ABSTRACT
In mice, the incisors grow throughout the animal's life, and this continuous renewal is driven by dental epithelial and mesenchymal stem cells. Sox2 is a principal marker of the epithelial stem cells that reside in the mouse incisor stem cell niche, called the labial cervical loop, but relatively little is known about the role of the Sox2+ stem cell population. In this study, we show that conditional deletion of Sox2 in the embryonic incisor epithelium leads to growth defects and impairment of ameloblast lineage commitment. Deletion of Sox2 specifically in Sox2+ cells during incisor renewal revealed cellular plasticity that leads to the relatively rapid restoration of a Sox2-expressing cell population. Furthermore, we show that Lgr5-expressing cells are a subpopulation of dental Sox2+ cells that also arise from Sox2+ cells during tooth formation. Finally, we show that the embryonic and adult Sox2+ populations are regulated by distinct signalling pathways, which is reflected in their distinct transcriptomic signatures. Together, our findings demonstrate that a Sox2+ stem cell population can be regenerated from Sox2− cells, reinforcing its importance for incisor homeostasis.
KEY WORDS: Incisor, Stem cells, Sox2, Lgr5, Hierarchy, Morphogenesis, Renewal
Summary: Sox2 plays a key role in incisor morphogenesis and renewal through initiating differentiation towards the enamel-secreting ameloblast fate, but is not required for establishing the dental epithelial stem cell niche.
INTRODUCTION
Renewing organs, such as hair, intestine and certain types of teeth, rely on the ability of stem cells (SCs) to self-renew and differentiate. To ensure tissue homeostasis, the number of SCs in a niche must be kept stable, and conditions such as tissue damage can trigger an SC population increase (Fuchs and Chen, 2012). When the damage is too great or when it affects SCs themselves, the early SC progeny or the niche cells can exhibit plasticity and de-differentiate in order to replenish the SC compartment (Rompolas et al., 2013; Tian et al., 2011). These capacities reflect the potential of the SC niche to control cell fate (Lane et al., 2014).
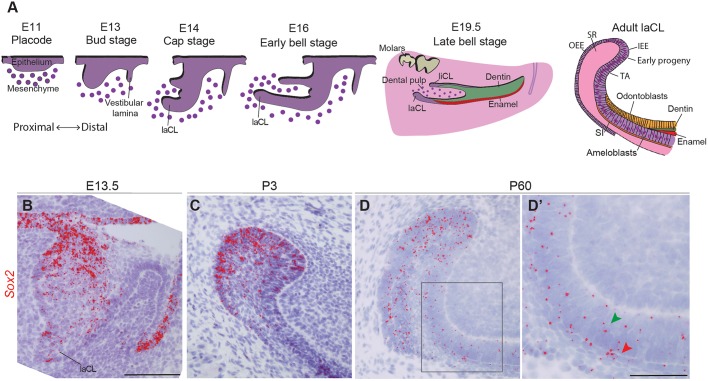
To compensate for its constant wear, the mouse incisor grows continuously. This life-long growth is fuelled by dental epithelial SCs located at the proximal end of the incisor, in a structure called the labial cervical loop (laCL). The laCL arises from the dental epithelium around embryonic day (E) 14, and its various cell types are well defined prior to birth (E19) (Fig. 1A). The stellate reticulum (SR) is a pool of epithelial cells located at the core of the laCL. It is surrounded posteriorly and labially by the columnar outer enamel epithelium (OEE), and anteriorly and lingually by the columnar inner enamel epithelium (IEE). The IEE houses the early SC progeny, namely the transient-amplifying (TA) cells and the stratum intermedium (SI) cells (Harada et al., 2006). The TA cells generate pre-ameloblasts, which then differentiate into enamel-secreting ameloblasts (Fig. 1A) (Thesleff and Tummers, 2008). Initial reports suggesting that dental epithelial SCs are present in the SR (Harada et al., 1999) were followed by in vivo genetic fate mapping experiments demonstrating that Gli1 (Seidel et al., 2010), Sox2 (Juuri et al., 2012), Bmi1 (Biehs et al., 2013), Lrig1 and Igfbp5 (Seidel et al., 2017) mark SCs in the laCL; a number of potential dental SC markers that have not yet been tested through lineage tracing were recently identified using gene co-expression analysis (Seidel et al., 2017). Moreover, the expression of some genes that mark SCs in other organs, such as Lgr5 (Suomalainen and Thesleff, 2009), Abcg2, Oct3/4 (Pou5f1), Tbx1, Pitx2 and Yap (Yap1) (Cao et al., 2013; Gao et al., 2015; Hu et al., 2017; Li et al., 2011), has also been detected in the incisor SC niche.
Fig. 1.
Sox2 expression during incisor morphogenesis. (A) Illustration of mouse incisor development, representing the morphological steps from placode stage to adult. At E14, the dental lingual epithelium gives rise to the lingual cervical loop (liCL), while the labial side originates a larger structure: the labial cervical loop (laCL). The adult laCL is composed of the stellate reticulum (SR), the outer enamel epithelium (OEE) and inner enamel epithelium (IEE). The latter gives rise to the stem cell early progeny, namely the transient-amplifying cells (TA), the stratum intermedium (SI) and the ameloblasts. (B) Sox2 expression (red) is present throughout the entire dental epithelium at E13.5, at highest levels in the lingual side. (C) At P3, Sox2 expression is more sparse, and restricted to the laCL. (D,D′) In adult mice (P60), Sox2 is expressed in the laCL (SR, IEE, OEE and TA cells), as well as in the pre-ameloblasts, ameloblasts (green arrowhead) and SI (red arrowhead). The boxed region in D is magnified in D′. Scale bars: B-D, 100 µm; D′, 50 µm.
SOX2, the focus of this study, is an important transcription factor in the maintenance of pluripotency (Takahashi et al., 2006), formation of endodermal organs (Que et al., 2007; Xie et al., 2014) and development of ectodermal tissues (Arnold et al., 2011; Clavel et al., 2012). We previously reported that SOX2 is a marker for dental epithelial SCs in the mouse incisor and that it is not expressed in the mesenchyme (Juuri et al., 2012). Recently, we showed that deletion of Sox2 in the dental epithelium at E10.5 (Pitx2Cre/+;Sox2fl/fl) drastically impairs incisor formation and leads to disappearance of the organ by E18. We also showed that Sox2 deletion using a ubiquitous promoter during incisor renewal (Rosa26CreER/+;Sox2fl/fl) slowed down incisor growth (Sun et al., 2016).
Here, we deleted Sox2 in the epithelium at E11 using ShhGFP-Cre/+ (Dassule and McMahon, 1998) to analyse the effects on cell differentiation. We found that SOX2 is necessary for ameloblast lineage commitment. Also, we specifically deleted Sox2 expression from Sox2+ cells (Sox2CreER/fl) and assessed the consequences of short- and long-term deletion on the laCL. We analysed the effect on laCL shape and on the expression pattern of Sox2 as well as Lgr5, a marker that has been suggested to be expressed by SCs in the laCL (Chang et al., 2013; Suomalainen and Thesleff, 2009). We found that loss of Sox2 led to a change in laCL morphology and to the disappearance of Lgr5 expression. Moreover, our data suggest that SR cells are capable of re-establishing a cell population expressing Sox2 and Lgr5. Together, these data indicated the importance of maintaining a Sox2+ SC population within the adult laCL. Moreover, we have observed that the transcriptomic signature of the Sox2+ cells varies between embryonic and adult stages, reflecting their distinct potential. Our data reveal a complex hierarchy in the laCL, and a degree of cellular plasticity not previously identified in the incisor SC niche.
RESULTS
Sox2 expression pattern changes during the transition from embryonic to adult incisor
Our previous use of a reporter mouse strain (Sox2GFP), immunohistochemistry, and RNA in situ hybridisation pointed to distinct Sox2-expressing populations (Juuri et al., 2012), and thus the Sox2+ cell population in the mouse incisor had not yet been definitively identified. Therefore, we used the highly sensitive RNAscope single mRNA in situ hybridisation method (Wang et al., 2012) to investigate the expression pattern of Sox2 during tooth morphogenesis (Fig. 1). Consistent with previous reports (Juuri et al., 2012; Sun et al., 2016; Zhang et al., 2012), Sox2 was expressed throughout the dental epithelium at E13.5 (Fig. 1B) and became gradually restricted to the laCL perinatally (Fig. 1C). At postnatal day (P) 60, the Sox2 transcripts appeared more scattered than at P3, when most cells within the laCL are Sox2+ (Juuri et al., 2012). The use of this more sensitive method allowed the detection of Sox2 transcripts in several epithelial lineages of the P60 incisor (Fig. 1D). Although most of the Sox2+ cells were found in the SR and enamel epithelium (EE) of the laCL, we detected transcripts in the TA cells, pre-ameloblasts, ameloblasts, and the SI (Fig. 1D′). We have previously shown that Sox2 and its upstream regulator Fgf8 are regulated by miRNAs in the laCL (Juuri et al., 2012; Michon et al., 2010), and this miRNA regulation could be the cause of the more restricted SOX2 protein domain.
