Abstract
Background
The aim of this study was to assess the relatedness of molecular types of Staphylococcus aureus isolates colonizing cystic fibrosis (CF) patients with their antimicrobial resistance and prevalence of toxin genes.
Methods
A total of 215 isolates from the airways of 107 patients with CF were tested for spa and SCCmec type, antimicrobial resistance and carriage of toxin genes.
Results
t015, t084, t091, t700 and t002 were the largest group (approximately 25%) among all 69 identified spa types. Five new spa types, t14286, t14287, t14288, t14289 and t14290, were identified and registered. Isolates from CF patients were clustered into 11 multi-locus sequence typing clonal complexes, with CC30, CC22, CC97, CC45, CC15 and CC5 being the most frequent ones. Twelve (5.6%) methicillin-resistant S. aureus (MRSA) isolates and 102 (47.7%) multidrug-resistant isolates were identified, along with three SCCmec types (I, III and V). All isolates (both MRSA and methicillin-sensitive S. aureus) were Panton–Valentine leucocidin-negative, and 56.7% harbored egc genes. This was the first study documenting the presence of ST398-V-t571 livestock-associated MRSA in a European patient with CF.
Conclusion
These findings imply that individuals with CF can also be colonized with animal-related ST398 MRSA, and justify constant monitoring of staphylococcal colonization and identification of epidemic S. aureus clones in this group.
Keywords: Staphylococcus aureus, cystic fibrosis, ST398 MRSA, Panton–Valentine leukocidin, spa typing, MRSA
Introduction
Staphylococcus aureus is a primary pathogen colonizing the airways and causing respiratory infections in cystic fibrosis (CF) patients. It is isolated in approximately one-third of the patients, usually shortly after diagnosis. In some cases, staphylococcal colonization persists till the end of patient’s life. Respiratory colonization with S. aureus strains is common during the first decade of life and plays a vital role in morbidity and mortality in most CF patients. S. aureus strains may affect the respiratory function of the lungs in CF patients, which is associated with a high risk of life-threatening infections.1
The growing prevalence of S. aureus in patients with CF observed recently in Europe and the US may result from an increase in the number of isolates resistant to antimicrobial agents, in particular methicillin-resistant S. aureus (MRSA).2 Epidemiological surveillance of staphylococcal colonization is essential to reduce the incidence of epidemic S. aureus clones and to prevent the spread thereof. Molecular typing is a very helpful and important instrument that can be used to study the relatedness, genetic diversity and clonal distribution of S. aureus isolates. Although a number of various techniques have been used to identify S. aureus strains, spa typing and multi-locus sequence typing (MLST) are the most popular methods for the epidemiological analysis of staphylococcal isolates from different sources.3–6 Available evidence suggests that the results of spa typing correlate strongly with the outcomes of clonal grouping obtained with other methods.7
The incidence of staphylococcal colonization in CF patients is still poorly recognized, and only few previous studies have analyzed the clonal structure of S. aureus strains isolated from this group of subjects.
The aim of this study was to assess the relatedness of molecular types of S. aureus isolates colonizing CF patients with their antimicrobial resistance and prevalence of toxin genes.
Materials and methods
Bacterial isolates
The study included material from 107 CF patients (age 1 month to 47 years) treated at the Outpatient Cystic Fibrosis Clinic at “Polanki” Children Hospital in Gdansk between 2012 and 2014. The number of collected swabs varied depending on the course of the treatment, from at least one per patient to two or even five. Collection of swabs for laboratory testing was a part of routine clinical practices. A total of 215 S. aureus isolates from throat swabs, sputum and bronchoalveolar lavage were studied. Throat swabs were obtained after provoking a cough (deep swabs). The material was subcultured onto Columbia blood agar and incubated at 35°C for 24 hours. Suspected staphylococcal isolates were identified on the basis of colony characteristics, pigment production, Gram staining, hemolysis and Staphyslide agglutination test (BioMérieux, Marcy-l’Étoile, France). Additionally, all strains identified as S. aureus were examined for the presence of species-specific thermostable nuclease gene (nucSA), as described by Baron et al.8 The isolates were stored at −80°C in trypticase soy broth (Oxoid, Basingstoke, UK) supplemented with 15% glycerol.
spa typing
spa typing was performed according to Harmsen et al.9 Nucleotide sequencing of the repeat-containing region of the spa gene was conducted on both DNA strands of the PCR product by GenoMed (St Louis, MO, USA), using BigDye Terminator Ready Reaction Cycle Sequencing Kit. The spa types were identified with Ridom StaphType software v.2.1.1 (Ridom GmbH, Münster, Germany),9 and grouped using BURP, Ridom StaphType software. spa types were clustered if the cost between members of a given group was less than or equal to four. The BURP algorithm was used to assign spa types into spa clonal complexes (spa-CCs) with defined default parameters “exclude spa types shorter than five repeats” and “cluster spa types into the same group if cost distances are less than four”.10 Since the results of spa typing and MLST are highly concordant,7 the spa typing data could be easily mapped on the MLST types using SpaServer database (http://spaserver.ridom.de/).
Preparation of bacterial DNA
Total DNA of S. aureus isolates was purified using Genomic Mini DNA Kit (A&A Biotechnology, Gdynia, Poland), in line with the manufacturer’s instructions.
