ABSTRACT
ADP-ribosylation factors (ARF) GTPases are activated by guanine nucleotide exchange factors (GEFs) to support cellular homeostasis. Key to understanding spatio-temporal regulation of ARF signaling is the mechanism of GEF recruitment to membranes. Small GEFs are recruited through phosphoinositide (PIP) binding by a pleckstrin homology (PH) domain downstream from the catalytic Sec7 domain (Sec7d). The large GEFs lack PH domains, and their recruitment mechanisms are poorly understood. We probed Golgi recruitment of GBF1, a GEF catalyzing ARF activation required for Golgi homeostasis. We show that the homology downstream of Sec7d-1 (HDS1) regulates Golgi recruitment of GBF1. We document that GBF1 binds phosphoinositides, preferentially PI3P, PI4P and PI(4,5)P2, and that lipid binding requires the HDS1 domain. Mutations within HDS1 that reduce GBF1 binding to specific PIPs in vitro inhibit GBF1 targeting to Golgi membranes in cells. Our data imply that HDS1 and PH domains are functionally analogous in that each uses lipid-based membrane information to regulate GEF recruitment. Lipid-based recruitment of GBF1 extends the paradigm of lipid regulation to small and large GEFs and suggests that lipid-based mechanisms evolved early during GEF diversification.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: ADP-ribosylation factors, GBF1, Golgi, HDS1 domain, Sec7 guanine nucleotide exchange factor, Membrane trafficking
Summary: Our work represents the first report showing PIP binding by a large GEF GBF1 and the crucial importance of such binding in membrane recruitment.
INTRODUCTION
ADP-ribosylation factors (ARFs) are essential regulators of cellular physiology that facilitate processes as diverse as membrane traffic, actin dynamics and lipid metabolism (D'Souza-Schorey and Chavrier, 2006). Like all small GTPases, ARFs cycle between an inactive (GDP-bound) and an active (GTP-bound) form. ARFs have low intrinsic exchange rates, and require guanine exchange factors (GEFs) to increase the efficiency of activation in cells. All ARF GEFs contain a ∼200 amino acid catalytic Sec7 domain (Sec7d), so named because it was first identified in the yeast Sec7p. This region shows the highest sequence conservation across the phylogenetic spectrum (Bui et al., 2009). GEFs contain other shared motifs, and their presence or absence groups them into two subfamilies, large and small GEFs. The large GEFs, with molecular weights >200 kDa include GBF1 and BIG1/2; the small GEFs, with molecular weights <200 kDa, include EFA6, BRAG and cytohesin/ARNO. The large GEFs are present in all eukaryotes, suggesting an early appearance during evolution, whereas the small GEFs are present only in metazoans.
All GEFs are cytosolic proteins that undergo cycles of membrane association and dissociation, with the membrane-localized pool representing the functional fraction. Thus, GEFs initiate the downstream effector cascades only at the membrane and must be recruited to the appropriate membranes to activate ARFs. Understanding of GEF recruitment is uneven: a lot is known about membrane targeting of small ARF GEFs, but knowledge of how large GEFs are recruited is more limited. Membrane recruitment of the cytohesin, BRAG and EFA6 small GEFs is regulated by a phosphoinositide (PIP)-binding pleckstrin homology (PH) domain immediately downstream of Sec7d. Thus, there exists a general paradigm for these GEFs, in which lipid binding by PH provides molecular information for membrane targeting of the GEF. Large GEFs lack PH domains, and instead contain homology downstream of Sec7d-1 (HDS1) domains immediately downstream of Sec7d. Previous work on yeast Sec7p showed that its HDS1 domain also regulates membrane recruitment, but by binding to an activated form of ARF1 instead of to lipids (Richardson and Fromme, 2012; Richardson et al., 2012). However, this mechanism of recruitment is not utilized by other large GEFs (yeast Gea1p, Gea2p and GBF1) (Gustafson and Fromme, 2017; Richardson et al., 2012; Szul et al., 2005). Thus, we explored whether the HDS1 within GBF1 functions more like a PH domain and regulates membrane recruitment through PIP binding.
GBF1 is a large GEF that localizes to the Golgi (Kawamoto et al., 2002; Niu et al., 2005; Szul et al., 2005; Zhao et al., 2002). GBF1 is required for ARF activation, which sustains COPI recruitment, Golgi homeostasis and secretory traffic; inactivation or depletion of GBF1 inhibits these processes (Garcia-Mata et al., 2003; Szul et al., 2007; Zhao et al., 2006). Yet, despite its crucial importance, the mechanisms that regulate membrane recruitment of GBF1 are poorly defined. Here, we report that specific motifs within the HDS1 domain immediately downstream of the catalytic Sec7d are essential for GBF1 recruitment to Golgi membranes. We show that GBF1 binds specific PIPs, and that selective mutations within HDS1 that impair PIP binding in vitro also inhibit GBF1 targeting to Golgi membranes and cellular function in vivo. Our results extend the generality of a lipid-based regulatory mechanism in which a domain (PH or HDS1) immediately downstream from the catalytic Sec7d uses PIP binding to impart recruitment information to the GEF.
