Abstract
Background: The presence of Class 1, 2 and 3 integrons in clinical isolates of Pseudomonas aeruginosa with multi-drug resistance phenotype has rendered the organism as a new concern. Objective: This study aimed to investigate the prevalence of Class 1, 2 and 3 integrons in multi-drug resistant clinical isolates of Pseudomonas aeruginosa collected from hospitals in the city of Tabriz
Materials and Methods: A total of 200 P. aeruginosa non-duplicated clinical isolates were collected from inpatients and outpatients in different wards of hospitals from May to November 2016. The bacteria were identified by conventional microbiological methods. Antibiotic susceptibility test was performed by disk diffusion method and the presence of integrons was analyzed by polymerase chain reaction (PCR).
Results: Colistin was the most effective antibiotic, while 98% of the isolates were resistant to cefotaxime. Fifty-three percent of the isolates were recorded as multi-drug resistant (MDR) phenotype; however, 27.5% of the isolates were resistant to more than 8 antibiotics. In this study, 55 (27.5%), 51 (25.5%), and 30 (15%) clinical isolates of P. aeruginosa were positive for Class 1, 2 and 3 integrons, respectively. aac(6)II in Class I integrons and dfrA1 in ClassII and aacA7 in Class II integrons were the most prevalent genes. Resistance to aminoglycosides were the most common genes harbored by integrons.
Conclusion: The results of this study showed that the prevalence of Class 1, 2 and 3 in integron genes in most P. aeruginosa strains islated from different parts and equipment used in the hospital. The role of these transferable genetic agents has been proven in the creation of resistance. Therefore, it is essential to use management practices to optimize the use of antibiotics, preferably based on the results of antibiogram and trace coding genes for antibiotic resistance.
Keywords: Pseudomonas aeruginosa, Antibiotic resistance, Integrons, Iran
1. Introduction
Pseudomonas aeruginosa is a common environmental, Gramnegative, ubiquitous bacterium that causes a variety of infections in immunocompromised, hospitalized patients [1, 2]. This organism trends to increase resistance towards many antimicrobial agents and a high percentage of the P. aeruginosa clinical isolates show the multidrug resistance (MDR) phenotype [3, 4]. The most effective anti-pseudomonal agents are beta-lactams, aminoglycosides and fluoroquinolones [5, 6]. Mechanisms of resistance to antimicrobial agents include production of beta-lactamases, multidrug efflux pumps, presence of integrons and downregulation of outer membrane porins [5-7]. Many of the antibiotic resistance genes found on plasmids and transposons that are located at a unique site named integron [8, 9]. These elements mediate the integration of genes through the action of a DNA integrase (intI) and a specific recombination site (attI) that acts as a receptor of gene cassettes [10]. Approximately, There are 90 distinct integron Classes that most of them located on chromosomes, and about 10% of the sequenced bacterial genomes carry these elements [11]. The first of integrons that have been described are Classes 1, 2, and 3 that exhibit a number of features not typical of the more numerically dominant chromosomal integron Classes. In total, they are carried on transposons and/or plasmids and most commonly contain up to 6 cassettes drawn from a pool of about 100 cassettes and almost all of which encode antibiotic resistance determinants [12, 13]. Class 1, 2 and 3 are three main well characterized integrons [14, 15]. The Classe 1 integrons are the most common integrons that found in P. aeruginosa, Acinetobacter baumannii and in members of Enterobacteriaceae family [16, 17]. Prevalence of Class 2 and 3 integrons among these pathogens is not widely reported [18, 19]. The aim of this study is the analysis of Class 1, 2 and 3 integrons prevalence of and their association with drug resistance in clinical isolates of P. aeruginosa.
2. Materials and Methods
2.1. Bacterial isolates and clinical data
We collected 200 non-duplicated P. aeruginosa clinical isolates from inpatient and outpatients in three teaching and treatment hospitals of Tabriz, Iran from 2015 to 2016 (September to April). The isolates were obtained from blood, sputum, urine, respiratory tract, wound, and cerebral spinal fluid (CSF). The bacteria were identified by routine microbiological tests such as Gram stain, inability to fermentation of lactose, oxidation and fermentation test (O/F), oxidase test, growth on Cetrimide agar medium (Liofilchem, Italy), pigmentation test [20].
