Abstract
Background:
Current advancements in next-generation sequencing technology have made possible to sequence whole genome but assembling a large number of short sequence reads is still a big challenge. In this article, we present the comparative study of seven assemblers, namely, ABySS, Velvet, Edena, SGA, Ray, SSAKE, and Perga, using prokaryotic and eukaryotic paired-end as well as single-end data sets from Illumina platform.
Results:
Results showed that in case of single-end data sets, Velvet and ABySS outperformed in all the seven assemblers with comparatively low assembling time and high genome fraction. Velvet consumed the least amount of memory than any other assembler. In case of paired-end data sets, Velvet consumed least amount of time and produced high genome fraction after ABySS and Ray. In terms of low memory usage, SGA and Edena outperformed in all the assemblers. Ray also showed good genome fraction; however, extremely high assembling time consumed by the Ray might make it prohibitively slow on larger data sets of single and paired-end data.
Conclusions:
Our comparison study will provide assistance to the scientists for selecting the suitable assembler according to their data sets and will also assist the developers to upgrade or develop a new assembler for de novo assembling.
Keywords: NGS (next-generation sequencing), DBG (de Bruijn graph), OLC (overlap layout consensus), ENA (European Nucleotide Archive), bps (base pairs)
Introduction
DNA sequencing has revolutionized the current advancements in the field of science and technology. It has been widely used in applied field of medicine, genetic engineering, food science, etc.1 In current era, next-generation sequencing (NGS) is the most advanced technology of DNA sequencing, which provides more accuracy and speed than previously known Sanger sequencing.2 Paired-end sequencing in NGS, which involves the sequencing of both forward and reverse fragments of DNA, has further increased the accuracy and ability to detect indels which otherwise was not possible in single-end sequencing.3
Next-generation sequencing technique produces millions of short sequence reads and assembling these short sequence reads without a reference genome is one of the challenging task for de novo assemblers.4 In the past few years, several de novo sequence assembling algorithms have been developed to handle and assemble the large amount of short sequence reads to form longer fragments called contigs but choosing the appropriate assembler for paired-end or single-end data is still a challenging job.5
The currently available assembling algorithms include de Bruijn graph (DBG), overlap layout consensus (OLC), string graph, greedy, and hybrid algorithm.6
De Bruijn graph is the graph algorithm based on k-mers approach, which splits the short reads into smaller k-mers, and these k-mers overlap by k − 1 which is the next k-mer. Dividing the sequences into smaller sizes also helps improving the crisis of different initial read lengths, whereas OLC is also the graph-based algorithm which builds overlap graph by overlapping the similar sequences.7
Finding overlapping sequences is usually the slowest part of the assembly and these overlapped sequences then pack fragments of the overlap graph into contigs. The DBG algorithm is faster and OLC algorithm executes better for longer sequence reads. String graph algorithm is the variant of OLC algorithm, which performs global overlap graph by eliminating unnecessary sequences.8
Greedy algorithms start by joining the short sequence reads that are best overlapped to produce contigs. Most greedy assemblers use heuristic techniques that are designed to eliminate misassembling of recurring sequences.9
Hybrid assembling algorithm refers to the mixing various assembling algorithms. It is used to reduce the number of contigs and errors produced by other algorithms.10
There are many de novo assemblers available online which have been developed by applying one of these five assembling algorithms. Our study evaluated the de novo sequence assemblers for Illumina-based paired-end and single-end short reads data sets. This study provides guidance to the biologists and bioinformaticians in selecting the appropriate assembler according to their data sets and it also assists developers to upgrade or develop a new assembler for de novo assembling.
Materials and Methods
Data sets
To compare the performance of each assembler, Illumina HiSeq 2000–based short sequence reads were downloaded from publicly available database European Nucleotide Archive (ENA)11 (Tables 1 and 2). For the estimation of genome fraction, all the reference genomes were downloaded from National Center for Biotechnology Information (NCBI) genome database. Short sequence reads included 7 paired-end and 8 single-end prokaryotic data sets and also 5 paired-end and 5 single-end eukaryotic data sets. All the data sets have maximum read length of 100 bps.
