Abstract
Background
The efficacy and applicability of molecular testing to guide the selection of antibiotics in triple Helicobacter pylori (H. pylori) eradication regimens have not been reported. We tested a 7-day, genotypic resistance-guided triple H. pylori eradication therapy in a high-resistance setting.
Methods
Consecutive dyspeptic patients with H. pylori infection were prospectively enrolled. Genotypic resistances to clarithromycin (23SrRNA mutations) and fluoroquinolones (gyrA mutations) were determined from gastric biopsy specimens using a commercially available molecular assay (GenoTypeâ HelicoDR). A tailored genotypic resistance-guided 7-day triple therapy comprised esomeprazole, amoxicillin, and either clarithromycin (wild-type 23SrRNA), levofloxacin (23SrRNA mutated/wild-type gyrA) or rifabutin (both 23SrRNA/gyrA mutated). H. pylori eradication was confirmed by 13C-urea breath test.
Results
Of 148 subjects screened, 51 patients were enrolled (male/female: 27/24, mean age: 50.7±11.4 years, treatment-naïve/-experienced: 32/19). The molecular kit was easily implemented, allowing for rapid (within 24 h) and relatively inexpensive determination of H. pylori resistance (clarithromycin: 47.1%, fluoroquinolones: 15.7%, dual clarithromycin/fluoroquinolones: 7.8%). For patients who received clarithromycin-, levofloxacin- and rifabutin-containing triple therapy, the respective eradication rates were 24/27, 20/20, and 2/4 by intention-to-treat (ITT); and 24/24, 19/19 and 2/3 by per-protocol (PP) analysis. Overall eradication rates were 90.2% (95% confidence interval [CI] 77.8-96.3%) by ITT and 97.8% (95%CI 87-99.8%) by PP analysis, showing no significant difference between treatment-naïve and -experienced patients (ITT: 87.5% vs. 94.7%, P=0.64; PP: 96.4% vs. 100%, respectively, P=1.00).
Conclusions
Regardless of prior treatment history, a genotypic resistance-guided 7-day triple therapy, based on a simple molecular assay, achieved a high H. pylori eradication rate.
Keywords: Helicobacter pylori, tailored therapy, 23SrRNA, gyrA, triple Helicobacter pylori eradication therapy
Introduction
Despite decades of efforts, the optimal strategy to eradicate Helicobacter pylori (H. pylori) remains uncertain. Empirical triple therapies used to represent the backbone of eradicating H. pylori, but are now obsolete in most countries [1]. In high-resistance countries where bismuth and/or tetracycline are unavailable (e.g., Greece, with >27% prevalence of clarithromycin resistance [2,3]), non-bismuth quadruple regimens are currently recommended as first-line therapeutic options, whereas levofloxacin-containing regimens are reserved as salvage treatments [4]. However, even with these regimens, eradication rates >95% are infrequently achieved and even those >90% are disputed [5-8]. Increasing antibiotic resistance is the main reason for the failure of empirical treatments, characterized by significant geographic variation [9]. This precludes any realistic expectation that a universal anti-H. pylori empirical regimen, with stable effectiveness over time, will ever become available. Thus, as for any other bacterial infection, tailoring the selection of antibiotics based on the individual resistance pattern appears to be the viable alternative in order to maintain excellent (>95%) cure rates [10,11].
A critical factor hampering the individualization of H. pylori therapies is that conventional culture-based susceptibility testing methods (e.g., E-test) are not always available and have several shortcomings: firstly, they require endoscopy (which is invasive and costly); secondly, they are time-consuming; and thirdly, they do not completely reflect in vivo eradication, including inaccurate detection of the so-called heteroresistance status [12]. Recent consensuses recommend the use of molecular testing in clinical practice, as an alternative to traditional culture, for both diagnosis and evaluation of H. pylori antibiotic susceptibility [4,13]. Different polymerase chain reaction (PCR)-based approaches allow the measurement of 3 point mutations in the H. pylori 23SrRNA gene (namely A2143G, A2142G and A2142C) that account for >90% of primary clarithromycin resistance in western countries [14,15]. Similarly, a molecular approach is available to test for quinolone resistance by detecting gyrA mutations [16]. These methods can be directly applied to gastric specimens, offering rapid and highly accurate results (reportedly >80-90% sensitivity/specificity) and the possibility of noninvasive evaluation (e.g., using fecal specimens) [16-19].