2.2. Antimicrobial susceptibility testing
The antibiotic susceptibility test was done by Kirby-Bauer disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines. The antibiotic disks (MAST, England) included amoxicillin-clavulanate (20/10 μg), imipenem (10 μg), colistin (10 μg), amikacin (30 μg), cefepime (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), tobramycin (30 μg), gentamicin (30 μg), ciprofloxacin (5 μg), Polymyxin B (300 units), gatifloxacin (5 μg) and piperacillin (100 μg) [21, 22]. MDR was definite as acquired resistant to at least one agent in three or more antimicrobial Classification [23]. P. aeruginosa ATCC 27853 was used as quality control strain [21].
2.3. DNA extraction and detection of int genes
Genomic DNA was extracted by the tissue buffer boiling method. Precisely, similar colonies of the bacterial isolates were mixed with 20 μ1 of tissue buffer (0.25% SDS + 0.05 M NaOH) and the mixture was incubated for 15 minutes in 95°C [24]. The mixture was centrifuged for 1 minute in 13,000 g. After centrifugation, 180 ìl of Milli-Q water was added to the aqueous solution. The extracted DNA was frozen in -20°C until usage [25].
The int genes were amplified by the polymerase chain reaction (PCR) method. Each PCR reaction was done by CINNAGEN master mix (SinaClon, Tehran, Iran) and specific primers which are shown in Table 1. The amplification was carried out in a DNA thermal cycler (Eppendorf master cycler gradient, Germany) as follows: initial denaturation at 94°C for 10 min, followed by 30 to 40 repeated cycles of denaturation at 94°C for 40 s, 50 s for annealing at 57°C for intI-1, intI-2 and 59°C for intI-3, and 55 s for extension at 72°C, followed by 10 min at 72°C for final extension. The amplified products were analyzed by electrophoresis on 1% agarose gel and staining by the ethidium bromide [26].
Table 1.
List of primers were used for the PCR amplification and sequencing of integrons in the present study.
| Target region | Primer sequence (5́ → 3́) | Size of product | Annealing Temperature | References |
|---|---|---|---|---|
| intI-1F | TCATGGCTTGTTATGACTGT GTAGGGCTTATTATGCACGC |
600 bp* | 57̊C | 26 |
| intI-1R | ||||
| intI-2F | GATGCCATCGCAAGTACGAG CGGGATCCCGGACGGCATGCACGATTTGTA |
750 bp* | 57̊C | 27 |
| intI-2R | ||||
| intI-3F | GCCTCCGGCAGCGACTTTCAG ACGGATCTGCCAAACCTGACT |
650 bp* | 59̊C | 28 |
| intI-3R |
bp = base pair, F = forward sequence, R = reverse sequence.
2.4. Statistical analysis
SPSS Version 22 (IBM SPSS Statistics, New York, USA) was used for statistical analysis. Descriptive statistics, Chi-square or Fisher's exact test was used to evaluate the data. P-value below 0.05 was considered statistically significant.
3. Results
Among 200 P. aeruginosa isolates, 145 of them were collected from Imam Reza hospital, while 35 and 20 isolates were obtained from Sina and Pediatric hospital, respectively. Also, 115 isolates were from inpatients and others were achieved from outpatients. Fifty percent of the patients were men and the age ranges of patients were from new-born to 89 year-old. The clinical isolates were collected from sputum (3 isolates), CSF (7 isolates), purulent wound (57 isolates), blood (19 isolates), respiratory tracts (38 isolates), and urine (76 isolates).
The results of antibiotic susceptibility test were shown in Table 2. The isolates were most sensitive to polypeptide antibiotics such as Colistin. The results showed that the majority of the isolates (98%) were resistant to Cefotaxime (Table 2) and 53% of the isolates were multidrug resistance (MDR).
Table 2.