Table 1.
Prokaryotic data sets used in this study.
| S. no. | Data set | ENA run accession | Data set type | No. of reads |
|---|---|---|---|---|
| 1 | Staphylococcus aureus | ERR353143 | Paired-end | 137 022 |
| 2 | Streptococcus pneumoniae | ERR490828 | Paired-end | 321 004 |
| 3 | Escherichia coli | ERR490638 | Paired-end | 737 008 |
| 4 | Mycobacterium tuberculosis | ERR495003 | Paired-end | 770 994 |
| 5 | Neisseria flava | DRR015798 | Paired-end | 1 218 573 |
| 6 | Aeromonas salmonicida | DRR015726 | Paired-end | 2 267 875 |
| 7 | Rothia mucilaginosa | DRR015851 | Paired-end | 4 098 002 |
| 8 | Streptococcus suis | DRR015872 | Single-end | 113 512 |
| 9 | Streptococcus pyogenes | SRR1148216 | Single-end | 724 546 |
| 10 | Salmonella enterica | ERR233905 | Single-end | 1 490 584 |
| 11 | Neisseria gonorrhoeae | SRR969383 | Single-end | 1 840 438 |
| 12 | Chlamydia muridarum | SRR1736648 | Single-end | 3 099 636 |
| 13 | Clostridioides difficile | ERR465798 | Single-end | 5 094 314 |
| 14 | Bacillus anthracis | ERR1596542 | Single-end | 7 466 661 |
| 15 | Chlamydia trachomatis | SRR1038047 | Single-end | 9 129 274 |
Table 2.
Eukaryotic data sets used in this study.
| S. no. | Data set | ENA run accession | Data set type | No. of reads |
|---|---|---|---|---|
| 1 | Homo sapiens | DRR002191 | Paired-end | 126 605 856 |
| 2 | Drosophila melanogaster | DRR016722 | Paired-end | 95 461 377 |
| 3 | Arabidopsis thaliana | ERR1224454 | Paired-end | 30 841 688 |
| 4 | Saccharomyces cerevisiae | ERR052652 | Paired-end | 17 584 902 |
| 5 | Fungi | SRR1614243 | Paired-end | 22 344 195 |
| 6 | Homo sapiens | DRR002191 | Single-end | 126 605 856 |
| 7 | Drosophila melanogaster | DRR002191 | Single-end | 95 461 377 |
| 8 | Arabidopsis thaliana | ERR1224454 | Single-end | 30 841 688 |
| 9 | Saccharomyces cerevisiae | ERR052652 | Single-end | 17 584 902 |
| 10 | Fungi | SRR1614243 | Single-end | 22 344 195 |
Genome assemblers
Seven assemblers (Table 3), which represent 5 different assembly algorithm strategies, were selected to assemble paired-end and single-end data sets.
Table 3.
De novo assemblers selected for this study.
| S. no. | ASSEMBLER | Programming LANGUAGE | ALGORITHM | Input reads |
|---|---|---|---|---|
| 1 | ABySS14 | C++ | De Bruijn graph (DBG) | Paired-end and single-end |
| 2 | Velvet15 | C | De Bruijn graph (DBG) | Paired-end and single-end |
| 3 | Edena16 | C++ | Overlap/layout/consensus (OLC) | Paired-end and single-end |
| 4 | SGA17 | C++ | String graph | Paired-end |
| 5 | Ray18 | C++ | Hybrid | Paired-end and single-end |
| 6 | SSAKE19 | Perl | Greedy | Paired-end and single-end |
| 7 | Perga20 | C | Greedy | Paired-end and single-end |
All the selected assemblers were executed on the virtual machine, which was designed using Oracle VM VirtualBox with 2 VCPU, 4 GB of RAM memory and 64-bit Linux Ubuntu Server 14.04 operating system (supplementary file 1).
