ABSTRACT
The role of the zebrafish transcription factor Nanog has been controversial. It has been suggested that Nanog is primarily required for the proper formation of the extra-embryonic yolk syncytial layer (YSL) and only indirectly regulates gene expression in embryonic cells. In an alternative scenario, Nanog has been proposed to directly regulate transcription in embryonic cells during zygotic genome activation. To clarify the roles of Nanog, we performed a detailed analysis of zebrafish nanog mutants. Whereas zygotic nanog mutants survive to adulthood, maternal-zygotic (MZnanog) and maternal mutants exhibit developmental arrest at the blastula stage. In the absence of Nanog, YSL formation and epiboly are abnormal, embryonic tissue detaches from the yolk, and the expression of dozens of YSL and embryonic genes is reduced. Epiboly defects can be rescued by generating chimeric embryos of MZnanog embryonic tissue with wild-type vegetal tissue that includes the YSL and yolk cell. Notably, cells lacking Nanog readily respond to Nodal signals and when transplanted into wild-type hosts proliferate and contribute to embryonic tissues and adult organs from all germ layers. These results indicate that zebrafish Nanog is necessary for proper YSL development but is not directly required for embryonic cell differentiation.
KEY WORDS: Nanog, Zebrafish, Maternal-to-zygotic transition, MZT, Yolk syncytial layer, YSL, Zygotic genome activation, ZGA, Lineage tracing, GESTALT
Highlighted Article: Detailed analyses of zebrafish nanog mutants, including tissue-specific rescue and chimera analysis, indicate that Nanog is necessary for yolk syncytial layer development but is not directly required for embryonic cell differentiation.
INTRODUCTION
The transcription factor Nanog is part of the core circuitry that regulates mammalian pluripotency (reviewed by Theunissen and Jaenisch, 2014). In vitro studies have shown that removal of Nanog triggers differentiation of mouse and human embryonic stem cells (Chambers et al., 2007; Hyslop et al., 2005; Loh et al., 2006; Mitsui et al., 2003). However, a subset of Nanog mutant mouse embryonic stem cells are able to self-renew (Chambers et al., 2007). In vivo studies have revealed that Nanog is required for inner cell mass pluripotency and epiblast development (Mitsui et al., 2003). However, in chimeras with wild-type cells, Nanog mutant cells can give rise to tissues from all germ layers (Chambers et al., 2007). Thus, mouse Nanog is involved in, but not absolutely required for, the maintenance of the pluripotent state (Carter et al., 2014; Chambers et al., 2007; Schwarz et al., 2014).
The roles of zebrafish Nanog in pluripotency and differentiation are less well-understood. Xu et al. (2012) reported that nanog was provided maternally and present in all embryonic and extra-embryonic cells. Morpholino-mediated knockdown of nanog mRNA resulted in developmental arrest prior to gastrulation. Nanog morphants displayed defects in the formation of the yolk syncytial layer (YSL), the extra-embryonic tissue that attaches the embryo to the yolk and generates Nodal and BMP signals that pattern mesendoderm (Carvalho and Heisenberg, 2010; Chen and Kimelman, 2000; D'Amico and Cooper, 2001; Hong et al., 2011; Kimmel and Law, 1985; Mizuno et al., 1996; Xu et al., 2012). Gene expression analysis in nanog morphants revealed the absence of YSL markers such as mxtx2 and the misregulation of hundreds of embryonic genes, including Nodal and its target genes. Injecting mxtx2 mRNA into YSL precursors of nanog morphants partially rescued YSL formation and the expression of Nodal and several of its target genes. Although no cell-autonomy data were shown to determine whether Nanog was required in embryonic cells, the study suggested that the primary role of Nanog is to regulate the formation of the YSL (Xu et al., 2012).
Two subsequent studies analyzed potential roles of zebrafish Nanog in embryonic cells (Lee et al., 2013; Perez-Camps et al., 2016). Lee et al. (2013) defined a set of genes expressed at the maternal-to-zygotic transition (MZT), expression of which was reduced in nanog morphants. Chromatin immunoprecipitation experiments suggested that many of these genes were direct targets of Nanog (Bogdanovic et al., 2012; Lee et al., 2013; Leichsenring et al., 2013; Xu et al., 2012). Based on the reduced expression of genes in morphants and the Nanog binding data, the study concluded that Nanog, along with Pou5f1 (now known as Pou5f3 in zebrafish) and the SoxB1 family, was involved in the first wave of zygotic transcription in embryonic cells. Subsequent reviews have interpreted these results to conclude that Nanog is directly required for zygotic genome activation in embryonic cells (Langley et al., 2014; Lee et al., 2014; Onichtchouk and Driever, 2016; Paranjpe and Veenstra, 2015), even though the majority of zygotic genes are activated in nanog morphants (Lee et al., 2013; Xu et al., 2012). Perez-Camps et al. (2016) reported that morpholino knockdown of nanog caused defects in BMP signaling and target gene expression, and suggested that Nanog acts to promote ventral cell-fate specification. Surprisingly, neither study (Lee et al., 2013; Perez-Camps et al., 2016) mentioned the extra-embryonic YSL phenotype of nanog morphants (Xu et al., 2012) or tested the postulated direct roles of Nanog in embryonic cells.
Here, we clarify the embryonic and extra-embryonic requirements for Nanog using tissue-specific rescue and chimera analysis. Our results indicate that the primary role of zebrafish nanog is YSL formation and that it is not essential for embryonic cell differentiation.
RESULTS
Generation of nanog mutants
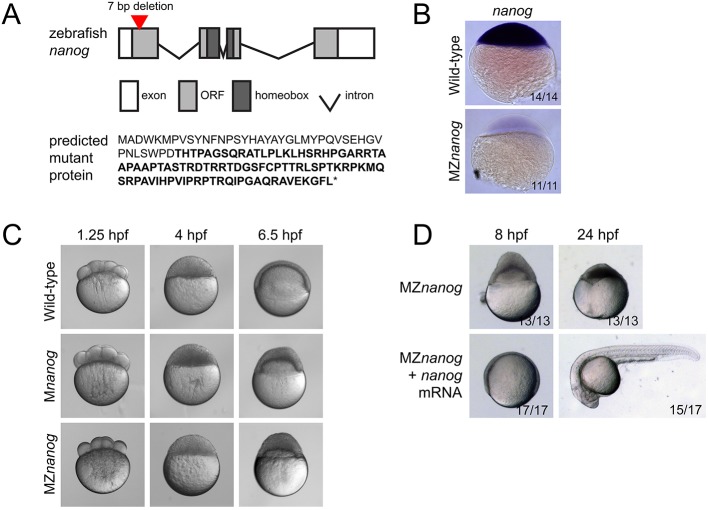
The interpretation of morpholino experiments can be complicated by potential partial loss-of-function phenotypes and the short half-life of morpholinos. To avoid these confounding effects in our studies, we generated nanog mutants using transcription activator-like effector nucleases (TALENs) (Carroll, 2014). We isolated an allele containing a 7 bp deletion predicted to cause a frameshift and premature termination codon before the homeodomain required for DNA binding (Fig. 1A). The mutant nanog mRNA was not detectable at sphere stage [4 hours post-fertilization (hpf)], presumably owing to nonsense-mediated decay (Fig. 1B). Homozygous zygotic nanog (Znanog) mutant embryos showed no phenotypic defects and could be raised to fertile adults. By contrast, maternal-zygotic nanog mutants (MZnanog) and maternal-only nanog mutants (Mnanog) arrested at sphere stage, did not undergo normal epiboly (Fig. 1C), and died when embryonic tissue detached from the yolk. The defects observed in MZnanog embryos were rescued by injection of nanog mRNA at the 1-cell stage (Fig. 1D). Rescued embryos could be raised to fertile adults, establishing that the observed phenotype is solely due to disruption of the nanog gene. The nanog mutant phenotype strongly resembles the previously reported nanog morphant phenotype (Xu et al., 2012). As Mnanog and MZnanog mutants had very similar or identical phenotypes, whereas Znanog mutants were viable, we conclude that maternal, but not zygotic, Nanog has essential roles during embryogenesis.
Fig. 1.