Detection of methicillin resistance and determination of SCCmec cassette type
The isolates were screened for their resistance to oxacillin on the basis of growth of blue colonies in the selective medium (ORSAB; Oxoid). Suspected MRSA isolates were further examined for the presence of S. aureus mecA gene, as described elsewhere.11 Each PCR contained mecA-positive (S. aureus ATCC 43300) and mecA-negative (S. aureus ATCC 29213) strains as controls. All mecA-negative S. aureus isolates able to grow on ORSA plates were tested for the carriage of mecC gene using primers described by Cuny et al.12 Typing of the staphylococcal chromosomal cassette mec (SCCmec) was carried out as described previously by Milheirico et al.13 The PCR products were electrophoretically resolved in 1.5% agarose gel containing 0.5 µg/mL ethidium bromide.
Antimicrobial resistance
The resistance of S. aureus isolates to antimicrobial agents was determined by disk diffusion and interpreted according to the CLSI document no. M02-A11.14 The following drugs were used for the test: penicillin, erythromycin, azithromycin, roxithromycin, clindamycin, lincomycin, ciprofloxacin, ofloxacin, levofloxacin, tetracycline, amikacin, tobramycin, netilmicin, gentamicin, fusidic acid, sulfamethoxazole/trimethoprim, chloramphenicol, vancomycin (all from Becton Dickinson, Franklin Lakes, NJ, USA) and mupirocin (Oxoid). Multidrug resistance was defined as resistance to antimicrobial agents from at least three various classes. For isolates identified as resistant to erythromycin but susceptible to clindamycin, D-test was performed to detect inducible clindamycin resistance. Minimal inhibitory concentration (MIC) for vancomycin was determined by E-tests, in line with the manufacturer’s instructions (AB Biodisk, Solna, Sweden).
Detection of toxin genes
Genes for enterotoxins (sea, seb, sec, sed, see, seg, seh, sei, sej, sek, sel, sem, sen, seo, seu), exfoliative toxins (eta, etb), toxic shock syndrome toxin-1 (tst) and Panton–Valentine leukocidin (lukS-PV/lukF-PV) were detected by means of multiplex PCR, as described elsewhere.15
Statistical analysis
Distributions of the study variables were presented as numbers and percentages, and compared between groups with Pearson’s chi-squared test or Fisher’s exact test. All calculations were carried out with Statistica 10 software (StatSoft, Tulsa, OK, USA), with the threshold of statistical significance set at p<0.001.
Results
spa typing
The 215 S. aureus isolates from CF patients represented 69 spa types, primarily t015 (n=14, 6.5%), t084 (n=11, 5.1%), t091 (n=11, 5.1%), t700 (n=11, 5.1%) and t002 (n=8, 3.7%), which constituted approximately one-fourth of all examined isolates (n=55, 25.6%). Only one isolate was spa non-typeable. The examined material included five new spa types: t14286 (n=2), t14287 (n=1), t14288 (n=2), t14289 (n=5) and t14290 (n=1); all of them had been registered in the international database, Ridom SpaServer (http://spaserver.ridom.de/) (Table 1). MRSA isolates were assigned to four spa types: t2029, t073, t151 and t571.
Table 1.
spa types and spa clonal complexes of Staphylococcus aureus isolated from cystic fibrosis patients
| Cluster | spa | spa-CC | MLST-CC | Predicted ST | N |
|---|---|---|---|---|---|
| 1 | t700 | spa-CC 021 | CC30 | ST30 | 11 |
| 1 | t012 | spa-CC 021 | CC30 | ST30 | 5 |
| 1 | t338 | spa-CC 021 | CC30 | ST30 | 4 |
| 1 | t2029 | spa-CC 021 | CC30 | ST30/ST239 | 4 |
| 1 | t019 | spa-CC 021 | CC30 | ST30 | 3 |
| 1 | t234 | spa-CC 021 | CC30 | ST30/ST239 | 2 |
| 1 | t913 | spa-CC 021 | CC30 | ST30 | 2 |
| 1 | t021 | spa-CC 021 | CC30 | ST30 | 1 |
| 1 | t342 | spa-CC 021 | CC30 | ST30 | 1 |