The relationship between the presence of integrons and resistance to antibiotics in clinical isolates of Pseudomonas aeruginosa in the present study.
| All isolates (n = 200) | Integron positives (n = 55) | Integron negatives (n = 145) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| Antibiotics | R* | I* | S* | R* | I* | S* | R* | I* | S* | P value** |
| No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | ||
| Amikacin | 110 | 9 | 81 | 34 | 2 | 19 | 79 | 3 | 63 | NS* |
| (55) | (4.5) | (40.5) | (61.8) | (3.6) | (34.5) | (54.4) | (2) | (43.4) | ||
| Cefepime | 125 | 0 | 75 | 36 | 0 | 19 | 89 | 0 | 56 | NS |
| (62.5) | (37.5) | (65.4) | (34.5) | (61.3) | (38.6) | |||||
| Ceftazidime | 113 | 0 | 67 | 40 | 0 | 15 | 93 | 0 | 52 | NS |
| (66.5) | (33.5) | (72.7) | (27.2) | (64.1) | (33.8) | |||||
| Tobramycin | 115 | 0 | 85 | 26 | 0 | 29 | 86 | 0 | 59 | NS |
| (57.5) | (42.5) | (47.2) | (52.7) | (59.3) | (40.6) | |||||
| Gentamicin | 124 | 0 | 76 | 39 | 0 | 16 | 85 | 0 | 60 | NS |
| (62) | (38) | (70.9) | (29) | (58.9) | (41.3) | |||||
| Imipenem | 93 | 12 | 95 | 25 | 3 | 17 | 73 | 5 | 67 | NS |
| (46.5) | (6) | (47.5) | (45.4) | (5.4) | (30.9) | (50.3) | (3.4) | (46.2) | ||
| Colistin | 6 | 0 | 194 | 2 | 0 | 53 | 4 | 0 | 141 | NS |
| (3) | (97) | (3.6) | (96.3) | (2.7) | (97.2) | |||||
| Ciprofloxacin | 125 | 0 | 75 | 33 | 0 | 22 | 88 | 0 | 57 | NS |
| (62.5) | (37.5) | (60) | (40) | (60.6) | (39.3) | |||||
| Amoxicillinclavulanate | 121 | 0 | 79 | 33 | 0 | 22 | 86 | 0 | 59 | NS |
| (60.5) | (39.5) | (60) | (40) | (59.3) | (40.6) | |||||
| Cefotaxime | 149 | 3 | 48 | 39 | 1 | 15 | 109 | 2 | 34 | NS |
| (74.5) | (1.5) | (24) | (70.9) | (1.8) | (27.2) | (75.1) | (1.3) | (23.4) | ||
| Ceftazidimeclavulanate | 110 | 0 | 90 | 33 | 0 | 22 | 77 | 0 | 68 | NS |
| (55) | (45) | (60) | (40) | (53.1) | (46.8) | |||||
R: resistant. I: intermediate. S: susceptible. NS: not statistically significant.
Statistical analysis was done by Chi-square method using SPSS software. All other statistical analysis were done by descriptive methods.
All isolates were evaluated with PCR analysis to detect integrons Classes. According to the PCR results, 55 (27.5%), 51 (25.5%), and 30 (15%) isolates were contained Class 1, 2, and 3 integron, respectively. The results of association between antibiotic resistance and presence of the integrons are shown in Table 3, 4 and 5. In this study, there was no significant association between antibiotic resistance and the presence of integron among the isolates.
Table 3.
Gene cassettes in the Class I integrons in clinical isolates of Pseudomonas aeruginosa.
| Length of variable region (s) (bp) | Gene cassette (s) | No. of isolates (%) | The name of hospital (s) |
|---|---|---|---|
| 750 | aadB | 3 (5.4%) | I,I,I |
| 1200 | aadA6-orfD | 4 (7%) | I,I,I,I |
| 1200 |
aadB-aadA1, blaoxa-10+ (aac(6)-II), blaoxa-10 |
13 (24%) | I,I,I,I,S,S,I,S,S,I,I,I,I |
| 1250 |
aac(6)-II, aacA4, blaoxa-10+blavlm-6, aac(6)-Ib |
20 (36%) | I,I,I,I,I,S,I,I,I,I,I,I,I,S,S,S,I,I,I,I |
| 1500 | aacA4-catB10 | 2 (3.5%) | I,I |
| 1700 | aacA4-blaOXA10 | 13 (24%) | I,I,I,I I,I,I,I,I,I,I,I,I,I |
| Total | 55 isolates |
* I: Imam, S: Sina. C: Kodakan.
