ABSTRACT
The chemosensory system in Sinorhizobium meliloti has several important deviations from the widely studied enterobacterial paradigm. To better understand the differences between the two systems and how they are optimally tuned, we determined the cellular stoichiometry of the methyl-accepting chemotaxis proteins (MCPs) and the histidine kinase CheA in S. meliloti. Quantitative immunoblotting was used to determine the total amount of MCPs and CheA per cell in S. meliloti. The MCPs are present in the cell in high abundance (McpV), low abundance (IcpA, McpU, McpX, and McpW), and very low abundance (McpY and McpZ), whereas McpT was below the detection limit. The approximate cellular ratio of these three receptor groups is 300:30:1. The chemoreceptor-to-CheA ratio is 23.5:1, highly similar to that seen in Bacillus subtilis (23:1) and about 10 times higher than that in Escherichia coli (3.4:1). Different from E. coli, the high-abundance receptors in S. meliloti are lacking the carboxy-terminal NWETF pentapeptide that binds the CheR methyltransferase and CheB methylesterase. Using transcriptional lacZ fusions, we showed that chemoreceptors are positively controlled by the master regulators of motility, VisNR and Rem. In addition, FlbT, a class IIA transcriptional regulator of flagellins, also positively regulates the expression of most chemoreceptors except for McpT and McpY, identifying chemoreceptors as class III genes. Taken together, these results demonstrate that the chemosensory complex and the adaptation system in S. meliloti deviates significantly from the established enterobacterial paradigm but shares some similarities with B. subtilis.
IMPORTANCE The symbiotic soil bacterium Sinorhizobium meliloti is of great agricultural importance because of its nitrogen-fixing properties, which enhances growth of its plant symbiont, alfalfa. Chemotaxis provides a competitive advantage for bacteria to sense their environment and interact with their eukaryotic hosts. For a better understanding of the role of chemotaxis in these processes, detailed knowledge on the regulation and composition of the chemosensory machinery is essential. Here, we show that chemoreceptor gene expression in S. meliloti is controlled through the main transcriptional regulators of motility. Chemoreceptor abundance is much lower in S. meliloti than in Escherichia coli and Bacillus subtilis. Moreover, the chemoreceptor-to-kinase CheA ratio is different from that of E. coli but similar to that of B. subtilis.
KEYWORDS: alfalfa, chemoreceptors, flagellar motor, plant symbiosis, transcriptional control
INTRODUCTION
Chemotaxis is a mechanism by which bacteria rapidly respond to their immediate environment, ultimately moving toward favorable niches and away from repellents (1–3). It has been implicated in various bacterial processes, like pathogenicity, nodulation, and biofilm production (4, 5). Chemotaxis has been most widely studied in the enterobacterium Escherichia coli, which swims in series of runs and tumbles through the rotation of peritrichous flagella. During runs, flagella rotate counterclockwise (CCW), leading to the formation of a flagellar bundle with synchronized rotation. In E. coli, tumbles are achieved by reverting the rotation of one or more flagella in the clockwise (CW) direction, which causes the flagellar bundle to splay apart and randomly reorients the cell in three-dimensional space (6). However, the flagellar motors of certain bacterial species exhibit only unidirectional rotation. In the case of Rhodobacter sphaeroides, pausing rotation of the single polar flagellum causes the cell to tumble, while resuming flagellar rotation results in a straight run (7). In Sinorhizobium meliloti, the asynchrony caused by the slowing down of one or more of the strictly CW rotating flagella results in a tumble (8–11).
Bacterial chemotaxis is accomplished through the sensing of environmental signals by chemoreceptors called methyl-accepting chemotaxis proteins (MCPs). E. coli has four transmembrane MCPs: Tap, Tar, Trg, and Tsr. While Tap senses dipeptides, Tar mediates taxis toward aspartate and maltose, Trg recognizes ribose and galactose, and Tsr responds to serine and 3,4-dihydroxymandelic acid, a catabolite of norepinephrine (12–14). A fifth receptor, Aer, which is anchored to the inside the cytoplasmic membrane, acts as an oxygen sensor (13, 14). The number of different chemoreceptors varies within species. While Mesorhizobium loti has only one putative chemoreceptor gene, Vibrio cholerae has 45 (15). Chemoreceptors form stable homodimers that are in turn arranged in trimers. These trimers of dimers, along with other cytoplasmic chemotaxis proteins, are packaged in large hexagonal arrays called chemoreceptor clusters. MCPs typically consist of a periplasmic ligand-binding domain, two membrane-spanning helices, and a cytoplasmic signaling domain. Binding of ligand to the periplasmic domain causes a piston-like movement through the transmembrane domains to the cytoplasmic domain, where it acts as a signal to the cytosolic chemotaxis proteins (14, 16). While the periplasmic domains of MCPs are vastly diverse to accommodate various ligands, the cytoplasmic signaling domains are highly conserved, even among different species (3). A two-component system using the histidine-aspartate phosphorelay mediates the chemotactic signal transduction from the chemoreceptor cluster to the flagellar motors. The first component, CheA, is the histidine kinase that binds to the cytoplasmic domain of the MCPs via a coupling protein, CheW. The second component, CheY, is the response regulator that interacts with the flagellar motor complex, thereby controlling its rotation. Binding of an attractant to the periplasmic domain of an MCP induces a conformational change in the cytoplasmic domain that inhibits the autophosphorylation of CheA. In E. coli, when CheA is inactive and no signal is being passed to the flagellar motors, continued CCW rotation results in a run. In the absence of a bound attractant or presence of a repellent, ATP-dependent CheA autophosphorylation is stimulated. Phosphorylated CheA (CheA-P) transfers the phosphate group to a conserved aspartate residue in CheY (2, 3). CheY-P interacts with FliM of the flagellar motor complex and signals the motor to switch to the CW direction, subsequently resulting in a tumble (17, 18). In E. coli, CheZ is a phosphatase which increases the dephosphorylation rate of CheY-P and thereby allows for signal termination (19–21). An adaptation system involving CheR and CheB is employed for increased sensitivity and real-time modulation of chemotactic activity based on the local environment. CheR is a methyltransferase that constitutively adds methyl groups to conserved sites on the cytoplasmic signaling domain of MCPs. CheB acts as a methylesterase and is activated through its phosphorylation by CheA-P (3, 15). In E. coli, only the highly abundant MCPs Tar and Tsr have a conserved pentapeptide, NWETF, at their carboxy termini, which serves as the site for CheR and CheB docking (22). The concerted addition and removal of methyl groups by CheR and CheB, respectively, brings about the conformational changes in MCPs required for the resetting and adaptation of the chemotaxis system (23, 24).
The importance of bacterial chemotaxis in establishing symbiosis with plant hosts has been well documented for members of the Rhizobiaceae family, including S. meliloti (25–28). Recent studies of S. meliloti motility and chemotaxis have uncovered marked deviations from the enterobacterial paradigm (9, 29). Unlike E. coli, S. meliloti not only has six transmembrane chemoreceptors (McpT to McpX and McpZ) but also has two soluble cytosolic receptors (McpY and IcpA). The size of the ligand-binding domains of S. meliloti chemoreceptors varies greatly between 160 and 390 amino acid (aa) residues (30). Our group has shown that McpU and McpX play a role in host interaction by sensing plant-derived amino acids and quaternary ammonium compounds, respectively (31–34). However, the function of the remaining six chemoreceptors is not known. Furthermore, S. meliloti does not utilize a CheZ phosphatase but employs an indirect phosphate sink mechanism for signal termination. Here, phosphate groups from the response regulator CheY2 are shuttled back via CheA to an additional response regulator protein, CheY1 (35). S. meliloti involves three additional proteins in chemotaxis not seen with E. coli, namely, CheD, CheS, and CheT (36). A CheD analog in Bacillus subtilis serves as an MCP deamidase and thus plays a role in adaptation (37). However, it is unclear whether S. meliloti CheD exerts a similar function. CheS and CheT have no homologs in enteric bacteria but display homology to some unassigned proteins in other alphaproteobacteria, like Caulobacter crescentus and Rhizobium leguminosarum. CheS enhances the phosphate flow from CheA-P to CheY1 by increasing the affinity between CheA-P and CheY1 by 100-fold (38). The one kinase-two response regulator system and presence of an auxiliary protein allows the implementation of a tunable switch-like signal processing (39). The function of CheT in chemotaxis is currently unknown.