Efficiency evaluation
The efficiency of each assembler was evaluated using various parameters, which include assembling total time, maximum memory usage, and maximum CPU usage.
Accuracy evaluation
The output of assemblers was decomposed into contigs. All these contig information were stored in contig files which were produced as an end result of assembling by an assembler. Contig files were used for the accuracy evaluation of each assembler using different parameters including the total number of contigs and N50 contig length. These parameters were collected using Assemblathon 2 script12 which is written in Perl language to calculate the metrics of each contig file. Genome fraction was calculated using QUAST tool13 to find the similarity between the contig sequences and the reference genome.
Statistical analyses
For data analysis, R (version 3.3.2) was used. The data were tested using Shapiro-Wilk normality to find whether data are normally distributed or not. To determine statistical significance, parametric and nonparametric tests were used according to the data. A 2-tailed P values less than .05 were considered as significant.
Results
Efficiency, as well as the accuracy of each assembler, was analyzed by generated contig files using various evaluation techniques. Our study involved evaluation of 7 different assemblers with alternative assembly algorithms such as ABySS and Velvet, the DBG-based assemblers; Edena which is an OLC-based assembler; SGA which uses string graph algorithm; SSAKE and Perga, the greedy-based assembler; and Ray which worked on hybrid algorithm (Table 3).
Total assembling time
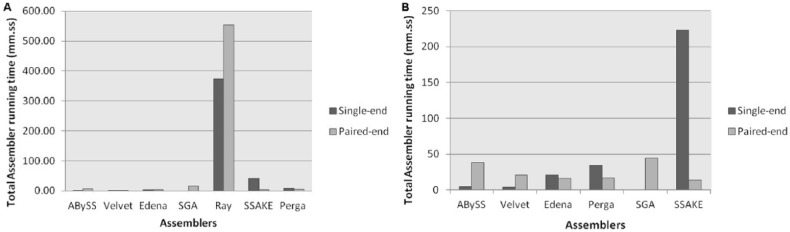
The total assembling time in minutes was calculated using Linux time command, and median of each assembler was compared using Mann-Whitney test. The results showed that Ray, the hybrid assembler, consumed more time on paired-end data sets with a median time of 553.95 minutes and single-end data sets with a median time of 373.15 minutes than any other assembler and reached very high level of significance with P < .05 in prokaryotic data sets, whereas in eukaryotic data set, SSAKE, the greedy assembler, consumed more time on single-end data sets with a median time of 223.10 minutes and lowest in paired-end data sets with a median time of 13.85 minutes.
Velvet, the DBG assembler, consumed lowest median time of 1.49 minutes on paired-end data sets and 1.26 minutes on single-end data sets, whereas in eukaryotic data sets, Velvet showed lowest time of 1.26 minutes on single-end data sets with median of 4.90 minutes. ABySS, which is also the DBG assembler, was second in consuming the lowest median time of 1.93 minutes on single-end prokaryotic and eukaryotic data sets. SSAKE was second lowest time-consuming tool (on paired-end prokaryotic data sets) with median time of 1.93 minutes (Figure 1).
Figure 1.
The comparison of total median assembling time of each assembler for (A) paired-end and single-end prokaryotic data sets and (B) paired-end and single-end eukaryotic data sets.
Memory and CPU usage
The maximum assembling memory usage in megabytes (MBs) and CPU usage in percentage (%) were also calculated using Linux command and the assemblers were compared using independent samples test. On prokaryotic paired-end data sets, the results showed that ABySS, the DBG assembler, and Perga, the greedy assembler, consumed the highest amount of memory than any other assembler with a significance of P < .05, whereas SGA, the string graph assembler, and Edena, the OLC assembler, used the least amount of memory than other assemblers. On eukaryotic paired-end data sets, ABySS and Velvet consumed the highest amount of memory, whereas SSAKE used the least amount of memory.
On prokaryotic single-end data sets, SSAKE and Perge, the greedy assemblers, consumed the highest amount of memory, whereas Edena consumed the lowest memory among all.