Despite the evidence of their benefits, there is still paucity of data concerning the applicability of molecular tests to guide the selection of antibiotics in clinical practice. Historically, triple regimens comprising a proton pump inhibitor (PPI) and amoxicillin, together with either clarithromycin (standard triple therapy; STT), levofloxacin (levofloxacin-based triple therapy; LTT) or rifabutin (rifabutin-based triple therapy; RTT) have represented popular treatment options, partly because of their relative simplicity and convenient schedule. Nevertheless, increasing antibiotic resistance rates narrow the potential for using triple therapies in an empirical fashion. Therefore, in this pilot trial, we aimed to evaluate a genotypic resistance-guided triple therapy for H. pylori eradication in a high-resistance setting in Greece.
Patients and methods
This was a pilot, open, prospective study for the treatment of patients with infection by H. pylori. The study was performed from February to March 2017 in the Gastroenterology Department of the “Konstantopouleio-Patission” General Hospital (Nea Ionia, Athens, Greece). The study protocol was in accordance with the principles of the Declaration of Helsinki and was approved by the Ethics Committee of our institution. Written informed consent was obtained from all participating subjects. Data in this pilot study do not overlap our previous evaluation (n=28) of genotypic resistance-guided triple therapy, published in abstract form [20].
Study population
Consecutive patients with dyspepsia, referred and scheduled for upper endoscopy, were prospectively enrolled. Eligible patients were those 18-70 years old who gave informed consent and who had documented H. pylori infection. The following were the exclusion criteria: a history of allergies to the medications used; treatment in the preceding 2 months with antibiotics, bismuth preparations, or non-steroid anti-inflammatory drugs, and in the preceding two weeks with PPIs; previous esophageal or gastric surgery; serious systemic disease; and pregnancy or lactation. For eligible patients, the demographic data, history of previous H. pylori treatment, smoking status, and the endoscopic findings were all recorded.
H. pylori detection and determination of 23SrRNA and gyrA mutations
All eligible patients underwent upper endoscopy at the time of study entry. A gastric biopsy specimen was taken from the antrum for rapid urease testing (HelicotecUT plus test, Strong Biotech Corp-Taiwan). For those who tested positive, two mucosal biopsy specimens (one from the corpus and one from the antrum) were obtained for molecular testing and stored at -20*C in a freezer until the extraction and processing of DNA. Those patients who tested positive on both rapid urease and molecular testing were considered as being infected with H. pylori.
A molecular genetic assay based on DNA-strip technology, the Geno Type® HelicoDR (Hain Lifescience GmbH, Nehren, Germany), was used for the detection of H. pylori and the evaluation of antimicrobial drug susceptibility. This method can be directly applied to gastric specimens, employing the simultaneous detection of resistance to clarithromycin (mutations A2146G, A2146C and A2147G in 23SrRNA gene; codons 2146 and 2147; GenBank accession number NC_000915) and fluoroquinolones (mutations N87K, D91N, D91G and D91Y in gyrA gene; codons 87 and 91). The method has been previously validated and described in detail elsewhere [16,21]. Briefly, the procedure may be schematically divided into three steps. Initially the DNA is extracted from biopsy samples using the validated QIAmp DNA Mini Kit (Qiagen GmbH, Germany). The next step is represented by a multiplex amplification of DNA regions of interest using the biotinylated primers supplied in the Geno TypeÒ HelicoDR kit. The last phase of the molecular test is a reverse hybridization, performed using a specific incubator (TwinCubator, Hain, Lifescience, Nehren, Germany). Hybridization is performed on DNA strips that had been coated at the Hain Lifescience factory (Nehren, Germany) with different specific oligonucleotides (DNA probes). Results concerning H. pylori detection and antibiotic susceptibility are obtained by the analysis of the positive and negative bands in the DNA strips, according to the manufacturer’s instructions.
A 13C-urea breath test (Helicobacter Test INFAI®, INFAI GmbH, Cologne, Germany) was performed 8-12 weeks after completion of treatment and a negative test result was considered indicative of successful H. pylori eradication.