Deletion of Sox2 leads to incisor defects during morphogenesis
To decipher the function of SOX2 during incisor morphogenesis, we conditionally deleted the gene in the dental epithelium. We have previously demonstrated that the timing of Cre-driven recombination can dramatically impact the dental phenotype (Cao et al., 2010; Michon et al., 2010; Seidel et al., 2010). As the Pitx2-driven Sox2cKO led to the absence of incisors at late stages of morphogenesis (Sun et al., 2016), we decided to use ShhCre-GFP/+ to delete Sox2. Shh is expressed later than Pitx2, and almost all dental epithelial cells derive from early Shh+ cells (Juuri et al., 2013b). The Shh-driven Sox2cKO mice have a hyperplastic dental epithelium in the second and third molars (Juuri et al., 2013a), but no incisor phenotype has been described. As the incisors of the Shh-Cre;Sox2fl/fl mice had a different phenotype to that previously reported in Pitx2-Cre;Sox2fl/fl mice (Sun et al., 2016), and the incisor was present until the end of embryogenesis, this gave us the opportunity to analyse the dental phenotype at later developmental stages.
We used RNAscope to determine the efficiency of Sox2 ablation in Shh-Cre;Sox2fl/fl mice. By E13.5, essentially no Sox2 transcripts were detected in the incisor epithelium (Fig. S1A,B). Moreover, the incisor shape was drastically affected in the mutants. At this stage, the control incisor had invaginated into the dental mesenchyme, and the forming laCL contained a large Sox2+ cell population (Fig. S1A). The incisor of the Sox2cKO littermates displayed a shallow laCL and a wider dental lamina (Fig. S1B). As previously reported (Sun et al., 2016), this phenotype was accompanied by an enlargement of the Shh+ population (Fig. S1C,D). At E13.5, both the basal (high P-cadherin) and suprabasal (low P-cadherin) (Jussila et al., 2015) cell compartments were present in the mutant incisors (Fig. S1E,F).
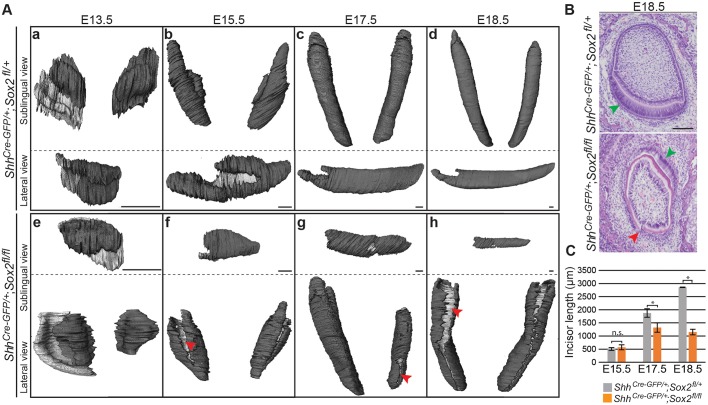
To determine the consequences of this early phenotype for subsequent dental morphogenesis, we reconstructed the dental epithelium from micro-computed tomography (micro-CT) scans of Sox2cKO embryos and their control littermates from E13.5 to E18.5. The three-dimensional (3D) renderings showed that the morphology of the Sox2cKO incisor differed from that of control littermates at all developmental stages (Fig. 2Aa-h), and the shape and length of the incisors varied both among and within individual embryos (Fig. 2Ae-h; data not shown). A recurrent trait in the Sox2cKO incisors was the presence of clefts in the labial epithelium (Fig. 2Af-h). These defects were visualised in histological sections as discontinuities in the epithelial tissue (Fig. 2B, red arrowhead). Histological sections also evidenced defective cell differentiation at E18.5, with ameloblast-like cells present in the lingual region (Fig. 2B, green arrowhead).
Fig. 2.
Mouse lower incisor shape and length are regulated by Sox2. (A) 3D reconstructions from micro-CT scans showing the dental epithelium in dark grey. The internal layer of the dental epithelium (b-d,f-h) or vestibular lamina (a,e) appears in light grey. Sox2cKO incisors exhibit an aberrant morphology at all embryonic stages. At E13.5, the tooth domain is broader, and at E15.5 clefts (red arrowhead) appear. The defects can vary within the same individual (g). (B) Histological staining of frontal sections shows that the well-organised ameloblast layer seen in control incisor (green arrowhead) is not visible in Sox2cKO individuals. A cleft is visible on the labial side of the mutant incisor (red arrowhead). (C) The length of the epithelial compartment is similar in the control and Sox2cKO at E15.5. The incisor length increases over 2 mm from E15.5 to E18.5 in controls, but only 0.6 mm in mutants. E17.5, *P=0.016; E18.5, *P=1×10−5; n.s., not significant. n=3. Scale bars: A, 50 µm; B, 100 µm.
We observed no significant differences in incisor length at E15.5 (Fig. 2C). However, Sox2cKO lower incisors were significantly shorter than those of control littermates at E17.5, and they did not grow further after this stage. The tooth size defect was not attributable to decreased cell proliferation, as we did not detect any significant difference in the density of phospho-histone H3 (pH-H3)+ cells in the dental epithelium at E13.5, nor in the laCL at E18.5 (Fig. 3A). These results are in line with the report on the molars of ShhCre-GFP/+;Sox2fl/fl embryos (Juuri et al., 2013a). Moreover, we did not observe any obvious increase in apoptotic cells by TUNEL assay in the Sox2cKO (Fig. S2), indicating that Sox2 loss does not affect cell death rate, in agreement with other Sox2 loss-of-function models (Sun et al., 2016). Taken together, our data indicated a role for SOX2 in incisor morphogenesis and dental epithelium cell differentiation.
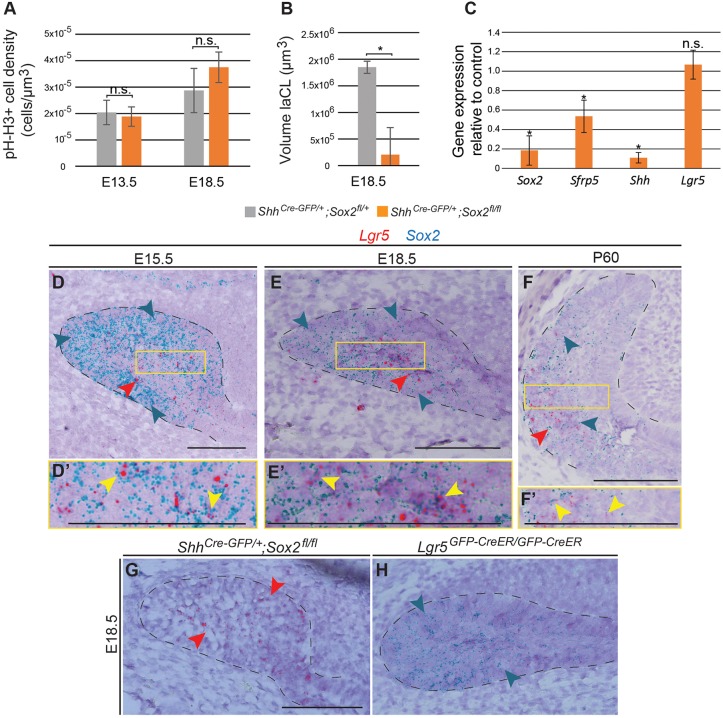
Fig. 3.
Sox2cKO impacts laCL volume and the expression of different differentiation markers. (A) Quantification of phospho-histone H3 (pH-H3)+ cell density in the dental epithelium reveals no significant defect in cell proliferation in Sox2cKO. (B) The volume of the E18.5 laCL is drastically decreased in Sox2cKO. *P=0.026. (C) qPCR analysis demonstrates that Sox2cKO induces a decrease in Sfrp5 and Shh expression. Lgr5 expression remains unaffected. *P<0.05. (A-C) n=3. (D) Sox2 transcripts (blue arrowheads in all images) at E15.5 are detected in all cells of the laCL (outlined). Lgr5 transcripts (red arrowheads in all images) mark a subset of the Sox2+ population in the SR. (E) At E18.5 the Sox2 expression domain is smaller than at previous stages. Lgr5 expression is localised to the SR. The laCL houses the expression of both Sox2 (EE and SR) and Lgr5 (SR). (F) In the adult incisor, Sox2 transcripts are localised in different areas of the laCL, while Lgr5 expression is confined to the most proximal part. (D′,E′,F′) Magnifications of the boxed regions in D,E,F. Yellow arrowheads indicate cells expressing both Lgr5 and Sox2 transcripts. (G,H) Sox2cKO shows no changes in Lgr5 expression, as Lgr5KO displays a normal Sox2 expression pattern. Scale bars: 100 µm. qPCR.
The laCL and its cell populations are established in Shh-Cre;Sox2fl/fl mice
Next, we investigated the structure of the laCL upon Sox2 deletion in Shh-Cre;Sox2fl/fl embryos. As expected from the epithelial reconstructions (Fig. 2A), laCL volume was drastically reduced in the Sox2cKO (Fig. 3B), but the structure was not completely absent. Hence, in addition to Sox2 we assessed via RT-qPCR the expression of Sfrp5, which is a marker of early Sox2+ cell progeny (Juuri et al., 2012), Shh, which is expressed in TA cells, pre-ameloblasts and immature ameloblasts (Seidel et al., 2010), and Lgr5, which is expressed by a minor cell population in the laCL and marks SCs in several adult organs (Chang et al., 2013; Suomalainen and Thesleff, 2009; Yang et al., 2015) (Fig. 3C). Sox2 expression was drastically reduced in the mutant incisors. Sfrp5 and Shh expression levels were also decreased, indicating defects in cell differentiation. Surprisingly, reduced SHH expression was found only in the lingual side of the incisor (Fig. S3). We did not detect an impact on Lgr5 expression, which has been reported at E14.5 in the molar bud (Kawasaki et al., 2014) but in the laCL incisor only after E16.5 (Suomalainen and Thesleff, 2009). However, the sensitivity of the RNAscope assay enabled the detection of Lgr5 transcripts already at E15.5 in the incisor laCL (Fig. 3D-F) and, interestingly, the expression patterns of Sox2 and Lgr5 overlap (Fig. 3D-F′, Fig. S4).