Table 4.
Gene cassettes in the Class II integrogens structure by PCR method in clinical isolates of Pseudomonas aeruginosa.
| Length of variable region (s) (bp) | Gene cassette (s) | No. of isolates (%) | The name of hospital (s) |
|---|---|---|---|
| 500 | dfrA1 | 15 (30%) | I,I,I,I,I,S,S,S,I,I,S,I,I,I,P |
| 600 | Hypothetical gene cassette | 15(30%) | I,I,I,I,I,I,I,S,S,P,P,S,S,I,I |
| Total | 30 isolates |
*I: Imam. S: Sina. C: Kodakan.
Table 5.
Gene cassettes in the Class III integrogens structure by PCR method in clinical isolates of Pseudomonas aeruginosa.
| Length of variable region (s) (bp) | Gene cassette (s) | No. of isolates (%) | The name of hospital (s) |
|---|---|---|---|
| 400 | aacA7+ aacA4-blaoxA2 | 30(59%) | I,I,I,I,I,I,I,S,S,S,I,I,I,I,I,I,I,S,I,S,I,I,I,I,P,I,I,I,I,I |
| 600 | Hypothetical gene cassette | 21(41%) | I,I,I,S,I,I,S,I,I,I,I,S,I,I,I,P,S,P,P,S,P |
| Total | 51 isolates |
* I: Imam. S: Sina. C: Kodakan.
Fig. 1.
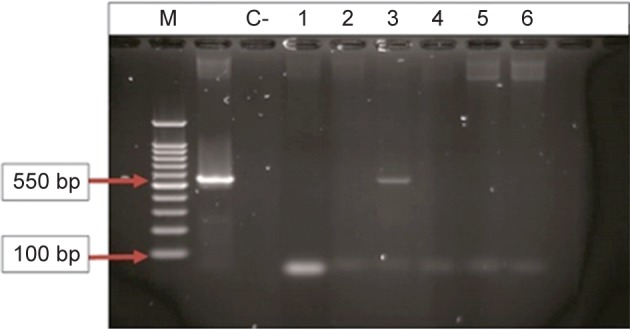
PCR product of the amplification of int-1after agarose electerophoresis. M: ladder (100 bp). C+: control positive of a gene (int-1). C-: control negative. 1-6: Pseudomonas aeruginosa isolates (sample number 3 was positive).
4. Discussion
P. aeruginosa is an opportunistic pathogen that encompasses a wide range of human infection [4], particularly resistant to many antibiotics that makes it hard to treat [27]. Currently it is known one of the most important nosocomial infections with high mortality. Recent studies have shown that transfer of resistance genes by integrons has important role in acquiring resistant in bacteria. Many resistance genes can be transferred by integrons. These genes can be originated by plasmids and transposons [18]. This study aimed to track presence of three main Classes of integrons including Class 1, 2, 3 integrons in P. aeruginosa isolated from main hospitals in northwest of Iran. The most prevalent integron in our isolates was Class 1 with presence in 55% of isolates. However, 78.4% of isolates with Class 2 integrons were MDR, which shows its importance in transfer of resistance genes. All identified integrons harbored gene cassettes and aad and aac genes were the most prevalent genes in our isolates. These genes are corresponding to resistance to aminoglicosides. Several recent studies reported high presence of integrons harboring resistance genes cassettes such as Ren et al. in 2012 from the United States[28], Kali et al. In 2011, from France [29], Tacon et al. In 2012 from Brooklyn [30], as well as Taghavi et al. in 2013 from Iran [31]. In our study, most of isolates were isolated from infected wounds (28.5%) and urinary tract infection (38%), respectively. In Babay study in Saudi Arabia, the most prevalence of isolates from wounds was reported [32]. Antibiotic susceptibility pattern of P. aeruginosa showed 53% of isolates were resistant to more than 5 antibiotics. In Thailand, Poonsuk et al. showed an increase in resistance of P. aeruginosa isolates to Amikacin (92.1%), Ceftazidime (96%), Gentamicin (99%) and Cipro-floxacin (95%)[33]. Fazeli et al. have shown that P. aeruginosa isolates were resistant to Ciprofloxacin (29%) and Gentamicin (32.2%) [34]. Ciprofloxacin is one of the best options available for the treatment of infections caused by P.aeruginosa, particularly in treatment of urinary tract infections [35]. In our isolates, 62.5% of isolates were resistant to Ciprofloxacin. In Latin America (26.8%) and Europe (32%) of isolates were reported to be resistant to Ciprofloxacin [36-38]. In our study, frequency of Class 1, 2 and 3 