ABSTRACT
Nisin is a lantibiotic, a member of a family of polypeptides containing lanthionine with antimicrobial activity. Nisin-producing microorganisms require immunity proteins for self-protection from nisin itself. Lactococcus lactis, a microorganism that synthesizes nisin, has an integral NisFEG ABC transporter and an NisI lipoprotein that function in nisin immunity. Here, we present the crystal structure of the full length of NisI22-C, a lipid-free form of NisI, determined at 1.9-Å resolution. As with the nuclear magnetic resonance (NMR) structures of the N- and C-terminal domains of NisI, NisI22-C is composed of N- and C-terminal domains, both of which display a fold similar to that found in SpaI, a lipoprotein with immunity against subtilin in Bacillus subtilis. The full-length structure of NisI22-c reveals a large, deep cleft by the interdomain association, one side of which is occupied by the residues important for immunity. Opposite the cleft, a shallow groove is found where nisin-interacting residues are distributed in the periphery composed of the C-terminal negative patch. Based on a sulfate ion found in the large and deep cleft, a model of NisI in complex with a farnesyl diphosphate backbone of lipid II is proposed, suggesting a mechanism for increasing the chances of encountering nisin.
KEYWORDS: nisin, lantibiotics, NisI, immunity protein, SpaI, Lactococcus lactis, crystal structure, X-ray crystallography
INTRODUCTION
Bacteriocins are proteinaceous antimicrobial agents produced naturally by bacteria and have been categorized as alternatives to traditional antibiotics (1). Lantibiotics belonging to the class I bacteriocins, which involve posttranslational modifications, have inhibitory and killing activity against clinically important strains, such as Streptococcus pneumoniae, methicillin-resistant Staphylococcus aureus (MRSA), and vancomycin-resistant enterococci (VRE) (2), and have been studied with the intent to generate derivatives with properties suitable for general medical applications (3, 4). Lantibiotics are synthesized through posttranslational modifications such as dehydration and cyclization, producing lanthionine or methyllanthionine, and are secreted to the extracellular space (5).
Nisin, the first identified lantibiotic, is produced by Gram-positive bacteria, such as Lactococcus lactis (6) and Streptococcus uberis (7). Nisin is produced from the posttranslational modification of an inactive precursor, NisA (8), and the cleavage of a leader sequence containing 21 amino acids, which is required for extracellular transportation (8). After synthesis of the NisA precursor, NisB and NisC catalyze the dehydration of serine and threonine residues and the cyclization of these dehydrated residues with a cysteine residue, which is limited to the region consisting of 34 amino acids, excluding the leader sequence (9, 10). The modified preprotein is then transported by NisT (11) and localized to the extracellular side of the cytoplasmic membrane, where the leader sequence is cleaved off by the NisP protease (12). The fully modified active nisin is characterized by five lanthionine- or methyllanthionine-based rings, which are designated rings A, B, C, D, and E from the N terminus to the C terminus, with the last two rings intertwined.
One of the mechanisms of antimicrobial activity by nisin is based on pore formation in the cytoplasmic membrane of Gram-positive bacteria (13). However, pore formation by nisin differs from that of other antimicrobial peptides in that nisin pore formation is targeted. Lipid II, the final intermediate precursor for peptidoglycan synthesis, provides docking sites for nisin on the membrane (14). It has been shown that after interaction with lipid II, the C-terminal tails of nisin molecules are inserted into the membrane and associate each other to form the pore (15). Nuclear magnetic resonance (NMR) studies of the nisin-lipid II complex have shown that the N-terminal A and B lanthionine rings of nisin form a structure like cage for specific binding to the pyrophosphate moiety of lipid II, corroborating the specific binding of lipid II by nisin (16). In addition to antimicrobial pore formation, an alternative mode of antimicrobial activity of lantibiotics, including nisin, is the targeting lipid II. It has recently been shown that nisin kills bacteria by impairing the growth of bacterial compartments that require continuous synthesis of peptidoglycan, such as the septum, via the segregation of lipid II (17).