To further study the connection between Sox2 and Lgr5 expression, we analysed their expression patterns upon deletion of either gene (Fig. 3E,G,H). Lgr5 null mice (Lgr5KO) die neonatally due to gastrointestinal problems and ankyloglossia (Morita et al., 2004). These mice also exhibit cleft palate, but we did not identify morphological abnormalities in the laCL. In the control embryos, Sox2 transcripts were found in the EE and SR, and Lgr5 transcripts were mainly restricted to the SR. Interestingly, in the Sox2cKO laCL, Lgr5 expression was maintained, and the Sox2 expression pattern did not change in the Lgr5KO incisor (Fig. 3G,H). These observations highlighted the importance of Sox2 for ameloblast lineage commitment, but not for establishment of the laCL.
Sox2 expression is quick to recover after transient Sox2 deletion in the adult incisor
Having identified the importance of Sox2 for dental epithelial cell differentiation during embryogenesis, we next analysed the effects of Sox2 absence in the renewing incisor. As Sox2cKO mice die perinatally, we used K14-CreER mice (Huelsken et al., 2001; Järvinen et al., 2006; Vasioukhin et al., 1999) to delete Sox2 in the laCL. We investigated the morphology and Sox2 expression pattern at 2 days, 11 days and 1 month after Cre activation. No obvious differences were observed in histological sections (Fig. S5A) nor in the SOX2 pattern (by immunofluorescence staining; data not shown). We also analysed the expression levels of Sox2, Sfrp5, Lrg5 and Bmi1 24 h after induction via qPCR and found no statistically significant differences from controls (data not shown). This suggested a lack of recombination in the laCL, and our examination of Sox2 and keratin 14 (K14) expression in the adult laCL at the protein and transcript levels (Fig. S5B,C) showed minimal overlap. K14 was expressed in the SR region, but not in the most lingual part of the SR, nor in the IEE, where many of the Sox2+ cells reside; this is in line with the low recombination levels reported in the OEE with this Cre line (Hu et al., 2017).
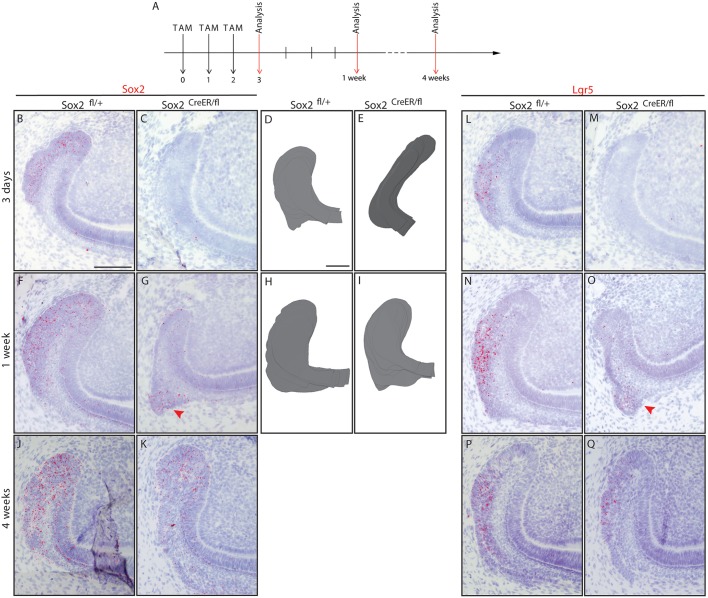
Therefore, we generated Sox2CreER/fl mice to specifically delete Sox2 in the Sox2+ cells upon Tamoxifen administration. Two-month-old Sox2CreER/fl mice were administered Tamoxifen for three consecutive days. We then examined the laCL at 3 days, 1 week and 4 weeks chase (Fig. 4A), and Sox2fl/+ littermates were used as controls. We also injected adult Sox2CreER/fl mice with corn oil and observed no aberrant phenotype (Fig. S6). After 3 days of chase, we observed a large decrease in the number of Sox2 transcripts (Fig. 4B,C). Moreover, the spherical shape of the laCL was lost in the mutants, which instead exhibited an elongated SC niche (Fig. 4D,E). However, the transient loss of Sox2 expression did not lead to cell death (Fig. S7), and after 5 days without Cre activation some Sox2 transcripts were detected, although the expression was fainter than in the control laCL (Fig. 4F,G). Interestingly, by this time point, laCL shape was restored (Fig. 4H,I). At 1 month chase, we did not detect any differences in laCL morphology or Sox2 expression pattern between control and mutant (Fig. 4J,K).
Fig. 4.
Sox2 and Lgr5 expression are lost then restored in Sox2CreER/fl laCL. (A) The experimental setup. (B,C) Sox2 and (L,M) Lgr5 expression are almost abolished after three Tamoxifen injections in Sox2CreER/fl mice. (D,E) The Sox2CreER/fl laCL is narrower than that of controls. One week after the first Tamoxifen administration, a faint Sox2 signal is detected in the laCL (F,G, arrowhead), while the Lgr5 expression pattern appears to be normal in the mutant (N,O, arrowhead). (H,I) At this stage, the morphology of the laCL appears to be normal. (J,K,P,Q) One month after Cre recombinase activation, the mouse incisor SC niche of Sox2CreER/fl mice is indistinguishable from that of control littermates. Scale bars: 100 µm.
Lgr5 marks a small cell population in the laCL (Suomalainen and Thesleff, 2009; Yang et al., 2015), and its expression pattern overlaps with that of Sox2 (Fig. 3D-F, Fig. S3). Therefore, we investigated the effect of Sox2 loss on Lgr5 expression. After 3 days of Tamoxifen administration, Lgr5 transcripts were greatly decreased (Fig. 4L,M), similarly to Sox2. Four days later, Lgr5 expression levels were similar to those of the control (Fig. 4N,O) and returned to normal after 1 month of chase (Fig. 4P,Q).
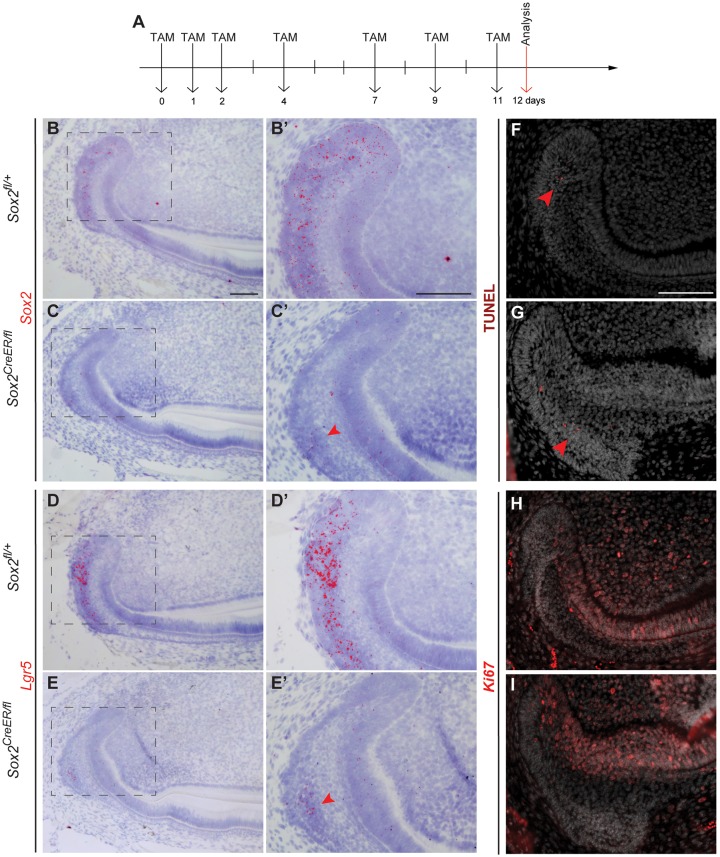
To evaluate the effect of long-term Sox2 loss, we administered Tamoxifen to Sox2CreER/fl mice seven times over 11 days. Incisors were collected 1 day after the last injection (Fig. 5A). As expected, very few Sox2 transcripts were detected in the laCL, whereas controls exhibited expression of Sox2 as previously reported (Fig. 5B-C′). Lgr5 expression was faint as well (Fig. 5D-E′). Very few Sox2 transcripts were detected in the area where Lgr5 expression was localised. Moreover, the morphology of the laCL was not affected, similar to results obtained with another Sox2cKO model Rosa26CreER/+;Sox2fl/fl (Sun et al., 2016). We further examined the cellular response to the deletion of Sox2 in the Sox2+ SCs and did not observe any obvious increase in cell death or aberrant cell proliferation (Fig. 5F-I).
Fig. 5.