integrons were 55 (27.5%), 51 (25.5%), and 30 (15%) of isolates, respectively. Other studies from our country reported Class 1 Integrons in 95% of the isolates and Class 2 in 54% and Class 3 in 10% of the isolates [39]. In a study by Shibata et al., integron 1 was the most common integron and integron 3 was observed sporadic in isolates from Japan [40]. Integrons 2 is reported from 9% of isolates from Zanjan- Iran [41] and was reported in 5.3% of isolates isolated from Malaysia [42]. In the present study, Colistin was the most effective antibiotic against P. aeruginosa (Table 2). Highest resistance was observed to cefotaxime (98%). All of the integron positive isolates in the present study Contained genetic cassettes. aad and aac genes family were the most common genes in cassettes. These genes are corresponding on resistance to aminoglycosides. In a same study conducted in neighbor region of our country (Turkey) the most common gene in cassettes was aad gene. In the present study aac (6)–II gene was the most common gene identified in the cassettes. This gene is the most common identified gene in the structure of Class 1 integrons, in clinical isolates of P.aeruginosa reported worldwide. This gene (aac (6) –II) is an enzyme encoding an aminoglycoside ('6) -N-acetyltransferase (II- (6) AAC), which causes resistance to Netilmicin, tobramycin and kanamycin. aadB gene was the second common genes in our studied cassettes. This gene codes the enzyme aminoglycoside ("2) - adenyltransferase (ANT (2") - Ia), which can cause resistance to kanamycin, gentamicin and tobramycin. The aadA1 and aacA4 genes were other identified genes and blaoxa-10 gene from Class D of bosh penicillinase, as a broad-spectrum beta-lactamase. This gene is corresponding to enzymes to hydrolyze the beta-lactam antibiotics such as penicillins. In addition, in several isolates we had co-presence of aacA4 and catB 10 genes ,which is only reported previously from isolates originated from Iran [43]. The catB 10 gene is a chloramphenicol acetyltransferase enzyme causes resistance to chloramphenicol. Blast of gene cassettes in our isolates showed presence of two new genes in cassettes including blaoxa-10+blavlm-6 and aac(6)-Ib which there is no previous report for them from Iran. Presence of resistance in isolates and their easy movement indicates importance of infection control and stewardship programs in hospitals [44]. In this study, there was no significant association between antibiotic resistance and the presence of integron among the isolates. It may be due to a high rate of resistance in our isolates and a low number of sensitive isolates or vis-versa. But future studies the same number of resistant and sensitive isolates can be more helpful to define they exact role on resistance. The finding of the present study is a comprehensive study on integron carriage in our study region and will help clinicians to define the best stewardship for controlling distribution of the resistance.
5. Conclusion
Results of the present study indicate increasing prevalence of integrons corresponding to antibiotic gene cassettes movement. These integrons had genes for resistance to different family members and caused multi drug resistance in our isolates. We had less prevalence of class 1 integrons but higher prevalence of class 3 integrons. Presence of different resistance genes indicates high risk of resistance transmission and distribution of MDR isolates in hospitals. Antibiotic consumption control and antibiotic stew-warship are necessary for reducing resistance in clinical isolates in this region.