Tailored treatment protocol
A 7-day tailored treatment consisted of choosing triple therapy according to 23SrRNA and gyrA mutational analyses, as follows:
Wild-type 23SrRNA: STT; comprising esomeprazole 40 mg b.i.d., amoxicillin 1 g b.i.d. and clarithromycin 500 mg b.i.d.
23SrRNA mutated/wild-type gyrA: LTT; comprising esomeprazole 40 mg b.i.d., amoxicillin 1 g b.i.d. and levofloxacin 500 mg b.i.d.
23SrRNA mutated/gyrA mutated: RTT; comprising esomeprazole 40 mg b.i.d., amoxicillin 1 g t.i.d. and rifabutin 150 mg b.i.d.
Tolerability and compliance
Adverse events were investigated by means of a structured clinical interview immediately after the completion of therapy and were categorized as nominal variables (i.e., present or absent). Drug compliance was determined by counting unused medication. For this purpose, any tablet that was not consumed was brought back to the clinic for pill count. Poor compliance was defined as taking less than 90% of the total medication prescribed.
Statistical analysis
Intergroup differences were evaluated using Student’s t test or analysis of variance for continuous data. For categorical data, the chi-square test or Fisher’s exact test were used, as appropriate. H. pylori eradication rates were evaluated by intention to treat (ITT) and per protocol (PP) analyses, and 95% confidence intervals (CI) were calculated using the Wilson score method with continuity correction [22]. In the ITT analysis, data from all included patients were evaluated, and patients who did not complete the study were counted as treatment failures. PP analysis excluded patients who violated the study protocol or who were noncompliant with the prescribed treatment. SPSS version 24 for Macintosh (IBM SPSS, Chicago, IL, USA) was used for the statistical analyses and a two-sided P-value of <0.05 was regarded as statistically significant.
Results
Patient data and antibiotic resistance rates
The study flowchart is shown in Fig. 1. A total of 148 subjects were initially screened for inclusion in the trial, of whom 97 were excluded. Thus, 51 patients were finally recruited (male/female: 27/24, mean age: 50.7±11.4 years); 32 were treatment-naïve and 19 (37.2%) had a prior history of H. pylori eradication. When the rapid urease test was positive, the concordance with molecular detection of H. pylori was 100% (51/51). The overall rates of genotypic resistance to clarithromycin and fluoroquinolones were 47.1% (24/51) and 15.7% (8/51) respectively. Dual clarithromycin/quinolone resistance was detected in 4/51 (7.8%). A trend towards higher clarithromycin resistance rates was noted for experienced compared to treatment-naïve patients: 63.2% (12/19) vs. 37.5% (12/32), P=0.09. Quinolone resistance was detected in 21.1% (4/19) of treatment-experienced and 12.5% (4/32) of treatment-naïve patients (P=0.45). Based on the individual genotypic susceptibility pattern, 27/51 patients received STT, 20/51 received LTT and 4/51 received RTT; there was no significant difference in the baseline characteristics of the three treatment groups (Table 1).
Figure 1.

Flow chart of the study
Wt, wild type; mut, mutated; CAM, clarithromycin; LEV, levofloxacin; RIF, rifabutin; ITT, intention to treat; PP, per protocol.
Table 1.
Baseline characteristics

Adverse events and compliance
All but two patients (one each from the STT and RTT groups) returned for confirmation of final eradication. A total of 13 (28.3%) patients complained of at least one side effect (Table 2). One patient in the STT group, who suffered an allergic skin rash, and another in the LTT group, who complained of nausea, interrupted the treatment early, at 4 and 6 days respectively, with H. pylori eradication being successful only in the second case. The overall compliance rate was 95.6% (44/46) and there was no significant difference between the three treatment subgroups (P=0.92).
Table 2.
Frequency of adverse events

Eradication rates
The overall eradication rates were 90.2% (95%CI 77.8-96.3%) by ITT analysis and 97.8% (95%CI 87-99.8%) by PP analysis (Table 3). There were no statistically significant differences in the eradication rates between treatment-naïve and -experienced patients (ITT analysis: 87.5% vs. 94.7%, P=0.64; PP analysis: 96.4% vs. 100%, P=1.00). For STT, LTT and RTT, the overall cure rates were 24/27, 20/20 and 2/4 respectively by ITT analysis, and 24/24, 19/19 and 2/3 by PP analysis (Table 4).