A day after an 11-day Sox2 ablation, the Sox2 expression pattern, proliferation and cell differentiation appear disturbed. (A) The experimental setup. (B-E′) A small amount of Sox2 transcripts is detected in the proximal area of the SR (arrowhead), where faint expression of Lgr5 is seen (arrowhead). (F,G) TUNEL assay confirms that there is no increase in apoptosis in the laCL, where only a few positive cells were found (arrowheads). (H,I) The domain of proliferating cells, visualised with Ki67 staining, appears similar to the control. Scale bars: 100 µm.
The Sox2+ population is regenerated from the SR
The restoration of a Sox2+ cell population days after deleting Sox2 suggested a high degree of plasticity within the laCL. To understand the origin of the newly generated Sox2+ population, we studied the dividing cells during laCL restoration using EdU incorporation. Under normal renewal conditions, dividing cells are localised to the lingual and distal part of the laCL, the IEE, the TA cells, and the distalmost SR cells (Hu et al., 2017) (Fig. 6A). In comparing this pattern with Sox2 expression, it is apparent that most of the proliferating cells in the laCL do not strongly express Sox2 (Fig. 6B). In addition, the cells in the proximal region of the laCL are quiescent (Seidel et al., 2010). We marked the proliferative cells 3 days after the first Tamoxifen administration and analysed their position and quantity 4 days later (Fig. 6C). In the control, the EdU+ cells were located in the central and proximal sections of the SR, where Sox2 is faintly expressed (Fig. 6D-E′). By contrast, in the Sox2CreER/fl mice, these cells were displaced to the proximalmost part, close to the Lgr5+ cell area (Fig. 6F-G′). Furthermore, the percentage of EdU+ cells was significantly increased in the mutant SR compared with the control (Fig. 6H). This reflected an increase in proliferation in the SR at the beginning of the rescue period.
Fig. 6.
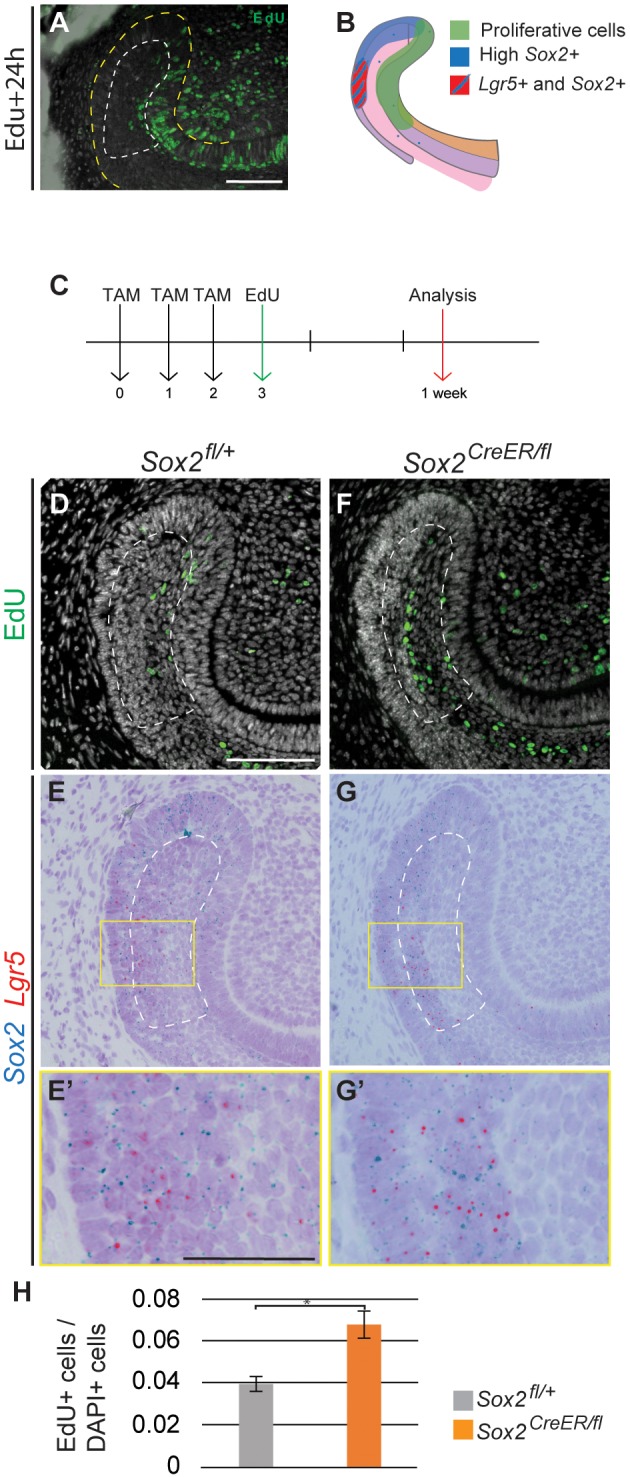
Expression of Sox2 and Lgr5 is rescued from the SR. (A) Pattern of proliferative cells (EdU+) 24 h after being administered to the mouse. (B) Schematic representation of the proliferative region and the Sox2+ and Lgr5+ domains. (C) The experimental setup. EdU+ cells (D,F) and Sox2 and Lgr5 mRNA detection (E,G) performed in identical sections. (E′,G′) Higher magnifications of the boxed regions in E,G. (H) Quantification of EdU+ cells in the SR of control and Sox2CreER/fl mice. The area quantified is delineated by the dashed line in D-G. Results are expressed as the fraction of EdU+ cells among total cells (number of nuclei, DAPI) compared with the control. *P=0.007. Scale bars: D-G, 100 µm; E′,G′, 50 µm.
Modulation of the Sox2+ cell signature in embryonic versus adult incisors
The observation that almost all laCL cells are Sox2+ at E15.5 (Fig. S3), when Lgr5 expression appears, suggested that embryonic Sox2+ cells might give rise to the Lgr5+ subpopulation during incisor formation. However, during laCL regeneration after Sox2 ablation, the Sox2+ cells seemed to partially arise from the subpopulation expressing both Sox2 and Lgr5 (Fig. 4F,G,N,O). This observation raised the question: how similar are the Sox2+ populations in the forming and renewing incisor? We therefore compared the transcriptome of the Sox2+ cells in the early incisor (E14.5) with that of the Sox2+ cells in the renewing incisor (P30) using gene expression microarrays. We first extracted the transcripts that were similarly expressed in embryonic and adult Sox2+ cells (−2<fold change<2) and compared them with the transcriptome of mouse embryonic stem cells (mESCs) as a naïve cell reference (Fig. 7A). Then, we compared the transcriptomes of the embryonic and adult Sox2+ cells (Fig. 7B). We selected the transcripts that were statistically significant (P<0.05, ANOVA) and exhibited a consequential fold change (fold change>2) (Table S1).
Fig. 7.
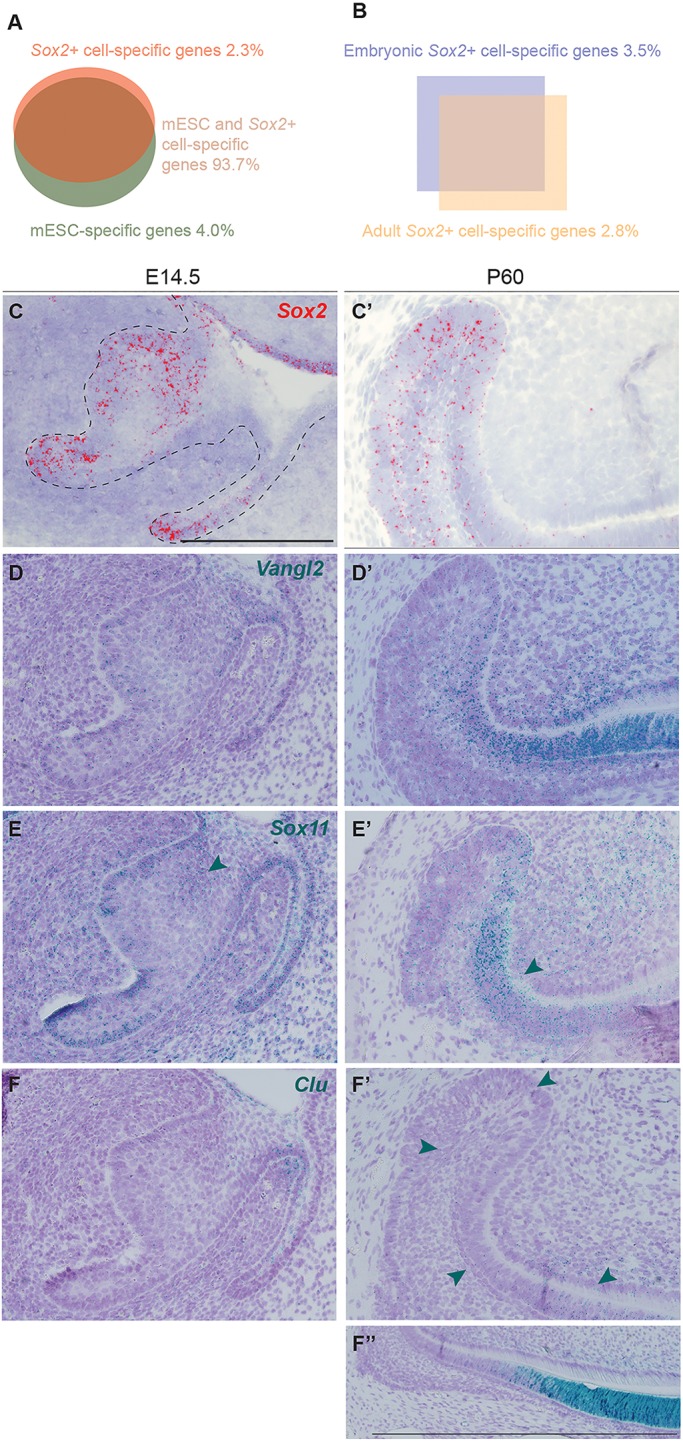
Transcriptomic changes between Sox2+ embryonic progenitors and Sox2+ dental SC. (A) Signatures of Sox2+ cells and mESCs overlap by 93.7%. (B) Comparison of embryonic and adult Sox2+ cells: 3.5% of genes are specific to embryonic Sox2+ cells, and 2.8% specific to the renewing incisor Sox2+ SCs. (C,C′) Sox2 expression pattern at E14.5 and adult stage. (D,D′) Vangl2 is enriched in Sox2+ cells compared with mESCs. It is expressed in the embryonic incisor and in the adult tooth. (E,E′) Sox11 is highly expressed in the embryonic incisor (arrowhead) and in the surrounding mesenchyme. In the adult, expression is mostly localised to the TA cells (arrowhead). (F,F′) Clu expression is seen in the adult incisor (arrowheads), with the majority of transcripts found in the differentiated epithelial cells (F″). Scale bars: 100 µm.