Acknowledgments
This study was supported by Drug Applied Research Center, Tabriz University of Medical Sciences, as the Master's thesis of Mr Mobaraki. We thank all hospital staff for their collaboration in sample collection.
References
- 1.Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2000; 2: 1051-60. [DOI] [PubMed] [Google Scholar]
- 2.Bialvaei AZ, Kafil HS, Asgharzadeh M, Yousef Memar M, Yousefi M. Current methods for the identification of carbapenemases. J Chemother. 2016; 28: 1-19. [DOI] [PubMed] [Google Scholar]
- 3.Zahedi Bialvaei A, Samadi Kafil H, Ebrahimzadeh Leylabadlo H, Asgharzadeh M, Aghazadeh M. Dissemination of carbapenemases producing Gram negative bacteria in the Middle East. Iran J Microbiol. 2015; 7: 226-46. [PMC free article] [PubMed] [Google Scholar]
- 4.Leylabadlo HE, Asgharzadeh M, Aghazadeh M. Dissemination of carbapenemases producing Gram negative bacteria in the Middle East. Iran J Microbiol. 2015; 7: 226. [PMC free article] [PubMed] [Google Scholar]
- 5.Rossolini G, Mantengoli E. Treatment and control of severe infections caused by multiresistant Pseudomonas aeruginosa. Clin Microbiol Infect. 2005; 11: 17-32. [DOI] [PubMed] [Google Scholar]
- 6.Hancock RE, Speert DP. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist Updat. 2000; 3: 247-55. [DOI] [PubMed] [Google Scholar]
- 7.Bialvaei AZ, Kafil HS, Asgharzadeh M, Yousefi M, Aghazadeh M. How Social Networks Can Affect Infectious Disease Control: An Experience From Northwest Iran. Infect Control Hosp Epidemiol. 2016; 37: 489. [DOI] [PubMed] [Google Scholar]
- 8.Goli HR, Nahaei MR, Rezaee MA, Hasani A, Kafil HS, Aghazadeh M, et al. Prevalence and molecular characterization of Class 1 integrons among clinical isolates of Pseudomonas aeruginosa in Northwest of Iran. Mol Gen Microbiol Virol. 2017; 32: 109-15. [Google Scholar]
- 9.Severino P, Magalhães VD. The role of integrons in the dissemination of antibiotic resistance among clinical isolates of Pseudomonas aeruginosa from an intensive care unit in Brazil. Res Microbiol. 2002; 153: 221-6. [DOI] [PubMed] [Google Scholar]
- 10.Martinez E, de la Cruz F. Genetic elements involved in Tn21 sitespecific integration, a novel mechanism for the dissemination of antibiotic resistance genes. EMRO J. 1990; 9: 1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillings M, Boucher Y, Labbate M, Holmes A, Krishnan S, Holley M, et al. The evolution of Class 1 integrons and the rise of antibiotic resistance. J Bacteriol. 2008; 190: 5095-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Recchia GD, Hall RM. Gene cassettes: a new Class of mobile element. Microbiology. 1995; 141: 3015-27. [DOI] [PubMed] [Google Scholar]
- 13.Stokes H, O'gorman D, Recchia GD, Parsekhian M, Hall RM. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol Microbiol. 1997; 26: 731-45. [DOI] [PubMed] [Google Scholar]
- 14.Mazel D, Dychinco B, Webb VA, Davies J. A Distinctive Class of Integron in the Vibrio Cholerae Genome. Science. 1998; 280: 605-8. [DOI] [PubMed] [Google Scholar]
- 15.Mazel D. Integrons and the origin of antibiotic resistance gene cassettes-super integrons with thousands of gene cassettes may have set the stage for pathogens to develop antibiotic resistance very rapidly. ASM News-American Society for Microbiology. 2004; 70: 520-5. [Google Scholar]
- 16.Ruiz-Martinez L, Lopez-Jimenez L, Fuste E, Vinuesa T, Martinez JP, Vinas M. Class 1 integrons in environmental and clinical isolates of Pseudomonas aeruginosa. Intern J Antimicrob Agents. 2011; 38: 398-402. [DOI] [PubMed] [Google Scholar]