Until now, two studies had investigated the cellular quantities of bacterial chemotaxis proteins, one in E. coli and one in B. subtilis (40, 41). While total protein amounts may change depending on growth conditions and nutrient availability, cellular ratios of chemotaxis proteins were fairly robust. Both studies also revealed that ratios between certain proteins, such as CheA and CheW, remained constant, while the ratio of others, such as CheA and the MCPs, differed greatly between species (40, 41). To gain a better understanding of deviations evolved in the S. meliloti chemotaxis system and to understand how the system is tuned for optimum performance and sensitivity, we determined the cellular amounts and ratios of CheA and all eight chemoreceptors in S. meliloti using quantitative immunoblotting. Furthermore, we explored the regulation of chemoreceptor gene expression within the flagellar gene hierarchy. Ultimately, computational models can be used to simulate the interactions of all chemotaxis proteins and to evaluate the chemotaxis system holistically under various physiologically relevant conditions (42).
RESULTS
The mcp genes are part of the flagellar regulon and transcribed as class III genes.
Previously, we have shown that all genes in the flagellar gene cluster are organized in a four-class hierarchy (43, 44). The LuxR-type VisNR and the OmpR-like Rem act as class IA and IB transcriptional regulators, respectively. They control the expression of class II (comprising flagellar assembly and motility genes) and class III (comprising flagellin and chemotaxis genes), which requires class IIA for expression (43). FlbT is a class IIA positive flagellar regulator (45). While icpA is the first gene of the che operon and therefore classified as a class III gene, the regulation of the remaining seven chemoreceptor genes is unknown. With the exception of mcpW, which is cotranscribed with a putative cheW, all other mcp genes are monocistronic and scattered throughout the genome (30, 46). To answer whether expression of mcp genes follows the same control mechanisms, we transferred vectors with translational fusions of six of the mcp promoters and of the promoter of the che operon as a control (30) to RU11/001 (WT), RU11/814 (ΔvisNR), RU11/555 (Δrem), and RU13/110 (ΔflbT) strains and assayed for ß-galactosidase activity as listed in Table 1. We found that all three genes were required for the transcription of chemoreceptor genes. Only two of the genes with weaker promoters, namely, mcpT and mcpY, exhibited some residual transcriptional activity in ΔvisNR and Δrem strains and about 80% activity in the ΔflbT strain compared to those measured in the wild type. In conclusion, mcp genes are part of the flagellar regulon and positively regulated by its master transcriptional regulators with a certain degree of decoupling for mcpT and mcpY.
TABLE 1.
In vivo mcp promoter activities in WT and ΔvisN/R, Δrem, and ΔflbT mutant strains
| Plasmida (lacZ fusion) | β-Galactosidase activityb (Miller units) for strain: |
|||
|---|---|---|---|---|
| RU11/001c (WT) | RU11/814 (ΔvisNR) | RU11/555 (Δrem) | RU13/110 (ΔflbT) | |
| pRU2728 (mcpT) | 42 | 7 | 7 | 35 |
| pRU2283 (mcpU) | 235 | 0 | 0 | 4 |
| pRU2784 (mcpW) | 127 | 2 | 0 | 6 |
| pRU2994 (mcpX) | 417 | 0 | 0 | 6 |
| pRU2898 (mcpY) | 29 | 13 | 12 | 23 |
| pRU2787 (mcpZ) | 154 | 0 | 0 | 17 |
| pRU2250 (icpA = che) | 156 | 0 | 0 | 25 |
Transcription from nine chemoreceptor promoters was assessed with plasmid-borne lacZ fusions in wild-type (RU11/001), ΔvisN/R (RU11/814), Δrem (RU11/555), and DflbT (RU13/110) strains during exponential growth. Cells diluted in RB were layered on Bromfield agar plates and grown to an OD600 of 0.15 to 0.25. The che operon (che) is composed of the genes icpA, orf2, cheY1, cheA, cheW, cheR, cheB, cheY2, cheD, and orf10.
β-Galactosidase activities (47) of three to five independent experiments were averaged. Standard deviations were between 0.5 and 6%.
Values for the wild type were taken from the work of Meier et al. (30).
Quantification of transmembrane chemoreceptors.
To quantify the six transmembrane chemoreceptors (McpT to McpX and McpZ), we purified and raised polyclonal antibodies against the periplasmic ligand-binding region of each MCP. This way, we avoided generating antibodies targeting the highly conserved cytosolic domains and obviate cross-reactivity with other chemoreceptors in the cell extracts. An important factor for attaining consistent results during quantitative immunoblot analysis is the choice of the appropriate growth phase for cell harvest. In S. meliloti, expression of flagellar and chemotaxis genes (including the che operon) is under tight transcriptional control through the activity of a class IB regulator, Rem (43). Furthermore, it has been shown previously that chemoreceptors in S. meliloti follow the expression pattern of Rem and are maximally expressed at mid-exponential phase (48). Thus, an OD600 of 0.25 was selected for harvesting cells for immunoblotting (30). Standard curves were established by adding various amounts of the purified periplasmic regions of each MCP to cell extracts of corresponding deletion strains. Signals from immunoblots were detected using X-ray films with different exposures, and band intensities were determined with ImageJ.
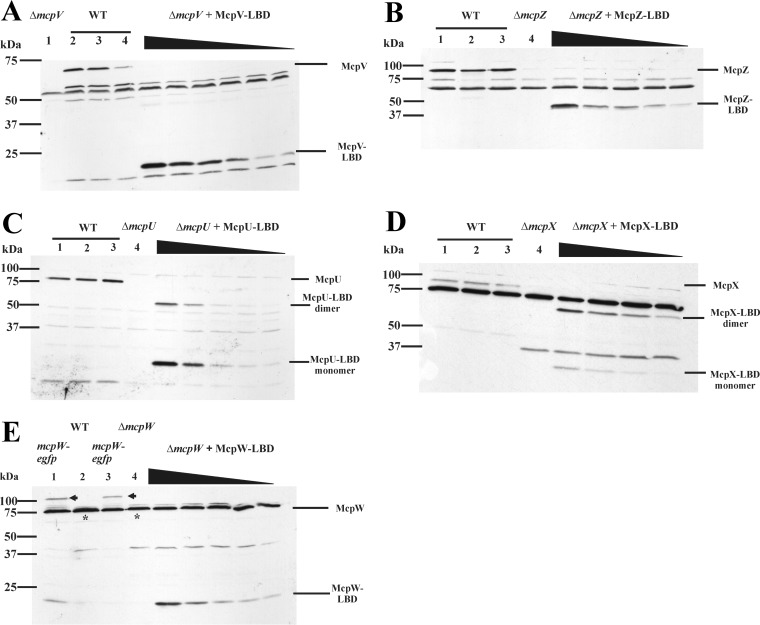
A representative blot for McpV (65.34 kDa) showed a distinct band below the 75-kDa marker in lanes 2 to 4, containing wild-type cell extracts, which is markedly absent from lane 1, containing the mcpV deletion cell extract (Fig. 1A). McpV-LBD (20.23 kDa), which was used to create a standard curve, can be seen below the 25-kDa marker in the lanes containing mcpV deletion cell extracts and purified McpV-LBD in decreasing amounts. A similar blot is seen for McpZ (90.15 kDa), with various amounts of McpZ-LBD (45.97 kDa) added to extracts from the mcpZ deletion strain (Fig. 1B).
FIG 1.
Representative immunoblots used to quantify transmembrane chemoreceptors. (A) McpV. Lane 1 (ΔmcpV) contains RU11/830 (mcpV deletion strain) cell lysate from 1 ml culture at an OD600 of 0.25. Lanes 2 to 4 contain RU11/001 (WT) cell lysates from 1 ml of culture at an OD600 of 0.25. ΔmcpV + McpV-LBD lanes contain purified McpV-LBD (7.5, 5.0, 4.0, 3.0, 2.0, and 1.0 ng) mixed with RU11/830 cell lysates. Representative immunoblots were used to quantify transmembrane chemoreceptors. (B) McpZ. Lanes 1 to 3 contain RU11/001 (WT) cell lysates from 1 ml of culture at an OD600 of 0.25. Lane 4 (ΔmcpZ) contains RU11/818 (mcpZ deletion strain) cell lysate from 1 ml culture at an OD600 of 0.25. ΔmcpZ + McpZ-LBD lanes contain purified McpZ-LBD (2.6, 1.3, 0.78, 0.52, and 0.26 ng) mixed with RU11/830 cell lysates. (C) McpU. Lanes 1 to 3 contain RU11/001 (WT) cell lysates from 1 ml of culture at an OD600 of 0.25. Lane 4 (ΔmcpU) contains RU11/828 (mcpU deletion strain) cell lysate from 1 ml culture at an OD600 of 0.25. ΔmcpU + McpU-LBD lanes contain purified McpU-LBD (2.0, 1.0, 0.5, 0.1, and 0.05 ng) mixed with RU11/828 cell lysates. McpU-LBD exists in monomeric and dimeric forms, as indicated. (D) McpX. Lanes 1 to 3 contain RU11/001 (WT) cell lysates from 1 ml of culture at an OD600 of 0.25. Lane 4 (ΔmcpX) contains RU11/805 (mcpX deletion strain) cell lysate from 1 ml culture at an OD600 of 0.25. ΔmcpX + McpX-LBD lanes contain purified McpX-LBD (1.0, 0.75, 0.5, and 0.25 ng) mixed with RU11/805 cell lysates. McpX-LBD exists in monomeric and dimeric forms, as indicated. (E) McpW. Lanes 1 and 3 contain RU13/143 (mcpW-egfp) cell lysates from 1 ml of culture at an OD600 of 0.25. Lane 2 contains RU11/001 (WT) cell lysates from 1 ml of culture at an OD600 of 0.25. Lane 4 (ΔmcpW) contains RU11/803 (mcpW deletion strain) cell lysate from 1 ml culture at an OD600 of 0.25. ΔmcpW + McpW-LBD lanes contain purified McpW-LBD (0.5, 0.25, 0.1, 0.075, and 0.05 ng) mixed with RU11/803 cell lysates. The intensity of the McpW-GFP band in lanes 1 and 3 (arrows) is equal to the difference in intensities between the band in lane 2 (asterisk) and its corresponding nonspecific band in lane 4.