Table 3.
Eradication efficacy of the tailored triple therapy protocol
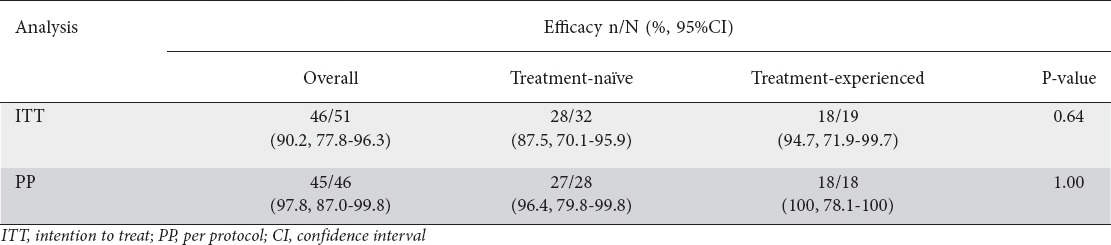
Table 4.
Eradication efficacies of three tailored therapy subgroups

Discussion
The present pilot study reveals several novel findings. First, it demonstrates that genotypic resistance-guided modified triple H. pylori eradication therapy is highly effective (ITT>90%, PP>95%). This was achieved in the face of high rates of antibiotic resistance: clarithromycin 47.1%, quinolones 15.7%, and dual clarithromycin/quinolone resistance 7.8%. As would be expected for a tailored strategy, the cure rates were shown to be irrespective of treatment history, with >95% (PP) efficacy achieved in both treatment-naïve and -experienced groups. Clarithromycin-resistant H. pylori strains were indeed more prevalent among patients with previous treatment failure (likely reflecting secondary resistance), but were successfully eradicated in 11/12 cases (ITT). Likewise, although not at a statistically significant level, the rate of quinolone resistance was higher in the treatment-experienced group (21.1% vs. 12.5%), calling into question the efficacy of an empirical second-line levofloxacin-containing regimen.
The overall (PP) efficacy of tailored triple therapy in eradicating single clarithromycin- and quinolone-resistant H. pylori strains was 100% (23/23). Even more strikingly, we obtained high efficacy using a relatively short (7-day) therapeutic course. This compares favorably to current empiric regimens, generally prescribed for 10-14 days, outlining a sparing effect of the molecular strategy with respect to treatment duration [23,24]. Notably, we used high-dose (1000 mg/day; 500 mg b.i.d.) levofloxacin in the tailored protocol, in contrast to the low dose (500 mg/day) recommended in empirical LTT regimens [4]. Moreover, we chose high-dose (40 mg b.i.d) esomeprazole, a PPI metabolized by a non-enzymatic pathway, aiming to optimize acid suppression [23]. In this context, an interesting approach has been to tailor H. pylori eradication according to the individual cytochrome P450 (CYP) 2C19 (CYP2C19) status [25], though this was not undertaken in the current study.
To date, the clinical value of genotypic resistance has seldom been appraised. Interestingly, preliminary data suggest a relative discordance between phenotypic (culture) and genotypic (PCR-based) determination of clarithromycin resistance [26,27]. In an Italian study, this discrepancy was mainly due to the detection of heteroresistant status by PCR, while genotypic resistance that failed to appear phenotypically was associated with H. pylori eradication in >80% of patients [26]. Moreover, specific point mutations (in particular A2143G) significantly lowered the eradication rate, further highlighting the potential of genotypic assessment. Accordingly, a relative association with therapeutic outcomes has been demonstrated for levofloxacin resistance [28].
From a practical standpoint, the GenoType® HelicoDR test was easily implemented, yielding several potential advantages. This commercially available molecular kit allows for rapid results (within 24 h) and simultaneous detection of both clarithromycin and fluoroquinolone resistances, at a relatively affordable price (approximately €53/single test). Although the test can be performed with only one patient sample, a cost-containing option is to run patient samples in batches. The optimum batch size is 10 patient samples, considering that in each run a positive and a negative control must be included, and the TwinCubator has a limit of 12 positions. The setup for carrying out the test requires a thermocycler, usually available in every molecular biology laboratory, and a specific incubator (TwinCubator, Hain, Lifescience, Nehren, Germany) for hybridization. As far as expertise is concerned, personnel who are skilled in molecular biology techniques should be capable of performing the test. Lastly, the molecular assay may successfully analyze biopsies without tissue transport/storage limitations, as no living cells are required [21].