Generation and phenotype of MZnanog mutants. (A) Top: TALENs were used to generate a 7 bp deletion within the first exon of nanog. Exons, introns, the open reading frame (ORF) and the homeobox domain are indicated. Bottom: The predicted mutant protein sequence, with intact amino acids (40 of 384 total) in normal font and frameshifted amino acids in bold font. Asterisk indicates a premature stop codon. (B) In situ hybridization for nanog expression in wild-type and MZnanog embryos at sphere stage. (C) Wild-type, Mnanog and MZnanog embryos imaged at 1.25 hpf (8-cell stage), 4 hpf (sphere stage) and 6.5 hpf (shield stage in wild type). Epiboly defects in Mnanog and MZnanog are apparent at 6.5 hpf. (D) MZnanog embryos are shown at 8 hpf and at 24 hpf, either uninjected or injected with 5 pg nanog mRNA at the 1-cell stage. In B and D, the number of embryos exhibiting the illustrated phenotype is shown in the bottom-right corner of each image as a proportion of total embryos examined.
nanog is required for proper formation of the YSL
YSL development and the expression of YSL markers are impaired in nanog morphants (Xu et al., 2012). Several lines of evidence indicate that YSL formation is similarly impacted in MZnanog mutants. First, yolk injection of SYTOX Green dye indicated that marginal blastomere nuclei form a normal syncytium in MZnanog at sphere stage (Fig. 2A). However, by 6.5 hpf (shield stage in wild type) these yolk syncytial nuclei are aberrantly clustered together and display an enlarged morphology in MZnanog, reminiscent of the phenotypes caused by microtubule defects in the YSL (Takesono et al., 2012). We next investigated the F-actin band, a network of actin filaments that forms in the YSL during gastrulation and is postulated to promote blastopore closure (Cheng et al., 2004; Köppen et al., 2006; Wilkins et al., 2008). We stained for actin using phalloidin and found that the F-actin band was absent in MZnanog at 8 hpf (Fig. 2B), consistent with the phenotypic arrest in the mutants before epiboly (Fig. 1C). Interestingly, MZnanog embryos still form the shield structure and occasionally exhibit dorsal cell internalization and marginal constriction, even in the absence of epiboly or the F-actin band. These results suggest that YSL formation is initiated in MZnanog mutants but the YSL fails to differentiate properly and maintain the attachment of embryo to yolk.
Fig. 2.
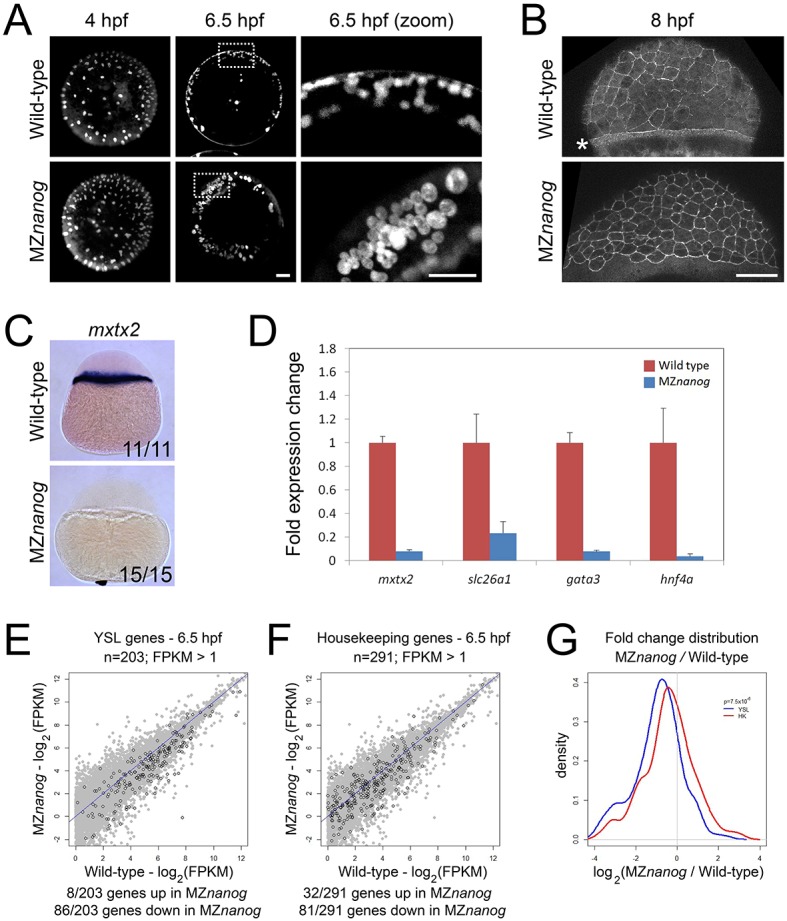
The YSL is impaired in MZnanog embryos. (A) Yolk syncytial nuclei (YSN) were labeled by SYTOX Green dye injection into the yolk of 128-cell wild-type and MZnanog embryos, and imaged at 4 hpf and 6.5 hpf; n=7 for all conditions. Images on the right are magnifications of the boxed areas to the left. (B) The F-actin band, indicated with an asterisk, was visualized using phalloidin staining in fixed wild-type and MZnanog embryos at 8 hpf; n=8 for both wild type and MZnanog. (C) In situ hybridization for mxtx2 expression in wild-type and MZnanog embryos at sphere stage. The number of embryos exhibiting the illustrated expression pattern is shown in the bottom-right corner of each image as a proportion of total embryos examined. (D) Fold expression change for mxtx2, slc26a1, gata3 and hnf4a comparing wild-type and MZnanog embryos at 4 hpf (sphere stage) using RT-qPCR. Error bars show s.d. for three technical replicates (ten embryos per replicate). (E) Differential expression of genes expressed in the YSL, comparing wild type and MZnanog at 6.5 hpf using RNAseq. All genes are in gray, YSL-expressed genes are in black and were previously defined (Xu et al., 2012), and filtered for zygotic expression (Rabani et al., 2014). (F) Differential expression of zygotically expressed housekeeping genes (Lee et al., 2013), comparing wild type and MZnanog at shield stage using RNAseq. All genes are in gray, housekeeping genes are in black. Only those genes with wild-type mRNA expression >1 fragments per kilobase of transcript per million mapped reads (FPKM) are plotted. Genes are categorized as up- or downregulated if their expression in MZnanog differs more than 2-fold from that in wild type. (G) Distribution of fold changes derived from RNAseq data comparing wild-type and MZnanog embryos for all YSL (blue) and housekeeping (HK; red) genes visualized using a kernel density estimation. The displayed P-value (P=7.5×106) comparing these two sets was calculated using a Student's two-tailed t-test. Scale bars: 100 μm.
We next investigated gene expression in the YSL. MZnanog embryos lacked expression of the key YSL determinant mxtx2 at sphere stage (Fig. 2C). Expression of other YSL marker genes, such as slc26a1, gata3 and hnf4a (Xu et al., 2012), was decreased or absent as determined by RT-qPCR (Fig. 2D). RNAseq experiments showed that the expression of many genes expressed in the YSL (Xu et al., 2012) was reduced in MZnanog mutants at 6.5 hpf (Fig. 2E). Expression of many of these genes was not completely eliminated, probably owing to their co-expression in embryonic cells. Compared with zygotically expressed housekeeping genes (Fig. 2F), YSL genes were significantly downregulated in MZnanog embryos (P=7.5×10−6; Fig. 2G). These results confirm and extend previous morphant studies that concluded that Nanog is required for YSL development (Xu et al., 2012).