Purified McpU-LBD (27.66 kDa) and McpX-LBD (32.55 kDa) were used to quantify the corresponding proteins (McpU, 74.38 kDa; McpX, 83.73 kDa) in S. meliloti wild-type cell extracts. Although the proteins were separated under denaturing conditions and a reducing agent was added to the loading buffer, both proteins existed in monomeric and dimeric forms (Fig. 1C and D). The McpU-LBD standard curve showed that the ratio of monomer to dimer was approximately 80:20. In contrast, McpX-LBD mainly existed as a dimer, with the ratio of monomer to dimer being approximately 20:80. Thus, the intensities of both bands, monomer and dimer, were added for the quantification of McpU and McpX.
For immunoblots probed with anti-McpW antibodies, McpW-LBD (15.96 kDa) was used as a standard. We observed that the McpW (72.50 kDa) band in wild-type cell extracts overlapped with a cross-reacting band of similar size, as indicated in Fig. 1E. This produced a more intense band, as marked by an asterisk, compared to lane 4, containing the mcpW deletion strain extract. To confirm that the additional band intensity was caused by McpW, we loaded extracts of strain RU13/143 expressing McpW-enhanced green fluorescent protein (eGFP) from its native chromosomal locus in lanes 1 and 3. McpW-eGFP (104.84 kDa) appeared above the 100-kDa marker band, and the band intensity of the 75-kDa band was decreased to that of the mcpW deletion strain (Fig. 1E, lane 4). We next subtracted the 75-kDa band appearing in the mcpW deletion strain extract from the 75-kDa band of the wild-type extract to quantify McpW. Additionally, we quantified the band intensity of McpW-eGFP. Both quantification methods yielded the same amount of McpW.
To quantify the number of MCP molecules per cell, we determined the number of S. meliloti cells in 2 ml of cell culture grown in minimal medium at an optical density at 600 nm (OD600) of 0.25. Using serial dilutions and spread plating, we determined that 1 ml cell culture of S. meliloti at an OD600 of 0.25 contained 2.56 × 108 ± 0.31 × 108 cells. For the six transmembrane chemoreceptors, the numbers ranged from a few molecules to several hundred per cell (Table 2). However, we were unable to quantify McpT. Neither crude serum nor affinity-purified antibodies raised against McpT-LBD allowed detection of a band corresponding to purified McpT-LBD or McpT in wild-type cell extracts. As an alternative strategy for quantifying McpT, extracts of a strain expressing McpT-eGFP from its native chromosomal locus (RU13/142) were probed using anti-eGFP antibodies. We were able to detect purified eGFP at amounts as small as 5 pg. No corresponding bands were detected in extracts of the McpT-eGFP strain. In conclusion, of the five transmembrane receptors quantified, McpV was the most abundant chemoreceptor, being present in more than 6-fold higher numbers than the next most abundant receptor, McpU (Table 2). The other receptors followed the order McpU > McpX > McpW > McpZ.
TABLE 2.
Cellular content of chemoreceptors and CheA chemotaxis protein contents in S. meliloti strain RU11/001
| Protein | No. of molecules/cella |
|---|---|
| McpT | BDL |
| McpU | 47 ± 6 |
| McpV | 299 ± 55 |
| McpW | 17 ± 4 |
| McpX | 39 ± 7 |
| McpY | 1 ± 1 |
| McpZ | 3 ± 1 |
| IcpA | 17 ± 6 |
| Receptor total | 423 ± 56 |
| CheA | 18 ± 5 |
Means ± standard deviations are reported. Values were obtained from six independent immunoblots. BDL, below detection limit.
Quantification of cytosolic chemoreceptors.
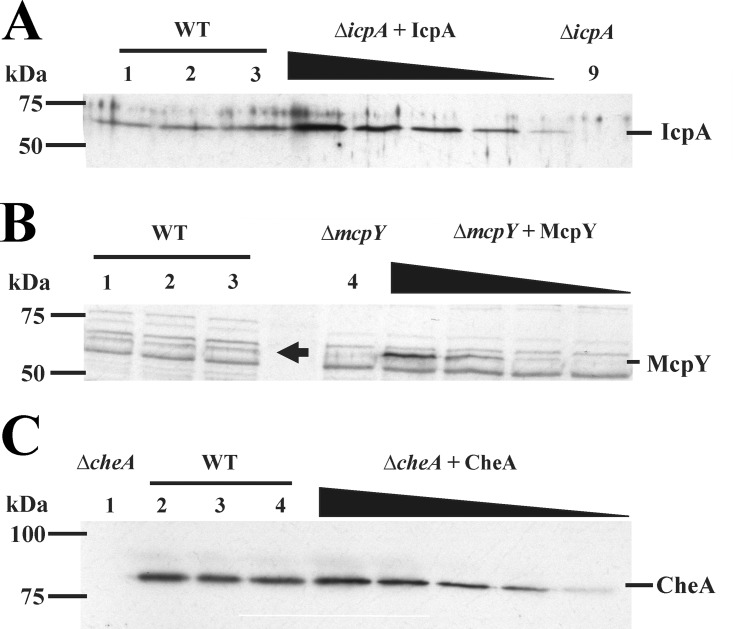
For the two cytosolic chemoreceptors, IcpA (57.54 kDa) and McpY (64.30 kDa), full-length proteins were used to serve as controls for a standard curve, because antibodies raised against these proteins have been generated previously (30). While the IcpA antibodies had few cross-reactivities with other proteins in the cell lysates, the McpY antibody reacted nonspecifically with a number of proteins in the lysates (Fig. 2A and B). IcpA was present at levels comparable to those of McpW. The amounts of McpY were the smallest of any chemoreceptor that could be quantified (Table 2).
FIG 2.
Representative immunoblot used to quantify cytosolic receptors and CheA. (A) McpY. Lanes 1 to 3 contain RU11/001 (WT) cell lysates from 1 ml of culture at an OD600 of 0.25. Lane 4 (ΔmcpY) contains RU11/804 (mcpY deletion strain) cell lysate from 1 ml culture at an OD600 of 0.25. ΔmcpY + McpY lanes contain purified McpY (0.1, 0.05, 0.01, and 0.005 ng) mixed with RU11/804 cell lysates. The arrow indicates the McpY protein in the WT cell lysates. (B) IcpA. Lanes 1 to 3 contain RU11/001 (WT) cell lysates from 1 ml of culture at an OD600 of 0.25. Lane 9 (ΔicpA) contains RU11/815 (icpA deletion) cell lysate from 1 ml culture at an OD600 of 0.25. ΔicpA + IcpA lanes contain purified IcpA (1.28, 0.96, 0.64, 0.32, and 0.16 ng) mixed with RU11/815 cell lysates. (C) CheA. Lane 1 (ΔcheA) contains RU11/310 (cheA deletion strain) cell lysate from 1 ml culture at an OD600 of 0.25. Lanes 2 to 4 contain RU11/001 (WT) cell lysates from 1 ml of culture at an OD600 of 0.25. ΔcheA + CheA lanes contain purified CheA (1.0, 0.8, 0.6, 0.4, and 0.2 ng) mixed with RU11/310 cell lysates.
Quantification of CheA.
To determine the stoichiometry of chemoreceptors to the kinase CheA and to compare our data with those from the E. coli and B. subtilis studies (40, 41), we quantified the cellular amounts of CheA. Purified full-length CheA (81.12 kDa) and existing polyclonal antibodies were used for quantification (35). A typical blot (Fig. 2C) exhibited a band above the 75-kDa marker in WT cell lysates, which was absent from the lane containing ΔcheA cell lysates. As before, a standard curve was established by adding increasing amounts of the purified full-length CheA protein. CheA was found to be present in low abundance and at levels similar to those of McpW and IcpA (Table 2).