Ours is the first attempt to characterize the efficacy of a triple genotypic resistance-guided therapy. A few previous studies, conducted in Asia, assessed the potential of tailoring quadruple therapies based on genotypic resistance. Using PCR-based evaluation of clarithromycin/quinolone susceptibility, Liou et al guided the selection of antibiotics (either clarithromycin, levofloxacin, or tetracycline) in a 14-day sequential regimen [29]. The overall eradication rate was 80.7% (ITT). However, only patients with multiple (at least two) treatment failures were included, exhibiting clarithromycin (>85%) and quinolone (>45%) resistance. Furthermore, a subset of patients was treated empirically (according to their medication history), precluding clear-cut conclusions regarding the efficacy of the molecular-based strategy. More recently, Chinese investigators evaluated a genotypic resistance-guided quadruple therapy for first-line H. pylori eradication [30]. Based on genotypic determination of clarithromycin resistance (rate 37.7%), the ITT/PP eradication rates with tailored clarithromycin- and furazolidone-containing quadruple therapies were 98%/100% and 92%/94% respectively.
Our tailored treatment protocol has several strengths. These include a predefined strategy for selecting antibiotics, administration of a convenient triple 7-day schedule, and use of an easily applicable molecular testing method. Nevertheless, a series of critical limitations should be acknowledged. Firstly, this pilot, non-controlled study was designed to generate preliminary data. Therefore, larger studies with a randomized design are needed to validate our molecular-based strategy, assessing its comparative efficacy (and cost-effectiveness) over current empiric treatments. Secondly, we only determined the choice of antibiotics according to clarithromycin and quinolone resistance, although RTT was prescribed empirically in patients harboring dual clarithromycin/quinolone resistant H. pylori strains. The only eradication failure recorded in PP analysis indeed corresponded to a patient receiving RTT, advancing the hypothesis that multidrug resistance may represent the “Achilles’ heel” of our tailored strategy. Few available data, however, suggest that the prevalence of H. pylori resistance to rifabutin remains low [31,32]. Nevertheless, studies evaluating a 7-day RTT have produced conflicting eradication rates (PP), ranging between 46.5% and 80.6% [33-35]. Based on the results of a well-designed randomized trial [35], we used a high PPI (esomeprazole 40 mg b.i.d.) and amoxicillin (1 g t.i.d.) dose, aiming to optimize the efficacy of 7-day RTT. Therefore, assuming a worst-case scenario of 50% eradication, and taking into account a less than 10% prevalence of dual clarithromycin/quinolone resistance in our region, the PP efficacy of tailored triple therapy should not be expected to fall below 95%. Likewise, a longer (for instance 12-day) treatment duration could have led to better eradication outcomes in the RTT group [36,37]. Thirdly, neither amoxicillin resistance nor CYP2C19 polymorphisms were determined. However, we would expect the impact of these factors, if any, to be minimal, as resistance of H. pylori to amoxicillin is uncommon [38] and the metabolism of esomeprazole has been reported to be independent of CYP2C19 status [39]. Lastly, we used gastric specimens for genotyping, which is an invasive and expensive technique, as it requires endoscopy. These limitations could have been avoided by using noninvasive determination methods (e.g. based on fecal specimens) [40].
In summary, this pilot study reveals that a simple molecular susceptibility testing method guiding 7-day triple therapy can achieve a high H. pylori eradication rate, regardless of prior treatment history. Our findings deserve further validation in randomized controlled trials and countries with different patterns of antibiotic resistance.
Summary Box.