- 17.Fluit AC, Schmitz FJ. Resistance integrons and super-integrons. Clin Microbiol Infect. 2004; 10: 272-88. [DOI] [PubMed] [Google Scholar]
- 18.Gu B, Tong M, Zhao W, Liu G, Ning M, Pan S, et al. Prevalence and characterization of Class I integrons among Pseudomonas aeruginosa and Acinetobacter baumannii isolates from patients in Nanjing, China. J Clin Microbiol. 2007; 45: 241-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Z, Li L, Shirtliff ME, Alam M, Yamasaki S, Shi L. Occurrence and characteristics of Class 1 and 2 integrons in Pseudomonas aeruginosa isolates from patients in southern China. J Clin Microbiol. 2009; 47: 230-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Najafi K, Kafil HS, Shokrian S, Azimi S, Asgharzadeh M, Yousefi M, et al. Virulence Genes and Antibiotic Resistance Profile of Pseudomonas aeruginosa Isolates in Northwest of Iran. J Pure Appl Microbiol. 2015; 9: 383-9. [Google Scholar]
- 21.Clinical, Institute LS. Performance standards for antimicrobial susceptibility testing. Twenty-fourth informational supplement M100-S24. CLSI Wayne, PA; 2014. [Google Scholar]
- 22.Fattahi S, Aghazadeh M, Nahaei MR, Asgharzadeh M, Kafil HS. Comparison of virulence factors fimA, papC, and hly among uropathogenic Escherichia coli isolates producing and nonproducing extended spectrum beta-lactamases. Ann Trop Med Public health. 2017; 10: 404. [Google Scholar]
- 23.Gholizadeh P, Maftoon H, Aghazadeh M, Asgharzadeh M, Kafil HS. Current opinions in the infection control of carbapenem-resistant Enterobacteriaceae species and Pseudomonas aeruginosa. Rev Med Microbiol. 2017; 28: 97-103. [Google Scholar]
- 24.Asgharzadeh M, Shahbabian K, Kafil HS, Rafi A. Use of DNA Fingerprinting in Identifying the Source Case of Tuberculosis in East Azarbaijan Province of Iran. J Med Sci. 2007; 7: 418-21. [Google Scholar]
- 25.Kafil HS, Mobarez AM. Spread of Enterococcal Surface Protein in Antibiotic Resistant Entero-coccus faecium and Enterococcus faecalis isolates from Urinary Tract Infections. Open Microbiol J. 2015; 9: 14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asgharzadeh M, Mazloumi A, Kafil HS, Ghazanchaei A. Mannosebinding lectin gene and promoter polymorphism in visceral leishmaniasis caused by Leishmania infantum. Pak J Biol Sci. 2007; 10: 1850-4. [DOI] [PubMed] [Google Scholar]
- 27.Bialvaei AZ, Samadi Kafil H. Colistin, mechanisms and prevalence of resistance. Curr Med Res Opin. 2015; 31: 707-21. [DOI] [PubMed] [Google Scholar]
- 28.Ren CL, Konstan MW, Yegin A, Rasouliyan L, Trzaskoma B, Morgan WJ, et al. Multiple antibiotic-resistant Pseudomonas aeruginosa and lung function decline in patients with cystic fibrosis. J Cystic Fibrosis. 2012; 11: 293-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cholley P, Thouverez M, Hocquet D, van der Mee-Marquet N, Talon D, Bertrand X. Most multidrug-resistant Pseudomonas aeruginosa isolates from hospitals in eastern France belong to a few clonal types. J Clin Microbiol. 2011; 49: 2578-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taccone FS, Cotton F, Roisin S, Vincent JL, Jacobs F. Optimal meropenem concentrations to treat multidrug-resistant Pseudomonas aeruginosa septic shock. Antimicrob Agents Chemother. 2012; 56: 2129-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taghvaee R, Shojapour M, Sadeghi A, Pourbabaie AA. The Study of Antibiotic Resistance Pattern and the Frequency of Extended-Spectrum Beta-Lactamases (ESBL) in Pseudomonas aeruginosa Strains Isolated from Medical Centers in Arak City, Iran. Qom Univ Med Sci J. 2013; 7: 36-41. [Google Scholar]