| 1 | t726 | spa-CC 021 | CC30 | ST30/ST1901 | 1 |
| 1 | t3285 | spa-CC 021 | CC30 | ST30 | 1 |
| 2 | t005 | spa-CC 005 | CC22 | ST22 | 7 |
| 2 | t14289 | spa-CC 005 | CC22 | ST22 | 5 |
| 2 | t709 | spa-CC 005 | CC22 | ST22 | 3 |
| 2 | t223 | spa-CC 005 | CC22 | ST22 | 1 |
| 2 | t474 | spa-CC 005 | CC22 | ST22 | 1 |
| 2 | t891 | spa-CC 005 | CC22 | ST22 | 1 |
| 2 | t2618 | spa-CC 005 | CC22 | ST22 | 1 |
| 2 | t4585 | spa-CC 005 | CC22 | ST22 | 1 |
| 3 | t359 | spa-CC 267/359 | CC97 | ST97 | 5 |
| 3 | t1236 | spa-CC 267/359 | CC97 | ST97 | 3 |
| 3 | t267 | spa-CC 267/359 | CC97 | ST97 | 2 |
| 3 | t521 | spa-CC 267/359 | CC97 | ST97 | 2 |
| 3 | t865 | spa-CC 267/359 | CC97 | ST97 | 2 |
| 4 | t880 | spa-CC 065 | CC45 | ST45/ST1914 | 7 |
| 4 | t330 | spa-CC 065 | CC45 | ST45 | 3 |
| 4 | t065 | spa-CC 065 | CC45 | ST45 | 2 |
| 4 | t715 | spa-CC 065 | CC45 | ST45 | 1 |
| 5 | t084 | spa-CC 4096/091 | CC15 | ST15 | 11 |
| 5 | t091 | spa-CC 4096/091 | CC7 | ST7 | 11 |
| 5 | t2932 | spa-CC 4096/091 | CC7/CC15 | ST7 | 2 |
| 5 | t4096 | spa-CC 4096/091 | CC15 | ST15 | 1 |
| 6 | t002 | spa-CC 5213 | CC5 | ST5 | 8 |
| 6 | t1228 | spa-CC 5213 | CC5 | ST5 | 3 |
| 6 | t6991 | spa-CC 5213 | CC5 | ST5 | 2 |
| 6 | t5213 | spa-CC 5213 | CC5 | ST5 | 1 |
| 7 | t078 | spa-CC 078 | CC25 | ST25 | 7 |
| 7 | t14286 | spa-CC 078 | CC25 | ST25 | 2 |
| 7 | t167 | spa-CC 078 | CC25 | ST25 | 1 |
| 8 | t094 | spa-CC 2636 | CC15 | ST15 | 2 |
| 8 | t547 | spa-CC 2636 | CC15 | ST15 | 1 |
| 8 | t2636 | spa-CC 2636 | CC15 | ST15/ST1905 | 1 |
| 9 | t11263 | spa-CCa | CC30 | ST34 | 3 |
| 9 | t166 | spa-CCa | CC30 | ST34 | 1 |
| 10 | t015 | spa-CCb | CC45 | ST45 | 14 |
| 10 | t073 | spa-CCb | CC45 | ST45 | 4 |
| 11 | t645 | spa-CCc | CC121 | ST121 | 2 |
| 11 | t14288 | spa-CCc | CC121 | ST121 | 2 |
| Singleton | t127 | Singleton | CC1 | ST1 | 6 |
| Singleton | t571 | Singleton | CC398 | ST398 | 6 |
| Singleton | t151 | Singleton | CC5 | ST5/ST225 | 5 |
| Singleton | t209 | Singleton | CC9 | ST109 | 5 |
| Singleton | t4771 | Singleton | – | – | 4 |
| Singleton | t056 | Singleton | CC101 | ST101 | 3 |
| Singleton | t156 | Singleton | CC12 | ST12 | 3 |
| Singleton | t008 | Singleton | CC8 | ST8 | 2 |
| Singleton | t230 | Singleton | CC45 | ST45 | 2 |
| Singleton | t148 | Singleton | – | ST72/ST1434/ST1723 | 1 |
| Singleton | t647 | Singleton | – | ST1027 | 1 |
| Singleton | t924 | Singleton | CC30 | ST30 | 1 |
| Singleton | t4428 | Singleton | – | – | 1 |
| Singleton | t4992 | Singleton | – | – | 1 |
| Singleton | t14287 | Singleton | – | – | 1 |
| Singleton | t14290 | Singleton | – | – | 1 |
| Excluded | t693 | Excluded | CC9 | ST9 | 4 |
| Excluded | t362 | Excluded | CC45 | ST45 | 2 |
| Excluded | t748 | Excluded | CC30 | ST30 | 2 |
| Excluded | t026 | Excluded | CC45 | ST45 | 1 |
| Excluded | t1509 | Excluded | CC15 | ST15 | 1 |
| NT | NT | NT | NT | NT | 1 |
Abbreviations: spa-CC, spa clonal complex; MLST, multi-locus sequence typing; NT, non-typeable.
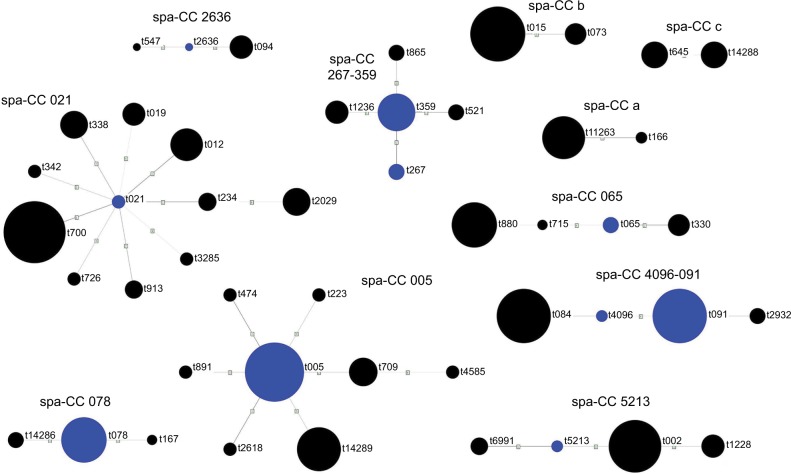
The identified spa types were clustered into 11 spa-CCs by BURP repeat analysis. Ten isolates (4.7%) represented spa types that were excluded from BURP cluster analysis due to the presence of less than five spa repeats. Another 43 isolates (20%) belonged to spa types with a repeat pattern not associated with other detected staphylococci (i.e. singletons). The majority of isolates represented five clusters, namely spa-CC 021 belonging to ST-CC30, spa-CC 005 belonging to ST-CC22, spa-CC 267/359 belonging to ST-CC97, spa-CC 065 belonging to ST-CC45 and spa-CC 4096/091 belonging to ST-CC15 and ST-CC7. The newly described spa types were distributed across various clusters and singletons (Figure 1; Table 1).
