exposed (0.125 μM of K2Cr2O7 for 25 weeks) shRNA vector control cells [pLKO.1-Control-shRNA-BEAS-2B-Cr(VI)] and each HMTase stable knockdown [pLKO.1-G9a-shRNA-BEAS-2B-Cr(VI), pLKO.1-SUV39H1-shRNA-BEAS-2B-Cr(VI), pLKO.1-EZH2-shRNA-BEAS-2B-Cr(VI)] cells with 2.5 μM of Cr(VI) (K2Cr2O7) for 12 h to induce acute DNA damage. Cells were then washed with PBS and allowed to recover for 36 h in culture media without Cr(VI). Cr(VI)-caused DNA damage was revealed as previously reported (Peterson-Roth et al., 2005) by IF staining of γH2AX, a sensitive molecular marker for monitoring DNA damage and repair (Kuo and Yang, 2008; Mah et al., 2010; Ivashkevich et al., 2012). It was found that similar acute DNA damages are detected in all group of cells treated with 2.5 μM of Cr(VI) for 12 h as evidenced by similar extensive positive IF staining of γH2AX (designated as Rest 0 h in Fig. 5A). After 36 h recovery in the absence of Cr(VI), the extensive γH2AX positive staining was still observed in chronic low dose Cr(VI)-exposed shRNA vector control [pLKO.1-Control-shRNA-BEAS-2B-Cr(VI)] cells (designated as Rest 36 h in Fig. 5A). In contrast, the γH2AX positive staining was significantly reduced in chronic low dose Cr(VI)-exposed each HMTase stable knockdown cells (Rest 36 h in Fig. 5A). The quantitation of the γH2AX positive staining in each group of cells is shown in Fig. 5B. These results suggest that chronic low dose Cr(VI) exposure-transformed cells [pLKO.1-Control-shRNA-BEAS-2B-Cr(VI)] display impaired DNA damage repair capacity. In contrast, stable knockdown of HMTases (G9a, SUV39H1 or EZH2) significantly reduces chronic Cr(VI) exposure-caused DNA damage repair deficiency, implying an important role of Cr(VI)-induced epigenetic changes in its genotoxic effect.
Fig. 5. Stable knockdown of HMTases significantly reduces Cr(VI) exposure -caused DNA damage.
(A) Representative images of immunofluorescence (IF) staining of γH2AX in chronic low dose Cr(VI)-exposed shRNA vector control and each HMTase shRNA stable knockdown BEAS-2B cells treated with 2.5 μM of Cr(VI) (K2Cr2O7). After exposure to 0.125 μM of Cr(VI) (K2Cr2O7) for 25 weeks, vector control and each HMTase stable knockdown cells were seeded in 6-well plates. After overnight culture, cells were treated with 2.5 μM of K 2Cr2O7 for 12 h. At the end of the treatment, one set of cells were used for IF staining of γH2AX (designated as Rest 0 h). The other set of cells were washed with PBS and cultured for additional 36 h in the absence of K2Cr2O7 and used for IF staining of γH2AX (designated as Rest 36 h) as described in Methods. Scale bar: 100 μm. (B) Quantitation of γH2AX positive staining in Cr(VI)-exposed cells described in (A). Results are expressed as percent (%) of γH2AX positive staining per field of view. Data are presented as mean ± SD (n=30 fields of view). *p< 0.05, compared to Cr(VI)-exposed shRNA vector control cells. Similar results were obtained in two additional experiments.
4. Discussion
Although Cr(VI) is a common and well-recognized environmental carcinogen causing lung and other cancer in humans, the mechanism of Cr(VI) carcinogenicity is not clearly understood. While Cr(VI) is generally considered as a genotoxic carcinogen, studies showed that Cr(VI) exposure also causes epigenetic changes. However, how Cr(VI) exposure causes epigenetic changes and whether Cr(VI)-caused epigenetic changes contributes significantly to its carcinogenicity are largely unknown. This study was designed to investigate these important unanswered questions.