Fluorescence microscopy of eGFP-expressing strains.
The small amounts of chemoreceptor proteins detected in immunoblots raised the question of whether only a subpopulation of cells displayed expression. We chose four representative proteins to assess their expression in individual cells via C-terminal eGFP fusions (48), namely, two transmembrane receptors (McpU and McpV), a cytosolic receptor (IcpA), and the autokinase CheA. We showed previously that all four fusion proteins localize to one or both cell poles (48). Cultures were grown as described for quantitative immunoblots, and cells from three biological replicates per strain were analyzed by fluorescence microscopy in conjunction with computerized image analysis to determine their fluorescent patterns (Table 3). For the four strains, between 24 and 45% of the cells displayed fluorescent foci predominantly at one cell pole. Representative images for each strain are depicted in Fig. 3. Thus, between one-half and three-quarters of cells in a population do not express the chemosensory cluster.
TABLE 3.
Proportion of S. meliloti cells with fluorescent polar foci expressing eGFP fusions from native chromosomal gene loci
| Strain | Protein fusion | % of cells with fluorescent polar focia |
|---|---|---|
| RU13/212 | McpV-eGFP | 24 ± 4 |
| RU13/243 | CheA-eGFP | 44 ± 5 |
| RU13/301 | McpU-eGFP | 28 ± 3 |
| RU13/303 | IcpA-eGFP | 42 ± 1 |
Results are means ± standard deviations. Values were obtained from 5,000 to 6,400 cells in three independent experiments. Wild-type cells had no detectable fluorescent signal.
FIG 3.
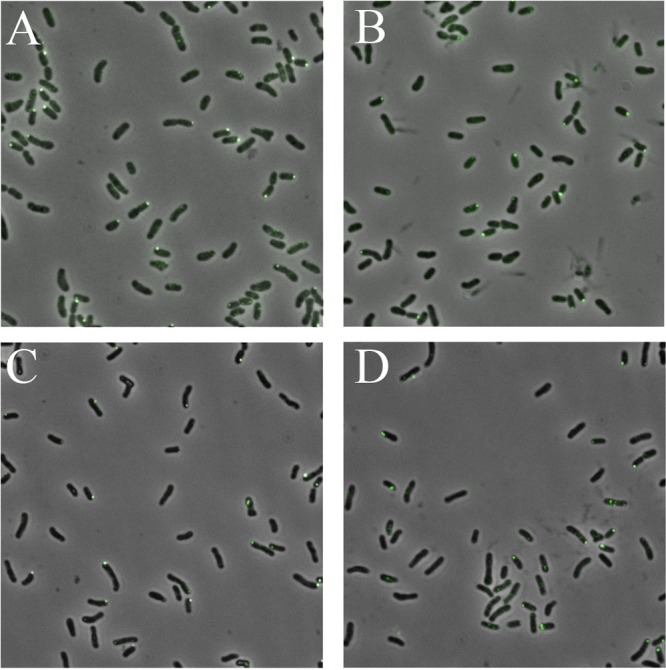
Localization of McpU, McpV, IcpA, and CheA fused to eGFP in S. meliloti cells by fluorescence microscopy. (A) McpU-eGFP; (B) McpV-eGFP; (C) IcpA-eGFP; (D) CheA-eGFP.
DISCUSSION
Chemotaxis is a complex process, which ultimately aids in the survival of bacteria in a rapidly changing environment. Since it consumes considerable amounts of the cell's energy, tight regulation of the expression of all components of the chemotaxis machinery is required (49). Our group has shown previously that motility and chemotaxis genes in the S. meliloti flagellar regulon are expressed in a transcriptional hierarchy (43). Our current studies expand the previously established scheme by including the mcp genes as class III genes and FlbT (class IIA) as a positive regulator for class III genes (with mcpT and mcpY as exceptions). Similar to the coordinated regulation in enterobacteria (50), class III gene expression is dependent on the completion of basal body structure and flagellar export. This suggests that a control mechanism comparable to the one operating in enterobacteria exists in S. meliloti. Expression of chemotaxis and motility genes not only is dependent on growth phase and temperature but also can vary based on the richness of the medium. While enterobacteria are typically motile in rich media (51), S. meliloti exerts higher motility under nutrient-poor conditions (52). Furthermore, it remains to be seen whether expression of chemoreceptors is induced by their respective chemoeffector, as shown for certain C and N sources in Pseudomonas putida (53).
A number of different chemotaxis proteins have to interact to bring about the desired changes in motility. The cellular amounts of these proteins must be at the exact levels tuned for optimal performance. We set out to quantify the amounts of chemoreceptors and the histidine kinase CheA in an S. meliloti cell. The total molecule amount of all eight chemoreceptors in an S. meliloti cell is 423 ± 56 (Table 2). This is extremely small compared to 59,960 ± 5,960 in B. subtilis and 26,000 ± 1,800 in E. coli (40, 41). From the viable cell counts and dry weight analyses, it is evident that an S. meliloti cell is about half the size of an E. coli cell (18). The smaller size may enable the S. meliloti cell to function efficiently with a smaller total amount of chemoreceptors. Additionally, not all cells expressed detectable quantities of eGFP fusion proteins under these culture conditions (Table 3). Therefore, it is possible that amounts for those cells expressing chemotaxis proteins are actually two to four times larger. We also estimated the percentage of motile cells in the population and found that about 30% of the cells are motile (data not shown), which correlates with the percentage of cells with fluorescent foci. However, this observation does not have an effect on the determined protein ratios.
In E. coli, the chemoreceptors are present in high (Tsr and Tar) and low (Trg, Tap, and Aer) abundances (22, 40). In S. meliloti, one chemoreceptor, McpV, is present at high levels, four are present in small amounts (McpU, McpW, McpX, and IcpA), two are present at extremely low abundance (McpY and McpZ), and one was not detectable (McpT). The approximate ratios of receptors are 1 (McpY and McpZ) to 15 (McpU, McpW, McpX, and IcpA) to 150 (McpV). The high abundance of McpV correlates with its CheA- and CheW-independent localization in S. meliloti (48). It can be hypothesized that the abundance of McpV enables its localization at the pole independently of other chemotaxis proteins and, in fact, that it may act as a scaffold receptor to recruit other receptors to localize at the poles. Interestingly, when observing the localization of McpV-eGFP through cell division, new clusters are formed in the midcell near the septum of the new daughter cells (54). A similar behavior has been described for E. coli chemoreceptors, which localize to future division sites (55). The function of McpV in sensing environmental cues is not known and is the subject of current investigations. However, we identified the low-abundance receptors McpU and McpX as general amino acid and quaternary ammonium compound sensors, respectively (31–33). In B. subtilis, the major receptors for taxis toward all amino acids and sugars, McpB and McpC, are also present in relatively low cellular abundance (41). This feature might be commonplace in soil bacteria, as it is not seen in E. coli, where the high-abundance receptor Tsr mediates taxis toward serine and Tar mediates taxis toward aspartate and maltose (22).
Finally, two of the three receptors that are present in very low numbers (McpT and McpY) appear to be regulated differently from the other receptors. Although their expression is partially dependent on VisNR and Rem, the transcription of their genes is not controlled by the class IIA regulator FlbT. Furthermore, we have preliminary data showing that both of their genes exhibit 30 to 40% residual expression in alfalfa root nodules while all other chemoreceptors are not expressed (data not shown). Due to these differences, we speculate that McpT and McpY play a role in planta, which could explain their extremely low expression levels under our assay conditions.
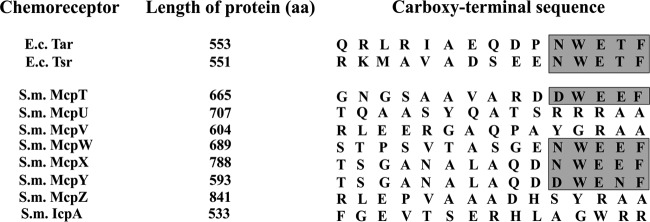
In E. coli, the presence of the carboxy-terminal pentapeptide, representing the CheR and CheB binding site, correlates with receptor abundance (56, 57). Despite the absence of this binding motif from low-abundance receptors of E. coli, receptor methylation and demethylation occur efficiently due to assistance by high-abundance receptors within chemoreceptor clusters (58). In S. meliloti, the most abundant receptor, McpV, is lacking the motif. Instead, only McpT, McpW, McpX, and McpY, which are low- or extremely-low-abundance chemoreceptors, possess the conserved carboxy-terminal pentapeptide (Fig. 4) (30). The adaptation process in S. meliloti has not been investigated, and the composition of chemoreceptor arrays is unknown. Therefore, the reason for the opposite correlation of receptor abundance and presence of the CheR/CheB binding motif remains to be elucidated.