Figure 1.
Cluster analysis of spa types of Staphylococcus aureus isolates from cystic fibrosis patients. Blue circles represent group founders and circle sizes are proportional to the frequency of a given spa type.
Abbreviation: spa-CC, spa clonal complex.
Antimicrobial resistance
The examined isolates showed resistance to penicillin (83.3%), azithromycin (50.7%), erythromycin (49.8%), roxithromycin (49.8%), clindamycin (43.7%), lincomycin (43.7%), ciprofloxacin (13%), ofloxacin (13%), levofloxacin (9.3%), tetracycline (7.9%), amikacin (8.8%), tobramycin (6.1%), netilmicin (6.1%) and gentamicin (5.6%). The D-test demonstrated that 10.7% of isolates from CF patients represented the inducible phenotype of clindamycin resistance (MLSBi). Resistance to fusidic acid, sulfamethoxazole/trimethoprim and chloramphenicol was found in 4.2%, 3.3% and 1.9% of the isolates, respectively (Table 2). All isolates were sensitive to vancomycin and mupirocin, with MICs for the former antimicrobial agent ranging between 0.25 µg/mL and 1 µg/mL.
Table 2.
Antimicrobial resistance profiles of Staphylococcus aureus isolated from cystic fibrosis patients
| Antibiotics | MRSA % (n=12) |
MSSA % (n=203) |
p-value | Total S. aureus % (n=215) |
|---|---|---|---|---|
| Penicillin | 100 (n=12) | 82.3 (n=167) | 0.110 | 83.3 (n=179) |
| Azithromycin | 100 (n=12) | 47.8 (n=97) | <0.001 | 50.7 (n=109) |
| Erythromycin | 100 (n=12) | 46.8 (n=95) | <0.001 | 49.8 (n=107) |
| Roxithromycin | 100 (n=12) | 46.8 (n=95) | <0.001 | 49.8 (n=107) |
| Clindamycin | 100 (n=12) | 40.4 (n=82) | <0.001 | 43.7 (n=94) |
| Clindamycinind* | 91.6 (n=11) | 5.9 (n=12) | <0.001 | 10.7 (n=23) |
| Lincomycin | 100 (n=12) | 40.4 (n=82) | <0.001 | 43.7 (n=94) |
| Ciprofloxacin | 50 (n=6) | 10.8 (n=22) | <0.001 | 13 (n=28) |
| Ofloxacin | 41.6 (n=5) | 11.3 (n=23) | 0.002 | 13 (n=28) |
| Levofloxacin | 41.6 (n=5) | 7.4 (n=15) | <0.001 | 9.3 (n=20) |
| Tobramycin | 50 (n=6) | 3 (n=6) | <0.001 | 6.1 (n=13) |
| Amikacin | 25 (n=3) | 6.4 (n=13) | 0.017 | 8.8 (n=19) |
| Netilmicin | 25 (n=3) | 4.9 (n=10) | 0.005 | 6.1 (n=13) |
| Gentamicin | 41.6 (n=5) | 4.4 (n=9) | <0.001 | 5.6 (n=12) |
| Tetracycline | 33.3 (n=4) | 6.4 (n=13) | 0.001 | 7.9 (n=17) |
| Fusidic acid | 16.6 (n=2) | 3.4 (n=7) | 0.026 | 4.2 (n=9) |
| Sulfamethoxazole/trimethoprim | 16.6 (n=2) | 2.5 (n=5) | 0.007 | 3.3 (n=7) |
| Chloramphenicol | 0 | 2 (n=4) | 0.623 | 1.9 (n=4) |
| Vancomycin | 0 | 0 | 0 | |
| Mupirocin | 0 | 0 | 0 | |
| Multidrug-resistant | 100 (n=12) | 41.9 (n=90) | <0.001 | 47.4 (n=102) |
Note:
Clindamycinind: inducible clindamycin resistance.
Abbreviations: MRSA, methicillin-resistant S. aureus; MSSA, methicillin-sensitive S. aureus.
Twelve (5.6%) S. aureus isolates were identified as MRSA. All these isolates were mecA-positive. All MRSA isolates displayed multidrug resistance. The prevalence of multidrug-resistant (MDR) staphylococci among MRSA isolates turned out to be significantly higher than among methicillin-sensitive S. aureus (MSSA) (p<0.001). All MRSA isolates were resistant to macrolides and lincosamides, and 91.6% showed inducible phenotype of clindamycin resistance (MLSBi). The latter proportion was significantly higher than the percentage of MSSA isolates with inducible resistance to clindamycin (p<0.001) (Table 2). MRSA isolates represented three distinct SCCmec types: SCCmec I, III and V. Isolates SCCmec V belonged to ST398-t571 (Table 3).
Table 3.