What is already known:
The prevalence of Helicobacter pylori (H. pylori) resistance to antibiotics is steadily increasing worldwide
H. pylori eradication rates with empirical therapies are rapidly declining
There is still paucity of data concerning the applicability of molecular tests to guide the treatment of H. pylori infection
What the new findings are:
A genotypic-resistance-guided modified triple regimen is highly effective (per protocol efficacy >95%) for H. pylori eradication
Tailored triple therapy, based on a simple and relatively affordable molecular assay, is an alternative approach, both convenient and highly efficacious, for the treatment of H. pylori infection
Biography
General Hospital of Nea Ionia “Konstantopouleio-Patission”, Athens, Greece
Footnotes
Conflict of Interest: None
References
- 1.Papastergiou V, Georgopoulos SD, Karatapanis S. Treatment ofHelicobacter pyloriinfection:past, present and future. World J Gastrointest Pathophysiol. 2014;5:392–399. doi: 10.4291/wjgp.v5.i4.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Georgopoulos SD, Xirouchakis E, Martinez-Gonzales B, et al. Randomized clinical trial comparing ten day concomitant and sequential therapies forHelicobacter pylorieradication in a high clarithromycin resistance area. Eur J Intern Med. 2016;32:84–90. doi: 10.1016/j.ejim.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 3.Karamanolis GP, Daikos GL, Xouris D, Goukos D, Delladetsima I, Ladas SD. The evolution ofHelicobacter pyloriantibiotics resistance over 10 years in Greece. Digestion. 2014;90:229–231. doi: 10.1159/000369898. [DOI] [PubMed] [Google Scholar]
- 4.Malfertheiner P, Megraud F, 'O'Morain CA, et al. European Helicobacter and Microbiota Study Group and Consensus panel. Management ofHelicobacter pyloriinfection-the Maastricht V/Florence Consensus Report. Gut. 2017;66:6–30. doi: 10.1136/gutjnl-2016-312288. [DOI] [PubMed] [Google Scholar]
- 5.Greenberg ER, Anderson GL, Morgan DR, et al. 14-day triple, 5-day concomitant, and 10-day sequential therapies forHelicobacter pyloriinfection in seven Latin American sites:a randomised trial. Lancet. 2011;378:507–514. doi: 10.1016/S0140-6736(11)60825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liou JM, Chen CC, Chen MJ, et al. Taiwan Helicobacter Consortium. Sequential versus triple therapy for the first-line treatment ofHelicobacter pylori:a multicentre, open-label, randomised trial. Lancet. 2013;381:205–213. doi: 10.1016/S0140-6736(12)61579-7. [DOI] [PubMed] [Google Scholar]
- 7.Vakil N, Vaira D. Treatment forH. pyloriinfection:new challenges with antimicrobial resistance. J Clin Gastroenterol. 2013;47:383–388. doi: 10.1097/MCG.0b013e318277577b. [DOI] [PubMed] [Google Scholar]
- 8.Thung I, Aramin H, Vavinskaya V, et al. Review article:the global emergence ofHelicobacter pyloriantibiotic resistance. Aliment Pharmacol Ther. 2016;43:514–533. doi: 10.1111/apt.13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papastergiou V, Georgopoulos SD, Karatapanis S. Treatment ofHelicobacter pyloriinfection:meeting the challenge of antimicrobial resistance. World J Gastroenterol. 2014;20:9898–9911. doi: 10.3748/wjg.v20.i29.9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papastergiou V, Georgopoulos SD, Karatapanis S. Current and future insights inH. pylorieradication regimens:the need of tailoring therapy. Curr Pharm Des. 2014;20:4521–4532. doi: 10.2174/13816128113196660726. [DOI] [PubMed] [Google Scholar]
- 11.Ierardi E, Giorgio F, Iannone A, et al. Noninvasive molecular analysis ofHelicobacter pylori:Is it time for tailored first-line therapy? World J Gastroenterol. 2017;23:2453–2458. doi: 10.3748/wjg.v23.i14.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alebouyeh M, Yadegar A, Farzi N, et al. Impacts ofH. pylorimixed-infection and heteroresistance on clinical outcomes. Gastroenterol Hepatol Bed Bench. 2015;8:S1–S5. [PMC free article] [PubMed] [Google Scholar]