nanog regulates the expression of a subset of early zygotic genes
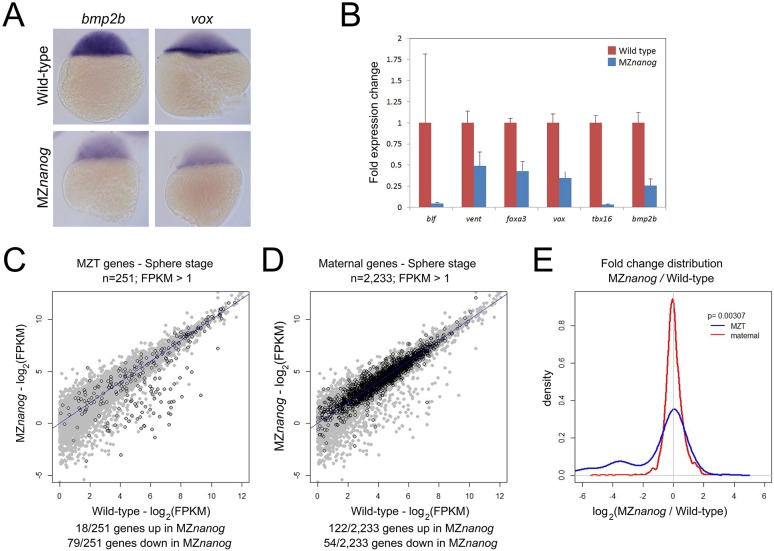
nanog morphants were reported to display a widespread reduction in zygotic gene expression (Lee et al., 2013). To determine whether similar defects are found in MZnanog mutants, we used in situ hybridization, RT-qPCR and RNAseq. Using in situ hybridization and RT-qPCR, we demonstrated that the expression of several early zygotic genes was decreased in MZnanog at sphere stage (Fig. 3A,B). RNAseq experiments confirmed that the expression of many of the previously defined early zygotic genes (Lee et al., 2013) was reduced in MZnanog embryos at sphere stage (Fig. 3C). Compared with maternally provided mRNAs, which are largely unaffected (Fig. 3D), the levels of early zygotic RNAs are significantly reduced in MZnanog mutants (P=0.00307; Fig. 3E). The expression of 79/251 early zygotic genes was reduced >2-fold in MZnanog embryos. Many of these genes are part of developmental signaling pathways (Table S1). Of the 79 genes reduced in MZnanog mutants, 68 were also significantly decreased in the nanog morphants (Lee et al., 2013). These results indicate that nanog mutants and morphants have similar gene expression defects.
Fig. 3.
Defects in early zygotic gene expression in MZnanog embryos. (A) In situ hybridization for bmp2b and vox expression in wild-type and MZnanog embryos at sphere (4 hpf) stage. (B) Fold expression change for the indicated genes comparing wild type and MZnanog at sphere stage using RT-qPCR. Error bars show s.d. for three technical replicates (ten embryos per replicate). (C) Differential expression of early zygotic genes, comparing wild type and MZnanog at sphere stage using RNAseq. All genes are in gray, early zygotic genes (MZT genes) in black were previously defined (Lee et al., 2013). (D) Differential expression of maternally provided genes, comparing wild type and MZnanog at sphere stage using RNAseq. All genes are in gray, maternally provided genes in black were previously defined (Rabani et al., 2014). Only those genes with wild-type mRNA expression >1 FPKM are plotted. Genes are categorized as up- or downregulated if their expression in MZnanog differs more than 2-fold from that in wild type. (E) Distribution of fold changes between wild-type and MZnanog embryos for all early zygotic (blue) and maternal (red) genes visualized using a kernel density estimation. The displayed P-value comparing these two sets was calculated using a Student's two-tailed t-test.
Expression of nanog mRNA in YSL precursors can rescue MZnanog embryos
To clarify the embryonic and extra-embryonic requirements for Nanog, we performed tissue-specific rescue experiments. We first attempted rescue by injection of nanog mRNA into the yolk at the 64-cell stage, or directly into the YSL at the 512-cell stage, but observed little to no rescue. We hypothesized that nanog expression is required early during embryogenesis, and that delayed expression by mRNA injection at later stages is insufficient to rescue the phenotype. We therefore performed YSL rescue experiments by depositing mRNA in the precursor cells of the YSL by vegetal yolk injection at the 4-cell stage (Xu et al., 2012). Co-injection of GFP mRNA was used to verify that expression was predominantly in the YSL, although we cannot exclude the possibility that low levels of injected mRNA are inherited and translated in embryonic cells (Fig. S1A,B). Similar to 1-cell nanog injection, 4-cell yolk injection of nanog mRNA was sufficient to rescue the epiboly defects and yolk detachment in most MZnanog mutants (Fig. 4A). Additionally, expression of mxtx2 and ndr2 was restored in 4-cell yolk-injected MZnanog embryos (Fig. S1C), indicating that YSL function was rescued. At 24 hpf, the majority of mutants displayed axis rescue, tail elongation, and differentiation of various cell types (Fig. 4B,C). In contrast, injection of nanog mRNA into one of 16 cells did not result in any rescue of MZnanog embryos (Fig. 4A,C). We next attempted to rescue MZnanog by mxtx2 mRNA injection (Xu et al., 2012). Four-cell yolk injection of mxtx2 mRNA rescued the epiboly defect and yolk detachment in MZnanog mutants (Fig. 4D,E), and some rescued embryos developed differentiated tissues, including somites, notochord, eyes, brain, melanocytes, otoliths and blood. Generally, rescue by nanog mRNA was more pronounced than rescue by mxtx2 mRNA. These results suggest that the primary role of Nanog is in YSL development, and that YSL defects are a major cause of the embryonic phenotypes observed in MZnanog mutants.
Fig. 4.
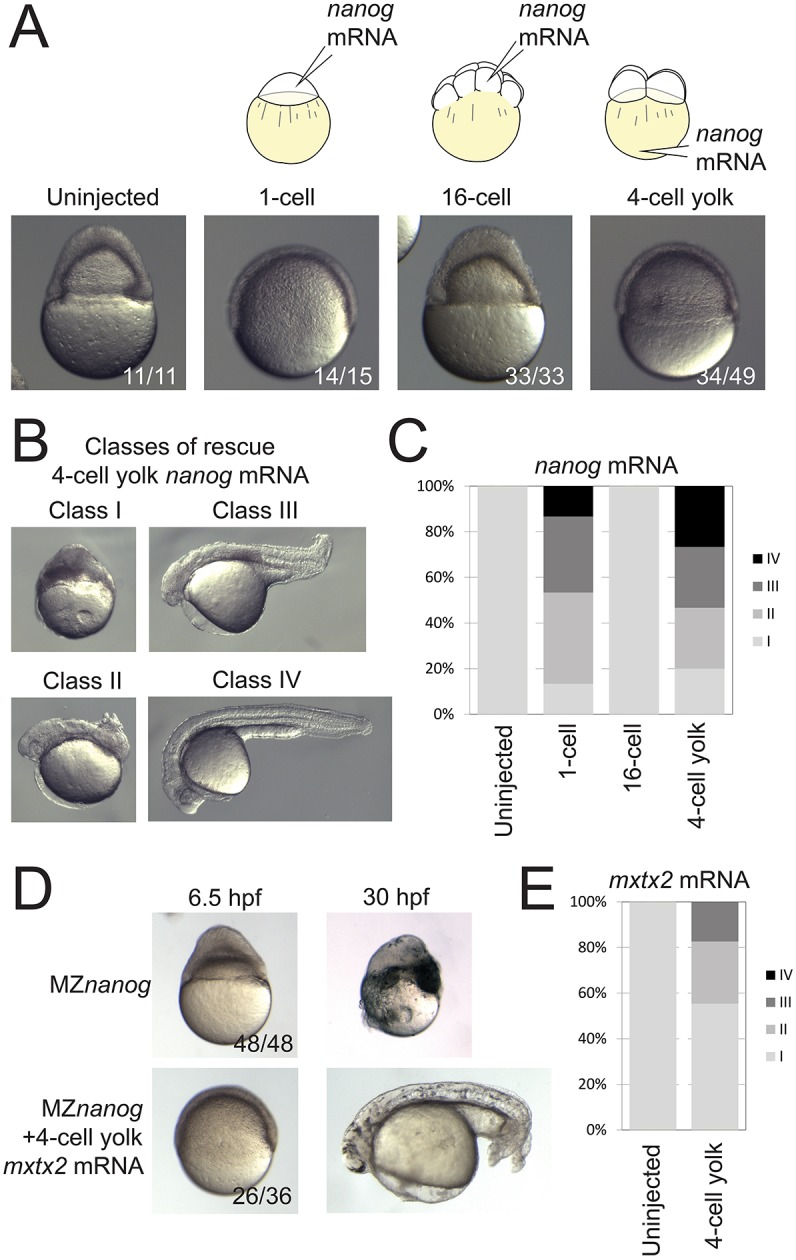
Rescue of MZnanog embryos by YSL expression of nanog or mxtx2 mRNA. (A) Uninjected MZnanog embryos, or MZnanog embryos injected with 33 pg GFP mRNA and 25 pg nanog mRNA into the cell at the 1-cell stage, into a cell at the 16-cell stage, or into the vegetal yolk at the 4-cell stage, were sorted for appropriate spatial GFP expression at sphere stage (as shown in Fig. S1A), and scored and imaged at 8 hpf. (B) Embryos were scored at 24 hpf into four rescue phenotype classes as described in Materials and Methods. (C) Quantification of rescue phenotype scores illustrated as stacked bar plots (uninjected n=11; 1-cell n=15; 16-cell n=33; 4-cell yolk n=30). (D,E) Control MZnanog embryos, or embryos injected into the vegetal yolk at the 4-cell stage with 33 pg GFP mRNA and 50 pg mxtx2 mRNA, were sorted for appropriate spatial GFP expression at sphere stage, scored as above, and imaged at 6.5 hpf and at 30 hpf (D). These scores were quantified and illustrated as stacked bar plots (E) (control n=48; 4-cell yolk n=92). In A and D, the number of embryos exhibiting the illustrated phenotype is shown in the bottom-right corner of each image as a proportion of total embryos examined.