Characteristics of methicillin-resistant Staphylococcus aureus isolates from cystic fibrosis patients
| Patient | Number of isolates | Cluster | spa | spa-CC | MLST- CC | Predicted ST | SCCmec | Toxin genes | Antibiotic resistance |
|---|---|---|---|---|---|---|---|---|---|
| A | 2 | 1 | t2029 | spa-CC 021 | CC30 | ST30/ST239 | III | sea | FOX, AZ, ERY, ROX, L, CLIN, TET |
| A | 1 | 1 | t2029 | spa-CC 021 | CC30 | ST30/ST239 | III | sea | FOX, AZ, ERY, ROX, L, CLIN, TET, GEN, AM, NET, TO, SXT |
| B | 1 | 10 | t073 | spa-CCb | CC45 | ST45 | II | seg, sei, sem, sen, seo | FOX, AZ, ERY, ROX, L, CLIN, CIP |
| C | 1 | Singleton | t151 | Singleton | CC5 | ST5/ST225 | II | None | FOX, AZ, ERY, ROX, L, CLIN, CIP, OF, LEV, GEN, TO |
| C | 2 | Singleton | t151 | Singleton | CC5 | ST5/ST225 | II | None | FOX, AZ, ERY, ROX, L, CLIN, CIP, OF, LEV, GEN, TO |
| C | 2 | Singleton | t151 | Singleton | CC5 | ST5/ST225 | II | None | FOX, AZ, ERY, ROX, L, CLIN, TET, CIP, OF, LEV, GEN, TO, NET, AM |
| D | 3 | Singleton | t571 | Singleton | CC398 | ST398 | V | None | FOX, AZ, ERY, ROX, L, CLIN, SXT |
Abbreviations: spa-CC, spa clonal complex; MLST, multi-locus sequence typing; FOX, cefoxitin; AZ: azithromycin; ERY, erythromycin; ROX, roxithromycin; L, lincomycin; CLIN, clindamycin; TET, tetracycline; GEN, gentamicin; AM, amikacin; NET, netilmicin; TO, tobramycin; SXT, sulfamethoxazole/trimethoprim; CIP, ciprofloxacin; OF, ofloxacin; LEV, levofloxacin.
Prevalence of toxin genes
The list of virulence factors harbored by the examined CF isolates included sea (16.7%), seb (3.7%), sec (15.8%), sed (0.5%), seg (56.7%), seh (4.7%), sei (57.6%), sej (2.8%), sek (36.7%), sel (27%), sem (56.7%), sen (57.2%), seo (57.6%), seu (28.8%), eta (4.7%) and tst (14.9%) (Table 4). Enterotoxin gene cluster (egc)-positive isolates (56.7%) turned out to be the largest group among all examined staphylococci.
Table 4.
Prevalence of toxin genes among Staphylococcus aureus isolated from cystic fibrosis patients
| Toxin genes* | Number of CF isolates (%) |
|---|---|
| sea | 36 (16.7) |
| seb | 8 (3.7) |
| sec | 34 (15.8) |
| sed | 1 (0.5) |
| see | 0 |
| seg | 122 (56.7) |
| seh | 10 (4.7) |
| sei | 124 (57.6) |
| sej | 6 (2.8) |
| sek | 79 (36.7) |
| sel | 58 (27) |
| sem | 122 (56.7) |
| sen | 123 (57.2) |
| seo | 124 (57.6) |
| seu | 62 (28.8) |
| eta | 10 (4.7) |
| etb | 0 |
| tst | 32 (14.9) |
| lukS-PV/lukF-PV | 0 |
Note:
Genes encoding staphylococcal enterotoxins (sea, seb, sec, sed, see, seg, seh, sei, sej, sek, sel, sem, sen, seo, seu), exfoliative toxins (eta, etb), toxic shock syndrome toxin-1 (tst) and Panton–Valentine leukocidin (lukS-PV/lukF-PV).
Abbreviation: CF, cystic fibrosis.
Discussion
Using spa typing, we demonstrated a substantial diversity of S. aureus isolates from the airways of CF patients (a total of 69 various spa types were isolated from 107 patients). Even greater diversity has been recently reported by Masoud-Landgraf et al16 who identified up to 48 spa types within only 58 isolates from Austrian patients with CF. Comparison of our hereby presented findings with recent data from other European countries shows that spa types isolated from CF patients are similar to those from individuals with other staphylococcal colonization or from the carriers.17,18 Thus, CF patients do not seem to be colonized with any unique spa type(s), but rather colonized with the types being currently spread in a given population.
spa types identified in this study formed clonal complexes with a pattern typical for Polish hospitals.19,20 Clonal complexes CC30 and CC22 from clusters 1 and 2 for many years have been predominating in many European countries, including Poland.21,22 It should be emphasized that the predominance of CC30 complex has also been recently reported by Masoud-Landgraf et al16 in Austrian CF patients. In contrast to studies that have been conducted a decade ago, cluster 1 (CC30) did not include any t037/ST239 isolates, previously widespread in Polish hospitals.19,20
Despite a huge diversity of spa types (eight out of 20 isolates), the second cluster with CC22 complex showed high homogeneity in terms of sequence type (ST22), which distinguished this cluster from other clusters (especially cluster 5) in which an opposite phenomenon was observed.
The third cluster, with CC97 complex, included less isolates than the remaining two and showed greater genetic stability, representing only one sequential type. This complex can be represented by isolates with CA-MRSA phenotype, originating from both humans and cattle.22,23 In our study, staphylococci belonging to CC97 complex turned out to be sensitive to methicillin (MSSA); likewise, more than 90% of S. aureus isolates from our material were sensitive to methicillin.