As a self-protection strategy against lantibiotics, genes of immunity proteins are found together with genes for the biosynthesis of lantibiotics (18–21). For example, nisin-producing L. lactis bacteria use an integral ATP-binding cassette (ABC) transporter, NisFEG, and a specific lantibiotic binding protein, NisI. It has been shown that NisFEG can function as an immunity protein by transporting nisin out of the cytoplasmic membrane, thereby removing nisin from the membrane (22). NisI carries out its immune activity by binding to nisin, thereby preventing nisin from reaching its target molecules (22–24). NisI is a lipoprotein and, as a preprotein, it consists of 245 amino acids. The N-terminal leader sequence containing a lipobox consensus sequence (16-GLSGCY-21), where the cysteine residue is modified with a diacylglycerol moiety, functions as a secretion signal, and the N-terminal 19 amino acids before the cysteine residue of the lipobox are removed during posttranslational modification (18, 25). The resulting NisI protein is anchored to the extracellular surface of the cytoplasmic membrane and confers immunity to nisin. NisI also exists as a lipid-free form (LFNisI) in the media, probably due to escape from lipid modification (26). A functional study with C-terminally truncated NisI mutants showed that the C-terminal fragment consisting of 21 amino acids is important for nisin immunity, specifically by inhibiting nisin-mediated pore formation (27, 28). Recently, structural studies of the isolated N- and C-terminal domains of NisI using NMR revealed that NisI consists of two structurally similar domains, both of which are structurally homologous to SpaI, an LanI immunity protein to subtilin in B. subtilis, but whereas the N-terminal domain is specific for the interaction with the membrane, the C-terminal domain is involved in the recognition of nisin (24, 29). Currently, the structure of the full length of NisI is not available, limiting the information about the organization of the N- and C-terminal domains of NisI.
A line of evidence suggests that NisI exerts its immunity by blocking nisin from reaching its target molecules, such as lipid II, through a direct interaction (22–24). However, the molecular mechanism by which NisI protects nisin-producing strains from nisin is poorly understood. One of the reason could be the absence of the 3D structure of the full length of NisI, which could provide a structural basis for biochemical and biophysical studies. In order to gain an insight into the organization of the N- and C-terminal SpaI folds of NisI, the crystal structure of the full length of NisI has been determined. Interestingly, the interdomain association of the N- and C-terminal domains of NisI produces a deep cleft and a groove formed by the N- and C-terminal domains of NisI, which might be an important binding site of nisin or other molecules.
RESULTS AND DISCUSSION
Overall structure of NisI22-C.
A truncated version of L. lactis NisI (NisI22-C; residues 22 to 245) without the N-terminal signal peptide leader sequence and the lipobox (LSGC; residues 17 to 20) was used for structural studies (Fig. 1). Initially, native NisI22-C was used for crystallization, but it did not produce any crystals. As a rescue strategy for crystallization, reductive methylation of the lysine residues of NisI22-C was carried out to generate methylated NisI22-C (30). Because of the simplicity of the steps for chemical modification and its direct applicability to purified proteins (30, 31), reductive methylation has been used for getting an initial crystal or crystals to diffract better. Importantly, several studies showed that native and methylated proteins have very similar structures and maintain their biochemical activity (32–34). Therefore, methylated NisI22-C was crystallized and was used for structural determination as described before (35). We then determined the X-ray crystal structure of NisI at a resolution of 1.9 Å with an Rwork of 17.6% and an Rfree of 20.9% using the single anomalous dispersion (SAD) method (Table 1). The N-terminal (residues 22 to 27), C-terminal (residues 244 to 245), and internal residues (residues 130 to 136 and residues 196 to 206) were not traced, due to the poor electron density map. There was one NisI22-C molecule in the asymmetry unit.
FIG 1.
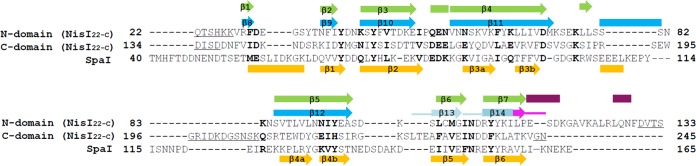
Structure-guided sequence alignment. Structure-guided sequence alignment of the N- and C-terminal domains of NisI22-C from L. lactis with those of SpaI from B. subtilis. Identical and similar residues are shown in bold, gaps are shown with hyphens, and internal disordered residues are underlined. The residue numbers are relative to those of the methionine residues in the prelipoproteins before processing of the N-terminal leader sequence. Secondary structures (β-strand, arrows; α-helix, rectangles) of NisI22-C are shown above the alignment and colored in the scheme as in Fig. 2A, and those of SpaI are shown in orange below the alignment. For the 21-amino-acid fragment of the C-terminal domain of NisI22-C, which includes the 5-amino-acid C-terminal fragment (pink), the loops of the secondary structures are also shown with thick lines. The numbers of the secondary structures are assigned only to β-strands comprising the β-barrel structure, and those of SpaI are based on the published report (29).