RESULTS AND DISCUSSION
GBF1 recruitment to Golgi membranes requires the LF motif in HDS1
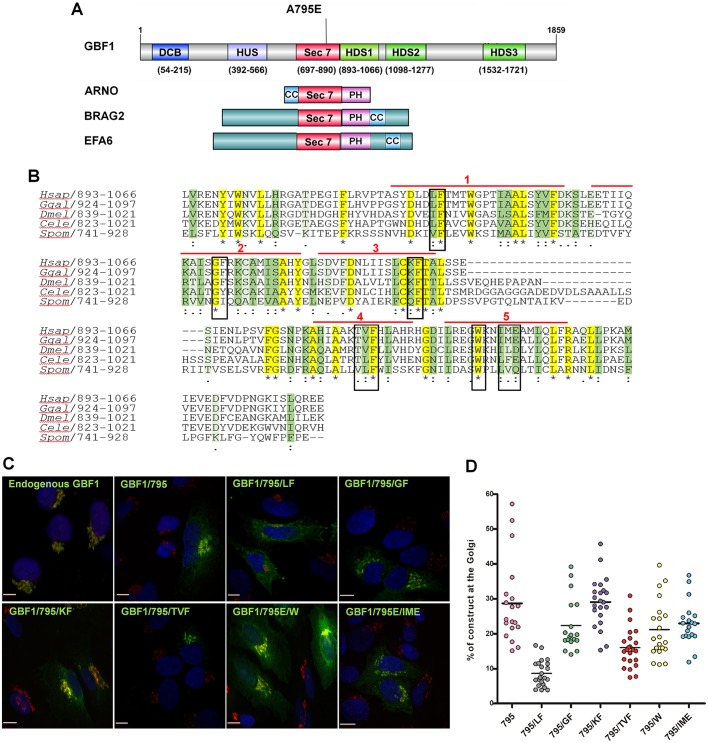
The PH domains of small GEFs that mediate their membrane recruitment are immediately downstream from the catalytic Sec7d (Fig. 1A). In the large GEFs, that position is occupied by HDS1, and we performed structure–function analyses of HDS1 to assess its participation in GBF1 recruitment to Golgi membranes. The ∼173 residue HDS1 domain is predicted to contain five α-helices. We targeted the highly conserved LF926,927 in α-helix 1, GF956,957 in α-helix 2, KF983,984 in α-helix 3, TVF1011-1013 in α-helix 4, and IME130-133 in α-helix 5 for mutagenesis (Fig. 1B). We chose alanine substitutions because our previous findings suggested that such modifications do not usually cause misfolding. Thus, we selected motifs within each α-helix that contained highly conserved amino acids, but did not contain an alanine residue. Importantly, all constructs were full-length, had N-terminal GFP (this tag does not interfere with Golgi targeting) (Szul et al., 2005) and contained the A795E mutation that confers brefeldin A (BFA) resistance to GBF1 (all constructs are designated 795). Targeting of each GBF1 mutant was assessed relative to the Golgi marker GM130 by immunofluorescence (IF) of fixed and permeabilized cells. Golgi recruitment was observed for all constructs, except the GBF1/795/LF mutant, which was impaired in Golgi association and was predominantly in a diffuse cellular pattern (Fig. 1C; Figs S1 and S2). The recruitment efficiency of each construct was quantified relative to GM130: approximately 30% of GBF1/795 or GBF1/795/KF was detected at the Golgi, whereas less GBF1/795/GF (∼22%), GBF1/795/TVF (∼15%), GBF1/795/W (∼22%) and GBF1/795/IME (∼25%) localized to the Golgi (Fig. 1D). However, less than 10% of GBF1/795/LF localized to the Golgi, suggesting that the LF motif within α-helix 1 provides essential information for membrane recruitment.
Fig. 1.
Specific motifs in HDS1 regulate Golgi recruitment of GBF1. (A) Domain organization of GBF1, ARNO, BRAG2 and EFA6. The A795E mutation conferring BFA resistance to GBF1 is marked. DCB, dimerization and cyclophilin-binding; HUS, homology upstream of Sec7; HDS1-3, homology downstream of Sec7; CC, coiled coil; PH, pleckstrin homology. (B) Alignment of HDS1 sequences from Homo sapiens GBF1 and orthologs from Gallus gallus (Ggal), Drosophila melanogaster (Dmel), Caenorhabditis elegans (Cele) and Schizosaccharomyces pombe (Spom) shows identical residues in yellow and marked by asterisks. Strongly conserved residues are in dark green and marked by a colon; weakly conserved residues are in light green and marked with a period. The mutated residues are boxed and α-helical regions are marked by red lines. (C) HeLa cells transfected with the indicated constructs were processed for IF with anti-GFP and anti-GM130. Scale bars: 7 µm. (D) Images analogous to those in C were used to quantitate the percentage of each construct co-localizing with GM130. Data from 20 cells expressing each construct are presented as a scatter plot.
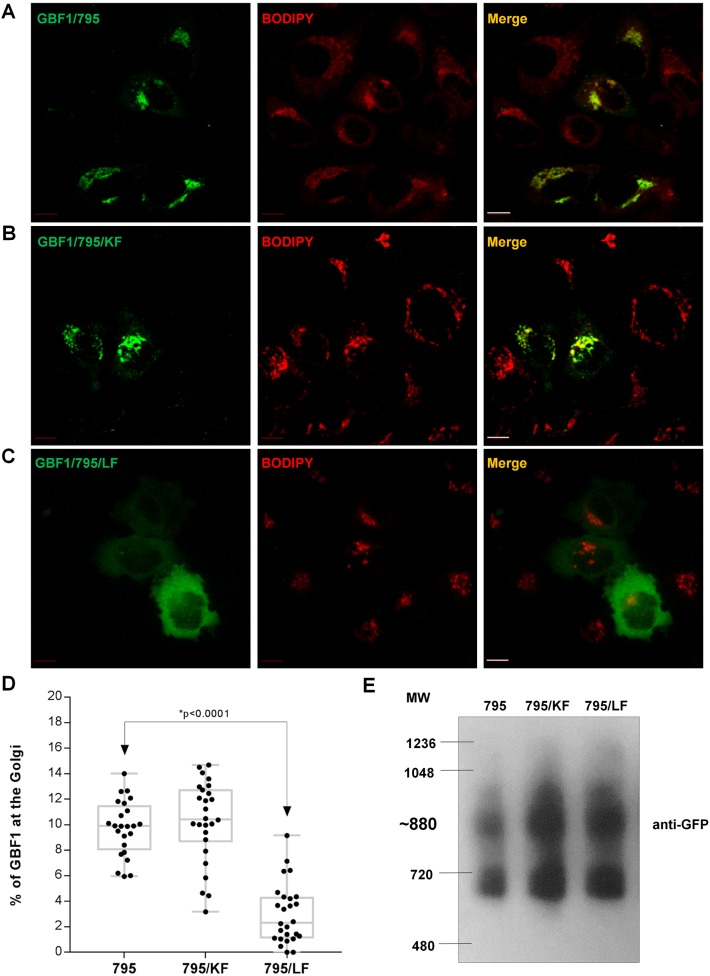
Live-cell imaging was used to confirm the targeting efficiency of GBF1/795, GBF1/795/KF and GBF1/795/LF (this method does not utilize permeabilization and, hence, allows the quantification of total cellular pool of GBF1) relative to the cell-permeant vital Golgi stain BODIPY TR C5-ceramide. GBF1/795 (Fig. 2A) and GBF1/795/KF (Fig. 2B) efficiently targeted to the Golgi, whereas GBF1/795/LF (Fig. 2C) showed diffuse cellular distribution. Quantification indicated approximately 8–11% of cellular GBF1/795 or GBF1/795/KF associating with Golgi membranes (Fig. 2D). This agrees with the ∼10% membrane association of endogenous GBF1 we reported using subcellular fractionation (Szul et al., 2005, 2007). In contrast, only ∼1–4% of GBF1/795/LF construct localized to the Golgi, indicating that HDS1 is crucial for GBF1 targeting to Golgi membranes, with the LF motif within α-helix 1 being essential for membrane recruitment.