nanog mutant cells can undergo epiboly
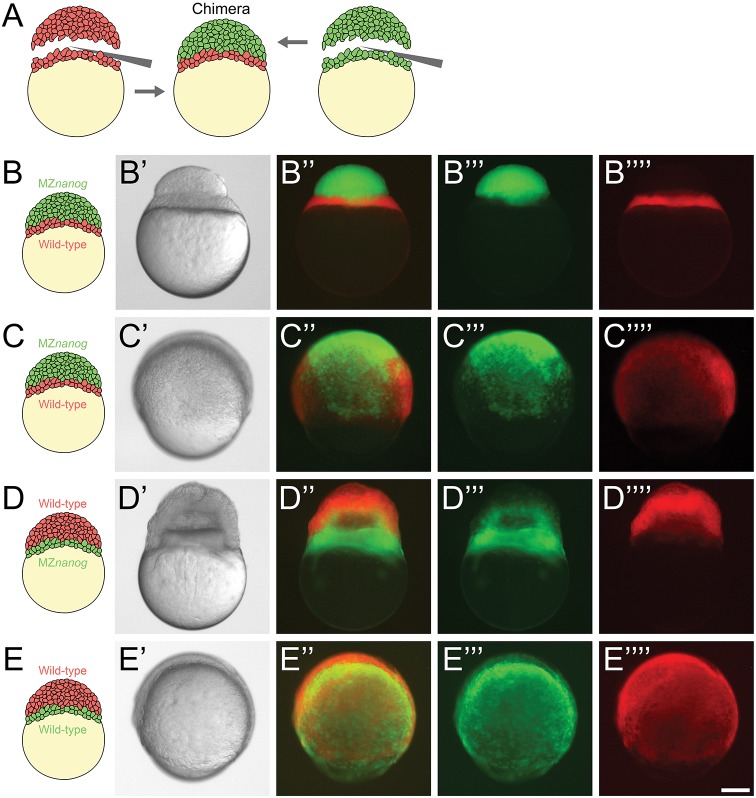
As an additional test to determine whether nanog acts primarily in the YSL, we performed whole blastoderm transplants (Holloway et al., 2009). We transplanted MZnanog blastoderm tissue onto wild-type embryos in which yolk cell was left intact but most of the blastoderm tissue was removed (Fig. 5A). Transplants were performed at the 256- to 512-cell stage, and donor and host tissues were labeled with different fluorescent dyes to track their contributions to the resulting chimera. Shortly after transplantation, we verified that the YSL was exclusively contributed by wild type and that embryonic cells were predominantly contributed by MZnanog tissue (Fig. 5B). Strikingly, these chimeras (n=9) underwent epiboly similar to wild-type embryos or wild-type-to-wild-type controls (n=10) (Fig. 5C,E). In the reciprocal experiment, we transplanted wild-type blastoderm tissue onto MZnanog embryos in which yolk cell was left intact but most of the blastoderm tissue was removed. These chimeras arrested during epiboly (n=5) and had a phenotype similar to MZnanog mutants (Fig. 5D). These experiments cannot rule out an additional role for Nanog in marginal cells but support the idea that Nanog is primarily required in the YSL.
Fig. 5.
Blastoderm transplants between wild-type and MZnanog embryos. (A) Diagram of the blastoderm transplant. Donor embryos were injected with dextran-Alexa-488 or dextran-Alexa-546. Blastoderms were separated from yolks and combined to generate chimera embryos. (B) A chimera imaged approximately 15 min after transplant of a wild-type yolk cell and YSL with MZnanog blastoderm (B-B″″), where MZnanog tissue is labeled in green (B‴) and wild-type tissue is labeled in red (B″″). (C) A chimera at 8 hpf of a wild-type yolk cell and YSL with MZnanog blastoderm (C-C″″), where MZnanog tissue is labeled in green (C‴) and wild-type tissue is labeled in red (C″″). (D) A reciprocal chimera at 8 hpf of wild-type blastoderm with MZnanog yolk cell (D-D″″), again with MZnanog tissue labeled in green (D‴) and wild-type tissue labeled in red (D″″). (E) A chimera at 8 hpf of a wild-type yolk cell and YSL with a wild-type blastoderm from a second embryo (E-E″″), where tissue derived from each donor embryo is distinctly labeled (E‴,E″″). Composites of the fluorescent channels are shown in B″, C″, D″ and E″. Scale bar: 100 μm.
nanog mutant cells can proliferate and differentiate in wild-type embryos
It has been proposed that Nanog is directly required in embryonic cells (Lee et al., 2013; Perez-Camps et al., 2016), but this assumption has not been tested rigorously. For example, mesendodermal marker gene expression is lost in nanog morphants (Lee et al., 2013; Xu et al., 2012) and MZnanog mutants (Fig. 3B, Table S1). Similarly, miR-430 expression and activity are abrogated in nanog morphants (Lee et al., 2013). Such defects may be caused directly by absence of Nanog in embryonic cells or indirectly by lack of Nanog in the YSL (Fig. S1C) (Xu et al., 2012). In support of the latter model, MZnanog embryos injected with Nodal (ndr1) mRNA can express mesendodermal marker genes at similar levels as in wild-type embryos (Fig. S2). This results reveals that Nanog is not required for Nodal-induced cell-type specification. However, supporting a direct role for Nanog in embryonic cells, we found that a GFP reporter normally silenced by miR-430 is active in MZnanog embryos and in transplanted MZnanog cells (Fig. S3). This result supports a cell-autonomous requirement for Nanog in miR-430 activity.