On eukaryotic single-end data sets, Velvet and Edena consumed the lowest memory among all assembler, whereas Perga and SSAKE consumed the highest amount of memory (Figure 2).
Figure 2.
The mean comparison of memory usage and CPU usage of each assembler for (A) paired-end and single-end prokaryotic data sets and (B) paired-end and single-end eukaryotic data sets.
In terms of CPU usage, ABySS, Velvet, and SGA, the graph-based assemblers, consumed a huge amount of CPU, whereas Edena and SSAKE consumed least amount of CPU on prokaryotic paired-end data sets, whereas SSAKE also consumed least amount of CPU on eukaryotic paired-end data sets.
On prokaryotic and eukaryotic single-end data sets, Ray, Perge, and Velvet consumed huge amount of CPU as compared with SSAKE and Edena which consumed least amount of CPU (Figure 2).
Total number of contigs
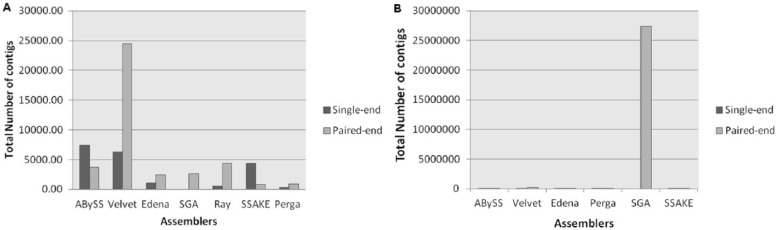
For further analysis of assembled contigs, the number of contigs was calculated by running Assemblathon script. In an ideal condition, the minimum number of contigs that matches the whole genome sequence could be generated from each assembly procedure. The results showed that on prokaryotic and eukaryotic paired-end data sets, the Velvet, the DBG assembler, assembled short reads into relatively short contigs and achieved significance of P < .05, whereas in case of single-end data sets, ABySS produced the high number of contigs followed by Velvet and SSAKE. However, SSAKE and Perga produced the low number of contigs on paired-end data sets (Figure 3).
Figure 3.
The comparison of the total number of contigs by median of each assembler for (A) paired-end and single-end prokaryotic data sets and (B) paired-end and single-end eukaryotic data sets.
N50 contig length
N50 contig length was calculated by running Assemblathon script on contig files produced by various assemblers. On prokaryotic and eukaryotic paired-end data sets, ABySS produced high N50 contig length, whereas Velvet produced low N50 contig length.
On prokaryotic single-end data sets, Velvet produced high N50 contig length with a median length of 1530.00 bp followed by ABySS with a median length of 1054.00 bp, whereas SSAKE and Perga produced low N50 contig length with a median length of 260.50 and 348.00 bp. On eukaryotic single-end data sets, Edena produced high N50 contig length with a median length of 57 252.00 bp, whereas Perga produced low N50 contig length with a median length of 12 654.50 bp (Figure 4).
Figure 4.
The comparison of the N50 contig length by median of each assembler for (A) paired-end and single-end prokaryotic data sets and (B) paired-end and single-end eukaryotic data sets.
Genome fraction
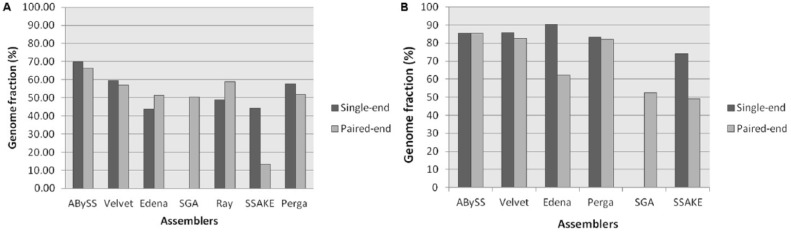
By mapping all the contigs onto the reference genomes using QUAST tool, we calculated the genome fraction of all the contigs generated by each assemblers which showed the percentage of aligned contig bases in the reference genome (Table 4). ABySS showed the high number of genome fraction with a mean of 66.3% on paired-end data sets and 69.8% in prokaryotic and eukaryotic single-end data sets. Ray showed second highest genome fraction with a mean of 58.8% followed by Velvet with third highest genome fraction with a mean of 57.1% on prokaryotic paired-end data sets, whereas Perga, Edena, and SGA showed average accuracy with a mean genome fraction of 51.9%, 51.4%, and 50.4% and SSAKE showed worst accuracy with mean genome fraction of 13.2%.