For the first analysis (Sox2+ versus mESCs), we observed that 927 transcripts (2.34% of the signature) were enriched in Sox2+ cells, independent of their stage, and 1583 transcripts (4% of the signature) were downregulated. This observation reflected that ∼6.34% of the signature was differentially regulated in Sox2+ cells compared with mESCs (Fig. 7A, Table S2). We compared the gene ontology processes (GOPs) between these samples and found that 187 were activated in Sox2+ cells, including those specific to ectodermal organ formation and regulation, and odontogenesis (Table S3). Also, both canonical and non-canonical Wnt signalling were activated. Among the enriched genes, we examined Vangl2 as a test case because it is a member of the planar cell polarity signalling pathway (non-canonical Wnt). Moreover, its expression has previously been reported in the ameloblasts and odontoblasts of embryonic molars, where it regulates cell alignment (Obara et al., 2017; Wu et al., 2016). We found Vangl2 transcripts in the incisor at E14.5 and in the adult laCL. However, in the adult stage, most of the transcripts were found in TA cells and ameloblasts, where there are fewer Sox2+ cells (Fig. 7C-D′).
When comparing embryonic and adult Sox2+ cells, we observed that 3.54% of the signature (1400 hits) was enriched in embryonic cells (Fig. 7B, Table S1). Among these, 143 GOPs were enriched over 2.5 fold, including cell division regulation processes (e.g. mitotic DNA replication, DNA replication initiation) (Table S4). We also found a number of genes important for the mineralisation of forming teeth, such as embigin (Xie et al., 2015), Six4 (Nonomura et al., 2010) and Cxcr4 (Juuri et al., 2013b), the last of which is thought to be important for the migration of epithelial progenitors in adult laCL (Yokohama-Tamaki et al., 2015). Also, Sox11, which is involved in palate development (Sock et al., 2004; Watanabe et al., 2016) and expressed in the mouse embryonic molar (Dy et al., 2008; Hargrave et al., 1997), was enriched by 4.40 fold. Sox11 transcripts were abundant in the mouse incisor epithelium at E14.5, with fewer transcripts in the adult laCL. In adults, we found Sox11 expression also in pre-ameloblasts and in the mesenchymal compartment (Fig. 7E,E′).
Similarly, 2.75% (1089 transcripts) of the signature was enriched in the adult Sox2+ population (Fig. 7B); 153 GOPs were enriched in adult Sox2+ cells, with those specific for the immune response well represented (e.g. antigen processing, macrophage activation, Toll-like receptor signalling) (Table S5). The adult Sox2+ population signature contained mineralisation markers (e.g. Dspp, enamelin, amelogenin, ameloblastin) and metalloproteinases (e.g. Mmp13, Mmp14, Mmp20) (Table S1). We also detected expression of Barx2, a gene involved in cell migration and differentiation (Juuri et al., 2013b). Moreover, clusterin (Clu), a stress-activated and apoptosis-associated chaperone, which has been found in embryonic and postnatal mouse molars (Chou et al., 2009; Khan et al., 2013; Shiota et al., 2012), was enriched in adult Sox2+ cells by a fold change of 187.08. In the embryonic (E14.5) incisor we found almost negligible amounts of Clu expression, and few transcripts were found in the vestibular lamina at this stage. In the adult, low Clu expression levels were found in the laCL; higher expression levels were evident in the pre-ameloblasts and ameloblasts (Fig. 7F-F″), where Sox2 transcripts were also found (Fig. 7C,C′).
DISCUSSION
We have previously demonstrated the role of Sox2+ SCs in incisor renewal (Juuri et al., 2012) and in successional tooth formation in the mouse (Juuri et al., 2013a). More recently, we showed that early deletion of Sox2 in the dental epithelium led to the absence of the incisor at E18 (Sun et al., 2016). As this prevented analysis during late time points of embryonic development, here we used the ShhGFPCre/+ allele to delete Sox2 at the dental placode stage. Sox2 deletion using this driver resulted in growth and shape irregularities, including a curved incisor phenotype, which, together with the drastic decrease of Shh and Sfrp5 expression, pointed to improper differentiation of cell lineages. Interestingly, SHH expression (Fig. S6) and ameloblast-like cells (Fig. 2B) were found on the lingual side of the incisor. SHH expression adjacent to the lingual cervical loop (liCL) has previously been linked to an expanded liCL and to ectopic lingual ameloblasts (Klein et al., 2008). However, the laCL structure was preserved, but its morphology and the cell arrangement were affected. These observations suggest an essential role for Sox2 in lineage commitment and early differentiation towards the enamel-secreting ameloblast fate. We have previously reported that the number of proliferative cells is reduced after early Sox2 deletion (Sun et al., 2016). However, we did not detect a significant reduction in proliferation in the Sox2cKO model used here; instead, we propose that slower renewal is caused by defects in cell differentiation. We conclude that Sox2 plays a key role in the maintenance of enamel organ morphology and proper cell differentiation during incisor morphogenesis.
Lgr5 marks intestinal and skin SCs (Barker et al., 2007; Haegebarth and Clevers, 2009; Jaks et al., 2008), and it also marks a small epithelial cell population in the mouse incisor laCL (Chang et al., 2013; Suomalainen and Thesleff, 2009; Yang et al., 2015). Our identification of Lgr5+ cells within the Sox2cKO laCL indicates that other potential laCL SC populations can be present in the absence of Sox2. Therefore, we propose that Sox2 expression is of importance for initiating differentiation towards ameloblast fate, but not for establishing the SC niche.
In the adult incisor, Sox2 deletion in the Sox2+ cells (Sox2CreER/fl) caused a drastic, but temporary change in laCL shape, and the loss of Lgr5 expression. We showed that Lgr5 and Sox2 expression overlap during embryonic and postnatal stages. Therefore, we conclude that Lgr5+ cells represent a subpopulation of Sox2+ cells in the developing and renewing laCL. This situation differs from other SC niches, such as the stomach, where Lgr5 and Sox2 mark distinct cell populations (Arnold et al., 2011).
To ensure tissue homeostasis, the number of SCs in a niche is kept stable, but tissue damage can trigger an SC increase (Fuchs and Chen, 2012). If the damage is too great, the niche cells can display signs of transient plasticity in order to replenish the SC compartment, as shown in skin (Rompolas et al., 2013) and intestine (Tian et al., 2011). Such intra-organ plasticity (Blanpain and Fuchs, 2014) requires cell de-differentiation or transdifferentiation to ensure the maintenance of organ integrity. While this mechanism is well studied in other SC niches, it has not yet been documented in the dental context. The loss of Sox2 expression in Sox2CreER/fl mice led to a morphologically thinner laCL depleted of Sox2+ and Lgr5+ cells. This phenotype was rapidly rescued, and the Lgr5+ subpopulation was the first to emerge, from the distal section of the laCL. Moreover, our EdU incorporation experiment demonstrated that some SR cells were plastic enough to regenerate the lost cell populations within the laCL.
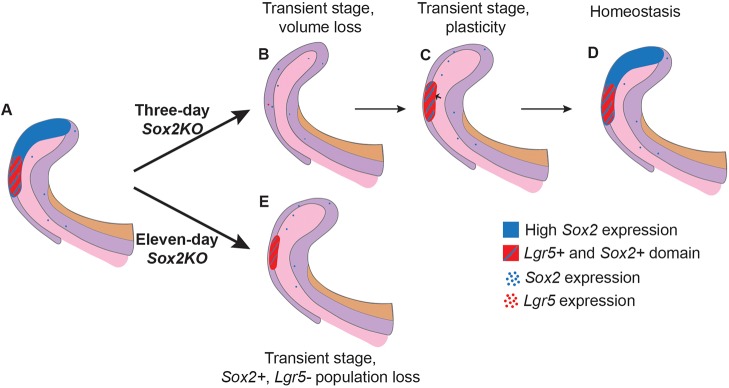
Taken together, our data suggest that after damage the SR cells regenerate first a Sox2+ Lgr5+ double-positive cell population, and then a Sox2+ Lgr5− population (Fig. 8A-D). Also, these results suggest that Sox2 marks a heterogeneous population, where different lineage specificities exist. However, long-term ablation of Sox2 did not lead to laCL shape malformation, and the laCL maintained a very small Sox2+ Lgr5+ cell population (Fig. 8E).