Fig. 2.
Intact LF motif in HDS1 is required for GBF1 recruitment to Golgi. (A–C) HeLa cells expressing the indicated constructs and stained with BODIPY TR C5-ceramide were imaged under live-cell conditions. Scale bars: 10 µm. (D) Quantification of the percentage of total cellular construct that co-localizes with BODIPY indicates a statistically significant difference in targeting of GBF1/795 and GBF1/795/LF (P<0.0001 by simple one-way ANOVA; P<0.05 by t-test). (E) Lysates from HeLa cells expressing the indicated constructs were separated on a blue native gel and western blotted with anti-GFP.
We ensured that the loss of membrane association of GBF1/795/LF was not due to misfolding by showing analogous migration patterns of GBF1/795/LF, GBF1/795 and GBF1/795/KF, similar to that previously reported for endogenous GBF1 (Bhatt et al., 2016) on blue native gel electrophoresis (Fig. 2E). Hence, the inability of GBF1/795/LF to associate with Golgi membranes is not caused by major structural rearrangements.
GBF1 binds PIPs, and PIP binding requires the LF motif in HDS1
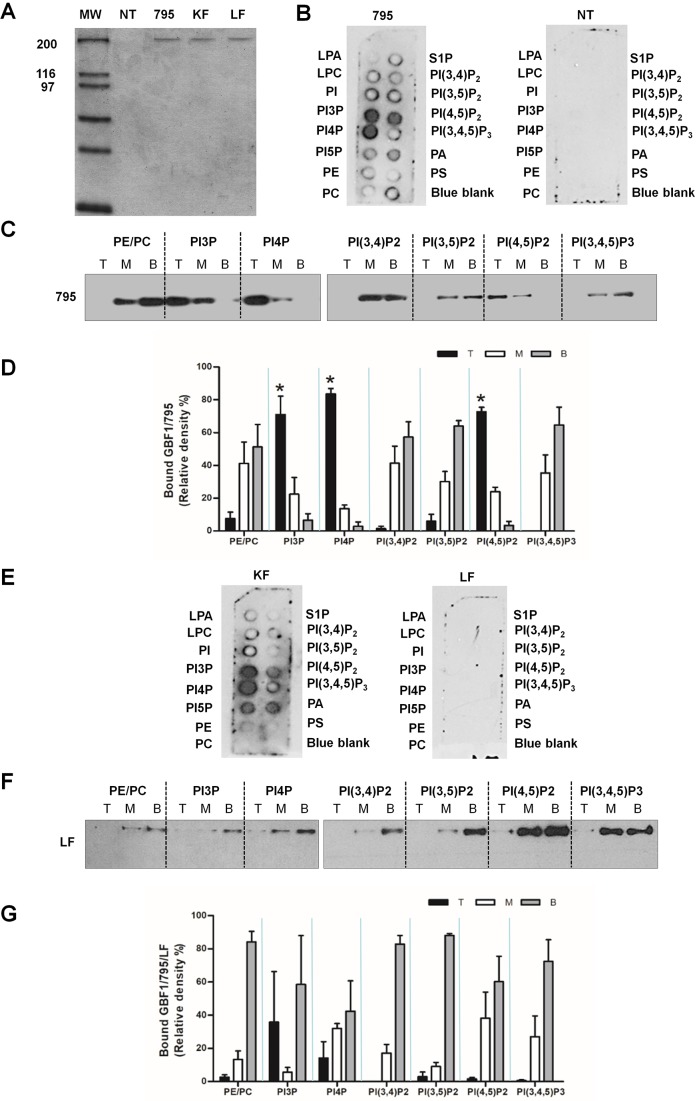
PH domains mediate membrane recruitment by binding PIPs, with PH from different proteins displaying binding specificity for different PIPs (Balla and Várnai, 2009; Macia et al., 2001, 2008; Stahelin et al., 2014; Stalder et al., 2011). We posited that HDS1 recruits GBF1 to membranes via direct binding to one or more lipids. We probed lipid binding of full-length GBF1 and assessed possible involvement of HDS1 in PIP binding by purifying GBF1/795, GBF1/795/KF and GBF1/795/LF from HEK cells overexpressing each recombinant protein and testing them in lipid strip and liposome flotation assays. As shown in Fig. 3A, equivalent amounts of each protein were purified, and GBF1 represented the only protein species detected in the gel (although we cannot exclude the possibility of extremely low levels of contaminants). A parallel purification was performed from mock-transfected (NT lane) cells to provide a negative control. Binding of purified GBF1/795 to nitrocellulose strips containing spotted lipids was detected by western blotting with anti-GBF1. GBF1/795 preferentially bound to PI4P, PI3P and PI(4,5)P2, and less to PA (Fig. 3B; 795 panel). The specificity of binding was assured by the lack of signal in strips incubated with material from untransfected cells (Fig. 3B; NT panel).
Fig. 3.
GBF1 binds PIPs, and intact LF motif in HDS1 is required for binding. (A) Coomassie Blue stained gel of purified His-myc-tagged GBF1/795, GBF1/795/KF and GBF1/795/LF. Material purified from mock-transfected cells (NT) is also shown. (B,E) Purified proteins from A were incubated with lipid strips, and bound protein detected by western blotting with anti-GBF1. Material from mock-transfected cells (NT) provides a negative control. (C,F) Purified proteins from A were incubated with liposomes containing the indicated PIPs and floated on Nycodenz gradients to generate top (T), middle (M) and bottom (B) fractions. Each fraction was western blotted with anti-GBF1. (D,G) Quantification of liposome flotation for GBF1/795 (D) and GBF1/795/LF (G). The graphs show the averages of the relative amounts of each construct in the top, middle and bottom fractions from three different experiments (n=3). Statistically significant differences in protein–liposome binding (P<0.05 by t-test) are marked by asterisks.
Lipid strips present the lipid to the protein in a restricted geometry, and we confirmed GBF1 lipid binding and specificity in the more stringent liposome flotation assay. Purified GBF1/795 was first incubated with liposomes of distinct PIP composition. Following flotation, the levels of GBF1/795 in the top layer (T), containing liposome-bound protein, and the medium (M) and bottom (B) layers, containing unbound protein, were assayed by western blotting with anti-GBF1. GBF1/795 was preferentially recovered in the liposome fraction when liposomes contained PI3P, PI4P or PI(4,5)P2 (Fig. 3C). GBF1/795 was not found with liposomes containing PE/PC, PI(3,4)P2, PI(3,5)P2 or PI(3,4,5)P3. This binding profile is identical to that obtained in the lipid strip assays, confirming GBF1 lipid-binding specificity. Quantification of lipid binding preference confirmed that GBF1/795 has highest binding preference for PI4P, followed by PI(4,5)P2 and then PI3P (Fig. 3D).