FIG 4.
Sequence comparison of the 15 C-terminal amino acid residues in the NWETF motif-containing E. coli (E.c.) receptors Tar and Tsr and all eight S. meliloti (S.m.) chemoreceptors. The conserved pentapeptide sequence is marked in gray.
The ratio of chemoreceptors to CheA in S. meliloti is approximately 23.5:1. This ratio is lower in E. coli (3.4:1) but is at the same level in B. subtilis (23:1) (40, 41). Presumably, this difference in chemoreceptor-to-CheA ratio reflects the more variable biotopes of soil bacteria (bulk soil versus rhizosphere) compared to those of gut bacteria. It remains to be investigated whether the structure of the CheA-CheW receptor complex is different in S. meliloti from the one described for E. coli (59). Apparently, ratios of chemotaxis proteins across genera are adapted and optimized according to their lifestyles. Our ongoing efforts to determine the amounts of cytosolic chemotaxis proteins would provide us with a snapshot of chemotaxis protein stoichiometry in S. meliloti. This in turn would shed more light on the various deviations of the S. meliloti chemosensory system from the enterobacterial paradigm and their corresponding benefits to the different (free-living versus symbiotic) life styles of S. meliloti.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Derivatives of E. coli K-12 and S. meliloti MV II-1 and the plasmids used are listed in Table 4. RU11/001 is a spontaneous streptomycin-resistant derivative of MVII-1 (65).
TABLE 4.
Bacterial strains and plasmids
| Strain/plasmid | Relevant characteristic | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm λ (DE3) | Novagen |
| ER2566 | lon ompT lacZ::T7 | NEB |
| S17-1 | recA endA thi hsdR RP4-2 Tc::Mu::Tn7 Tpr Smr | 60 |
| S. meliloti | ||
| RU11/001 | Smr, spontaneously streptomycin-resistant wild-type strain | 52 |
| RU11/310 | Smr ΔcheA | 61 |
| RU11/555 | Smr Δrem | 43 |
| RU11/803 | Smr ΔmcpW | 30 |
| RU11/804 | Smr ΔmcpY | 30 |
| RU11/805 | Smr ΔmcpX | 30 |
| RU11/814 | Smr ΔvisNR | 44 |
| RU11/815 | Smr ΔicpA | 30 |
| RU11/818 | Smr ΔmcpZ | 30 |
| RU11/828 | Smr ΔmcpU | 30 |
| RU11/830 | Smr ΔmcpV | 30 |
| RU11/838 | Smr ΔmcpT | 30 |
| RU13/142 | Smr mcpT-egfp | 48 |
| RU13/143 | Smr mcpW-egfp | 48 |
| RU13/243 | Smr cheA-egfp | 48 |
| RU13/301 | Smr mcpU-egfp | 48 |
| RU13/303 | Smr icpA-egfp | 48 |
| RU13/310 | Smr ΔflbT | This study |
| Plasmids | ||
| pHT28 | Apr, expression vector for E. coli fliM | 62 |
| pK18mobsacB | Kmr lacZ mob sacB | 63 |
| pKLD66 | Apr, expression vector | 64 |
| pTYB1 | Apr, expression vector | NEB |
| pTYB11 | Apr, expression vector | NEB |
| pBS352 | Apr, 858-bp NdeI/SapI PCR fragment containing periplasmic domain of mcpX cloned into pTYB1 | This study |
| pBS353 | Apr, 741-bp NdeI/SapI PCR fragment containing periplasmic domain of mcpU cloned into pTYB1 | This study |
| pBS409 | Apr, 474-bp SapI/PstI PCR fragment containing periplasmic domain of mcpV cloned into pTYB11 | This study |
| pBS426 | Apr, 1,218-bp SapI/SpeI PCR fragment containing periplasmic domain of mcpZ cloned into pTYB11 | This study |
| pBS487 | Apr, 1,602-bp KpnI/HindIII PCR fragment containing icpA cloned into pKLD66 | This study |
| pBS1030 | Apr, 444-bp NdeI/PstI PCR fragment containing periplasmic domain of mcpT cloned into pTYB1 | This study |
| pBS1031 | Apr, 423-bp NdeI/PstI PCR fragment containing periplasmic domain of mcpW cloned into pTYB1 | This study |
| pRU2250 | Tcr, icpA (1,974 bp)-lacZ (che) fusion cloned into pPHU236 | 30 |
| pRU2782 | Tcr, mcpT (320 bp)-lacZ fusion cloned into pPHU235 | 30 |
| pRU2783 | Tcr, mcpU (456 bp)-lacZ fusion cloned into pPHU236 | 30 |
| pRU2784 | Tcr, mcpW (303 bp)-lacZ fusion cloned into pPHU236 | 30 |
| pRU2787 | Tcr, mcpZ (409 bp)-lacZ fusion cloned into pPHU236 | 30 |
| pRU2790 | Apr, 1,779-bp KpnI/PstI PCR fragment containing mcpY replacing E. coli fliM in pHT28 | 30 |
| pRU2898 | Tcr, mcpY (786 bp)-lacZ fusion cloned into pPHU236 | 30 |
| pRU2994 | Tcr, mcpX (590 bp)-lacZ fusion cloned into pPHU236 | 30 |
Media and growth conditions.
E. coli strains were grown in lysogeny broth (LB) (66) at the indicated temperatures. S. meliloti strains were grown in TYC (0.5% tryptone, 0.3% yeast extract, 0.13% CaCl2 · 6H2O [pH 7.0]) (67) or SMM (Sinorhizobium motility medium; RB [6.1 mM K2HPO4, 3.9 mM KH2PO4, 1 mM MgSO4, 1 mM (NH4)2SO4, 0.1 mM CaCl2, 0.1 mM NaCl, 0.01 mM Na2MoO4, 0.001 mM FeSO4, 2 μg/liter biotin] [68], 0.2% mannitol, 2% TY) (43). Motile cells for immunoblotting and fluorescence microscopy were grown in SMM for 2 days, diluted to an OD600 of 0.02, and incubated at 30°C to an OD600 of 0.25. The following antibiotics were used in their final concentrations: for E. coli, ampicillin at 100 μg/ml, kanamycin at 50 μg/ml, and tetracycline at 10 μg/ml; for S. meliloti, neomycin at 120 μg/ml, streptomycin at 600 μg/ml, and tetracycline at 10 μg/ml.
Genetic and DNA manipulations.
S. meliloti DNA was isolated and purified as described previously (61). Plasmid DNA, DNA fragments, or PCR products were purified according to the manufacturers' instructions, and PCR amplification of chromosomal DNA was carried out according to published protocols (61). The flbT strain was generated in vitro by overlap extension PCR as described previously (69). Constructs containing the mutations were cloned into the mobilizable suicide vector pK18mobsacB, used to transform E. coli S17-1, and conjugally transferred to S. meliloti by filter mating (60, 70). Allelic replacement was achieved by sequential selections on neomycin and 10 or 15% sucrose as described previously (61). Confirmation of allelic replacement and elimination of the vector was obtained by gene-specific primer PCR and DNA sequencing. Derivatives of the broad-host-range plasmids pPHU235 and pPHU236 were used to transform E. coli S17-1 and conjugally transferred to S. meliloti by streptomycin-tetracycline double selection as described above (71).
β-Galactosidase assays.
Cultures of S. meliloti containing lacZ fusions grown on overlayered Bromfield agar plates were sampled, diluted 1:1 in Z buffer (47), permeabilized with 1 drop of toluene, and assayed for β-galactosidase activity by the method of Miller (47) as previously described (30).
Purification of recombinant proteins.
McpT-LBD (ligand-binding domain) (aa 30 to 178) was expressed from pBS1030, McpU-LBD (aa 40 to 287) from pBS353, McpV-LBD (aa 31 to 189) from pBS0409, McpW-LBD (aa 39 to 180) from pBS1031, McpX-LBD (aa 34 to 320) from pBS352, McpZ-LBD (aa 39 to 445) from pBS426, and CheA from pBS57 in E. coli ER2566 (Table 4), as described by Riepl et al. (72). Briefly, cells were grown to an OD600 of 0.6 to 0.8 at 37°C in LB, and expression was induced by 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 16°C for 16 h. Cells were harvested, suspended in IMPACT buffer (500 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], 20 mM Tris-HCl, pH 8.0), and lysed by three passages through a French pressure cell at 20,000 lb/in2 (SLM Aminco, Silver Spring, MD). A modified IMPACT buffer (2 M NaCl, 1 mM EDTA, 1 mM PMSF, 20 mM Tris-HCl, pH 8.0) along with Halt protease inhibitor cocktail (Life Technologies) was used for CheA purification. The soluble fraction was loaded on a chitin-agarose (New England Biolabs [NEB], Beverly, MA) column (6 cm by 5 cm), and intein-mediated cleavage was induced by equilibration of the column with IMPACT buffer containing 50 mM dithiothreitol (DTT) and incubation at 4°C for 2 to 3 days. Proteins were eluted with IMPACT buffer, and pooled fractions of each protein were further purified by fast-performance liquid chromatography (FPLC; Äktaprime plus; GE Healthcare) gel filtration on HiPrep 26/60 Sephacryl S-200 HR (GE Healthcare). The column was equilibrated and developed using 100 mM NaCl, 5% (vol/vol) glycerol, 80 mM Na2HPO4, 20 mM NaH2PO4, pH 7.5, at 0.5 ml/min, and protein-containing fractions were combined.