Fig. 8.
Model for the effects of short- and long-term Sox2 ablation. (A) Summary of Sox2 and Lgr5 expression domains in normal conditions within the laCL. (B) Upon conditional deletion of Sox2 in Sox2-expressing cells for 3 days, the laCL becomes narrower, and almost all Sox2 and Lgr5 transcripts are lost. (C) Shortly after, the volume of the laCL is back to normal due to an increase in cell proliferation in the SR (small arrow). Overlapping expression of Sox2 and Lgr5 is found in the distal side of the laCL. (D) Eventually, the laCL reaches homeostasis and returns to its original state. (E) In the event of Sox2 ablation for 11 days, the laCL maintains a small Lgr5+ Sox2+ cell population.
We have previously reported that the global deletion of Sox2 (Rosa26CreER/+;Sox2fl/fl) in adult mice leads to a reduction in incisor renewal rate (Sun et al., 2016). In the Sox2Cre/fl mice, the proliferation pattern in the laCL is maintained after prolonged Sox2 ablation. Therefore, we hypothesize that the incisor growth defect reported earlier (Sun et al., 2016) is caused by defective cell differentiation, similar to the embryonic scenario.
Finally, an important question that remains largely unanswered in the stem cell field is that of the origin of adult SCs. We found that the transcriptomic signatures of embryonic and adult Sox2+ cells are very similar. Moreover, they express a number of genes differently compared with naïve mESCs. From this observation, we propose that dental SC identity is represented either in the embryonic and adult Sox2+ cell overlapping transcriptomes or in the genes enriched in adult Sox2+ cells. Although our results alone are not enough to distinguish between these possibilities, an early generation of the dental SC signature would require the formation of the niche microenvironment early on. Interestingly, Notch1, a marker of the dental epithelial SC niche, is expressed in the mouse incisor at E14.5 (Felszeghy et al., 2010; Mucchielli and Mitsiadis, 2000). Therefore, we postulate that SC niche and Sox2+ cell dental fate are already established by E14.5 in the mouse incisor. The corollary of such a conclusion would be that the genes enriched in embryonic or adult Sox2+ cells should be related to the role of cells within the organ at this stage. For instance, Sox11, a transcription factor involved in epithelial-mesenchymal interactions (Hargrave et al., 1997), was enriched in the forming incisor. On the other hand, we found an enrichment of Clu in adult Sox2+ cells. This chaperone has been reported to play a role in secretory odontogenesis, an important function in the adult incisor (Khan et al., 2013), and was highly enriched in the incisor ameloblasts. These observations strengthen our hypothesis that the differences between embryonic and adult Sox2+ transcriptomes are inherent to the temporal role of the cell population (morphogenesis versus ameloblast lineage renewal). From these data, we conclude that the differences in gene expression that we observed reflect cues from the microenvironment and minor changes in the role of the Sox2+ cells.
Collectively, our data demonstrate the importance of a Sox2+ cell population for incisor renewal and cell differentiation. We also observed an impressive cellular plasticity in the laCL that acts to maintain the Sox2+ population. We propose that Lgr5+ cells are a subpopulation of the Sox2+ cells, and are the first cells to reappear in the event of transient Sox2 loss. This indicates the existence of a complex relationship between the Lgr5-expressing and Sox2-expressing cells.
MATERIALS AND METHODS
Mouse lines
The stage of the embryos was determined according to morphological criteria and noon of plug day was counted as E0.5. All animals are available from The Jackson Laboratory. ShhGFP-Cre/+;Sox2fl/fl were used as Sox2cKO (Juuri et al., 2013a). K14-CreER males [Tg(KRT14-cre/ERT)20Efu/J, stock 005107] were crossed with Sox2fl/fl females (Sox2tm1.1Lan/J, stock 013093) to generate K14-CreER;Sox2fl/fl mice. To generate the Sox2CreER/fl mice (inducible Sox2 cKO), Sox2fl/fl females were crossed with Sox2CreER/+ males [Sox2tm1(cre/ERT2)Hoch/J, stock 017593]. Lgr5GFP-CreER/+ [B6.129P2-Lgr5tm1(cre/ERT2)Cle/J, stock 008875] animals were crossed to generate Lgr5GFP-CreER/GFP-CreER embryos (Lgr5KO) embryos. Sox2GFP males (B6;129S-Sox2tm2Hoch/J, stock 017592) were crossed with NMRI females to produce Sox2GFP embryos. Mice were genotyped using the primers listed in Table S6. All aspects of mouse care and experimental protocols were approved by the Finnish National Board of Animal Experimentation.
Tamoxifen administration
A working solution of Tamoxifen (Sigma-Aldrich, T5648) in corn oil (Sigma-Aldrich, 47112-U) was prepared at 50 mg/ml. Tamoxifen solution was sonicated for 15 min and kept at −20°C. Mice were administered 10 mg Tamoxifen solution via oral gavage.
Tissue processing, histology, immunofluorescence, RNAscope, and TUNEL assay
For histology, tissues were fixed in 4% paraformaldehyde (PFA) at 4°C overnight, dehydrated, and embedded in paraffin. Adult samples were decalcified for 2 weeks in 0.5 M EDTA pH 7.5 after fixation. Samples were processed into 5 µm-thick sagittal sections. Hematoxylin-Eosin (H&E) staining was performed as previously described (Juuri et al., 2012).
For RNAscope in situ hybridisation (Advanced Cell Diagnostics, ACDbio), we used both the red channel and duplex kits. Mouse tissues were processed into 5 µm sections as described above. Sections were processed using an optimised protocol (as detailed in the supplementary Materials and Methods). Probes were purchased from ACDbio.
TUNEL assay was performed using the In Situ Cell Death Detection Kit, Fluorescein (Roche, 11684795910) according to the manufacturer's protocol.
We used the following antibodies in immunofluorescence assays: SOX2 (goat, Santa Cruz, SC-17320; 1:200), keratin 14 (rabbit, NeoMarkers, RB-9020-P; 1:200), P-cadherin (cadherin 3) (goat, R&D Systems, AF761; 1:500), phospho-histone H3 (rabbit, Abcam, ab5176; 1:200), Ki67 (rabbit, Abcam, ab16667; 1:200) and GFP (chicken, Abcam, ab13970; 1:200). As secondary antibodies, we used Alexa 488 goat anti-rabbit (Life Technologies, A11057; 1:500) and Alexa 568 donkey anti-goat (Life Technologies, A11008; 1:500). Nuclei were stained with Hoechst 33342 (Thermo Fisher Scientific, H3570). For DAB immunostaining we used SHH primary antibody (mouse, R&D Systems, AF464) with HRP anti-goat secondary antibody (rabbit, Jackson ImmunoResearch, 305-035-003). Detailed protocols are provided in the supplementary Materials and Methods.
EdU labelling assay
Mice were intraperitoneally injected with EdU (25 µg/g body weight; Thermo Fisher Scientific, A10044) 3 days after the first Tamoxifen administration. Samples were collected 4 days later and processed into 5 µm paraffin sections. Detection was performed using the Click-iT EdU Alexa Fluor 488 Imaging Kit (Thermo Fisher Scientific, C10337) according to the manufacturer's instructions.
Data acquisition and processing
Samples were imaged with a Zeiss Axio Imager M2 microscope and further processed with Adobe Photoshop. RNAscope signal was enhanced by selecting the red or blue-green pixels of the image using the ʻselect color range tool'. The selected areas were false-coloured using the ʻbrush tool'.
pH-H3+ cell, EdU+ cell and laCL volume quantifications
Samples were imaged with a Zeiss Axio Imager M2 microscope and processed with Adobe Photoshop. For embryonic stages, pH-H3+ cells were quantified for every second 5 µm-thick section, and the incisor area was drawn by hand using Zen 2011 software (Zeiss). For adult stages, EdU+ cells and laCL area were quantified from every second 5 µm-thick section from the central region of the incisor (a total of 70 µm). Volumes were calculated by taking into account the thickness of the sections.
Multiplex quantitative real-time (qRT) PCR
The proximal end of the incisor was dissected out from E18 embryos (approximately one-third of the total length) and stored at −80°C. RNA was extracted using the RNeasy Micro kit (Qiagen, 74004) and reverse transcribed using the QuantiTect Reverse Transcription Kit (Qiagen, 205310).
Multiplex qRT-PCR (CFX96 Touch Real-Time PCR Detection System, Bio-Rad) was performed using iTaq Universal Probe Super Mix (Bio-Rad, 1725130) and 10 ng cDNA per reaction. Probe combinations (PrimePCR Probe Assay, Bio-Rad) are provided in the supplementary Materials and Methods.
Micro-CT and 3D reconstructions
Samples were fixed in 4% PFA, rinsed with PBS, dehydrated in an ethanol series, and stained for 2 weeks in 0.1% phosphotungstic acid (PTA) in 70% ethanol. Samples were scanned with a micro-CT scanner (Phoenix Nanotom). Three mutant and two control lower jaws were scanned for each stage. 3D reconstructions were prepared with Avizo software (Thermo Fisher Scientific) from micro-CT scans (embryonic stages) and from H&E-stained sagittal sections (adult stages).