The preference for PI4P and PI(4,5)P2 is consistent with previous work implicating the Golgi-localized PI4 kinase (PI4KIIIα) in regulating GBF1 membrane association. Inactivating or depleting PI4KIIIα reduced membrane association of GBF1, whereas increased generation of PI4P by overexpressing the constitutively active form of Rab1b increased GBF1 recruitment (Dumaresq-Doiron et al., 2010). Furthermore, the GBF1 preference for PI4P and PI(4,5)P2 is consistent with the known distribution of these PIPs within the endo-membrane system, as both are detected at the organelles at the ER–Golgi interface (Balla, 2007; Balla and Várnai, 2009; D'Angelo et al., 2012; De Matteis et al., 2005, 2013; Godi et al., 2004; Várnai and Balla, 2008; Watt et al., 2002).
To determine whether lipid binding by GBF1/795 requires intact HDS1, we tested the GBF1/795/KF and GBF1/795/LF mutants in lipid strip binding. GBF1/795/KF bound to lipids in a manner analogous to GBF1/795, with strongest binding to PI3P and PI4P, and lower binding to PI(4,5)P2 and PA (Fig. 3E; KF panel). In some strips, binding to PI5P was observed, but this was not reproducible. In contrast, GBF1/795/LF did not bind lipids (Fig. 3E; LF panel). We confirmed that an intact LF motif is required for lipid binding in the liposome flotation assay. As shown in Fig. 3F and quantified in Fig. 3G, GBF1/795/LF is always recovered in the unbound fractions, indicating lack of lipid binding.
Our results with full-length GBF1 expressed in mammalian cells differ from the lipid-binding properties reported for the HDS1 domain alone or for GBF1 fragments containing the HDS1 domain (Bouvet et al., 2013; Mazaki et al., 2012). The mCherry-tagged HDS1 domain, expressed in cells treated with oleic acid to promote lipid droplet formation, associated with the surface of the lipid droplets but did not target Golgi membranes (Bouvet et al., 2013). Lipid droplets do not contain PIPs and it is possible that the binding of mCherry–HDS1 within this system reflects its hydrophobic propensity, rather than specific targeting. Similarly, bacterially expressed GST–HDS1 was shown to bind to PC (100%) and Golgi mix (50% PC, 20% PE, 10% PI, 5% PS, 16% cholesterol) liposomes, leading the authors to conclude that HDS1 does not have a requirement for charged and rare lipids such as PIPs (Bouvet et al., 2013). By contrast, a study showing the recruitment of endogenous GBF1 to the plasma membrane in stimulated neutrophils documented that a bacterially expressed fragment of GBF1 containing HDS1 did bind PIPs, but with a preference for the plasma membrane PI(3,5)P2 and PI(3,4,5)P3 (Mazaki et al., 2012). However, the PIP binding specificity of the GBF1 fragment was not confirmed by a liposome flotation assay. Both studies also probed the localization of small GBF1 fragments expressed in cells without assessing their folding or functionality. In contrast, our study assessed lipid binding of full-length GBF1 from mammalian cells that was shown to be correctly folded and fully functional when assayed for its ability to support Golgi homeostasis (discussed below).
Our results strongly support the conclusion that GBF1 binds a specific subset of PIPs [PI3P, PI4P and PI(4,5)P2] and that the LF motif within HDS1 is required for this binding. Our results show a strict correlation between GBF1 ability to be recruited to the Golgi and its ability to bind PIPs. Thus, our findings suggest that HDS1 acts in a manner similar to the PH domain in small GEFs and regulates GBF1 recruitment to Golgi membranes by binding specific PIPs.
Cellular function of GBF1 is dependent on the LF motif in HDS1
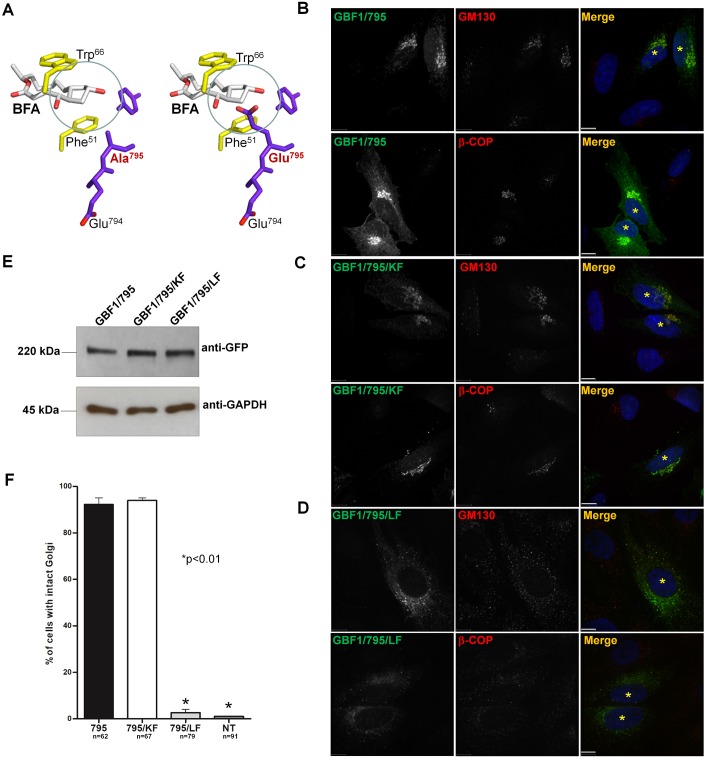
Considering that GBF1 activates ARF when associated with Golgi membranes, we tested the role of PIP binding in GBF1 cellular function. We used a ‘replacement’ assay in which GBF1 constructs containing the A795E mutation that makes them BFA resistant (Belov et al., 2008) are the sole functional GBF1 species within cells because the endogenous GBF1 had been inhibited with BFA. The A795E substitution within the Sec7d of GBF1 obstructs the BFA-binding site, but does not affect catalytic activity (Fig. 4A). Cells expressing GBF1/795, GBF1/795/KF or GBF1/795/LF were treated with BFA to inactivate endogenous GBF1 and their ability to support Golgi homeostasis then assessed. As expected, cells expressing GBF1/795 (but not untransfected cells) contained morphologically recognizable perinuclear Golgi and Golgi-associated β-COP, a marker of ARF activity at the Golgi (Fig. 4B). Cells expressing GBF1/795/KF also showed normal Golgi architecture and supported β-COP recruitment to Golgi membranes (Fig. 4C). By contrast, cells expressing GBF1/795/LF had disrupted Golgi and diffusely distributed β-COP (Fig. 4D). The difference in rescue phenotypes was not due to differences in expression levels of the constructs, as shown by the western blot of lysates from transfected cells (Fig. 4E). Quantification of rescue phenotypes indicated that GBF1/795 supports Golgi architecture in ∼90% of transfected cells, and that the KF mutation does not significantly affect the functionality of GBF1/795/KF (Fig. 4F). By contrast, GBF1/795/LF does not support Golgi homeostasis. Thus, an intact LF motif within the α-helix 1 of HDS1 in full-length GBF1 is required for PIP binding and is essential for GBF1 cellular function.