TABLE 1.
Data collection and refinement statistics for NisI22-C
| Statistic | Se-Met NisI22-Ca |
|---|---|
| Data collection | |
| Space group | P2(1)2(1)2(1) |
| Cell dimensions | |
| a, b, c (Å) | 45.98, 76.86, 76.66 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 30.0–1.9 (1.97–1.9) |
| Rmergeb (%) | 7.4 (32.3) |
| <I/σ (I)> | 11.1 (9.7) |
| Completeness (%) | 98.3 (95.7) |
| Redundancy | 13.2 (12.4) |
| Structure refinement | |
| Resolution (Å) | 30.0–1.9 |
| Reflections, total/test set | 21,483/1,971 |
| Rwork/Rfreec | 17.6/20.9 |
| No. of atoms, protein/sulfate ion/water | 1,604/5/188 |
| RMSDd | |
| Bond lengths (Å) | 0.007 |
| Angles (°) | 1.02 |
| Average B-factors (Å2) for: | |
| Protein | 24.2 |
| Ion | 19.8 |
| Water | 34.5 |
| Ramachandran plots (%) | |
| Favored region | 96.9 |
| Allowed region | 3.1 |
The numbers in parentheses are statistics from the highest-resolution shell.
Rmerge = Σ |Iobs - Iavg|/Iobs, where Iobs is the observed intensity of individual reflection and Iavg is average over symmetry equivalents.
Rwork = Σ ||Fo| − |Fc||/Σ |Fo|, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated with 9.1% of the data.
RMSD, root mean square deviation.
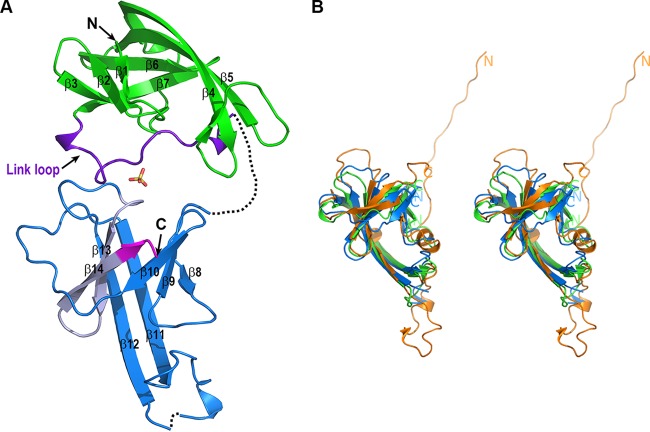
The structure of NisI22-C is composed mainly of β-strands (Fig. 2). NisI22-C consists of an N-terminal domain (residues 22 to 133) and a C-terminal domain (residues 134 to 245), which are structurally homologous despite the sequence identity being less than 14% (Fig. 1). Therefore, NisI22-C is arranged in a tandem repeat of structurally similar domains. This is consistent with the NMR structures of the isolated N- and C-terminal domains (24). As with the structures of the isolated N- and C-terminal domains, the structures of the N- and C-terminal domains display a similar fold that was first reported in the structure of SpaI, a lipoprotein with immunity function against subtilin lantibiotic from B. subtilis (Fig. 2B). The N-terminal domain has a β-barrel structure consisting of seven antiparallel twisted β-strands within one β-sheet with the strand order β1, β2, β3, β7, β6, β5, and β4. The anti-parallel β-sheet composed of β4 and β5 is relatively longer than the other strands, so that approximately half of the β-sheet protrudes from the core of the twisted β-sheet. The C-terminal domain has a similar β-barrel structure compared to the N-terminal domain, which is arranged in the strand order β8, β9, β10, β14, β13, β12, and β11. The β11 and β12, which are structurally equivalent to β4 and β5 in the N-terminal domain, also protruded from the core of the twisted β-sheet (Fig. 1 and 2A). The N- and C-terminal domains are connected by a link loop (residues 115 to 136), which is an extra region of the N-terminal domain excluding the core of the twisted β-sheet. Part of the link loop (residues 130 to 136) is not modeled due to the poor electron density map. Rather than just connecting the structurally homologous domains, the link loop is folded together around the core of the twisted β-sheet of the N-terminal domain on the side of the β-sheet consisting of β3 and β7 (Fig. 2A). In the NMR structure of the isolated C-terminal domain, the link loop was not definable, although the loop was included in the N-terminal region of the isolated C-terminal domain (24). Finally, the C-terminal domain is packed against the link loop region of the N-terminal domain using the side of the twisted β-sheet containing the β12–β13 loop and β9–β10 loop, resulting in a tightly compact association. One sulfate ion molecule resides in a pocket formed by packing between the N- and C-terminal domains, mediating the interactions between the link loops of the N-terminal domain and the C-terminal domain using hydrophobic and hydrogen bonds. It is notable that the β-barrel region of the N-terminal domain is not involved in interdomain interactions with the C-terminal domain (Fig. 2A).