Although some recently published papers documented a dramatic increase in the proportion of MRSA isolated from CF patients, even up to 25%,24 we found MRSA in only four study subjects; thus, MRSA isolates represented merely 5.6% of all staphylococci examined in this study. Low prevalence of MRSA documented in this study is generally consistent with the results of previous studies of CF patients from other European countries,16,25 but stays in opposition to frequent occurrence of these isolates reported from the US.24
It is colonization with MDR isolates, rather than resistance to methicillin, which seems to be a primary concern in CF patients; MDR isolates constituted nearly a half of staphylococci identified in this study, including all MRSA isolates. Aside from beta-lactams, MDR strains most often showed resistance to macrolides, lincosamides and fluoroquinolones, which is not surprising owing to current standards of antibacterial treatment in CF patients.
MRSA isolates identified in this study belonged to four different clonal complexes: CC30, CC45, CC5 and CC398. SCCmec types I–III carry additional genes that provide resistance to antibiotics other than beta-lactams; in contrast, SCCmec types IV and V usually do not carry additional drug resistance genes. ST5-t151 MRSA isolates identified in our study showed a pattern band characteristic for SCC-mec type II, and were characterized by the highest rate of antimicrobial resistance. Previously, this type was associated with an spa-CC found primarily in a hospital setting, and corresponded to one of the most prevalent hospital clones, ST5, or to its single-locus variant, Rhine-Hesse clone ST225. Both these STs have been recently identified among epidemic hospital clones from Poland and neighboring countries.20,26 The first CC5-ST5 pediatric clone was described in Portugal.27
Isolation of a single MRSA isolate ST45, with SCCmec type II, was previously reported in Poland, the US, Hong Kong and Finland.20,28,29 This isolate was resistant to macrolides, lincosamides and quinolones, and harbored egc.
SCCmec type III was related to ST239-III-t2029 type which involved isolates resistant to many antibiotics (macrolides, lincosamides, aminoglycosides, cotrimoxazole) and carried sea gene. This MRSA type has been previously isolated in a Malaysian hospital, also from the respiratory tract, and aside from the antibiotics mentioned above, also showed resistance to rifampicin and fusidic acid.30
Six of our isolates represented ST-389 type which may cause various human infections. A number of patients with soft tissue and skin infections caused by this type were reported.31 However currently, ST-398 is considered to be a noteworthy zoonotic pathogen commonly found in livestock and persons being in close contact with ST398-positive animals. This livestock-associated MRSA (LA-MRSA) clone is widespread in Europe, Asia and North America.5 LA-MRSA have been isolated from various human infections, including folliculitis, osteomyelitis, endocarditis and skin and soft tissue infections.31 To the best of our knowledge, the only published report documenting isolation of this clone from a CF patient originates from Brazil.5 Moreover, Chinese authors reported isolation of S. aureus ST398-V-t571 from a student’s nasal cavity.3 Our isolate originated from a 1-year-old boy whose mother practiced horse riding. According to the most recent report published in 2017, ST398 is a predominant staphylococcal isolate from equine infections caused by MRSA.4 In a recent study conducted by Mroczkowska et al,4 ST398 turned out to be a predominant clone isolated from animals in Poland. To the best of our knowledge, our current study is the first to demonstrate the colonization with LA-MRSA CC398 clone in a CF patient from Europe.
ST398 is commonly associated with the presence of Panton–Valentine leukocidin.32 However, both ST398-t571 and other isolates found in our patients were PVL-negative. The role of PVL in staphylococcal infections is still a matter of discussion.33,34 On the one hand, presence of PVL genes in MRSA isolates from CF patients was shown to be associated with invasive lung infection.35 On the other hand, either in our present study or in previous studies conducted in Europe and on other continents, all staphylococci from individuals with CF turned out to be PVL-negative.5,16,36 This implies that PVL is not necessarily a key virulence factor involved in staphylococcal infections in CF patients.
According to some previously published reports, CC22 may also include PVL-harboring S. aureus.37 However, although we have isolated staphylococci belonging to this complex, none of them harbored PVL-encoding genes. This implies that isolates belonging to the same clonal lineage may display diverse toxin gene profiles.
Enterotoxin genes seg, sei, sem, sen, seo and seu are organized within the egc. This cluster is located on the genomic island nSAb and considered to be the “nursery” of staphylococcal enterotoxins.38 Many staphylococcal isolates from our CF patients were egc-positive, which is consistent with previous reports.2,39 No relatedness with any specific molecular type was found in egc-positive isolates. In line with these findings, egc may be a virulence factor promoting colonization with S. aureus in CF patients.40
Staphylococcal superantigens were shown to nonspecifically activate proliferation of T lymphocytes, binding to VbTCR-2 on these cells and to MHC class II molecules. Indeed, some S. aureus isolates from our CF patients harbored genes for superantigens, including among them enterotoxins sea and sec, and tst. Also, Liu et al2 detected diverse superantigens genes in more than 50% of staphylococcal isolates from patients with CF. Taken all together, these findings imply that presence of superantigens genes may be linked with the pathogenesis of staphylococcal infections in this group.
To summarize, this study demonstrated that S. aureus isolates from CF patients vary considerably in terms of spa types, clonal complexes and carriage of toxin genes. We did not find any PVL-positive isolates, which implies that this toxin is not necessarily a key virulence factor involved in staphylococcal infections in CF patients. To the best of our knowledge, this is the first Polish study documenting genetic structure of S. aureus isolates from patients with CF, as well as the first European report describing the presence of ST398-V-t571 LA-MRSA in a person with this condition. This implies that patients with CF can also be colonized with ST398 MRSA, and justifies constant monitoring of staphylococcal colonization and identification of epidemic S. aureus clones in this group.