Fig. 4.
Intact LF motif in HDS1 is required for GBF1 activity in Golgi homeostasis. (A) Stick diagram of the BFA-binding pocket at the interface of ARF (yellow) and Sec7d (purple) showing the position of the wild-type A795E (red). The A795E mutation blocks BFA intercalation (circled). (B–D) HeLa cells expressing the indicated construct were treated with BFA (0.5 µg/ml, 30 min) and processed for IF with anti-GFP and either anti-GM130 or anti-β-COP. Transfected cells are marked with asterisks. Scale bars: 6 µm. (E) Western blot of lysates from cells expressing the indicated construct shows analogous level of expression of each protein (anti-GFP panel) relative to endogenous GAPDH (anti-GAPDH panel). (F) Quantification of the percentage of cells expressing each construct and containing intact Golgi after BFA treatment. Values represent means of three independent experiments; *P<0.01, calculated by Student's t-test.
Our results identify HDS1-mediated PIP binding as a novel regulator of GBF1 membrane recruitment. Currently, only the interaction of the N-terminal domain (amino acids 1–384) with the active form of Rab1b (Alvarez et al., 2003; Monetta et al., 2007) is known to promote GBF1 membrane association (Monetta et al., 2007). Membrane recruitment of GBF1 also appears to be regulated by homology downstream of Sec7d-2 (HDS2). Deletion of HDS2 (Bouvet et al., 2013; Ellong et al., 2011), and the L1246R mutation in HDS2 (Chen et al., 2017) inhibit membrane association, but the molecular mechanism of HDS2 action is unknown.
Our results document that multiple, structurally separate domains of GBF1 (N-terminus and HDS1) interact with distinct membrane components (Rab1b and PIPs) to regulate recruitment of GBF1 to membranes and thereby regulate its cellular functionality. Such dual coincidence detection of PIPs (PI3P) and Rabs (Rab5) has been observed for the recruitment of early endosome antigen 1 (EEA1) to endosomes (Simonsen et al., 1998) and could represent a general mechanism to ensure faithful membrane positioning of crucial cell components in the face of the intrinsic complexity of membrane networks.
MATERIALS AND METHODS
Materials
PC (L-α-phosphatidylcholine; egg, chicken); PE (L-α-phosphatidylethanolamine; brain, porcine); 18:1 PtdIns3P [(1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-3′-phosphate)]; PtdIns4P (L-α-phosphatidylinositol-4-phosphate ammonium salt, brain, porcine); 18:1 PtdIns5P [1,2-dio-leoyl-sn-glycero-3-phospho-(1′-myoinositol-5′-phosphate, ammonium salt)]; 18:1 PtdIns(3,4)P2 [1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-3′,4′-bisphosphate, ammonium salt)]; 18:1 PtdIns(4,5)P2 [(1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-4′,5′-bisphosphate ammonium salt)]; 18:1 PtdIns(3,5)P2 [(1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-3′,5′-bisphosphate, ammonium salt)]; 18:1 PtdIns (3,4,5)P3 [(1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-3′,4′,5′-trisphosphate, ammonium salt)] were purchased from Avanti Polar Lipids, Inc. USA. Texas-Red-PE (1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine, triethyl-ammonium salt) was bought from Life Technologies. Other reagents used were as follows: chloroform (MP Biomedicals, USA); sodium cholate (Alfa Aesar, USA); brefeldin A (Sigma-Aldrich, MO, USA); Pierce Glutathione Agarose (Thermo Scientific, IL, USA); Tween-20 (Fisher Scientific, USA); Ni-NTA agarose (Qiagen, CA, USA); Sephadex G-50 (Sigma-Aldrich, MO, USA); SuperSignal® West Femto Maximum Sensitivity Substrate (Thermo Scientific, IL, USA); Complete Protease Inhibitors Cocktail, EDTA-free (Santa Cruz Biotechnology, CA, USA); PIP strips (Echelon Biosciences Incorporated, USA); Nycodenz (Accurate Chemical and Scientific Corp., NY, USA); and BODIPY TR (Texas Red) C5-ceramide complexed with BSA (ThermoFisher Scientific, Grand Island, NY) Polyclonal and monoclonal GFP antibodies (Abcam, MA, USA); Monoclonal GBF1 antibodies (BD Bioscience, CA, USA); Polyclonal GBF1 antibodies (Thermo Scientific, IL, USA); Monoclonal GM130 (BD Transduction Laboratories, ON, USA); Polyclonal GM130 (Thermo Scientific, IL, USA); Monoclonal β COP antibodies (Roche, IN, USA); Monoclonal His (Thermo Scientific, IL, USA); Monoclonal Myc (Invitrogen, WI, USA); Secondary, anti-rabbit and anti-mouse antibodies conjugated with horseradish peroxidase (Pierce/Thermo Fisher Scientific Inc., IL, USA); Secondary antibodies conjugated with Alexa 488 or Alexa 594 (Invitrogen, WI, USA and Thermo Scientific, IL, USA).
Cell culture and transfection
Human HeLa (CCL-2) cell line was obtained from ATCC (The Global Bioresource Center, USA). HEK (GripTite™ 293 MSR, R79507) cell line was purchased from ThermoFisher Scientific (NY, USA). Cells were cultured in vitro in Eagle's minimum essential medium (MEM) supplemented with L-glutamine, 10% fetal bovine serum, 100 units/ml penicillin, 100 mg/ml streptomycin and 1 mM sodium pyruvate (Cellgro, Manassas, VA) at 37°C in humidified atmosphere and transfected with Mirus TransIT-LT1 Transfection Reagent (Mirus Bio Corporation, Madison, WI), according to the manufacturer's instructions.