Table 4.
List of all assemblers with their mean genome fraction.
| Assembler | Prokaryotic single-end | Prokaryotic paired-end |
|---|---|---|
| ABySS | 69.8 | 66.3 |
| Velvet | 59.6 | 57.1 |
| Edena | 43.8 | 51.4 |
| SGA | — | 50.4 |
| Ray | 48.7 | 58.8 |
| SSAKE | 44.3 | 13.2 |
| Perga | 57.6 | 51.9 |
| Assembler | Eukaryotic single-end | Eukaryotic paired-end |
| ABySS | 85.4 | 82.4 |
| Velvet | 82.6 | 85.6 |
| Edena | 62.2 | 90.4 |
| Perga | 82.0 | 83.2 |
| SGA | — | 52.4 |
| SSAKE | 49.2 | 74.0 |
On single-end prokaryotic data sets, Velvet showed second highest genome fraction with mean of 59.6% followed by Perga with third highest genome fraction with mean of 57.6%, whereas Ray, SSAKE, and Edena showed average accuracy with mean genome fraction of 48.7%, 44.3%, and 43.8%. On eukaryotic paired-end data sets, Edena showed highest genome fraction with a mean of 90.4% and the second highest Velvet with a mean of 85.6% whereas SSAKE showed lowest genome fraction with a mean of 49.2% and 74.0% in single-end and paired-end data sets (Figure 5). Practically, an assembler which produces the fewer number of contigs, with high N50 and high genome fraction, is considered to be ideal.
Figure 5.
The comparison of mean genome fraction of each assembler for (A) paired-end and single-end prokaryotic data sets and (B) paired-end and single-end eukaryotic data sets.
Discussion
We evaluated the selected assemblers with prokaryotic and eukaryotic paired-end and single-end Illumina-based short reads on a Linux-based server. Our results showed that Ray, the hybrid assembler, takes the highest time to complete the whole genome assembling on prokaryotic paired-end and single-end data sets21 but Ray was unable to run on eukaryotic paired-end and single-end data sets because Ray required huge RAM and multiple CPUs for assembling large number of reads. However, the DBG assemblers, Velvet and ABySS, are the best options for both types of data sets because of tremendous assembling speed by consuming the lowest assembling time among all other assemblers, whereas Velvet and ABySS are the best options only for eukaryotic single-end data sets.22 Edena, the OLC assembler, consumed lowest memory on both prokaryotic paired-end and single-end data sets; Velvet and Edena consumed lowest memory on eukaryotic single-end data sets; and SSAKE on eukaryotic paired-end data sets. SGA, the string graph assembler, was also a good choice to assemble paired-end data sets consuming low memory,23 but in terms of assembler data transformation, SGA consumed more time in indexing, correction, duplication removal, and overlapping steps before assembling that made SGA more complex than Edena, which needs only overlapping step to be performed before assembling. Velvet also consumed less memory on single-end data sets after Edena. In terms of high memory usage, ABySS and SSAKE were on the top on paired-end and single-end data sets, respectively.
In summary, in case of paired-end and single-end prokaryotic genomes, ABySS efficiently produced genome assembly and consumed less amount of time but consumed high amount of memory,24 whereas Velvet proved to be a time-efficient and memory-efficient program for only single-end data sets. Edena was a memory-efficient program for both types of data sets, and SGA was also a memory-efficient program, but it is only available for paired-end data.