McpY protein was overproduced in inclusion bodies from plasmid pRU2790 in E. coli BL21(DE3) (Table 4). Cells were grown at 37°C in LB at 300 rpm to an OD600 of 0.6 to 0.8, and expression was induced by 1 mM IPTG. Cells were harvested after 4 h of incubation at 37°C and suspended in 20 ml 0.5 mM EDTA, 20 mM Tris-HCl, pH 7.5, and cell lysates were prepared as described before. The lysate was centrifuged at 55,000 × g and 4°C for 20 min, the soluble fraction was discarded, and the pellet was washed twice with 1% (vol/vol) Triton X-100, 1 mM EDTA, 20 mM Tris-HCl, pH 7.5. Inclusion bodies were suspended in 10 ml denaturation buffer (8 M urea, 5 mM DTT, 50 mM Tris-HCl, pH 8.0) and centrifuged, and the supernatant was filtered through a 0.2-μm cellulose acetate syringe filter. Ten-milliliter samples were subjected to FPLC (Äktaprime plus; GE Healthcare) gel filtration on HiPrep 26/60 Sephacryl S-200 HR (GE Healthcare). The column was equilibrated and developed in denaturation buffer at 0.5 ml/min, and protein-containing fractions were combined. The protein was then refolded by dialysis against a 30-fold volume of 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 20% (vol/vol) glycerol, 50 mM Tris-HCl, pH 8.0, for 24 h at 4°C. Subsequently, dialysis was performed with 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 10% (vol/vol) glycerol, 50 mM Tris-HCl, pH 8.0, for 24 h at 4°C. Lastly, the protein was dialyzed in phosphate-buffered saline (PBS; 100 mM NaCl, 80 mM Na2HPO4, 20 mM NaH2PO4, pH 7.5) for 24 h at 4°C and stored in 5% glycerol at −80°C.
IcpA was overproduced as a fusion protein with 6-histidine-tagged maltose-binding protein (MBP) from pBS487 in E. coli BL21(DE3) (Table 4). Cells were grown to an OD600 of 0.6 to 0.8 at 37°C in LB, and expression was induced by 0.3 mM IPTG. Cells were harvested after 4 h of incubation at 25°C and then suspended in nickel-nitrilotriacetic acid (Ni-NTA) column buffer (500 mM NaCl, 25 mM imidazole, 1 mM PMSF, 20 mM NaPO4, pH 7.4). Cells were lysed by three passages through a French pressure cell at 20,000 lb/in2 (SLM Aminco, Silver Spring, MD). The soluble fraction was loaded onto a 5-ml NTA column (GE Healthcare Life Sciences) charged with Ni2+. Protein was eluted from the column using a linear gradient in Ni-NTA column buffer and elution buffer (500 mM NaCl, 350 mM imidazole, 1 mM PMSF, 20 mM NaPO4, pH 7.0). Fusion protein-containing fractions were pooled, and tobacco etch virus nuclear-inclusion-a endopeptidase (TEV protease) was added to a final concentration of 0.2 mg/ml. After incubation at room temperature for 24 h, the solution was centrifuged at 55,000 × g and 4°C for 30 min. Precipitated IcpA in the pellet fraction was solubilized at room temperature for 25 min in Ni-NTA column buffer containing 50 mM sodium cholate and filtered through a 0.22-μm cellulose acetate syringe filter. To remove any remaining MBP, the filtrate was loaded onto a 5 ml Ni-NTA column, and the IcpA-containing flowthrough fractions were pooled, concentrated, dialyzed against 300 mM NaCl, 50 mM sodium cholate, 5% glycerol, 40 mM Tris-HCl, pH 7.4, and stored at −80°C.
Immunoblotting.
Polyclonal antibodies raised against purified ligand-binding domains of McpT-McpX and McpZ were purified as described previously (67). Briefly, 1 mg of purified protein was separated on a 12.5% acrylamide gel and transferred to a nitrocellulose membrane (Amersham Protran 0.45 NC; GE Healthcare). Proteins were stained on the membrane using 1% Ponceau S, and the protein-containing membrane was cut into pieces (1 cm by 0.5 cm). Membrane pieces then were incubated for 16 h at 4°C with 2 ml of crude serum. The blots were washed three times with PBS–0.1% bovine serum albumin (BSA), twice with PBS–0.1% BSA–0.1% Nonidet P-40, and three times with PBS–0.1% BSA for 5 min per wash step. The specific antibodies were eluted from the membrane by incubating with 750 μl of 0.2 M glycine–HCl, pH 2.5, for 1 min, followed by neutralization with 375 μl of prechilled 1 M potassium phosphate, pH 9.0. The elution was repeated once, and the combined eluates were dialyzed three times against PBS and stored at −80°C.
Samples for immunoblots were prepared as follows. For whole-cell extracts, 1 ml cell culture of RU11/001 at an OD600 of 0.250 ± 0.002 was pelleted and suspended in 15 μl of supernatant and 15 μl of Laemmli buffer (4.5% SDS, 18.75 mM Tris-HCl, pH 6.5, 43.5% glycerin, 0.0125% bromophenol blue, and 5% β-mercaptoethanol). Samples then were boiled at 100°C for 10 min and stored at −30°C. Control samples comprised cell extracts from appropriate deletion strains treated identically. Standard curves were made by adding defined quantities of protein to appropriate deletion strain lysates. Immunoblot analyses were carried out as described previously (44). Briefly, proteins from cell extracts were separated on 12.5% acrylamide gels and then transferred to a 0.45-μm nitrocellulose membrane. The membrane was blocked overnight with 5% nonfat dry milk solution made in PBS–0.1% Tween 20. The blots were probed with a 1:200 dilution of purified antibodies or 1:5,000 dilution of crude serum. Mouse monoclonal anti-GFP was used at a 1:5,000 dilution to detect McpW-eGFP fusion protein. Blots were washed three times with PBS–0.1% Tween 20 and then probed with a 1:5,000 dilution of donkey anti-rabbit horseradish peroxidase-linked whole antibody. The blots were washed three times with PBS–0.1% Tween 20. Detection was performed by chemiluminescence (Amersham ECL Western blotting detection kit or SuperSignal West Femto maximum-sensitivity substrate for McpY) using Hyperfilm ECL (GE Healthcare). Images were captured by using an Epson Perfection 1640SU scanner, and intensities were quantified using ImageJ. Variations caused by improper blotting of proteins and manual error were minimized by only using those blots for quantification that had a standard curve with an R2 value of >0.95.
Protein concentrations were initially determined by the standard Bradford assay using quick-start Bradford 1× dye reagent (Bio-Rad) and a bovine serum albumin standard curve in accordance with the manufacturer's protocol. Amounts of proteins stated in the figure legends were calculated using this method. Accurate protein concentrations were obtained by quantitative amino acid analyses after total acid hydrolysis performed at the Protein Chemistry Laboratory, Texas A&M University.
Fluorescence microscopy.
Motile cells were pelleted and suspended in 15 μl of PBS. Five microliters was placed on a slide coated with poly-l-lysine, a coverslip was placed on top of the cell suspension droplet, and the edges were sealed with acrylic polymer to prevent drying. Images were taken with an Olympus IX71 microscope using a 100×, 1.4-numeric-aperture UPlanSApo objective lens equipped with a charge-coupled-device camera (Photometrics CoolSNAP HQ2CCD) and processed using SoftWorx software (Applied Precision). Fluorescence images of eGFP (excitation, 470 nm) were detected using a fluorescein isothiocyanate (525 nm) filter. Images were analyzed with MicrobeTracker, a MATLAB (MathWorks)-based software package (73).
Determination of dry weight.