Imaging
HeLa cells were seeded overnight on glass coverslips (12 mm diameter), transfected and processed for IF 24 h later. For the BFA replacement assay, transfected cells were washed in phosphate-buffered saline (PBS) and incubated with 0.5 µg/ml BFA for 30 min before fixation and processing for IF. For standard IF, cells were washed in PBS, fixed in 3% paraformaldehyde for 10 min, quenched with 10 mm ammonium chloride, permeabilized with 0.1% Triton X-100 in PBS, washed with PBS and blocked in PBS containing 2.5% goat serum, 0.2% Tween 20 for 5 min followed by blocking in PBS containing 0.4% fish skin gelatin and 0.2% Tween 20. Cells were incubated with primary antibodies for 1 h at room temperature. Coverslips were washed with PBS containing 0.2% Tween 20 and incubated with secondary antibodies for 45 min. Nuclei were stained with Hoechst dye. Coverslips were washed as above and mounted on slides in ProLong Gold antifade reagent (Invitrogen, Madison, WI).
Fluorescence patterns were visualized using a Leitz Wetlzar microscope with epifluorescence and Hoffman Modulation Contrast optics by Chroma Technology, Inc. (Bellows Falls, VT, USA). Images were captured with a 12-bit CCD camera from QImaging (Surrey, BC, Canada) and processed with iVision-Mac software. Confocal imaging was on a Perkin Elmer Ultraview ERS 6FE spinning disk attached to a Nikon TE 2000-U microscope. The system was equipped with laser and filter sets to visualize and image FITC, TRITC and DAPI fluorescence. Images were captured using a Hamamatsu C9100-50 EMCCD camera (Hamamatsu Photonics K.K., Hamamatsu city, Japan) and 60× or 100× Plan APO oil-immersion objectives. The imaging system was operated by Volocity 6.2 software (Perkin Elmer, Shelton, CT, USA).
For live imaging, transfected cells on coverslips were washed three times with imaging medium buffered with HEPES, pH 7.4 (Live Cell Imaging Solution; Molecular Probes, Grand Island, NY). Cells were incubated with 1 µM BODIPY TR (Texas Red) C5-ceramide complexed with BSA (ThermoFisher Scientific, Grand Island, NY) for 30 min at 37°C to label the Golgi apparatus. Coverslips were washed five times with HEPES imaging media and incubated in fresh MEM at 37°C in humidified atmosphere for 30 min. Coverslips were maintained on a thermostage at 37°C, 5% CO2 and 70% relative humidity in HEPES imaging media. To eliminate bias, the images were analyzed in an automated manner by Volocity software. To identify the ceramide-stained Golgi region (red), the threshold in the red channel was set to the average intensity across the entire image plus five standard deviations. The software could identify the Golgi region most consistently under these settings. To eliminate computational errors, all the identified Golgi regions were verified by visual inspection. The outer boundary of cells was delineated by using the average green intensity across the entire image. The total intensity of green within the cell (total cellular GFP–GBF1 levels) and the intensity of green that co-localized with red (Golgi-resident GFP–GBF1) was then determined and the percentage of total cellular GBF1 that resides at the Golgi was calculated.
Plasmids
All mutations were introduced into GFP-GBF1/A795E pcDNA4/To/Myc-His B (Invitrogen) using QuikChange XL site-directed mutagenesis kit from Agilent Technology. LF and KF mutations were introduced into GBF1/A795E-Venus vector using QuikChange XL site-directed mutagenesis kit from Agilent Technology.
Protein purification
His-tagged full-length human GBF1/795, GBF1/795/LF and GBF1/795/KF were expressed in and purified from HEK cells 48 h after transfection. Cells were lysed in lysis buffer (50 mM HEPES pH 7.4, 100 mM NaCl, protease inhibitor tablet) by passing five times through a 21G needle (BD Bioscience, CA, USA) and twice through a 27G needle (BD Biosciences, CA, USA). Cell debris was removed by centrifugation at 14,000 rpm for 15 min at 4°C. Supernatants were pre-cleared using Pierce Glutathione Agarose (Thermo Scientific, IL, USA) at 4°C for 1 h, and centrifuged at 1000 rpm for 2 min. Proteins were purified using Ni-NTA Agarose beads (Qiagen, CA, USA) (50% slurry) for 3 h at 4°C. Beads were recovered by centrifugation at 1000 rpm for 1 min and washed with buffer (20 mM HEPES pH 7.4, 100 mM NaCl, 20 mM imidazole) five times for 5 min at 4°C and then centrifuged at 1000 rpm for 2 min. Proteins were eluted from the beads with buffer (25 mM HEPES pH 7.4, 100 mM NaCl, 250 mM imidazole) three times for 5 min at 4°C and then centrifuged at 2000 rpm for 1 min. Purified proteins were analyzed by 8% SDS-PAGE gels and Coomassie Blue staining and stored at −80°C.
Lipid strip-binding assay
Filter strips (Echelon Biosciences Inc., USA) spotted with different lipids were blocked for 1 h in 3% fatty acid-free BSA in PBST (PBS containing 0.1% v/v Tween-20) and then incubated with 0.5 μg/ml of purified full-length GBF1/795, GBF1/795/LF or GBF1/795/KF for 1 h at room temperature. Filters were washed three times with PBST and incubated with mouse monoclonal anti-His antibody at room temperature for 60 min, followed by incubation with anti-mouse IgG conjugated with horseradish peroxidase. Filters were processed using SuperSignal® West Femto Maximum Sensitivity Substrate (Thermo Scientific, Rockford, IL, USA) and exposed to X-ray film.
Liposome preparation and flotation
Liposomes were prepared using a method based on that of Busse et al. (2013). Chloroform solutions of lipids PIP:PC:PE:TR-PE (2:73:23:2 ratio by weight) or for neutral liposomes lipids PC:PE:TR-PE (75:23:2 ratio) were premixed and dried under vacuum for 3 h to completely remove the solvent. The dried lipid film was then hydrated in 150 μl 3% (wt/vol) sodium cholate in HP150 buffer (20 mM HEPES pH 7.4, 150 mM KCl) and vortexed for 1 min. Cholate was removed from the liposome preparation by Sephadex G-50 chromatography in Bio-Rad 0.5 cm×15 cm columns.