While previous studies showed that acute Cr(VI) exposure triggers various epigenetic changes (Klein et al., 2002; Schnekenburger et al., 2007; Sun et al., 2009; Zhou et al., 2009), whether epigenetic dysregulations also occurs during chronic low dose Cr(VI) exposure-induced cell malignant transformation is unknown. Our findings that the levels of histone H3 repressive methylation marks (H3K9me2 and H3K27me3) are increased in chronic low dose Cr(VI) exposure-transformed cells indicate that chronic low dose Cr(VI) exposure is also capable of causing epigenetic dysregulation. We then further determined the underlying mechanism by which chronic Cr(VI) expose causes epigenetic changes. Increased levels of H3K9me2 and H3K27me3 could be caused by increased expression or activity of the related H3K9 methyltransferases (MTases) (GLP, G9a, SUV39H1, etc.) and the H3K27 MTase (EZH2); or by the downregulation of histone lysine demethylases. HMTases are important epigenetic regulators functioning as critical epigenome modifiers via changing histone tail methylation status. While differential histone tail posttranslational modifications have distinct effects on chromatin structure and gene expression, increased H3K9me2 and H3K27me3 silence gene expression and are believed to play critical roles in cancer (Yamagishi and Uchimaru, 2017; Rao et al., 2017; Morera et al., 2016; Casciello et al., 2015; Liu et al., 2014). We found that the expression levels of HMTases (GLP, G9a, SUV39H1 and EZH2) are increased Cr(VI)-transformed cells; and pharmacological inhibition or knockdown of these HMTases abolish the increases of H3 repressive methylation marks (H3K9me2 and H3K27me3). These findings indicate that chronic low dose Cr(VI) exposure causes epigenetic dysregulation (increases of H3K9me2 and H3K27me3) through upregulating the expression of several HMTases.
Whether Cr(VI)-caused epigenetic changes contribute significantly to Cr(VI) carcinogenicity remains to be determined. We attacked this important question by determining the role of Cr(VI)-caused epigenetic dysregulation in chronic Cr(VI) exposure-induced cancer stem cell (CSC)-like property and cell transformation. Our following findings strongly support such an idea that Cr(VI)-caused epigenetic dysregulation mediated by the upregulation of H3K9 and H3K27 MTases may play an important role in Cr(VI) carcinogenicity: (i) upregulation of these HMTases are also observed in human lung cancer resulting from Cr(VI) exposure; (ii) pharmacological inhibition or knockdown of these HMTases significantly reduces the malignant phenotypes of Cr(VI)-transformed cells; and (iii) stable knockdown the expression of these HMTases significantly decreases chronic Cr(VI) exposure-induced CSC-like property and cell transformation. Together, these findings not only suggest an important role of these HMTases in Cr(VI) carcinogenicity, they also open new avenues for additional mechanistic studies: further studies on the mechanism of chronic Cr(VI) exposure upregulating these HMTases expression are warrantied. Cr(VI) is known as a potent oxidative stress inducer, it will be interesting to explore whether Cr(VI)-mediated oxidative stress plays a role in HMTase up-regulation by chronic Cr(VI) exposure.
After demonstrating a critical role of epigenetic dysregulation in Cr(VI)-induced CSC-like property and cell transformation, we further explored the potential mechanism by which Cr(VI)-caused epigenetic dysregulation may contribute to its carcinogenicity. Although cancer has been traditionally viewed as a genetic disease, many studies showed that epigenetic dysregulation also play pivotal roles in cancer initiation and progression (Avgustinova and Benitah, 2016; Baylin and Jones, 2016). Exciting findings from the application of advanced sequencing technologies indicate that genetic and epigenetic mechanisms of cancer are not isolated events and the crosstalk between these two mechanisms exists extensively during the carcinogenic process (You and Jones, 2012). On one hand, the findings from whole exome sequencing of large numbers of human cancer cells and tissues revealed frequent mutations of genes that are epigenetic modifiers such as DNMTs (DNA methyltransferases) and HMTases, indicating the genetic regulation of cancer epigenetics. On the other hand, epigenetic silencing of key tumor suppressor genes by increased DNA methylation and/or histone repressive methylation modifications predisposes to increased DNA damage and genetic mutations, indicating the epigenetic regulation of cancer genetics. Our results showing that knockdown of HMTases significantly reduces Cr(VI) exposure-induced DNA damage strongly suggest that Cr(VI)-caused epigenetic dysregulation plays an important role in its genotoxic effect. While it is currently unknown whether Cr(VI) exposure causes gene mutations among key epigenetic regulators, our findings from this study provide novel evidence showing that the crosstalk between Cr(VI) epigenetic and genetic effects exists. It is thus reasonable to speculate that one potential mechanism of Cr(VI) epigenetic effect promoting CSC-like property and cell transformation is to silence the expression of tumor suppressors such as DNA repair genes via the increased histone H3 repressive methylation modifications. Further studies are needed to determine whether the expressions of certain DNA repair genes are downregulated by Cr(VI) exposure and the underlying mechanism.