Dry weight was determined as previously described (18). Five 25-ml samples were harvested by centrifugation, resuspended in 77 mM ammonium acetate, pH 7.0, transferred to tared Sarstedt tubes, centrifuged, washed in the same buffer, and lyophilized for 3 days. Medium and buffer were prefiltered (0.2 μm). A value of 0.053 ± 0.008 mg/ml of culture (means ± standard deviations [SD] for five determinations) was obtained. The cytoplasmic volume/milligram of dry weight of 1.4 μl was taken from Stock et al. (74).
ACKNOWLEDGMENTS
This study was supported by grant Scha914/2-1/2 from the Deutsche Forschungsgemeinschaft and NSF grant MCB-1253234.
We are indebted to Amanda Sebastian for performing the dry weight measurements, Jorge Escalante-Semerena for providing plasmid pKLD66, Jordan Mancl and Manisha Shrestha for providing TEV protease, Jinny Johnson for her help with the amino acid analyses, and members of the Scharf laboratory for critical reading of the manuscript.
REFERENCES
- 1.Bren A, Eisenbach M. 2000. How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory signal propagation. J Bacteriol 182:6865–6873. doi: 10.1128/JB.182.24.6865-6873.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porter SL, Wadhams GH, Armitage JP. 2011. Signal processing in complex chemotaxis pathways. Nat Rev Microbiol 9:153–165. doi: 10.1038/nrmicro2505. [DOI] [PubMed] [Google Scholar]
- 3.Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol 5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 4.Duan Q, Zhou M, Zhu L, Zhu G. 2013. Flagella and bacterial pathogenicity. J Basic Microbiol 53:1–8. doi: 10.1002/jobm.201100335. [DOI] [PubMed] [Google Scholar]
- 5.Pratt LA, Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 6.Berg HC. 2000. Constraints on models for the flagellar rotary motor. Philos Trans R Soc Lond 355:491–501. doi: 10.1098/rstb.2000.0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armitage JP, Macnab RM. 1987. Unidirectional, intermittent rotation of the flagellum of Rhodobacter sphaeroides. J Bacteriol 169:514–518. doi: 10.1128/jb.169.2.514-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Platzer J, Sterr W, Hausmann M, Schmitt R. 1997. Three genes of a motility operon and their role in flagellar rotary speed variation in Rhizobium meliloti. J Bacteriol 179:6391–6399. doi: 10.1128/jb.179.20.6391-6399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scharf B, Schmitt R. 2002. Sensory transduction to the flagellar motor of Sinorhizobium meliloti. J Mol Microbiol Biotechnol 4:183–186. [PubMed] [Google Scholar]
- 10.Scharf B. 2002. Real-time imaging of fluorescent flagellar filaments of Rhizobium lupini H13-3: flagellar rotation and pH-induced polymorphic transitions. J Bacteriol 184:5979–5986. doi: 10.1128/JB.184.21.5979-5986.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Attmannspacher U, Scharf B, Schmitt R. 2005. Control of speed modulation (chemokinesis) in the unidirectional rotary motor of Sinorhizobium meliloti. Mol Microbiol 56:708–718. doi: 10.1111/j.1365-2958.2005.04565.x. [DOI] [PubMed] [Google Scholar]
- 12.Pasupuleti S, Sule N, Cohn WB, MacKenzie DS, Jayaraman A, Manson MD. 2014. Chemotaxis of Escherichia coli to norepinephrine (NE) requires conversion of NE to 3,4-dihydroxymandelic acid. J Bacteriol 196:3992–4000. doi: 10.1128/JB.02065-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parkinson JS. 2003. Bacterial chemotaxis: a new player in response regulator dephosphorylation. J Bacteriol 185:1492–1494. doi: 10.1128/JB.185.5.1492-1494.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falke JJ, Hazelbauer GL. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci 26:257–265. doi: 10.1016/S0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szurmant H, Ordal GW. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev 68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hazelbauer GL, Falke JJ, Parkinson JS. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci 33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bren A, Eisenbach M. 1998. The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J Mol Biol 278:507–514. doi: 10.1006/jmbi.1998.1730. [DOI] [PubMed] [Google Scholar]
- 18.Scharf BE, Fahrner KA, Turner L, Berg HC. 1998. Control of direction of flagellar rotation in bacterial chemotaxis. Proc Natl Acad Sci U S A 95:201–206. doi: 10.1073/pnas.95.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scharf BE, Fahrner KA, Berg HC. 1998. CheZ has no effect on flagellar motors activated by CheY13DK106YW. J Bacteriol 180:5123–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blat Y, Eisenbach M. 1994. Phosphorylation-dependent binding of the chemotaxis signal molecule CheY to its phosphatase, CheZ. Biochemistry 33:902–906. doi: 10.1021/bi00170a008. [DOI] [PubMed] [Google Scholar]
- 21.McEvoy MM, Bren A, Eisenbach M, Dahlquist FW. 1999. Identification of the binding interfaces on CheY for two of its targets, the phosphatase CheZ and the flagellar switch protein FliM. J Mol Biol 289:1423–1433. doi: 10.1006/jmbi.1999.2830. [DOI] [PubMed] [Google Scholar]
- 22.Feng X, Baumgartner JW, Hazelbauer GL. 1997. High- and low-abundance chemoreceptors in Escherichia coli: differential activities associated with closely related cytoplasmic domains. J Bacteriol 179:6714–6720. doi: 10.1128/jb.179.21.6714-6720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Djordjevic S, Goudreau PN, Xu Q, Stock AM, West AH. 1998. Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain. Proc Natl Acad Sci U S A 95:1381–1386. doi: 10.1073/pnas.95.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anand GS, Goudreau PN, Stock AM. 1998. Activation of methylesterase CheB: evidence of a dual role for the regulatory domain. Biochemistry 37:14038–14047. doi: 10.1021/bi980865d. [DOI] [PubMed] [Google Scholar]
- 25.Caetano-Anolles G, Wall LG, De Micheli AT, Macchi EM, Bauer WD, Favelukes G. 1988. Role of motility and chemotaxis in efficiency of nodulation by Rhizobium meliloti. Plant Physiol 86:1228–1235. doi: 10.1104/pp.86.4.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller LD, Yost CK, Hynes MF, Alexandre G. 2007. The major chemotaxis gene cluster of Rhizobium leguminosarum bv. viciae is essential for competitive nodulation. Mol Microbiol 63:348–362. doi: 10.1111/j.1365-2958.2006.05515.x. [DOI] [PubMed] [Google Scholar]
- 27.Gulash M, Ames P, Larosiliere RC, Bergman K. 1984. Rhizobia are attracted to localized sites on legume roots. Appl Environ Microbiol 48:149–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scharf BE, Hynes MF, Alexandre GM. 2016. Chemotaxis signaling systems in model beneficial plant-bacteria associations. Plant Mol Biol 90:549–559. doi: 10.1007/s11103-016-0432-4. [DOI] [PubMed] [Google Scholar]
- 29.Schmitt R. 2002. Sinorhizobial chemotaxis: a departure from the enterobacterial paradigm. Microbiology 148:627–631. doi: 10.1099/00221287-148-3-627. [DOI] [PubMed] [Google Scholar]
- 30.Meier VM, Muschler P, Scharf BE. 2007. Functional analysis of nine putative chemoreceptor proteins in Sinorhizobium meliloti. J Bacteriol 189:1816–1826. doi: 10.1128/JB.00883-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webb BA, Compton KK, Castaneda Saldana R, Arapov T, Ray WK, Helm RF, Scharf BE. 2017. Sinorhizobium meliloti chemotaxis to quaternary ammonium compounds is mediated by the chemoreceptor McpX. Mol Microbiol 103:333–346. doi: 10.1111/mmi.13561. [DOI] [PubMed] [Google Scholar]
- 32.Webb BA, Helm RF, Scharf BE. 2016. Contribution of individual chemoreceptors to Sinorhizobium meliloti chemotaxis towards amino acids of host and nonhost seed exudates. Mol Plant Microbe Interact 29:231–239. doi: 10.1094/MPMI-12-15-0264-R. [DOI] [PubMed] [Google Scholar]
- 33.Webb BA, Hildreth S, Helm RF, Scharf BE. 2014. Sinorhizobium meliloti chemoreceptor McpU mediates chemotaxis toward host plant exudates through direct proline sensing. Appl Environ Microbiol 80:3404–3415. doi: 10.1128/AEM.00115-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webb BA, Compton KK, Del Campo JSM, Taylor D, Sobrado P, Scharf BE. 2017. Sinorhizobium meliloti chemotaxis to multiple amino acids is mediated by the chemoreceptor McpU. Mol Plant Microbe Interact 30:770–777. doi: 10.1094/MPMI-04-17-0096-R. [DOI] [PubMed] [Google Scholar]
- 35.Sourjik V, Schmitt R. 1998. Phosphotransfer between CheA, CheY1, and CheY2 in the chemotaxis signal transduction chain of Rhizobium meliloti. Biochemistry 37:2327–2335. doi: 10.1021/bi972330a. [DOI] [PubMed] [Google Scholar]