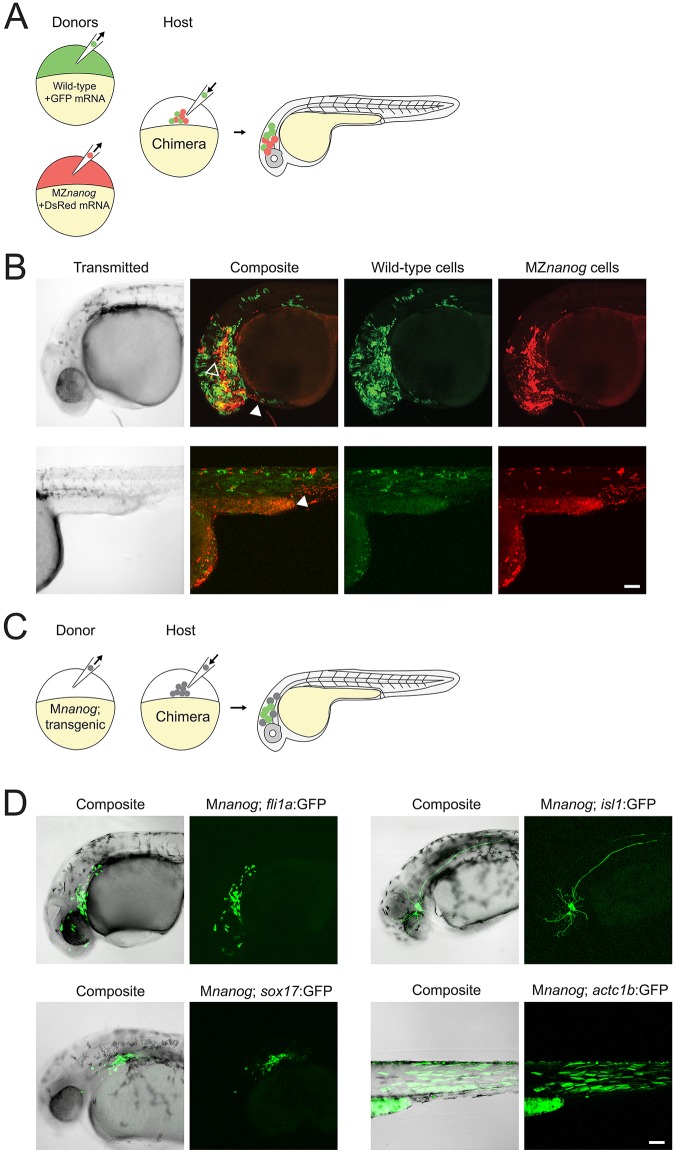
To test directly the role of Nanog in embryonic cell proliferation and differentiation, we co-transplanted fluorescently labeled MZnanog mutant and wild-type cells into wild-type hosts at sphere stage, and tracked their contributions to the host embryos (Fig. 6A). By 30 hpf, transplanted wild-type and MZnanog mutant cells had proliferated to a similar extent in the host embryo and contributed to many tissues, including brain (ectodermal derivative), muscle (mesodermal derivative) and hatching gland (axial mesodermal derivative) (Fig. 6B). To further test whether cells lacking Nanog can differentiate and contribute to tissues from all three germ layers, we used transgenic markers to follow donor cell differentiation (Fig. 6C), including fli1a:GFP (vasculature, a derivative of the mesodermal germ layer), actc1b:GFP (trunk muscle, mesodermal germ layer), isl1:GFP (trigeminal sensory neurons, ectodermal germ layer) and sox17:GFP (gastrointestinal tract, endodermal germ layer) (Higashijima et al., 1997; Lawson and Weinstein, 2002; Sagasti et al., 2005; Sakaguchi et al., 2006) (Fig. S4). We transplanted cells from transgenic Mnanog embryos into wild-type hosts and found that donor cells proliferated in the host embryo and activated each of the four marker transgenes (Fig. 6D). We additionally found that germ cells were specified in MZnanog embryos, and MZnanog germ cells migrated correctly to the host gonad when transplanted into wild-type host embryos (Fig. S5).
Fig. 6.
Transplantation of cells lacking Nanog into wild-type host embryos. (A) Diagram of co-transplantation of wild-type and MZnanog cells into wild-type host embryos, indicating their contribution to the host embryo. (B) Approximately 20 cells were transplanted from donor embryos (at 3-4 hpf) injected at the 1-cell stage with GFP mRNA (wild type) or DsRed mRNA (MZnanog) together into uninjected wild-type host embryos (n=27 across three independent trials). At 30 hpf, embryos were anesthetized, mounted and imaged by confocal microscopy. Two representative embryos are pictured, with arrowheads indicating contributions to eye (black arrowhead, upper row), hatching gland (white arrowhead, upper row), and muscle fibers (white arrowhead, lower row). (C) A diagram of transplantation of Mnanog; transgenic cells into a wild-type host embryo, with green cells in the host embryo indicating activation of the transgene in transplanted cells. Donor embryos (‘Mnanog; transgenic’) were the progeny of a Znanog female crossed to a transgenic male. (D) Approximately 20 cells were transplanted from donor embryos into uninjected wild-type host embryos. At 30 hpf, embryos were anesthetized, mounted and imaged by confocal microscopy. Representative embryos in all panels are displayed as maximum projections from a subset of a z-stack, with transgene or tracer expression overlaid onto the other channel or transmitted light channel for context (‘Composite’). Transgenic lines used were Tg(fli1a:GFP)y1 (n=11; Lawson and Weinstein, 2002), Tg(sox17:GFP)s870 (n=4, Sakaguchi et al., 2006), Tg(isl1:Gal4-VP16;UAS:GFP)zf154 (abbreviated isl1:GFP) (n=16; Sagasti et al., 2005) and Tg(actc1b:GFP)zf13 (n=5; Higashijima et al., 1997). Wild-type expression patterns for each transgene are shown in Fig. S4 for comparison. Scale bars: 100 μm.
Finally, we tested whether transplanted cells lacking Nanog can make long-term contributions to the adult host. We turned to GESTALT (genome editing of synthetic target arrays for lineage tracing), a long-term lineage-tracing method that uses CRISPR-Cas9 genome editing to introduce permanent mutations in a DNA barcode (McKenna et al., 2016). We injected Cas9 protein and sgRNAs into Mnanog; GESTALT transgenic embryos to edit the barcode and transplanted cells into wild-type hosts (Fig. 7A). Chimeric embryos were grown to adulthood, and intestine, heart, eyes and brain tissues were isolated. To detect the contributions of Mnanog; GESTALT cells, GESTALT barcodes were amplified and sequenced from each sample. Across all organs, we found hundreds of barcodes containing different combinations of edits, demonstrating that Mnanog cells can contribute to adult organs (Fig. 7B). These results reveal that zebrafish Nanog is not required for the proliferation, differentiation or survival of embryonic cells.
Fig. 7.
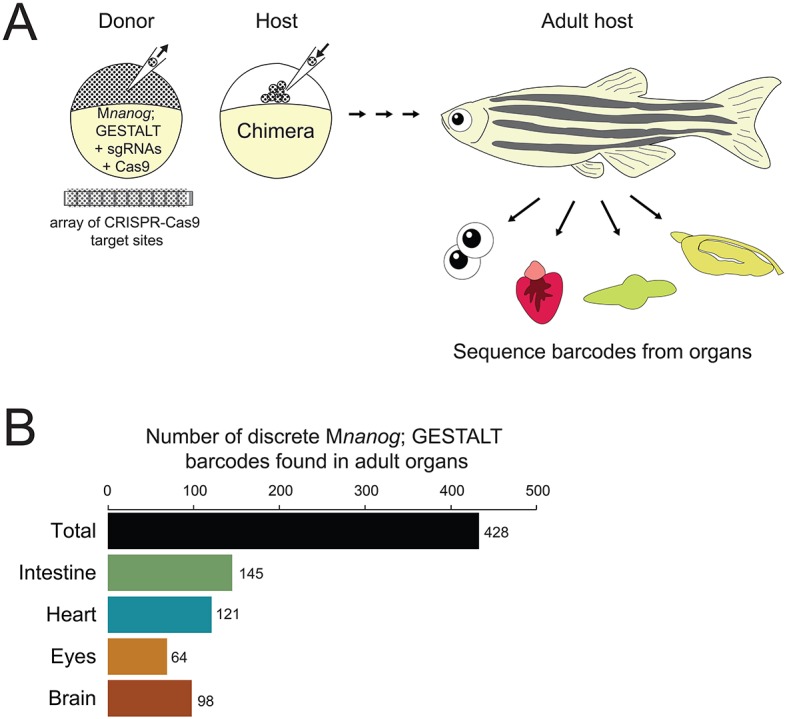
Long-term GESTALT fate mapping of transplanted cells lacking Nanog. (A) Diagram of cell transplantation at sphere stage from a Mnanog; GESTALT barcoded donor embryo into a wild-type host embryo. The donor embryo was injected with sgRNAs targeting CRISPR-Cas9 sites in the GESTALT barcode array. Host animals were grown to 90 days post-fertilization, when intestine, heart, eyes and brain were dissected (n=20 animals across two independent trials). Genomic DNA was prepared and barcodes (corresponding to surviving descendants of Mnanog; GESTALT transplanted cells) were sequenced from each organ. (B) After sequence processing of libraries from each organ across all 20 adults, distinct barcodes corresponding to different clones of transplanted Mnanog; GESTALT cells were counted. Shown is a summary of all 428 clones of Mnanog; GESTALT cells found in host animal organs.