5. Conclusions
In this study, we found that epigenetic dysregulations are detected in chronic low dose Cr(VI) exposure-transformed human bronchial epithelial cells as evidenced by the increased levels of histone H3 repressive methylation marks (H3K9me2 and H3K27me3) and the related HMTases (GLP, G9a, SUV39H1 and EZH2). Moreover, we also found that upregulation of HMTases are observed in Cr(VI) exposure-caused human lung cancer tissues. Further mechanistic studies using pharmacological inhibitors and shRNA gene knockdown approach reveal that: (i) chronic low dose Cr(VI) exposure increases histone H3 repressive methylation modifications through upregulating the expression of the related HMTases; (ii) upregulation of HMTases plays a causal role in chronic low dose Cr(VI) exposure-induced cancer stem cell (CSC)-like property and cell transformation; and (iii) knockdown of HMTases reduces Cr(VI) exposure-caused DNA damage suggesting potential epigenetic regulations of Cr(VI) genotoxic effects.
Supplementary Material
Highlights.
Chronic low dose Cr(VI) exposure causes epigenetic changes by upregulating HMTases
HMTases upregulation plays a causal role in Cr(VI)-induced CSC-like property
HMTases upregulation plays a causal role in Cr(VI)-induced cell transformation
Knockdown of HMTases reduces Cr(VI)-caused DNA damage
Acknowledgments
This research was supported by National Institutes of Health (5R01ES026151-03 and 1P30ES026529-01A1). The authors would like to thank Drs. Kathryn G. Severin and Xuefei Huang (Department of Chemistry, Michigan State University) for their excellent technical assistance in measuring intracellular chromium levels.
Footnotes
Disclosure of potential conflicts of interest: The authors declare that there are no conflicts of interest.
The authors declare they have no actual or potential competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agency for Toxic Substances and Disease Registry. Toxicological Profile for Chromium. Atlanta, GA: Agency for Toxic Substances and Disease Registry; 2000. [Google Scholar]
- Ali AH, Kondo K, Namura T, Senba Y, Takizawa H, Nakagawa Y, Toba H, Kenzaki K, Sakiyama S, Tangoku A. Aberrant DNA methylation of some tumor suppressor genes in lung cancers from workers with chromate exposure. Mol Carcinog. 2011;5:89–99. doi: 10.1002/mc.20697. [DOI] [PubMed] [Google Scholar]
- Arita A, Costa M. Epigenetics in metal carcinogenesis: nickel, arsenic, chromium and cadmium. Metallomics. 2009;1:222–8. doi: 10.1039/b903049b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avgustinova A, Benitah SA. The epigenetics of tumour initiation: cancer stem cells and their chromatin. Curr Opin Genet Dev. 2016;36:8–15. doi: 10.1016/j.gde.2016.01.003. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. Epigenetic Determinants of Cancer. Cold Spring Harb Perspect Biol. 2016;8 doi: 10.1101/cshperspect.a019505. pii: a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocato J, Costa M. Basic mechanics of DNA methylation and the unique landscape of the DNA methylome in metal-induced carcinogenesis. Crit Rev Toxicol. 2013;43:493–514. doi: 10.3109/10408444.2013.794769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casciello F, Windloch K, Gannon F, Lee JS. Functional Role of G9a Histone Methyltransferase in Cancer. Front Immunol. 2015;6:487. doi: 10.3389/fimmu.2015.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, Woodward WA, Hsu JM, Hortobagyi GN, Hung MC. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervona Y, Arita A, Costa M. Carcinogenic metals and the epigenome: understanding the effect of nickel, arsenic, and chromium. Metallomics. 2012;4:619–27. doi: 10.1039/c2mt20033c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–70. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–57. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries B, Wang Z, Oom AL, Fisher T, Tan D, Cui Y, Jiang Y, Yang C. MicroRNA-200b targets protein kinase Cα and suppresses triple-negative breast cancer metastasis. Carcinogenesis. 2014;35:2254–63. doi: 10.1093/carcin/bgu133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Agency for Research on Cancer. Chromium. IARC Monogr Eval Carcinog Risks Hum. 1990;49:49–256. [PMC free article] [PubMed] [Google Scholar]