FIG 2.
Structure of NisI22-C. (A) Ribbon diagram of NisI22-C. The region comprising the β-barrel structure (residues 22 to 114) and the link loop region (residues 115 to 136) of NisI22-C are displayed in green and purple, respectively. The C-terminal domain (residues 134 to 245) of NisI22-C is colored dark blue, except for the 21-amino-acid fragment, which is light gray, and the C-terminal 5 amino acids, which are magenta. The sulfate ion, which is derived from the crystallization solution, is shown in the stick model (oxygen in red and sulfur in yellow). Each secondary structure as described in Fig. 1 and the N- and C termini, are labeled. (B) Stereo view of the structural superposition of the N- and C-terminal domains of NisI22-C and SpaI (PDB accession number 2LVL). For clarity, the link loop region of the N-terminal domain of NisI22-C is not included. The ribbon diagrams of the N- and C-terminal domains of NisI22-C and SpaI are displayed in green, blue, and orange, respectively. The N- and C-terminal domains can be superimposed on those of SpaI with RMSDs of 3.1 Å and 2.8 Å over 87 and 87 Cα atoms, respectively. The domain boundaries of the N- and C-terminal domains of NisI22-C and the N- and C termini of SpaI are indicated with the N and C labels.
The secondary structure assignments of NisI22-C are consistent with the results from recent studies of the prereport on NMR resonance assignments of NisI21-C, except for breaking of the long continuous β-strand into two β-strands and the conversion of loops to a 310 α-helix, which might account for by the dynamic properties of NisI21-C in solution (36). The β3, β5, β10, and β11 are broken into two β-strands, and the loops ranging from Asp166 to Glu168 and from Lys223 to Ser224 form 310 α-helix and β-strand in solution (24).
Comparison of the N- and C-terminal domains.
A sequence alignment using the N-terminal 112 residues and the C-terminal 112 residues shows limited sequence conservation with approximately 14% identity, demonstrating that the N- and C-terminal domains are not related to each other (Fig. 1). However, a structural superposition reveals that the two domains have a similar structural fold (Fig. 2B). The β-barrel structures (residues 28 to 115 of the N-terminal domain; residues 137 to 243 of the C-terminal domain) of the N- and C-terminal domains are overlapped with a root mean square deviation (RMSD) of 2.5 Å over 86 Cα atoms calculated using the DaliLite program (37) (Fig. 2B). This deviation is comparable with the RMSD of 2.5 Å resulting from the superposition of the NMR structures of the isolated N- and C-terminal domains over 108 Cα atoms (24). The β-barrel structures of the N-terminal domain determined by NMR and X-ray crystallography are almost identical with those of an RMSD of 1.4 Å over 88 Cα atoms (see Fig. S1 in the supplemental material). Similarly, the β-barrel structures of the C-terminal domain determined by different methods are also almost identical with those of an RMSD of 1.2 Å over 95 Cα atoms (see Fig. S1 in the supplemental material). The β-barrel structures of both domains have nearly identical number of residues for each structurally equivalent strand. However, there are considerable differences when comparing loops, especially in the loop linking β4 with β5 in the N-terminal domain and the structurally equivalent β11–β12 loop in the C-terminal domain. The β11–β12 loop is relatively long, although half of the loop is structurally disordered (Fig. 1 and 2A).
Comparison with other structures.
Similarly to results obtained with the NMR structure of the isolated N- or C-terminal domain (NMR), searches of similar folds using the Dali server with the N-terminal domain containing the β-barrel and the link loop resulted in the highest Z score, 7.4, with SpaI, the lipoprotein with immunity against subtilin from B. subtilis (PDB accession number 2LVL) (29). The next most similar structure was a putative adenylate cyclase from Bacillus halodurans (PDB accession number 2GFG) with a Z score of 2.8, which was due to a similar fold characterized by the antiparallel β-sheets.