DISCUSSION
Our results support the conclusion that the primary role for zebrafish Nanog is in the specification of the YSL (Xu et al., 2012) and that it has no absolutely essential autonomous functions in embryonic cells.
Four lines of evidence support an essential role of Nanog in YSL formation. First, both MZnanog mutants (this study) and nanog morphants (Xu et al., 2012) lack expression of mxtx2, the master regulator of YSL maturation, as well as expression of several other YSL markers. Second, absence of Nanog blocks formation of the F-actin band within the YSL (Xu et al., 2012) and leads to epiboly defects and embryo detachment (Xu et al., 2012; this study). Interestingly, MZnanog embryos occasionally exhibit marginal constriction, even in the absence of epiboly or the F-actin band. Given the importance of the actin ring for marginal constriction after 50% epiboly (Behrndt et al., 2012; Holloway et al., 2009; Köppen et al., 2006; Popgeorgiev et al., 2011; Wilkins et al., 2008), MZnanog embryos seem to constrict off the yolk by a fundamentally different mechanism. Third, expression of nanog mRNA (this study) or mxtx2 mRNA (Xu et al., 2012; this study) predominantly in YSL precursors partially rescues the YSL gene expression, epiboly and embryonic patterning defects caused by the loss of nanog. Fourth, MZnanog mutant yolk and YSL do not support the epiboly movements of wild-type embryonic cells upon blastoderm transplants. These observations strengthen and extend the previously reached conclusion (Xu et al., 2012) that Nanog is essential for proper YSL development.
Four lines of evidence indicate that Nanog is not directly required in embryonic cells. First, nanog (this study) or mxtx2 (Xu et al., 2012; this study) expression predominantly in YSL precursors is sufficient to rescue many aspects of the phenotype, including epiboly, embryo attachment and embryonic cell differentiation. Second, in blastoderm transplants, MZnanog mutant cells undergo epiboly when combined with a wild-type yolk and YSL (this study). Third, MZnanog embryos injected with Nodal (ndr1) mRNA can express mesendodermal marker genes to levels similar to wild-type embryos (this study). Fourth, and most unexpectedly, embryonic cells that lack Nanog and are transplanted into wild-type hosts proliferate and differentiate into derivatives of all germ layers and into germ cells (this study). Strikingly, GESTALT-mediated long-term lineage tracing revealed that these cells survive to adulthood in a wild-type host. Together, these observations indicate that Nanog has no absolutely essential autonomous role in embryonic cell differentiation and survival.
Although our results cast doubt on the previously held assumption that Nanog is directly required in embryonic cells for zygotic gene activation (Lee et al., 2013), we did find that MZnanog embryos exhibit embryonic gene expression defects during MZT and that MZnanog mutant cells display reduced miR-430 activity. Seventy-nine genes expressed at sphere stage displayed a 2-fold or greater reduction of transcript levels in MZnanog mutants. Many of these genes encode components of developmental signaling pathways. The gene set reduced in nanog mutants broadly overlapped with genes downregulated in nanog morphants (Lee et al., 2013). Thus, previously observed gene regulation defects (Lee et al., 2013; Xu et al., 2012) were not simply a consequence of delayed development in morphants. Moreover, maternally provided nanog is present throughout the embryo (Fig. 1) (Xu et al., 2012), and Nanog binds promoters of many developmental regulators in the early embryo (Bogdanovic et al., 2012; Lee et al., 2013; Leichsenring et al., 2013; Xu et al., 2012). In agreement with our study, Veil et al. (2018) found that MZnanog cells can contribute to all categorized lineages. By quantification of cell contributions, they concluded that MZnanog cells show reduced potential for contribution to the host. It is not clear whether this reduced potential is due to proliferation or differentiation defects, or an increase in cell death. Although our study shows that Nanog is not strictly required in embryonic cell differentiation, these observations suggest a non-essential contribution of Nanog to the expression of some genes during MZT and in potentiating cell differentiation.
How can the seemingly contradictory observations on the roles of Nanog be reconciled (Lee et al., 2013; Perez-Camps et al., 2016; Xu et al., 2012; Veil et al., 2018; this study)? We suggest the following model. Nanog is primarily required for the proper development of the YSL and the activation of the YSL gene expression program. These genes include patterning signals that in turn regulate gene expression in embryonic cells. Nanog is not required for zygotic genome activation and cellular differentiation but binds in conjunction with other pluripotency factors to cis-regulatory regions of embryonic genes, including miR-430, and contributes to their transcription. In this scenario, zebrafish Nanog primarily acts extra-embryonically, and its embryonic requirements might be redundant with other factors and are similar to those in mammals (Carter et al., 2014; Chambers et al., 2007; Schwarz et al., 2014), i.e. involved in but not essential for the acquisition and maintenance of pluripotency and differentiation.
MATERIALS AND METHODS
Animal care
TL/AB strain zebrafish (Danio rerio) were used in this study. All vertebrate animal work was performed at the facilities of Harvard University, Faculty of Arts & Sciences (HU/FAS). The HU/FAS animal care and use program maintains full Association for Assessment and Accreditation of Laboratory Animal Care International accreditation, is assured with the Office of Laboratory Animal Welfare (A3593-01), and is currently registered with the United States Department of Agriculture. This study was approved by the Harvard University/Faculty of Arts & Sciences Standing Committee on the Use of Animals in Research & Teaching under Protocol 25–08.
Generation of nanog mutants
A TALEN pair targeting the first exon of nanog (TALEN L: TCCCGAATCTGAGCTGGC; TALEN R: TGTGACCCCGCCGGAGTGT) was generated using the TALE Toolbox (Sanjana et al., 2012). Wild-type embryos were injected at the 1-cell stage with 450 pg TALEN pair mRNA. Mutations were verified in injected embryos, and from clutches of outcrossed putative founder adults, by PCR from genomic DNA, followed by T7 Endonuclease I assay (NEB). Mutations within individual founder fish were identified by cloning PCR products followed by Sanger sequencing. An allele containing a 7 bp deletion was isolated and used for all further experiments (a166). These fish were genotyped using a PCR strategy – two allele-specific primers, in combination with a constant primer, separately identify wild-type and mutant alleles. Allele description, and primer and vector sequences can be found in Table S2.
Molecular cloning and in situ hybridization
Total RNA was isolated from embryos using EZNA Total RNA kits (Omega Biotek). cDNA was generated using iScript cDNA Synthesis kit (Bio-Rad). Open reading frames for nanog and mxtx2 were amplified by PCR from embryonic cDNA and cloned into the pCS2 vector for generation of mRNA and probes. Open reading frames for vox and bmp2b were amplified by PCR and cloned using the Strataclone kit (Agilent) for generation of probes. Plasmids encoding GFP, DsRed, ndr1, mxtx2, nanog, GFP-3xPT-miR-430 (Giraldez et al., 2005) and GFP-nanos1 (Köprunner et al., 2001) mRNAs were transcribed using mMessage mMachine kits (Thermo Fisher). Vector sequences can be found in Table S2. Antisense probes for in situ hybridization were transcribed using the DIG RNA labeling kit (Roche). All RNAs were purified using EZNA Total RNA kits (Omega Biotek). In situ hybridization was performed as previously described (Thisse and Thisse, 2008); stained embryos were cleared and imaged with a Zeiss Axio Imager.Z1 microscope.
RT-qPCR
Total RNA and cDNA were generated as above. qPCR was conducted using iTaq (Bio-Rad) on a CFX96 (Bio-Rad). Primer sequences are listed in Table S2.
RNAseq
Total RNA was isolated from MZnanog and wild-type embryos at 4 hpf (sphere stage) and 6.5 hpf (shield stage) (n=40 embryos each condition) following a previously published protocol (Pauli et al., 2012). RNA quality was confirmed by Bioanalyzer (Agilent). RNAseq libraries were generated using the TruSeq RNA Library Prep Kit v2 (Illumina) and sequenced on a HiSeq 2500, generating single-end 51 bp reads. Reads were aligned for each sample using TopHat v2.0.13 (Trapnell et al., 2009) with the following command ‘tophat -o <output directory> -p 16 --no-novel-juncs -G <gene table> <Bowtie2 genome index> <fastq reads>’. Transcript abundance and differential expression were determined using Cufflinks v2.2.1 (Trapnell et al., 2012) with the following command for each developmental stage ‘cuffdiff -p 16 -b <genome.fa -u -L <labels> -o <output directory> <gene table> <wild-type aligned reads .bam file> <mutant aligned reads .bam file>’. Differential gene expression plots were generated in Rstudio. Raw and processed data are available at Gene Expression Omnibus under accession number GSE89245.