- Ivashkevich A, Redon CE, Nakamura AJ, Martin RF, Martin OA. Use of the γ-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Lett. 2012;327:123–33. doi: 10.1016/j.canlet.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein CB, Su L, Bowser D, Leszczynska J. Chromate-induced epimutations in mammalian cells. Environ Health Perspect. 2002;110(Suppl 5):739–43. doi: 10.1289/ehp.02110s5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, Takahashi Y, Hirose Y, Nagao T, Tsuyuguchi M, Hashimoto M, Ochiai A, Monden Y, Tangoku A. The reduced expression and aberrant methylation of p16(INK4a) in chromate workers with lung cancer. Lung Cancer. 2006;53:295–302. doi: 10.1016/j.lungcan.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Kondo Y. Targeting histone methyltransferase EZH2 as cancer treatment. J Biochem. 2014;156:249–57. doi: 10.1093/jb/mvu054. [DOI] [PubMed] [Google Scholar]
- Kuo LJ, Yang LX. Gamma-H2AX - a novel biomarker for DNA double-strand breaks. In Vivo. 2008;22:305–9. [PubMed] [Google Scholar]
- Labra M, Grassi F, Imazio S, Di Fabio T, Citterio S, Sgorbati S, Agradi E. Genetic and DNA-methylation changes induced by potassium dichromate in Brassica napus L. Chemosphere. 2004;54:1049–58. doi: 10.1016/j.chemosphere.2003.10.024. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu K, Qin S, Xu C, Min J. Epigenetic targets and drug discovery: Part 1: Histone methylation. Pharmacol Ther. 2014;143:275–294. doi: 10.1016/j.pharmthera.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Mah LJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24:679–86. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- Martinez-Zamudio R, Ha HC. Environmental epigenetics in metal exposure. Epigenetics. 2011;6:820–7. doi: 10.4161/epi.6.7.16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer J, Reynolds M, Stoddard L, Zhitkovich A. Causes of DNA single-strand breaks during reduction of chromate by glutathione in vitro and in cells. Free Radic Biol Med. 2006;40:1981–92. doi: 10.1016/j.freeradbiomed.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Morera L, Lübbert M, Jung M. Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin Epigenetics. 2016;8:57. doi: 10.1186/s13148-016-0223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12:133–43. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- Ott I, Kircher B, Gust R. Investigations on the effects of cobalt-alkyne complexes on leukemia and lymphoma cells: cytotoxicity and cellular uptake. J Inorg Biochem. 2004;98:485–9. doi: 10.1016/j.jinorgbio.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Ovesen JL, Fan Y, Chen J, Medvedovic M, Xia Y, Puga A. Long-term exposure to low-concentrations of Cr(VI) induce DNA damage and disrupt the transcriptional response to benzo[a]pyrene. Toxicology. 2014;316:14–24. doi: 10.1016/j.tox.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson-Roth E, Reynolds M, Quievryn G, Zhitkovich A. Mismatch repair proteins are activators of toxic responses to chromium-DNA damage. Mol Cell Biol. 2005;25:3596–607. doi: 10.1128/MCB.25.9.3596-3607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Wang Z, Li Y, Miao Y, Ren Y, Luan Y. Characterization of sphere-forming cells with stem-like properties from the small cell lung cancer cell line H446. Cancer Lett. 2012;323:161–70. doi: 10.1016/j.canlet.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, Peyton M, Zou Y, Kurie JM, Dimaio JM, Milchgrub S, Smith AL, Souza RF, Gilbey L, Zhang X, Gandia K, Vaughan MB, Wright WE, Gazdar AF, Shay JW, Minna JD. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–34. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- Rao VK, Pal A, Taneja R. A drive in SUVs: From development to disease. Epigenetics. 2017;12:177–186. doi: 10.1080/15592294.2017.1281502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikow K, Zhitkovich A. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: nickel, arsenic, and chromium. Chem Res Toxicol. 2008;21:28–44. doi: 10.1021/tx700198a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnekenburger M, Talaska G, Puga A. Chromium cross-links histone deacetylase 1-DNA methyltransferase 1 complexes to chromatin, inhibiting histone-remodeling marks critical for transcriptional activation. Mol Cell Biol. 2007;27:7089–101. doi: 10.1128/MCB.00838-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen S, Ahmed M, Lorenzi F, Nateri AS. Spheroid-Formation (Colonosphere) Assay for in Vitro Assessment and Expansion of Stem Cells in Colon Cancer. Stem Cell Reviews. 