Microarray
Incisor buds and laCL were microdissected from Sox2GFP animals and then incubated with 0.2 U/ml dispase for 10 min at room temperature and the mesenchymal tissue removed. The epithelial explants were incubated with StemPro accutase (Thermo Fisher Scientific, A1110501) for 45 min at 37°C. Samples were passed through a cell strainer (pore size 35 µm) and 7AAD (Thermo Fisher Scientific, A1310) was added to mark dead cells. 7AAD-negative and GFP-positive cells were collected by fluorescence-activated cell sorting (FACS). RNA was extracted using the RNeasy Micro Kit (Qiagen, 74004). Total RNAs were provided to the Functional Genomics Unit, University of Helsinki, Finland, for hybridisation on MTA Affymetrix Arrays. Samples were amplified and labelled using the Affymetrix WT Pico Reagent Kit (Thermo Fisher Scientific, 902622) (see the supplementary Materials and Methods). Three replicates were prepared for each sample, and mESCs were used as a non-dental cell reference. For further analysis, we discarded the non-significant hits (P>0.05, ANOVA).
Statistical analysis
For each experiment n=3, except for gene expression analysis in K14-CreER;Sox2fl/fl mice where n=6 (data not shown). Each incisor is considered one biological replicate (only one incisor was analysed per animal), except for gene expression in Sox2cKO, where one litter was considered as a biological replicate (n=3). Data are shown as mean±s.d. (equal variances not assumed). Unpaired, two-tailed t-test was used to test statistical significance. P<0.05 was used as significance threshold.
Supplementary Material
Acknowledgements
We thank Kaisa Ikkala, Solja Kalha, Raija Savolainen, Jhon Alves and Merja Mäkinen for excellent technical help, and Irma Thesleff and Anamaria Balic for critical reading of the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.S.-N., Z.S., B.A.A., O.D.K., F.M.; Methodology: M.S.-N., K.S., O.D.K., F.M.; Validation: M.S.-N., K.S., F.M.; Formal analysis: M.S.-N., O.D.K., F.M.; Investigation: M.S.-N., K.S., L.B.-B.; Resources: M.S.-N., O.D.K., F.M.; Data curation: M.S.-N., F.M.; Writing - original draft: M.S.-N., O.D.K., F.M.; Writing - review & editing: M.S.-N., K.S., Z.S., L.B.-B., B.A.A., O.D.K., F.M.; Visualization: M.S.-N., F.M.; Supervision: F.M.; Project administration: F.M.; Funding acquisition: M.S.-N., O.D.K., F.M.
Funding
This work was supported by the Finnish Doctoral Program in Oral Sciences (M.S.-N.), the Academy of Finland (Suomen Akatemia) (F.M.), and by National Institutes of Health grant R35-DE026602 (O.D.K). Deposited in PMC for release after 12 months.
Data availability
The microarray data have been deposited at Gene Expression Omnibus under accession number GSE104808.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.155929.supplemental
References
- Arnold K., Sarkar A., Yram M. A., Polo J. M., Bronson R., Sengupta S., Seandel M., Geijsen N. and Hochedlinger K. (2011). Sox2+ adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 9, 317-329. 10.1016/j.stem.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N., van Es J. H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P. J. et al. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003-1007. 10.1038/nature06196 [DOI] [PubMed] [Google Scholar]
- Biehs B., Hu J. K.-H., Strauli N. B., Sangiorgi E., Jung H., Heber R.-P., Ho S., Goodwin A. F., Dasen J. S., Capecchi M. R. et al. (2013). BMI1 represses Ink4a/Arf and Hox genes to regulate stem cells in the rodent incisor. Nat. Cell Biol. 15, 846-852. 10.1038/ncb2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C. and Fuchs E. (2014). Plasticity of epithelial stem cells in tissue regeneration. Science 344, 1242281-1242281 10.1126/science.1242281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Wang J., Li X., Florez S., Huang Z., Venugopalan S. R., Elangovan S., Skobe Z., Margolis H. C., Martin J. F. et al. (2010). MicroRNAs play a critical role in tooth development. J. Dent. Res. 89, 779-784. 10.1177/0022034510369304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Jheon A., Li X., Sun Z., Wang J., Florez S., Zhang Z., McManus M. T., Klein O. D. and Amendt B. A. (2013). The Pitx2:miR-200c/141:noggin pathway regulates Bmp signaling and ameloblast differentiation. Development 140, 3348-3359. 10.1242/dev.089193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. Y. F., Wang C., Jin C., Yang C., Huang Y., Liu J., McKeehan W. L., D'Souza R. N. and Wang F. (2013). Self-renewal and multilineage differentiation of mouse dental epithelial stem cells. Stem Cell Res. 11, 990-1002. 10.1016/j.scr.2013.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.-Y., Chen W.-C., Lee A.-C., Hung S.-M., Shih N.-Y. and Chen M.-Y. (2009). Clusterin silencing in human lung adenocarcinoma cells induces a mesenchymal-to-epithelial transition through modulating the ERK/Slug pathway. Cell Signal. 21, 704-711. 10.1016/j.cellsig.2009.01.008 [DOI] [PubMed] [Google Scholar]
- Clavel C., Grisanti L., Zemla R., Rezza A., Barros R., Sennett R., Mazloom A. R., Chung C.-Y., Cai X., Cai C.-L. et al. (2012). Sox2 in the dermal papilla niche controls hair growth by fine-tuning BMP signaling in differentiating hair shaft progenitors. Dev. Cell 23, 981-994. 10.1016/j.devcel.2012.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassule H. R. and McMahon A. P. (1998). Analysis of epithelial–mesenchymal interactions in the initial morphogenesis of the mammalian tooth. Dev. Biol. 202, 215-227. 10.1006/dbio.1998.8992 [DOI] [PubMed] [Google Scholar]
- Dy P., Penzo-Méndez A., Wang H., Pedraza C. E., Macklin W. B. and Lefebvre V. (2008). The three SoxC proteins – Sox4, Sox11 and Sox12 – exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 36, 3101-3117. 10.1093/nar/gkn162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felszeghy S., Suomalainen M. and Thesleff I. (2010). Notch signalling is required for the survival of epithelial stem cells in the continuously growing mouse incisor. Differentiation 80, 241-248. 10.1016/j.diff.2010.06.004 [DOI] [PubMed] [Google Scholar]
- Fuchs E. and Chen T. (2012). A matter of life and death: self-renewal in stem cells. EMBO Rep. 14, 39-48. 10.1038/embor.2012.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Moreno M., Eliason S., Cao H., Li X., Yu W., Bidlack F. B., Margolis H. C., Baldini A. and Amendt B. A. (2015). TBX1 protein interactions and microRNA-96-5p regulation controls cell proliferation during craniofacial and dental development: implications for 22q11.2 deletion syndrome. Hum. Mol. Genet. 24, 2330-2348. 10.1093/hmg/ddu750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegebarth A. and Clevers H. (2009). Wnt signaling, lgr5, and stem cells in the intestine and skin. Am. J. Pathol. 174, 715-721. 10.2353/ajpath.2009.080758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H., Kettunen P., Jung H.-S., Mustonen T., Wang Y. A. and Thesleff I. (1999). Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J. Cell Biol. 147, 105-120. 10.1083/jcb.147.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H., Ichimori Y., Yokohama-Tamaki T., Ohshima H., Kawano S., Katsube K.-I. and Wakisaka S. (2006). Stratum intermedium lineage diverges from ameloblast lineage via Notch signaling. Biochem. Biophys. Res. Commun. 340, 611-616. 10.1016/j.bbrc.2005.12.053 [DOI] [PubMed] [Google Scholar]
- Hargrave M., Wright E., Kun J., Emery J., Cooper L. and Koopman P. (1997). Expression of theSox11 gene in mouse embryos suggests roles in neuronal maturation and epithelio-mesenchymal induction. Dev. Dyn. 210, 79-86. [DOI] [PubMed] [Google Scholar]
- Hu J. K.-H., Du W., Shelton S. J., Oldham M. C., DiPersio C. M. and Klein O. D. (2017). An FAK-YAP-mTOR signaling axis regulates stem cell-based tissue renewal in mice. Cell Stem Cell 21, 91-106. 10.1016/j.stem.2017.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsken J., Vogel R., Erdmann B., Cotsarelis G. and Birchmeier W. (2001). beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell 105, 533-545. 10.1016/S0092-8674(01)00336-1 [DOI] [PubMed] [Google Scholar]
- Jaks V., Barker N., Kasper M., van Es J. H., Snippert H. J., Clevers H. and Toftgård R. (2008). Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 40, 1291-1299. 10.1038/ng.239 [DOI] [PubMed] [Google Scholar]