YSL staining, expression and phenotypic scoring
YSL nuclei were labeled by injection of 1 nl of 0.5 mM SYTOX Green solution (Thermo Fisher) into the yolk cell at the 128-cell stage, as previously described (D'Amico and Cooper, 2001). F-actin was labeled using Alexa-647-phalloidin (Thermo Fisher), as previously described (Wilkins et al., 2008). Labeled embryos were mounted in 1% low melt agarose and imaged with either a Zeiss Pascal or Zeiss 880 inverted confocal microscope. YSL expression was performed through injection of mRNAs at the 4-cell stage into the vegetal yolk (Xu et al., 2012). Embryos with expression restricted to the YSL were scored and sorted at sphere stage using fluorescence from co-injected GFP mRNA. Phenotypes for all injected embryos were scored during gastrulation and at 24-30 hpf. At 24-30 hpf, classification used the following category definitions (shown in Fig. 4B): Class I, no rescue, ball of necrotic cells or exploded; Class II, axis rescue; Class III, axis rescue and tail extension; Class IV, similar to wild type.
Blastoderm transplantation
Blastoderm transplants were performed as previously described with minor modifications (Holloway et al., 2009). Dechorionated wild-type and MZnanog embryos were injected with 250 pg of either Dextran-Alexa-488 or Dextran-Alexa-546 (Thermo Fisher). Embryos (512-cell stage) were placed in an agarose-coated Petri dish containing 1× Ringer's solution supplemented with 1.6% cleared egg whites, which aids blastoderm adherence. A pulled glass knife was used to remove the blastoderm from the yolk cell of one embryo. This blastoderm was placed onto the yolk cell of a recipient embryo using a flamed glass knife. Pressure was applied for a few seconds to adhere the donor blastoderm onto the recipient yolk cell. Embryos were allowed to recover for approximately 10 min before transfer to 1/3× Ringer's solution, and subsequent monitoring and imaging.
Transplantation and imaging
Non-transplanted embryos were anesthetized when necessary, mounted in 3% methylcellulose, and imaged with a Leica MZ 16 F microscope. For transplantation, donor embryos were injected, as appropriate, with either GFP mRNA or DsRed mRNA (50 pg each). Approximately 20-40 cells from a donor embryo were transplanted at sphere stage to a host embryo. Transplant host embryos were screened for fluorescence at 30 hpf with a Leica MZ 16 F microscope, then mounted in 1% low melt agarose and imaged with a Zeiss Pascal confocal microscope.
GESTALT lineage tracing
Mnanog; GESTALT donor embryos were generated from crosses of homozygous nanog−/− female fish to homozygous Tg(ubb:DsRed-barcodev7,myl7:EGFP) male fish (McKenna et al., 2016). These donor embryos were injected with Cas9 protein (NEB) and sgRNAs targeting the v7 barcode (sequence in Table S2) (Gagnon et al., 2014). Transplantation was conducted as described above. Host embryos were grown to adulthood. Intestine, heart, eyes and brain were dissected from euthanized adults and frozen on dry ice. Adult organ gDNA was prepared using the Qiagen DNeasy kit. Sequencing libraries were prepared from each sample using two rounds of PCR to amplify barcodes and attach sample indexes and sequencing adapters (primer sequences in Table S2). Libraries were pooled and subjected to paired-end sequencing on an Illumina MiSeq, using a 500-cycle kit. After demultiplexing, reads were aligned and parsed to determine edited barcode sequences as described (McKenna et al., 2016). To filter sequencing noise, rare barcodes (present at <0.5% abundance per sample) were removed. The number of distinct edited barcodes (containing different combinations of CRISPR-Cas9 edits) present in each sample was counted. Each edited barcode was considered to be derived from the descendants of distinct clones of transplanted cells.
Supplementary Material
Acknowledgements
We thank members of the Zon lab for the Tg(fli1a:GFP) line and helpful discussions; Hazel Sive for a blastoderm transplantation protocol posted on her website; Adam Carte, Jeff Farrell, Jessica Leslie, Nathan Lord, Megan Norris, Andrea Pauli, Michal Rabani, Katherine Rogers and Maxwell Shafer for technical assistance; Marina Veil and Daria Onichtchouk for discussions and sharing their manuscript before publication; Miler Lee and Antonio Giraldez for helpful discussions; and the Harvard Bauer Core Facility, the Biopolymers Facility at Harvard Medical School and the Harvard Center for Biological Imaging.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.A.G., A.F.S.; Methodology: J.A.G.; Formal analysis: J.A.G., K.O.; Investigation: J.A.G., K.O.; Writing - original draft: J.A.G.; Writing - review & editing: J.A.G., A.F.S.; Supervision: A.F.S.; Project administration: A.F.S.; Funding acquisition: A.F.S.
Funding
This work was supported by a fellowship from the American Cancer Society (J.A.G.), the Harvard College Research Program (K.O.) and a grant from the National Institutes of Health (HD085905 to A.F.S). Deposited in PMC for release after 12 months.
Data availability
RNAseq data have been deposited in GEO under accession number GSE89245. GESTALT barcode data have been uploaded to the BioProject database with ID PRJNA416917.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.147793.supplemental
References
- Behrndt M., Salbreux G., Campinho P., Hauschild R., Oswald F., Roensch J., Grill S. W. and Heisenberg C.-P. (2012). Forces driving epithelial spreading in zebrafish gastrulation. Science 338, 257-260. 10.1126/science.1224143 [DOI] [PubMed] [Google Scholar]
- Bogdanovic O., Fernandez-Miñán A., Tena J. J., de la Calle-Mustienes E., Hidalgo C., van Kruysbergen I., van Heeringen S. J., Veenstra G. J. C. and Gómez-Skarmeta J. L. (2012). Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res. 22, 2043-2053. 10.1101/gr.134833.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. (2014). Genome engineering with targetable nucleases. Annu. Rev. Biochem. 83, 409-439. 10.1146/annurev-biochem-060713-035418 [DOI] [PubMed] [Google Scholar]
- Carter A. C., Davis-Dusenbery B. N., Koszka K., Ichida J. K. and Eggan K. (2014). Nanog-independent reprogramming to iPSCs with canonical factors. Stem Cell Rep. 2, 119-126. 10.1016/j.stemcr.2013.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho L. and Heisenberg C.-P. (2010). The yolk syncytial layer in early zebrafish development. Trends Cell Biol. 20, 586-592. 10.1016/j.tcb.2010.06.009 [DOI] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L. and Smith A. (2007). Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230-1234. 10.1038/nature06403 [DOI] [PubMed] [Google Scholar]
- Chen S. and Kimelman D. (2000). The role of the yolk syncytial layer in germ layer patterning in zebrafish. Development 127, 4681-4689. [DOI] [PubMed] [Google Scholar]
- Cheng J. C., Miller A. L. and Webb S. E. (2004). Organization and function of microfilaments during late epiboly in zebrafish embryos. Dev. Dyn. 231, 313-323. 10.1002/dvdy.20144 [DOI] [PubMed] [Google Scholar]
- D'Amico L. A. and Cooper M. S. (2001). Morphogenetic domains in the yolk syncytial layer of axiating zebrafish embryos. Dev. Dyn. 222, 611-624. 10.1002/dvdy.1216 [DOI] [PubMed] [Google Scholar]
- Gagnon J. A., Valen E., Thyme S. B., Huang P., Ahkmetova L., Pauli A., Montague T. G., Zimmerman S., Richter C. and Schier A. F. (2014). Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE 9, e106396 10.1371/journal.pone.0106396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A. J., Cinalli R. M., Glasner M. E., Enright A. J., Thomson J. M., Baskerville S., Hammond S. M., Bartel D. P. and Schier A. F. (2005). MicroRNAs regulate brain morphogenesis in zebrafish. Science 308, 833-838. 10.1126/science.1109020 [DOI] [PubMed] [Google Scholar]
- Higashijima S.-I., Okamoto H., Ueno N., Hotta Y. and Eguchi G. (1997). High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of Zebrafish origin. Dev. Biol. 192, 289-299. 10.1006/dbio.1997.8779 [DOI] [PubMed] [Google Scholar]