ABySS and Velvet also provided high scalability to handle a large amount of data than rest of the assemblers.
In terms of total number of contigs, we found that on paired-end data set, the Velvet, produced the greater number of contigs but low N50 value, whereas ABySS produced the greater number of contigs on single-end data sets and showed high N50 value on both data sets.25 This contrasted with the contigs produced from Edena, SGA, Ray, SSAKE, and Perga that produced the low number of contigs and low N50.
Ideally, contigs with high N50 and high genome fraction were our expectation but Velvet and ABySS worked more conservatively than others when it came to merging small contigs into larger contigs, which gave an assembly with a larger number of contigs.26 There could be a number of different things that might have led to this result such as k-mer size for the assembly, quality of the single-end vs paired-end data, and a bunch of other parameters that could have been used to build the assemblies.
To check the accuracy of genome assembly, the contigs were aligned to their related reference genomes using QUAST tool. ABySS showed high number of genome fraction on both paired-end and single-end data sets followed by Ray on paired-end data sets, and Velvet showed second highest genome fraction on single-end data sets.
Velvet and ABySS could be the best choice for both paired-end and single-end prokaryotic data sets with highest genome fraction among all selected assemblers27 but still there are some improvements needed to be incorporated into ABySS. There are several ways in which ABySS can be improved. ABySS consumption of memory and CPU on paired-end data sets is much higher than single-end data sets. ABySS mostly relies on mate pairs to assemble their contigs. This approach may perform poorly in case of lack of coverage and it has a known issue with deadlocking when using higher k values. So, tackling these issues and decreasing the memory and CPU usage make ABySS to be best in all other assemblers. Many research groups worldwide are working on building better genome assemblers. A group of researchers at the European Bioinformatics Institute28 developed the DBG-based genome assembler Velvet. Canada’s Michael Smith Genome Sciences Centre29 developed ABySS. These research groups are still working on improving their assemblers and they periodically release latest versions of their assemblers. De Bruijn graph–based genome assemblers are considered as the best genome assemblers.30
Conclusions
Our study evaluated 7 de novo sequence assemblers in terms of memory, time, and accuracy. We found that each assembler is capable of assembling whole prokaryotic or whole eukaryotic genome but the hybrid assembler Ray is not capable of assembling whole eukaryotic genome if you have about 4 GB of RAM or less. The selection of the best assembler is dependent on the uniqueness of the data sets and the user requirements. On single-end data sets, Velvet and ABySS, produced generally the best results among all 7 assemblers with comparatively low assembling time and high prokaryotic and eukaryotic genome fractions. Velvet also consumed the lowest memory usage on both single-end data sets. Some improvements are needed in ABySS including reduction in memory and CPU usage. On paired-end data sets, when a large amount of memory is not available, SGA and Edena might be a good choice. The hybrid approach, Ray, also showed high genome fraction; however, extremely high assembling time used by the Ray might make it prohibitively slow on larger data sets.
Supplementary Material
Acknowledgments
The authors thank Higher Education Commission and Virtual University of Pakistan for their generous support for conducting this research work.
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: MTP conceived the idea and guided overall design of experiments and drafting the manuscript. ARK conducted experiments and drafted the manuscript. NN and MEB collected data and analyzed the results. All authors read and approved the final manuscript. MS helped in responding peer reviewers’ comments.
Availability of Data and Materials: All the raw data sets can be downloaded from ENA server; links are available at https://github.com/alrafaykhan/DeNovoGenome/blob/master/Datasets.md.
All the testing codes are available at https://github.com/alrafaykhan/DeNovoGenome/blob/master/Commands.md.