- 13.Chey WD, Leontiadis GI, Howden CW, Moss SF. ACG Clinical Guideline:Treatment ofHelicobacter pyloriinfection. Am J Gastroenterol. 2017;112:212–239. doi: 10.1038/ajg.2016.563. [DOI] [PubMed] [Google Scholar]
- 14.De Francesco V, Margiotta M, Zullo A, et al. Clarithromycin-resistant genotypes and eradication ofHelicobacter pylori. Ann Intern Med. 2006;144:94–100. doi: 10.7326/0003-4819-144-2-200601170-00006. [DOI] [PubMed] [Google Scholar]
- 15.De Francesco V, Zullo A, Giorgio F, et al. Change of point mutations inHelicobacter pylorirRNA associated with clarithromycin resistance in Italy. J Med Microbiol. 2014;63:453–457. doi: 10.1099/jmm.0.067942-0. [DOI] [PubMed] [Google Scholar]
- 16.Cambau E, Allerheiligen V, Coulon C, et al. Evaluation of a new test, genotype HelicoDR, for molecular detection of antibiotic resistance inHelicobacter pylori. J Clin Microbiol. 2009;47:3600–3607. doi: 10.1128/JCM.00744-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baba S, Oishi Y, Watanabe Y, et al. Gastric wash-based molecular testing for antibiotic resistance inHelicobacter pylori. Digestion. 2011;84:299–305. doi: 10.1159/000332570. [DOI] [PubMed] [Google Scholar]
- 18.Graham DY, Kudo M, Reddy R, Opekun AR. Practical rapid, minimally invasive, reliable nonendoscopic method to obtainHelicobacter pylorifor culture. Helicobacter. 2005;10:1–3. doi: 10.1111/j.1523-5378.2005.00285.x. [DOI] [PubMed] [Google Scholar]
- 19.Kawai T, Yamagishi T, Yagi K, et al. Tailored eradication therapy based on fecalHelicobacter pyloriclarithromycin sensitivities. J Gastroenterol Hepatol. 2008;23(Suppl 2):S171–S174. doi: 10.1111/j.1440-1746.2008.05408.x. [DOI] [PubMed] [Google Scholar]
- 20.Mathou N, Lycousi S, Papastergiou V, et al. Efficacy of a 7-day genotypic resistance-guided triple therapy forHelicobacter pylorieradication. Helicobacter. 2016;21:82. doi: 10.20524/aog.2017.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miendje Deyi VY, Burette A, Bentatou Z, et al. Practical use of GenoType®HelicoDR, a molecular test forHelicobacter pyloridetection and susceptibility testing. Diagn Microbiol Infect Dis. 2011;70:557–560. doi: 10.1016/j.diagmicrobio.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Newcombe RG. Two-sided confidence intervals for the single proportion:comparison of seven methods. Stat Med. 1998;17:857–872. doi: 10.1002/(sici)1097-0258(19980430)17:8<857::aid-sim777>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 23.Georgopoulos SD, Papastergiou V, Karatapanis S. Treatment ofHelicobacter pyloriinfection:optimization strategies in a high resistance era. Expert Opin Pharmacother. 2015;16:2307–2317. doi: 10.1517/14656566.2015.1084503. [DOI] [PubMed] [Google Scholar]
- 24.Yuan Y, Ford AC, Khan KJ, et al. Optimum duration of regimens forHelicobacter pylorieradication. Cochrane Database Syst Rev. 2013;12:CD008337. doi: 10.1002/14651858.CD008337.pub2. [DOI] [PubMed] [Google Scholar]
- 25.Furuta T, Shirai N, Kodaira M, et al. Pharmacogenomics-based tailored versus standard therapeutic regimen for eradication ofH. pylori. Clin Pharmacol Ther. 2007;81:521–528. doi: 10.1038/sj.clpt.6100043. [DOI] [PubMed] [Google Scholar]
- 26.De Francesco V, Zullo A, Ierardi E, et al. Phenotypic and genotypicHelicobacter pyloriclarithromycin resistance and therapeutic outcome:benefits and limits. J Antimicrob Chemother. 2010;65:327–332. doi: 10.1093/jac/dkp445. [DOI] [PubMed] [Google Scholar]