- Järvinen E., Salazar-Ciudad I., Birchmeier W., Taketo M. M., Jernvall J. and Thesleff I. (2006). Continuous tooth generation in mouse is induced by activated epithelial Wnt/beta-catenin signaling. Proc. Natl. Acad. Sci. USA 103, 18627-18632. 10.1073/pnas.0607289103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jussila M., Aalto A. J., Sanz Navarro M., Shirokova V., Balic A., Kallonen A., Ohyama T., Groves A. K., Mikkola M. L. and Thesleff I. (2015). Suppression of epithelial differentiation by Foxi3 is essential for molar crown patterning. Development 142, 3954-3963. 10.1242/dev.124172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juuri E., Saito K., Ahtiainen L., Seidel K., Tummers M., Hochedlinger K., Klein O. D., Thesleff I. and Michon F. (2012). Sox2+ stem cells contribute to all epithelial lineages of the tooth via Sfrp5+ progenitors. Dev. Cell 23, 317-328. 10.1016/j.devcel.2012.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juuri E., Jussila M., Seidel K., Holmes S., Wu P., Richman J., Heikinheimo K., Chuong C.-M., Arnold K., Hochedlinger K. et al. (2013a). Sox2 marks epithelial competence to generate teeth in mammals and reptiles. Development 140, 1424-1432. 10.1242/dev.089599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juuri E., Saito K., Lefebvre S. and Michon F. (2013b). Establishment of crown–root domain borders in mouse incisor. Gene Expr. Patterns 13, 255-264. 10.1016/j.gep.2013.05.001 [DOI] [PubMed] [Google Scholar]
- Kawasaki M., Porntaveetus T., Kawasaki K., Oommen S., Otsuka-Tanaka Y., Hishinuma M., Nomoto T., Maeda T., Takubo K., Suda T. et al. (2014). R-spondins/Lgrs expression in tooth development. Dev. Dyn. 243, 844-851. 10.1002/dvdy.24124 [DOI] [PubMed] [Google Scholar]
- Khan Q.-E.-S., Sehic A., Khuu C., Risnes S. and Osmundsen H. (2013). Expression of Clu and Tgfb1 during murine tooth development: effects of in-vivo transfection with anti-miR-214. Eur. J. Oral Sci. 121, 303-312. 10.1111/eos.12056 [DOI] [PubMed] [Google Scholar]
- Klein O. D., Lyons D. B., Balooch G., Marshall G. W., Basson M. A., Peterka M., Boran T., Peterkova R. and Martin G. R. (2008). An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development 135, 377-385. 10.1242/dev.015081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane S. W., Williams D. A. and Watt F. M. (2014). Modulating the stem cell niche for tissue regeneration. Nat. Biotechnol. 32, 795-803. 10.1038/nbt.2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Kwon H.-J., Harada H., Ohshima H., Cho S.-W. and Jung H.-S. (2011). Expression patterns of ABCG2, Bmi-1, Oct-3/4, and Yap in the developing mouse incisor. Gene Expr. Patterns 11, 163-170. 10.1016/j.gep.2010.11.001 [DOI] [PubMed] [Google Scholar]
- Michon F., Tummers M., Kyyrönen M., Frilander M. J. and Thesleff I. (2010). Tooth morphogenesis and ameloblast differentiation are regulated by micro-RNAs. Dev. Biol. 340, 355-368. 10.1016/j.ydbio.2010.01.019 [DOI] [PubMed] [Google Scholar]
- Morita H., Mazerbourg S., Bouley D. M., Luo C.-W., Kawamura K., Kuwabara Y., Baribault H., Tian H. and Hsueh A. J. W. (2004). Neonatal lethality of LGR5 null mice is associated with ankyloglossia and gastrointestinal distension. Mol. Cell. Biol. 24, 9736-9743. 10.1128/MCB.24.22.9736-9743.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucchielli M.-L. and Mitsiadis T. A. (2000). Correlation of asymmetric Notch2 expression and mouse incisor rotation. Mech. Dev. 91, 379-382. 10.1016/S0925-4773(99)00293-2 [DOI] [PubMed] [Google Scholar]
- Nonomura K., Takahashi M., Wakamatsu Y., Takano-Yamamoto T. and Osumi N. (2010). Dynamic expression of Six family genes in the dental mesenchyme and the epithelial ameloblast stem/progenitor cells during murine tooth development. J. Anat. 216, 80-91. 10.1111/j.1469-7580.2009.01167.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara N., Suzuki Y., Irie K. and Shibata S. (2017). Expression of planar cell polarity genes during mouse tooth development. Arch. Oral Biol. 83, 85-91. 10.1016/j.archoralbio.2017.07.008 [DOI] [PubMed] [Google Scholar]
- Que J., Okubo T., Goldenring J. R., Nam K.-T., Kurotani R., Morrisey E. E., Taranova O., Pevny L. H. and Hogan B. L. M. (2007). Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development 134, 2521-2531. 10.1242/dev.003855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P., Mesa K. R. and Greco V. (2013). Spatial organization within a niche as a determinant of stem-cell fate. Nature 502, 513-518. 10.1038/nature12602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel K., Ahn C. P., Lyons D., Nee A., Ting K., Brownell I., Cao T., Carano R. A. D., Curran T., Schober M. et al. (2010). Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development 137, 3753-3761. 10.1242/dev.056358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel K., Marangoni P., Tang C., Houshmand B., Du W., Maas R. L., Murray S., Oldham M. C. and Klein O. D. (2017). Resolving stem and progenitor cells in the adult mouse incisor through gene co-expression analysis. Elife 6, e24712 10.7554/eLife.24712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota M., Zardan A., Takeuchi A., Kumano M., Beraldi E., Naito S., Zoubeidi A. and Gleave M. E. (2012). Clusterin mediates TGF-β-induced epithelial-mesenchymal transition and metastasis via twist1 in prostate cancer cells. Cancer Res. 72, 5261-5272. 10.1158/0008-5472.CAN-12-0254 [DOI] [PubMed] [Google Scholar]
- Sock E., Rettig S. D., Enderich J., Bosl M. R., Tamm E. R. and Wegner M. (2004). Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol. Cell. Biol. 24, 6635-6644. 10.1128/MCB.24.15.6635-6644.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Yu W., Sanz Navarro M., Sweat M., Eliason S., Sharp T., Liu H., Seidel K., Zhang L., Moreno M. et al. (2016). Sox2 and Lef-1 interact with Pitx2 to regulate incisor development and stem cell renewal. Development 143, 4115-4126. 10.1242/dev.138883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen M. and Thesleff I. (2009). Patterns of Wnt pathway activity in the mouse incisor indicate absence of Wnt/β-catenin signaling in the epithelial stem cells. Dev. Dyn. 239, 364.-. 10.1002/dvdy.22106 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S., Randle D. H., Kamijo T., Cleveland J. L., Sherr C. J., Roussel M. F., Kraft A. S., Yang V. W., Farese R. V. et al. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663-676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Thesleff I. and Tummers M. (2008). Tooth Organogenesis and Regeneration. StemBook (ed. The Stem Cell Research Community), doi/10.3824/stembook.1.37.1. [PubMed] [Google Scholar]
- Tian H., Biehs B., Warming S., Leong K. G., Rangell L., Klein O. D. and de Sauvage F. J. (2011). A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478, 255-259. 10.1038/nature10408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V., Degenstein L., Wise B. and Fuchs E. (1999). The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl. Acad. Sci. USA 96, 8551-8556. 10.1073/pnas.96.15.8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Flanagan J., Su N., Wang L.-C., Bui S., Nielson A., Wu X., Vo H.-T., Ma X.-J. and Luo Y. (2012). RNAscope. J. Mol. Diagnostics 14, 22-29. 10.1016/j.jmoldx.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Kawasaki K., Kawasaki M., Portaveetus T., Oommen S., Blackburn J., Nagai T., Kitamura A., Nishikawa A., Kodama Y. et al. (2016). Spatio-temporal expression of Sox genes in murine palatogenesis. Gene Expr. Patterns 21, 111-118. 10.1016/j.gep.2016.05.002 [DOI] [PubMed] [Google Scholar]
- Wu Z., Epasinghe D. J., He J., Li L., Green D. W., Lee M.-J. and Jung H.-S. (2016). Altered tooth morphogenesis after silencing the planar cell polarity core component, Vangl2. Cell Tissue Res. 366, 617-621. 10.1007/s00441-016-2489-4 [DOI] [PubMed] [Google Scholar]
- Xie W., Lynch T. J., Liu X., Tyler S. R., Yu S., Zhou X., Luo M., Kusner D. M., Sun X., Yi Y. et al. (2014). Sox2 modulates Lef-1 expression during airway submucosal gland development. AJP Lung Cell. Mol. Physiol. 306, L645-L660. 10.1152/ajplung.00157.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M., Xing G., Hou L., Bao J., Chen Y., Jiao T. and Zhang F. (2015). Functional role of EMMPRIN in the formation and mineralisation of dental matrix in mouse molars. J. Mol. Histol. 46, 21-32. 10.1007/s10735-014-9603-2 [DOI] [PubMed] [Google Scholar]
- Yang Z., Balic A., Michon F., Juuri E. and Thesleff I. (2015). Mesenchymal Wnt/β-catenin signaling controls epithelial stem cell homeostasis in teeth by inhibiting the antiapoptotic effect of Fgf10. Stem Cells 33, 1670-1681. 10.1002/stem.1972 [DOI] [PubMed] [Google Scholar]
- Yokohama-Tamaki T., Otsu K., Harada H., Shibata S., Obara N., Irie K., Taniguchi A., Nagasawa T., Aoki K., Caliari S. R. et al. (2015). CXCR4/CXCL12 signaling impacts enamel progenitor cell proliferation and motility in the dental stem cell niche. Cell Tissue Res. 362, 633-642. 10.1007/s00441-015-2248-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Yuan G., Liu H., Lin H., Wan C. and Chen Z. (2012). Expression pattern of Sox2 during mouse tooth development. Gene Expr. Patterns 12, 273-281. 10.1016/j.gep.2012.07.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.