Acknowledgments
This study was supported by a research grant (no. DEC-2011/03/B/NZ5/04276) from the National Science Centre, and partly by a specific subsidy of Ministry of Science and Higher Education for Medical University of Gdansk (grant nos. ST02-0099/07/402 and ST02-0543/07/289).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ratjen FA. Cystic fibrosis: pathogenesis and future treatment strategies. Respir Care. 2009;54(5):595–605. doi: 10.4187/aarc0427. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y, Zhang J, Zhong D, et al. Characterization of Staphylococcus aureus isolates from pediatric patients with cystic fibrosis. World J Microbiol Biotechnol. 2016;32(10):162. doi: 10.1007/s11274-016-2122-4. [DOI] [PubMed] [Google Scholar]
- 3.Du J, Chen C, Ding B, et al. Molecular characterization and antimicrobial susceptibility of nasal Staphylococcus aureus isolates from a Chinese medical college campus. PLoS One. 2011;6(11):17. doi: 10.1371/journal.pone.0027328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerin F, Fines-Guyon M, Meignen P, et al. Nationwide molecular epidemiology of methicillin-resistant Staphylococcus aureus responsible for horse infections in France. BMC Microbiol. 2017;17(1):104. doi: 10.1186/s12866-016-0924-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lima DF, Cohen RW, Rocha GA, Albano RM, Marques EA, Leao RS. Genomic information on multidrug-resistant livestock-associated methicillin-resistant Staphylococcus aureus ST398 isolated from a Brazilian patient with cystic fibrosis. Mem Inst Oswaldo Cruz. 2017;112(1):79–80. doi: 10.1590/0074-02760160342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mroczkowska A, Zmudzki J, Marszalek N, et al. Livestock-associated Staphylococcus aureus on Polish pig farms. PLoS One. 2017;12(2):e0170745. doi: 10.1371/journal.pone.0170745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strommenger B, Kettlitz C, Weniger T, Harmsen D, Friedrich AW, Witte W. Assignment of Staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J Clin Microbiol. 2006;44(7):2533–2540. doi: 10.1128/JCM.00420-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baron F, Cochet MF, Pellerin JL, et al. Development of a PCR test to differentiate between Staphylococcus aureus and Staphylococcus intermedius. J Food Prot. 2004;67(10):2302–2305. doi: 10.4315/0362-028x-67.10.2302. [DOI] [PubMed] [Google Scholar]
- 9.Harmsen D, Claus H, Witte W, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41(12):5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mellmann A, Weniger T, Berssenbrugge C, et al. Based Upon Repeat Pattern (BURP): an algorithm to characterize the long-term evolution of Staphylococcus aureus populations based on spa polymorphisms. BMC Microbiol. 2007;7:98. doi: 10.1186/1471-2180-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29(10):2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuny C, Layer F, Strommenger B, Witte W. Rare occurrence of methicillin-resistant Staphylococcus aureus CC130 with a novel mecA homologue in humans in Germany. PLoS One. 2011;6(9):8. doi: 10.1371/journal.pone.0024360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milheirico C, Oliveira DC, de Lencastre H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51(9):3374–3377. doi: 10.1128/AAC.00275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute . Performance standards for antimicrobial susceptibility testing; twenty-second informational supplement. Document M02-A11. Wayne, PA: Clinical and Laboratory Standards Institute; 2008. [Google Scholar]
- 15.Becker K, Friedrich AW, Lubritz G, Weilert M, Peters G, Von Eiff C. Prevalence of genes encoding pyrogenic toxin superantigens and exfoliative toxins among strains of Staphylococcus aureus isolated from blood and nasal specimens. J Clin Microbiol. 2003;41(4):1434–1439. doi: 10.1128/JCM.41.4.1434-1439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masoud-Landgraf L, Johler S, Badura A, et al. Genetic and phenotypic characteristics of Staphylococcus aureus isolates from cystic fibrosis patients in Austria. Respiration. 2015;89(5):390–395. doi: 10.1159/000377707. [DOI] [PubMed] [Google Scholar]
- 17.Becker K, Schaumburg F, Fegeler C, Friedrich AW, Kock R. Staphylococcus aureus from the German general population is highly diverse. Int J Med Microbiol. 2017;307(1):21–27. doi: 10.1016/j.ijmm.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, Friedrich AW. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 2010;7(1):1000215. doi: 10.1371/journal.pmed.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krzyszton-Russjan J, Empel J, Leski T, Gniadkowski M, Hryniewicz W. Clonal structure of the methicillin-resistant Staphylococcus aureus (MRSA) population in Poland: revision and update. Microb Drug Resist. 2005;11(2):127–136. doi: 10.1089/mdr.2005.11.127. [DOI] [PubMed] [Google Scholar]
- 20.Luczak-Kadlubowska A, Sulikowska A, Empel J, et al. Countrywide molecular survey of methicillin-resistant Staphylococcus aureus strains in Poland. J Clin Microbiol. 2008;46(9):2930–2937. doi: 10.1128/JCM.00869-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grundmann H, Schouls LM, Aanensen DM, et al. The dynamic changes of dominant clones of Staphylococcus aureus causing bloodstream infections in the European region: results of a second structured survey. Euro Surveill. 2014;19(49):20987. doi: 10.2807/1560-7917.es2014.19.49.20987. [DOI] [PubMed] [Google Scholar]