- 32.Babay HA. Antimicrobial resistance among clinical isolates of Pseudomonas aeruginosa from patients in a teaching hospital, Riyadh, Saudi Arabia, 2001-2005. Japan J infect Dis. 2007; 60: 123-5. [PubMed] [Google Scholar]
- 33.Poonsuk K, Tribuddharat C, Chuanchuen R. Class 1 integrons in Pseudomonas aeruginosa and Acinetobacter baumannii isolated from clinical isolates. Southern Asia J Trop Med Public Health. 2012; 43: 376-84. [PubMed] [Google Scholar]
- 34.Fazeli H, Akbari R, Moghim S, Narimani T, Arabestani MR, Ghoddousi AR. Pseudomonas aeruginosa infections in patients, hospital means, and personnel's specimens. J Res Med Sci. 2012; 17: 332-7. [PMC free article] [PubMed] [Google Scholar]
- 35.Gales AC, Jones RN, Turnidge J, Rennie R, Ramphal R. Characterization of Pseudomonas aeruginosa isolates: occurrence rates, antimicrobial susceptibility patterns, and molecular typing in the global SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin Infect Dis. 2001; 32 Suppl 2: S146-55. [DOI] [PubMed] [Google Scholar]
- 36.Bonfiglio G, Carciotto V, Russo G, Stefani S, Schito GC, Debbia E, et al. Antibiotic resistance in Pseudomonas aeruginosa: an Italian survey. J Antimicrob Chemother. 1998; 41: 307-10. [DOI] [PubMed] [Google Scholar]
- 37.Bouza E, Garcia-Garrote F, Cercenado E, Marin M, Diaz MS. Pseudomonas aeruginosa: a survey of resistance in 136 hospitals in Spain. The Spanish Pseudomonas aeruginosa Study Group. Antimicrob Agents Chemother. 1999; 43: 981-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown PD, Izundu A. Antibiotic resistance in clinical isolates of Pseudomonas aeruginosa in Jamaica. Pan Am J Public Health. 2004; 16: 125-30. [DOI] [PubMed] [Google Scholar]
- 39.Yousefi S, Nahaei M, Farajnia S, Ghojazadeh M, Akhi M, Sharifi Y, et al. Class 1 integron and Imipenem Resistance in Clinical Isolates of Pseudomonas aeruginosa: Prevalence and Antibiotic Susceptibility. Iran J Microbiol. 2010; 2: 115-21. [PMC free article] [PubMed] [Google Scholar]
- 40.Shibata N, Doi Y, Yamane K, Yagi T, Kurokawa H, Shibayama K, et al. PCR typing of genetic determinants for metallo-beta-lactamases and integrases carried by gram-negative bacteria isolated in Japan, with focus on the Class 3 integron. J Clin Microbiol. 2003; 41: 5407-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yayan J, Ghebremedhin B, Rasche K. Antibiotic Resistance of Pseudomonas aeruginosa in Pneumonia at a Single University Hospital Center in Germany over a 10-Year Period. PloS one. 2015; 10: e0139836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khosravi Y, Tay ST, Vadivelu J. Analysis of integrons and associated gene cassettes of metallo-beta-lactamase-positive Pseudomonas aeruginosa in Malaysia. J Med Microbiol. 2011; 60: 988-94. [DOI] [PubMed] [Google Scholar]
- 43.Nikokar I, Tishayar A, Flakiyan Z, Alijani K, Rehana-Banisaeed S, Hossinpour M, et al. Antibiotic resistance and frequency of Class 1 integrons among Pseudomonas aeruginosa, isolated from burn patients in Guilan, Iran. Iran J Microbiol. 2013; 5: 36-41. [PMC free article] [PubMed] [Google Scholar]
- 44.Sartelli M, Labricciosa FM, Barbadoro P, Pagani L, Ansaloni L, Brink AJ, et al. The Global Alliance for Infections in Surgery: defining a model for antimicrobial stewardship-results from an international cross-sectional survey. World J Emerg Surg. 2017; 12: 017-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]