- 36.Sourjik V, Sterr W, Platzer J, Bos I, Haslbeck M, Schmitt R. 1998. Mapping of 41 chemotaxis, flagellar and motility genes to a single region of the Sinorhizobium meliloti chromosome. Gene 223:283–290. doi: 10.1016/S0378-1119(98)00160-7. [DOI] [PubMed] [Google Scholar]
- 37.Glekas GD, Mulhern BJ, Kroc A, Duelfer KA, Lei V, Rao CV, Ordal GW. 2012. The Bacillus subtilis chemoreceptor McpC senses multiple ligands using two discrete mechanisms. J Biol Chem 287:39412–39418. doi: 10.1074/jbc.M112.413518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dogra G, Purschke FG, Wagner V, Haslbeck M, Kriehuber T, Hughes JG, Van Tassell ML, Gilbert C, Niemeyer M, Ray WK, Helm RF, Scharf BE. 2012. Sinorhizobium meliloti CheA complexed with CheS exhibits enhanced binding to CheY1, resulting in accelerated CheY1 dephosphorylation. J Bacteriol 194:1075–1087. doi: 10.1128/JB.06505-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amin M, Kothamachu VB, Feliu E, Scharf BE, Porter SL, Soyer OS. 2014. Phosphate sink containing two-component signaling systems as tunable threshold devices. PLoS Comput Biol 10:e1003890. doi: 10.1371/journal.pcbi.1003890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li M, Hazelbauer GL. 2004. Cellular stoichiometry of the components of the chemotaxis signaling complex. J Bacteriol 186:3687–3694. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cannistraro VJ, Glekas GD, Rao CV, Ordal GW. 2011. Cellular stoichiometry of the chemotaxis proteins in Bacillus subtilis. J Bacteriol 193:3220–3227. doi: 10.1128/JB.01255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levin MD, Morton-Firth CJ, Abouhamad WN, Bourret RB, Bray D. 1998. Origins of individual swimming behavior in bacteria. Biophys J 74:175–181. doi: 10.1016/S0006-3495(98)77777-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rotter C, Mühlbacher S, Salamon D, Schmitt R, Scharf B. 2006. Rem, a new transcriptional activator of motility and chemotaxis in Sinorhizobium meliloti. J Bacteriol 188:6932–6942. doi: 10.1128/JB.01902-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sourjik V, Muschler P, Scharf B, Schmitt R. 2000. VisN and VisR are global regulators of chemotaxis, flagellar, and motility genes in Sinorhizobium (Rhizobium) meliloti. J Bacteriol 182:782–788. doi: 10.1128/JB.182.3.782-788.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoang HH, Gurich N, González JE. 2008. Regulation of motility by the ExpR/Sin quorum-sensing system in Sinorhizobium meliloti. J Bacteriol 190:861–871. doi: 10.1128/JB.01310-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galibert F, Finan TM, Long SR, Pühler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P, Bothe G, Boutry M, Bowser L, Buhrmester J, Cadieu E, Capela D, Chain P, Cowie A, Davis RW, Dreano S, Federspiel NA, Fisher RF, Gloux S, Godrie T, Goffeau A, Golding B, Gouzy J, Gurjal M, Hernandez-Lucas I, Hong A, Huizar L, Hyman RW, Jones T, Kahn D, Kahn ML, Kalman S, Keating DH, Kiss E, Komp C, Lelaure V, Masuy D, Palm C, Peck MC, Pohl TM, Portetelle D, Purnelle B, Ramsperger U, Surzycki R, Thebault P, Vandenbol M, Vorholter FJ, Weidner S, Wells DH, Wong K, Yeh KC, Batut J. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 47.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 48.Meier VM, Scharf BE. 2009. Cellular localization of predicted transmembrane and soluble chemoreceptors in Sinorhizobium meliloti. J Bacteriol 191:5724–5733. doi: 10.1128/JB.01286-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soutourina O, Bertin PN. 2003. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev 27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- 50.Aldridge P, Hughes KT. 2002. Regulation of flagellar assembly. Curr Opin Microbiol 5:160–165. doi: 10.1016/S1369-5274(02)00302-8. [DOI] [PubMed] [Google Scholar]
- 51.Adler J, Templeton B. 1967. The effect of environmental conditions on the motility of Escherichia coli. J Gen Microbiol 46:175–184. doi: 10.1099/00221287-46-2-175. [DOI] [PubMed] [Google Scholar]
- 52.Pleier E, Schmitt R. 1991. Expression of two Rhizobium meliloti flagellin genes and their contribution to the complex filament structure. J Bacteriol 173:2077–2085. doi: 10.1128/jb.173.6.2077-2085.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lopez-Farfan D, Reyes-Darias JA, Krell T. 2017. The expression of many chemoreceptor genes depends on the cognate chemoeffector as well as on the growth medium and phase. Curr Genet 63:457–470. doi: 10.1007/s00294-016-0646-7. [DOI] [PubMed] [Google Scholar]
- 54.Meier VM. 2007. Funktionsanalyse und zellulaere lokalisierung der neun chemorezeptoren von Sinorhizobium meliloti. PhD thesis University of Regensburg, Regensburg, Germany. [Google Scholar]
- 55.Thiem S, Kentner D, Sourjik V. 2007. Positioning of chemosensory clusters in E. coli and its relation to cell division. EMBO J 26:1615–1623. doi: 10.1038/sj.emboj.7601610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu J, Li J, Li G, Long DG, Weis RM. 1996. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry 35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- 57.Barnakov AN, Barnakova LA, Hazelbauer GL. 1999. Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation. Proc Natl Acad Sci U S A 96:10667–10672. doi: 10.1073/pnas.96.19.10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li M, Hazelbauer GL. 2005. Adaptational assistance in clusters of bacterial chemoreceptors. Mol Microbiol 56:1617–1626. doi: 10.1111/j.1365-2958.2005.04641.x. [DOI] [PubMed] [Google Scholar]
- 59.Gegner JA, Graham DR, Roth AF, Dahlquist FW. 1992. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell 70:975–982. doi: 10.1016/0092-8674(92)90247-A. [DOI] [PubMed] [Google Scholar]
- 60.Simon R, O'Connell M, Labes M, Pühler A. 1986. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol 118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- 61.Sourjik V, Schmitt R. 1996. Different roles of CheY1 and CheY2 in the chemotaxis of Rhizobium meliloti. Mol Microbiol 22:427–436. doi: 10.1046/j.1365-2958.1996.1291489.x. [DOI] [PubMed] [Google Scholar]
- 62.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 63.Schäfer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 64.Rocco CJ, Dennison KL, Klenchin VA, Rayment I, Escalante-Semerena JC. 2008. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid 59:231–237. doi: 10.1016/j.plasmid.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krupski G, Götz R, Ober K, Pleier E, Schmitt R. 1985. Structure of complex flagellar filaments in Rhizobium meliloti. J Bacteriol 162:361–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scharf B, Schuster-Wolff-Bühring H, Rachel R, Schmitt R. 2001. Mutational analysis of Rhizobium lupini H13-3 and Sinorhizobium meliloti flagellin genes: importance of flagellin A for flagellar filament structure and transcriptional regulation. J Bacteriol 183:5334–5342. doi: 10.1128/JB.183.18.5334-5342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Götz R, Limmer N, Ober K, Schmitt R. 1982. Motility and chemotaxis in two strains of Rhizobium with complex flagella. J Gen Microbiol 128:789–798. [Google Scholar]
- 69.Higuchi R. 1989. Using PCR to engineer DNA, p 61–70. In Erlich HA. (ed), PCR technology principles and applications for DNA amplification. Stockton Press, New York, NY. [Google Scholar]
- 70.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology (NY) 1:783–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 71.Labes M, Pühler A, Simon R. 1990. A new family of RSF1010-derived expression and lac-fusion broad-host-range vectors for gram-negative bacteria. Gene 89:37–46. doi: 10.1016/0378-1119(90)90203-4. [DOI] [PubMed] [Google Scholar]
- 72.Riepl H, Maurer T, Kalbitzer HR, Meier VM, Haslbeck M, Schmitt R, Scharf B. 2008. Interaction of CheY2 and CheY2-P with the cognate CheA kinase in the chemosensory-signalling chain of Sinorhizobium meliloti. Mol Microbiol 69:1373–1384. doi: 10.1111/j.1365-2958.2008.06342.x. [DOI] [PubMed] [Google Scholar]
- 73.Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. 2011. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol 80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stock JB, Rauch B, Roseman S. 1977. Periplasmic space in Salmonella typhimurium and Escherichia coli. J Biol Chem 252:7850–7861. [PubMed] [Google Scholar]