- 27.De Francesco V, Zullo A, Ierardi E, Vaira D. Minimal inhibitory concentration (MIC) values and different point mutations in the23S rRNAgene for clarithromycin resistance in Helicobacter pylori. Dig Liver Dis. 2009;41:610–611. doi: 10.1016/j.dld.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Liou JM, Chang CY, Sheng WH, et al. Genotypic resistance inHelicobacter pyloristrains correlates with susceptibility test and treatment outcomes after levofloxacin- and clarithromycin-based therapies. Antimicrob Agents Chemother. 2011;55:1123–1129. doi: 10.1128/AAC.01131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liou JM, Chen CC, Chang CY, et al. Taiwan Helicobacter Consortium. Efficacy of genotypic resistance-guided sequential therapy in the third-line treatment of refractoryHelicobacter pyloriinfection:a multicentre clinical trial. J Antimicrob Chemother. 2013;68:450–456. doi: 10.1093/jac/dks407. [DOI] [PubMed] [Google Scholar]
- 30.Liu Q, Qi D, Kang J, et al. Efficacy of real-time PCR-based detection ofHelicobacter pyloriinfection and genotypic resistance-guided quadruple therapy as the first-line treatment for functional dyspepsia withHelicobacter pyloriinfection. Eur J Gastroenterol Hepatol. 2015;27:221–225. doi: 10.1097/MEG.0000000000000186. [DOI] [PubMed] [Google Scholar]
- 31.Glocker E, Bogdan C, Kist M. Characterization of rifampicin-resistant clinicalHelicobacter pyloriisolates from Germany. J Antimicrob Chemother. 2007;59:874–879. doi: 10.1093/jac/dkm039. [DOI] [PubMed] [Google Scholar]
- 32.Nishizawa T, Suzuki H, Matsuzaki J, et al. Helicobacter pyloriresistance to rifabutin in the last 7 years. Antimicrob Agents Chemother. 2011;55:5374–5375. doi: 10.1128/AAC.05437-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miehlke S, Hansky K, Schneider-Brachert W, et al. Randomized trial of rifabutin-based triple therapy and high-dose dual therapy for rescue treatment ofHelicobacter pyloriresistant to both metronidazole and clarithromycin. Aliment Pharmacol Ther. 2006;24:395–403. doi: 10.1111/j.1365-2036.2006.02993.x. [DOI] [PubMed] [Google Scholar]
- 34.Navarro-Jarabo JM, Fernández N, Sousa FL, et al. Efficacy of rifabutin-based triple therapy as second-line treatment to eradicateHelicobacter pyloriinfection. BMC Gastroenterol. 2007;7:31. doi: 10.1186/1471-230X-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim HC, Lee YJ, An B, Lee SW, Lee YC, Moon BS. Rifabutin-based high-dose proton-pump inhibitor and amoxicillin triple regimen as the rescue treatment forHelicobacter pylori. Helicobacter. 2014;19:455–461. doi: 10.1111/hel.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fiorini G, Vakil N, Zullo A, et al. Culture-based selection therapy for patients who did not respond to previous treatment forHelicobacter pyloriinfection. Clin Gastroenterol Hepatol. 2013;11:507–510. doi: 10.1016/j.cgh.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 37.Fiorini G, Zullo A, Vakil N, et al. Rifabutin triple therapy is effective in patients with multidrug-resistant strains ofHelicobacter pylori . J Clin Gastroenterol. 2018;52:137–140. doi: 10.1097/MCG.0000000000000540. [DOI] [PubMed] [Google Scholar]
- 38.De Francesco V, Giorgio F, Hassan C, et al. WorldwideH. pyloriantibiotic resistance:a systematic review. J Gastrointestin Liver Dis. 2010;19:409–414. [PubMed] [Google Scholar]
- 39.Tang HL, Li Y, Hu YF, Xie HG, Zhai SD. Effects of CYP2C19 loss-of-function variants on the eradication ofH. pyloriinfection in patients treated with proton pump inhibitor-based triple therapy regimens:a meta-analysis of randomized clinical trials. PLoS One. 2013;8:e62162. doi: 10.1371/journal.pone.0062162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Angol DC, Ocama P, Ayazika Kirabo T, Okeng A, Najjingo I, Bwanga F. Helicobacter pylorifrom peptic ulcer patients in Uganda is highly resistant to Clarithromycin and Fluoroquinolones:results of the genotype HelicoDR test directly applied on stool. Biomed Res Int. 2017;2017:5430723. doi: 10.1155/2017/5430723. [DOI] [PMC free article] [PubMed] [Google Scholar]