- Holloway B. A., Gomez de la Torre Canny S., Ye Y., Slusarski D. C., Freisinger C. M., Dosch R., Chou M. M., Wagner D. S. and Mullins M. C. (2009). A novel role for MAPKAPK2 in morphogenesis during zebrafish development. PLoS Genet. 5, e1000413 10.1371/journal.pgen.1000413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.-K., Jang M. K., Brown J. L., McBride A. A. and Feldman B. (2011). Embryonic mesoderm and endoderm induction requires the actions of non-embryonic Nodal-related ligands and Mxtx2. Development 138, 787-795. 10.1242/dev.058974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyslop L., Stojkovic M., Armstrong L., Walter T., Stojkovic P., Przyborski S., Herbert M., Murdoch A., Strachan T. and Lako M. (2005). Downregulation of NANOG induces differentiation of human embryonic stem cells to extraembryonic lineages. Stem Cells 23, 1035-1043. 10.1634/stemcells.2005-0080 [DOI] [PubMed] [Google Scholar]
- Kimmel C. B. and Law R. D. (1985). Cell lineage of zebrafish blastomeres: II. Formation of the yolk syncytial layer. Dev. Biol. 108, 86-93. 10.1016/0012-1606(85)90011-9 [DOI] [PubMed] [Google Scholar]
- Köppen M., Fernández B. G., Carvalho L., Jacinto A. and Heisenberg C.-P. (2006). Coordinated cell-shape changes control epithelial movement in zebrafish and Drosophila. Development 133, 2671-2681. 10.1242/dev.02439 [DOI] [PubMed] [Google Scholar]
- Köprunner M., Thisse C., Thisse B. and Raz E. (2001). A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 15, 2877-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley A. R., Smith J. C., Stemple D. L. and Harvey S. A. (2014). New insights into the maternal to zygotic transition. Development 141, 3834-3841. 10.1242/dev.102368 [DOI] [PubMed] [Google Scholar]
- Lawson N. D. and Weinstein B. M. (2002). In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307-318. 10.1006/dbio.2002.0711 [DOI] [PubMed] [Google Scholar]
- Lee M. T., Bonneau A. R., Takacs C. M., Bazzini A. A., DiVito K. R., Fleming E. S. and Giraldez A. J. (2013). Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360-364. 10.1038/nature12632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. T., Bonneau A. R. and Giraldez A. J. (2014). Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 30, 581-613. 10.1146/annurev-cellbio-100913-013027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichsenring M., Maes J., Mössner R., Driever W. and Onichtchouk D. (2013). Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science 341, 1005-1009. 10.1126/science.1242527 [DOI] [PubMed] [Google Scholar]
- Loh Y.-H., Wu Q., Chew J.-L., Vega V. B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J. et al. (2006). The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38, 431-440. 10.1038/ng1760 [DOI] [PubMed] [Google Scholar]
- McKenna A., Findlay G. M., Gagnon J. A., Horwitz M. S., Schier A. F. and Shendure J. (2016). Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 10.1126/science.aaf7907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M. and Yamanaka S. (2003). The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631-642. 10.1016/S0092-8674(03)00393-3 [DOI] [PubMed] [Google Scholar]
- Mizuno T., Yamaha E., Wakahara M., Kuroiwa A. and Takeda H. (1996). Mesoderm induction in zebrafish. Nature 383, 131-132. 10.1038/383131a0 [DOI] [Google Scholar]
- Onichtchouk D. and Driever W. (2016). Zygotic genome activators, developmental timing, and pluripotency. Curr. Top. Dev. Biol. 116, 273-297. 10.1016/bs.ctdb.2015.12.004 [DOI] [PubMed] [Google Scholar]
- Paranjpe S. S. and Veenstra G. J. C. (2015). Establishing pluripotency in early development. Biochim. Biophys. Acta 1849, 626-636. 10.1016/j.bbagrm.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A., Valen E., Lin M. F., Garber M., Vastenhouw N. L., Levin J. Z., Fan L., Sandelin A., Rinn J. L., Regev A. et al. (2012). Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 22, 577-591. 10.1101/gr.133009.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Camps M., Tian J., Chng S. C., Sem K. P., Sudhaharan T., Teh C., Wachsmuth M., Korzh V., Ahmed S. and Reversade B. (2016). Quantitative imaging reveals real-time Pou5f3-Nanog complexes driving dorsoventral mesendoderm patterning in zebrafish. Elife 5, e11475 10.7554/eLife.11475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popgeorgiev N., Bonneau B., Ferri K. F., Prudent J., Thibaut J. and Gillet G. (2011). The apoptotic regulator Nrz controls cytoskeletal dynamics via the regulation of Ca2+ trafficking in the Zebrafish blastula. Dev. Cell 20, 663-676. 10.1016/j.devcel.2011.03.016 [DOI] [PubMed] [Google Scholar]
- Rabani M., Raychowdhury R., Jovanovic M., Rooney M., Stumpo D. J., Pauli A., Hacohen N., Schier A. F., Blackshear P. J., Friedman N. et al. (2014). High-resolution sequencing and modeling identifies distinct dynamic RNA regulatory strategies. Cell 159, 1698-1710. 10.1016/j.cell.2014.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagasti A., Guido M. R., Raible D. W. and Schier A. F. (2005). Repulsive interactions shape the morphologies and functional arrangement of zebrafish peripheral sensory arbors. Curr. Biol. 15, 804-814. 10.1016/j.cub.2005.03.048 [DOI] [PubMed] [Google Scholar]
- Sakaguchi T., Kikuchi Y., Kuroiwa A., Takeda H. and Stainier D. Y. R. (2006). The yolk syncytial layer regulates myocardial migration by influencing extracellular matrix assembly in zebrafish. Development 133, 4063-4072. 10.1242/dev.02581 [DOI] [PubMed] [Google Scholar]
- Sanjana N. E., Cong L., Zhou Y., Cunniff M. M., Feng G. and Zhang F. (2012). A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 7, 171-192. 10.1038/nprot.2011.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz B. A., Bar-Nur O., Silva J. C. R. and Hochedlinger K. (2014). Nanog is dispensable for the generation of induced pluripotent stem cells. Curr. Biol. 24, 347-350. 10.1016/j.cub.2013.12.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takesono A., Moger J., Farooq S., Faroq S., Cartwright E., Dawid I. B., Wilson S. W. and Kudoh T. (2012). Solute carrier family 3 member 2 (Slc3a2) controls yolk syncytial layer (YSL) formation by regulating microtubule networks in the zebrafish embryo. Proc. Natl. Acad. Sci. USA 109, 3371-3376. 10.1073/pnas.1200642109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T. W. and Jaenisch R. (2014). Molecular control of induced pluripotency. Cell Stem Cell 14, 720-734. 10.1016/j.stem.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C. and Thisse B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59-69. 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L. and Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105-1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L. and Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562-578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veil, M. Schaechtle, M. A., Gao, M., Kirner, V. Buryanova, L., Grethen, R. and Onichtchouk, D. (2018). Maternal Nanog is required for zebrafish embryo architecture and for cell viability during gastrulation. Development 145, dev155366. 10.1242/dev.155366 [DOI] [PubMed] [Google Scholar]
- Wilkins S. J., Yoong S., Verkade H., Mizoguchi T., Plowman S. J., Hancock J. F., Kikuchi Y., Heath J. K. and Perkins A. C. (2008). Mtx2 directs zebrafish morphogenetic movements during epiboly by regulating microfilament formation. Dev. Biol. 314, 12-22. 10.1016/j.ydbio.2007.10.050 [DOI] [PubMed] [Google Scholar]
- Xu C., Fan Z. P., Müller P., Fogley R., DiBiase A., Trompouki E., Unternaehrer J., Xiong F., Torregroza I., Evans T. et al. (2012). Nanog-like regulates endoderm formation through the Mxtx2-Nodal pathway. Dev. Cell 22, 625-638. 10.1016/j.devcel.2012.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.