2016;12:492–499. doi: 10.1007/s12015-016-9664-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Hudson LG, Liu KJ. Oxidative stress and apoptosis in metal ion-induced carcinogenesis. Free Radic Biol Med. 2004;37:582–93. doi: 10.1016/j.freeradbiomed.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–8. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Meeran SM. Epigenetics of cancer stem cells: Pathways and therapeutics. Biochim Biophys Acta. 2014;1840:3494–502. doi: 10.1016/j.bbagen.2014.09.017. [DOI] [PubMed] [Google Scholar]
- Smolle M, Workman JL. Transcription-associated histone modifications and cryptic transcription. Biochim Biophys Acta. 2013;1829:84–97. doi: 10.1016/j.bbagrm.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout MD, Herbert RA, Kissling GE, Collins BJ, Travlos GS, Witt KL, Melnick RL, Abdo KM, Malarkey DE, Hooth MJ. Hexavalent chromium is carcinogenic to F344/N rats and B6C3F1 mice after chronic oral exposure. Environ Health Perspect. 2009;117:716–22. doi: 10.1289/ehp.0800208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Zhou X, Chen H, Li Q, Costa M. Modulation of histone methylation and MLH1 gene silencing by hexavalent chromium. Toxicol Appl Pharmacol. 2009;237:258–66. doi: 10.1016/j.taap.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann T, Schneider R. Targeting histone modifications--epigenetics in cancer. Curr Opin Cell Biol. 2013;25:184–9. doi: 10.1016/j.ceb.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Wang Z, Humphries B, Xiao H, Jiang Y, Yang C. MicroRNA-200b suppresses arsenic-transformed cell migration by targeting protein kinase Cα and Wnt5b-protein kinase Cα positive feedback loop and inhibiting Rac1 activation. J Biol Chem. 2014;289:18373–86. doi: 10.1074/jbc.M114.554246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhao Y, Smith E, Goodall GJ, Drew PA, Brabletz T, Yang C. Reversal and prevention of arsenic-induced human bronchial epithelial cell malignant transformation by microRNA-200b. Toxicol Sci. 2011;121:110–22. doi: 10.1093/toxsci/kfr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise SS, Holmes AL, Wise JP., Sr Hexavalent chromium- induced DNA damage and repair mechanisms. Rev Environ Health. 2008;23:39–57. doi: 10.1515/reveh.2008.23.1.39. [DOI] [PubMed] [Google Scholar]
- Yamagishi M, Uchimaru K. Targeting EZH2 in cancer therapy. Curr Opin Oncol. 2017;29:375–381. doi: 10.1097/CCO.0000000000000390. [DOI] [PubMed] [Google Scholar]
- Yang C, Liu Y, Lemmon MA, Kazanietz MG. Essential role for Rac in heregulin beta1 mitogenic signaling: a mechanism that involves epidermal growth factor receptor and is independent of ErbB4. Mol Cell Biol. 2006;26:831–42. doi: 10.1128/MCB.26.3.831-842.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Wu J, Zhang R, Zhang P, Eckard J, Yusuf R, Huang X, Rossman TG, Frenkel K. Caffeic acid phenethyl ester (CAPE) prevents transformation of human cells by arsenite (As) and suppresses growth of As-transformed cells. Toxicology. 2005;213:81–96. doi: 10.1016/j.tox.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Yao H, Guo L, Jiang BH, Luo J, Shi X. Oxidative stress and chromium(VI) carcinogenesis. J Environ Pathol Toxicol Oncol. 2008;27:77–88. doi: 10.1615/jenvironpatholtoxicoloncol.v27.i2.10. [DOI] [PubMed] [Google Scholar]
- You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Tan YS, Haslam SZ, Yang C. Perfluorooctanoic acid effects on steroid hormone and growth factor levels mediate stimulation of peripubertal mammary gland development in C57BL/6 mice. Toxicol Sci. 2010;115:214–24. doi: 10.1093/toxsci/kfq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wang Z, Jiang Y, Yang C. Inactivation of Rac1 reduces Trastuzumab resistance in PTEN deficient and insulin- like growth factor I receptor overexpressing human breast cancer SKBR3 cells. Cancer Lett. 2011;313:54–63. doi: 10.1016/j.canlet.2011.08.023. [DOI] [PubMed] [Google Scholar]
- Zhou X, Li Q, Arita A, Sun H, Costa M. Effects of nickel, chromate, and arsenite on histone 3 lysine methylation. Toxicol Appl Pharmacol. 2009;236:78–84. doi: 10.1016/j.taap.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.