Liposome suspension (70 µl) containing 1.8 mol% of different PIPs was incubated with 0.25 μg of purified protein diluted in HP150 buffer for 30 min at room temperature. Then, 150 μl of Nycodenz (80% wt/vol) was mixed with the sample, and the samples overlaid with 100 μl of 30% Nycodenz (wt/vol) and subsequently with 60 μl of HP150 buffer. The Nycodenz gradients were centrifuged in a TLA-100.2 rotor (Beckman) for 90 min at 75,000 rpm (245,000×g) at 4°C in an Optima TLX ultracentrifuge (Beckman Coulter). After centrifugation, 100 μl fractions were collected from the top, middle and bottom of the gradient. Fractions were analyzed by SDS-PAGE and western blotting using mouse anti-GBF1 antibody (dilution 1:500, overnight at 4°C) and secondary anti-mouse IgG labeled with horseradish peroxidase (1:5000, 1 h room temperature). Membranes were developed using SuperSignal® West Femto Maximum Sensitivity Substrate (Thermo Scientific, Rockford, IL, USA) and exposed to X-ray film.
Sequence alignments
Protein sequences for Homo sapiens GBF1 (Hsap; AAI17683.1) and orthologs from Gallus gallus (Ggal; XP_015143944.1), Drosophila melanogaster (Dmel; NP_725133.1), Caenorhabditis elegans (Cele; CAB03915.3), and Schizosaccharomyces pombe (Spom; NP_596613.1) were retrieved from GenBank. HDS1 domains were aligned using Clustal Omega (Lin et al., 2005; Simossis and Heringa, 2005; Simossis et al., 2005) alignments (Li et al., 2015; Squizzato et al., 2015). YASPIN (Lin et al., 2005) was used to predict the secondary structure of the HDS1 domain.
Supplementary Material
Acknowledgements
We thank Jacqueline Cherfils for providing the stick diagram, and Tomasz Szul, Paul Randazzo and Cathy Fuller for critical, yet supportive, comments.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.M.M., E.S.; Methodology: J.M.M., J.M.B., E.S.; Software: J.M.M., E.S.; Validation: J.M.M., J.M.B., E.S.; Formal analysis: J.M.M., J.M.B., E.S.; Investigation: J.M.M., E.L., A.A.I.; Resources: J.M.M., E.S.; Data curation: J.M.M., J.M.B., E.S.; Writing - original draft: J.M.M., M.L.S., E.S.; Writing - review & editing: J.M.M., M.L.S., R.A.K., E.S.; Visualization: J.M.M., E.S.; Supervision: J.M.M., R.A.K., E.S.; Project administration: J.M.M., E.S.; Funding acquisition: E.S.
Funding
This research was supported by grants from the National Science Foundation [MCB13-510] and from the National Institutes of Health [AI125561]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.210245.supplemental
References
- Alvarez C., Garcia-Mata R., Brandon E. and Sztul E. (2003). COPI recruitment is modulated by a Rab1b-dependent mechanism. Mol. Biol. Cell 14, 2116-2127. 10.1091/mbc.E02-09-0625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. (2007). Imaging and manipulating phosphoinositides in living cells. J. Physiol. 582, 927-937. 10.1113/jphysiol.2007.132795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. and Várnai P. (2009). Visualization of cellular phosphoinositide pools with GFP-fused protein-domains. Curr. Protoc. Cell Biol. Chapter 24, Unit 24 4 10.1002/0471143030.cb2404s42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov G. A., Feng Q., Nikovics K., Jackson C. L. and Ehrenfeld E. (2008). A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog. 4, e1000216 10.1371/journal.ppat.1000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt J. M., Viktorova E. G., Busby T., Wyrozumska P., Newman L. E., Lin H., Lee E., Wright J., Belov G. A., Kahn R. A. et al. (2016). Oligomerization of the Sec7 domain Arf guanine nucleotide exchange factor GBF1 is dispensable for Golgi localization and function but regulates degradation. Am. J. Physiol. Cell Physiol. 310, C456-C469. 10.1152/ajpcell.00185.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet S., Golinelli-Cohen M.-P., Contremoulins V. and Jackson C. L. (2013). Targeting of the Arf-GEF GBF1 to lipid droplets and Golgi membranes. J. Cell Sci. 126, 4794-4805. 10.1242/jcs.134254 [DOI] [PubMed] [Google Scholar]
- Bui Q. T., Golinelli-Cohen M.-P. and Jackson C. L. (2009). Large Arf1 guanine nucleotide exchange factors: evolution, domain structure, and roles in membrane trafficking and human disease. Mol. Genet. Genomics 282, 329-350. 10.1007/s00438-009-0473-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse R. A., Scacioc A., Hernandez J. M., Krick R., Stephan M., Janshoff A., Thumm M. and Kühnel K. (2013). Qualitative and quantitative characterization of protein-phosphoinositide interactions with liposome-based methods. Autophagy 9, 770-777. 10.4161/auto.23978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wu X., Yao L., Yan L., Zhang L., Qiu J., Liu X., Jia S. and Meng A. (2017). Impairment of cargo transportation caused by gbf1 mutation disrupts vascular integrity and causes hemorrhage in zebrafish embryos. J. Biol. Chem. 292, 2315-2327. 10.1074/jbc.M116.767608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo G., Vicinanza M., Wilson C. and De Matteis M. A. (2012). Phosphoinositides in Golgi complex function. Subcell. Biochem. 59, 255-270. 10.1007/978-94-007-3015-1_8 [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey C. and Chavrier P. (2006). ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, 347-358. 10.1038/nrm1910 [DOI] [PubMed] [Google Scholar]
- De Matteis M. A., Di Campli A. and Godi A. (2005). The role of the phosphoinositides at the Golgi complex. Biochim. Biophys. Acta 1744, 396-405. 10.1016/j.bbamcr.2005.04.013 [DOI] [PubMed] [Google Scholar]
- De Matteis M. A., Wilson C. and D'Angelo G. (2013). Phosphatidylinositol-4-phosphate: the Golgi and beyond. BioEssays 35, 612-622. 10.1002/bies.201200180 [DOI] [PubMed] [Google Scholar]
- Dumaresq-Doiron K., Savard M.-F., Akam S., Costantino S. and Lefrancois S. (2010). The phosphatidylinositol 4-kinase PI4KIIIalpha is required for the recruitment of GBF1 to Golgi membranes. J. Cell Sci. 123, 2273-2280. 10.1242/jcs.055798 [DOI] [PubMed] [Google Scholar]
- Ellong E. N., Soni K. G., Bui Q.-T., Sougrat R., Golinelli-Cohen M.-P. and Jackson C. L. (2011). Interaction between the triglyceride lipase ATGL and the Arf1 activator GBF1. PLoS ONE 6, e21889 10.1371/journal.pone.0021889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata R., Szul T., Alvarez C. and Sztul E. (2003). ADP-ribosylation factor/COPI-dependent events at the endoplasmic reticulum-Golgi interface are regulated by the guanine nucleotide exchange factor GBF1. Mol. Biol. Cell 14, 2250-2261. 10.1091/mbc.E02-11-0730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godi A., Di Campli A., Konstantakopoulos A., Di Tullio G., Alessi D. R., Kular G. S., Daniele T., Marra P., Lucocq J. M. and De Matteis M. A. (2004). FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 6, 393-404. 10.1038/ncb1119 [DOI] [PubMed] [Google Scholar]