- 22.Holtfreter S, Grumann D, Balau V, et al. Molecular epidemiology of Staphylococcus aureus in the general population in Northeast Germany: results of the Study of Health in Pomerania (SHIP-TREND-0) J Clin Microbiol. 2016;54(11):2774–2785. doi: 10.1128/JCM.00312-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Locatelli C, Cremonesi P, Caprioli A, et al. Occurrence of methicillin-resistant Staphylococcus aureus in dairy cattle herds, related swine farms, and humans in contact with herds. J Dairy Sci. 2017;100(1):608–619. doi: 10.3168/jds.2016-11797. [DOI] [PubMed] [Google Scholar]
- 24.Rosenthal VD, Bijie H, Maki DG, et al. International Nosocomial Infection Control Consortium (INICC) report, data summary of 36 countries, for 2004-2009. Am J Infect Control. 2012;40(5):396–407. doi: 10.1016/j.ajic.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 25.Campana S, Cocchi P, Döring G, Taccetti G, Moroney SM. Emergence of an epidemic clone of community-associated methicillin-resistant Panton-Valentine leucocidin-negative Staphylococcus aureus in cystic fibrosis patient populations. J Clin Microbiol. 2007;45(9):3146. doi: 10.1128/JCM.00132-07. author reply 3146–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasprzyk J, Piechowicz L, Wisniewska K, Dziewit L, Bronk M, Swiec K. Differentiation of spa types and staphylococcal cassette chromosome mec (SCCmec) in clinical methicillin-resistant Staphylococcus aureus isolated in medical sites of Gdansk region. Med Dosw Mikrobiol. 2015;67(2):79–88. Polish [with English abstract] [PubMed] [Google Scholar]
- 27.Oliveira DC, Tomasz A, de Lencastre H. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb Drug Resist. 2001;7(4):349–361. doi: 10.1089/10766290152773365. [DOI] [PubMed] [Google Scholar]
- 28.Ip M, Yung RW, Ng TK, et al. Contemporary methicillin-resistant Staphylococcus aureus clones in Hong Kong. J Clin Microbiol. 2005;43(10):5069–5073. doi: 10.1128/JCM.43.10.5069-5073.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson DA, Enright MC. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2003;47(12):3926–3934. doi: 10.1128/AAC.47.12.3926-3934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim KT, Teh CS, Yusof MY, Thong KL. Mutations in rpoB and fusA cause resistance to rifampicin and fusidic acid in methicillin-resistant Staphylococcus aureus strains from a tertiary hospital in Malaysia. Trans R Soc Trop Med Hyg. 2014;108(2):112–118. doi: 10.1093/trstmh/trt111. [DOI] [PubMed] [Google Scholar]
- 31.van Belkum A, Melles DC, Peeters JK, et al. Methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerg Infect Dis. 2008;14(3):479–483. doi: 10.3201/eid1403.0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao C, Liu Y, Zhao M, et al. Characterization of community acquired Staphylococcus aureus associated with skin and soft tissue infection in Beijing: high prevalence of PVL+ ST398. PLoS One. 2012;7(6):6. doi: 10.1371/journal.pone.0038577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bubeck Wardenburg J, Palazzolo-Ballance AM, Otto M, Schneewind O, DeLeo FR. Panton-Valentine leukocidin is not a virulence determinant in murine models of community-associated methicillin-resistant Staphylococcus aureus disease. J Infect Dis. 2008;198(8):1166–1170. doi: 10.1086/592053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Labandeira-Rey M, Couzon F, Boisset S, et al. Panton-Valentine leukocidin causes necrotizing pneumonia. Science. 2007;315(5815):1130–1133. doi: 10.1126/science.1137165. [DOI] [PubMed] [Google Scholar]
- 35.Elizur A, Orscheln RC, Ferkol TW, et al. Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus lung infection in patients with cystic fibrosis. Chest. 2007;131(6):1718–1725. doi: 10.1378/chest.06-2756. [DOI] [PubMed] [Google Scholar]
- 36.Mimica MJ, Berezin EN, Damaceno N, Carvalho RB. SCCmec type IV, PVL-negative, methicillin-resistant Staphylococcus aureus in cystic fibrosis patients from Brazil. Curr Microbiol. 2011;62(2):388–390. doi: 10.1007/s00284-010-9718-y. [DOI] [PubMed] [Google Scholar]
- 37.Masiuk H, Kopron K, Grumann D, et al. Association of recurrent furunculosis with Panton-Valentine leukocidin and the genetic background of Staphylococcus aureus. J Clin Microbiol. 2010;48(5):1527–1535. doi: 10.1128/JCM.02094-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monday SR, Bohach GA. Genes encoding staphylococcal enterotoxins G and I are linked and separated by DNA related to other staphylococcal enterotoxins. J Nat Toxins. 2001;10(1):1–8. [PubMed] [Google Scholar]
- 39.Ormerod KL, George NM, Fraser JA, Wainwright C, Hugenholtz P. Comparative genomics of non-pseudomonal bacterial species colonising paediatric cystic fibrosis patients. PeerJ. 2015;3:e1223. doi: 10.7717/peerj.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nowrouzian FL, Dauwalder O, Meugnier H, et al. Adhesin and superantigen genes and the capacity of Staphylococcus aureus to colonize the infantile gut. J Infect Dis. 2011;204(5):714–721. doi: 10.1093/infdis/jir388. [DOI] [PubMed] [Google Scholar]