References
- 1. Sperber GH, Sperber SM. Thompson and Thompson Genetics in Medicine. Baltimore, MD: Elsevier; 2008. [Google Scholar]
- 2. Buermans HPJ, Den Dunnen JT. Next generation sequencing technology: advances and applications. Biochimica et Biophysica Acta (BBA). 2014;1842:1932–1941. [DOI] [PubMed] [Google Scholar]
- 3. Grimm D, Hagmann J, Koenig D, Weigel D, Borgwardt K. Accurate indel prediction using paired-end short reads. BMC Genom. 2013;14:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. [DOI] [PubMed] [Google Scholar]
- 5. Baker M. De novo genome assembly: what every biologist should know. Nat Methods. 2012;9:333. [Google Scholar]
- 6. Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95:315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kang X, Tang S, Ma Y, Liu R, Wang Y. De Bruijn graph-based genomic sequence assembly algorithms and applications. Paper presented at: Green Computing and Communications (GreenCom), 2013 IEEE and Internet of Things (iThings/CPSCom), IEEE International Conference on and IEEE Cyber, Physical and Social Computing; August 20-23, 2013; Beijing, China, pp. 2094–2097. New York, NY: IEEE. [Google Scholar]
- 8. Li Z, Chen Y, Mu D, et al. Comparison of the two major classes of assembly algorithms: overlap–layout–consensus and de-Bruijn-graph. Brief Func Genom. 2012;11:25–37. [DOI] [PubMed] [Google Scholar]
- 9. Pop M, Salzberg SL, Shumway M. Genome sequence assembly: algorithms and issues. Computer. 2002;35:47–54. [Google Scholar]
- 10. Koren S, Schatz MC, Walenz BP, et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 2012;30:693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leinonen R, Akhtar R, Birney E, et al. The European Nucleotide Archive. Nuc Acids Res. 2010;39:D28–D31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bradnam KR, Fass JN, Alexandrov A, et al. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. GigaScience. 2013;2:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hernandez D, François P, Farinelli L, Østerås M, Schrenzel J. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res. 2008;18:802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Simpson JT, Durbin R. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 2012;22:549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boisvert S, Laviolette F, Corbeil J. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol. 2010;17:1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Warren RL, Sutton GG, Jones SJ, Holt RA. Assembling millions of short DNA sequences using SSAKE. Bioinformatics. 2007;23:500–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu X, Leung HC, Chin FY, et al. PERGA: a paired-end read guided de novo assembler for extending contigs using SVM and look ahead approach. PLoS ONE. 2014;9:e114253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sato K, Sakakibara Y. SL: an extension of the Velvet assembler to a de novo metagenomic assembler utilizing supervised learning. DNA Res. 2015;22:69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Y, Schmidt B, Maskell DL. Parallelized short read assembly of large genomes using de Bruijn graphs. BMC Bioinformatics. 2011;12:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dinh H, Rajasekaran S. A memory-efficient data structure representing exact-match overlap graphs with application for next-generation DNA assembly. Bioinformatics. 2011;27:1901–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haiminen N, Kuhn DN, Parida L, Rigoutsos I. Evaluation of methods for de novo genome assembly from high-throughput sequencing reads reveals dependencies that affect the quality of the results. PLoS ONE. 2011;6:e24182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. [DOI] [PubMed] [Google Scholar]
- 26. Pell J, Hintze A, Canino-Koning R, Howe A, Tiedje JM, Brown CT. Scaling metagenome sequence assembly with probabilistic de Bruijn graphs. Proc Nat Acad Sci. 2012;109:13272–13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peng Y, Leung HC, Yiu SM, Chin FY. IDBA—a practical iterative de Bruijn graph de novo assembler. Paper presented at: Annual International Conference on Research in Computational Molecular Biology; April 25-28, 2010; Lisbon, Portugal, pp. 426–440. Berlin, Germany; Heidelberg, Germany: Springer. [Google Scholar]
- 28. European Bioinformatics Institute Cambridgeshire UK. http://www.ebi.ac.uk; September 1994.
- 29. Canada’s Michael Smith Genome Sciences Centre, Vancouver, BC, Canada. http://www.bcgsc.ca; 1999.
- 30. Compeau PE, Pevzner PA, Tesler G. How to apply de Bruijn graphs to genome assembly. Nat Biotechnol. 2011;29:987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.