- Gustafson M. A. and Fromme J. C. (2017). Regulation of Arf activation occurs via distinct mechanisms at early and late Golgi compartments. Mol. Biol. Cell 28, 3660-3671. 10.1091/mbc.E17-06-0370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto K., Yoshida Y., Tamaki H., Torii S., Shinotsuka C., Yamashina S. and Nakayama K. (2002). GBF1, a guanine nucleotide exchange factor for ADP-ribosylation factors, is localized to the cis-Golgi and involved in membrane association of the COPI coat. Traffic 3, 483-495. 10.1034/j.1600-0854.2002.30705.x [DOI] [PubMed] [Google Scholar]
- Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y. M., Buso N. and Lopez R. (2015). The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43, W580-W584. 10.1093/nar/gkv279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K., Simossis V. A., Taylor W. R. and Heringa J. (2005). A simple and fast secondary structure prediction method using hidden neural networks. Bioinformatics 21, 152-159. 10.1093/bioinformatics/bth487 [DOI] [PubMed] [Google Scholar]
- Macia E., Chabre M. and Franco M. (2001). Specificities for the small G proteins ARF1 and ARF6 of the guanine nucleotide exchange factors ARNO and EFA6. J. Biol. Chem. 276, 24925-24930. 10.1074/jbc.M103284200 [DOI] [PubMed] [Google Scholar]
- Macia E., Partisani M., Favard C., Mortier E., Zimmermann P., Carlier M.-F., Gounon P., Luton F. and Franco M. (2008). The pleckstrin homology domain of the Arf6-specific exchange factor EFA6 localizes to the plasma membrane by interacting with phosphatidylinositol 4,5-bisphosphate and F-actin. J. Biol. Chem. 283, 19836-19844. 10.1074/jbc.M800781200 [DOI] [PubMed] [Google Scholar]
- Mazaki Y., Nishimura Y. and Sabe H. (2012). GBF1 bears a novel phosphatidylinositol-phosphate binding module, BP3K, to link PI3Kgamma activity with Arf1 activation involved in GPCR-mediated neutrophil chemotaxis and superoxide production. Mol. Biol. Cell 23, 2457-2467. 10.1091/mbc.E12-01-0062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monetta P., Slavin I., Romero N. and Alvarez C. (2007). Rab1b interacts with GBF1, modulates both ARF1 dynamics and COPI association. Mol. Biol. Cell 18, 2400-2010. 10.1091/mbc.E06-11-1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu T.-K., Pfeifer A. C., Lippincott-Schwartz J. and Jackson C. L. (2005). Dynamics of GBF1, a brefeldin A-sensitive Arf1 exchange factor at the golgi. Mol. Biol. Cell 16, 1213-1222. 10.1091/mbc.E04-07-0599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson B. C. and Fromme J. C. (2012). Autoregulation of Sec7 Arf-GEF activity and localization by positive feedback. Small GTPases 3, 240-243. 10.4161/sgtp.21828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson B. C., McDonold C. M. and Fromme J. C. (2012). The Sec7 Arf-GEF is recruited to the trans-Golgi network by positive feedback. Dev. Cell 22, 799-810. 10.1016/j.devcel.2012.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A., Lippe R., Christoforidis S., Gaullier J.-M., Brech A., Callaghan J., Toh B.-H., Murphy C., Zerial M. and Stenmark H. (1998). EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 394, 494-498. 10.1038/28879 [DOI] [PubMed] [Google Scholar]
- Simossis V. A. and Heringa J. (2005). PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 33, W289-W294. 10.1093/nar/gki390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simossis V. A., Kleinjung J. and Heringa J. (2005). Homology-extended sequence alignment. Nucleic Acids Res. 33, 816-824. 10.1093/nar/gki233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squizzato S., Park Y. M., Buso N., Gur T., Cowley A., Li W., Uludag M., Pundir S., Cham J. A., McWilliam H. et al. (2015). The EBI Search engine: providing search and retrieval functionality for biological data from EMBL-EBI. Nucleic Acids Res. 43, W585-W588. 10.1093/nar/gkv316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahelin R. V., Scott J. L. and Frick C. T. (2014). Cellular and molecular interactions of phosphoinositides and peripheral proteins. Chem. Phys. Lipids. 182, 3-18. 10.1016/j.chemphyslip.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder D., Barelli H., Gautier R., Macia E., Jackson C. L. and Antonny B. (2011). Kinetic studies of the Arf activator Arno on model membranes in the presence of Arf effectors suggest control by a positive feedback loop. J. Biol. Chem. 286, 3873-3883. 10.1074/jbc.M110.145532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szul T., Garcia-Mata R., Brandon E., Shestopal S., Alvarez C. and Sztul E. (2005). Dissection of membrane dynamics of the ARF-guanine nucleotide exchange factor GBF1. Traffic 6, 374-385. 10.1111/j.1600-0854.2005.00282.x [DOI] [PubMed] [Google Scholar]
- Szul T., Grabski R., Lyons S., Morohashi Y., Shestopal S., Lowe M. and Sztul E. (2007). Dissecting the role of the ARF guanine nucleotide exchange factor GBF1 in Golgi biogenesis and protein trafficking. J. Cell Sci. 120, 3929-3940. 10.1242/jcs.010769 [DOI] [PubMed] [Google Scholar]
- Várnai P. and Balla T. (2008). Live cell imaging of phosphoinositides with expressed inositide binding protein domains. Methods 46, 167-176. 10.1016/j.ymeth.2008.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt S. A., Kular G., Fleming I. N., Downes C. P. and Lucocq J. M. (2002). Subcellular localization of phosphatidylinositol 4,5-bisphosphate using the pleckstrin homology domain of phospholipase C delta1. Biochem. J. 363, 657-666. 10.1042/bj3630657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Lasell T. K. and Melancon P. (2002). Localization of large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi compartments: evidence for distinct functions in protein traffic. Mol. Biol. Cell 13, 119-133. 10.1091/mbc.01-08-0420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Claude A., Chun J., Shields D. J., Presley J. F. and Melancon P. (2006). GBF1, a cis-Golgi and VTCs-localized ARF-GEF, is implicated in ER-to-Golgi protein traffic. J. Cell Sci. 119, 3743-3753. 10.1242/jcs.03173 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.