NisI22-C contains two structural modules that have previously been shown to be associated with lantibiotic immunity through the NMR structure of SpaI (29). It is well known that SpaI functions in immunity against subtilin in B. subtilis (38). The sequences of the N- and C-terminal domains have approximately 15% and 16% sequence identities with SpaI, respectively. However, the antiparallel twisted β-barrel structures of the N- and C-terminal domains overlap SpaI by RMSDs of 3.1 Å and 2.8 Å over 87 and 87 Cα atoms, respectively (Fig. 2B). The β-barrel structures of the N- and C-terminal domains consist of seven strands, whereas those of SpaI consist of six strands, and the region of SpaI that corresponds to β1 of the N- and C-terminal domains of NisI22-C folds into an α-helix (Fig. 1). It is also worth noting that, while the β-sheets β4/β5 and β11/β12 in the β-barrel structures of the N- and C-terminal domains, respectively, form continuous long antiparallel β-sheets, there is a break in the structurally corresponding β-sheets β3a–β3b/β4a–β4b in the β-barrel structure of SpaI (Fig. 1). In SpaI, flexible loop 1 contains approximately 26 residues between β3a–β3b and β4a–β4b and is extended out from the β-barrel structure. In NisI22-C, loop 1 between β11 and β12 in the C-terminal domain has a similar residue number, and the N-terminal domain is composed of 10 residues, suggesting that the structure of the C-terminal domain is more similar to that of SpaI.
Nisin-binding site.
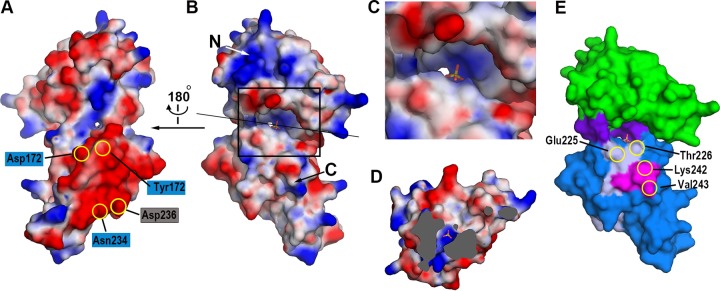
C-terminal truncation studies were previously performed to identify the nisin-binding site of NisI (27). A deletion mutant lacking a 5-amino-acid C-terminal fragment (residues 241 to 245) showed a 78% loss in immunity, indicating that this region is very important for immunity. Deleting 21 amino acids (residues 225 to 245) from the C terminus, which contains the 5-amino-acid C-terminal fragment, further decreased immunity. However, deletion of 74 amino acids (residues 172 to 245) from the C terminus, which contains 21 amino acids including the 5-amino-acid C-terminal fragment, did not further affect immunity, suggesting that the C-terminal 21-amino-acid fragment of NisI provides specificity to nisin and, thus, might be critical for the nisin binding (27). In the structure of NisI22-C, the C-terminal 5-amino-acid fragment comprises half of β14, and the C-terminal 21-amino-acid fragment comprises the β13/β14 antiparallel β-sheet (Fig. 1 and 2A), suggesting its important role as a structural frame. The C-terminal 5-amino-acid fragment mediates extensive interactions with the residues, such as Ile156, Tyr157, and Ser158 in β10 and Glu227 and Ala228 in β13 (Fig. 3). By analogy, the loss of immunity observed in the 21-amino-acid deletion, an approximately 10% immunity decrease compared with the 5-amino-acid deletion, might also be due to a collapse of the β-barrel structure. Importantly, it was shown that the deletion of the 21 amino acids results in unfolding of the C-terminal domain (24). However, the C-terminal 21-amino-acid fragment contains a secondary structural element (β13 and β14) conferring structural integrity, as well as residues residing in the loop region (β13–β14 and β12–β13 loops). Thus, it cannot be ruled out that this fragment might be involved in direct interactions with nisin. Recently, NMR titration experiments showed that Ile233 and Asn234, which reside in the β13–β14 loop, are involved in the nisin interaction (24). It is worth noting that among the C-terminal 21 amino acids, Asp236 contributes a negative surface patch (Fig. 4A), and Leu225, Thr226, Lys242, and Val243 contribute to the surface of the cleft formed by the N- and C-terminal domains (Fig. 4E).
FIG 3.
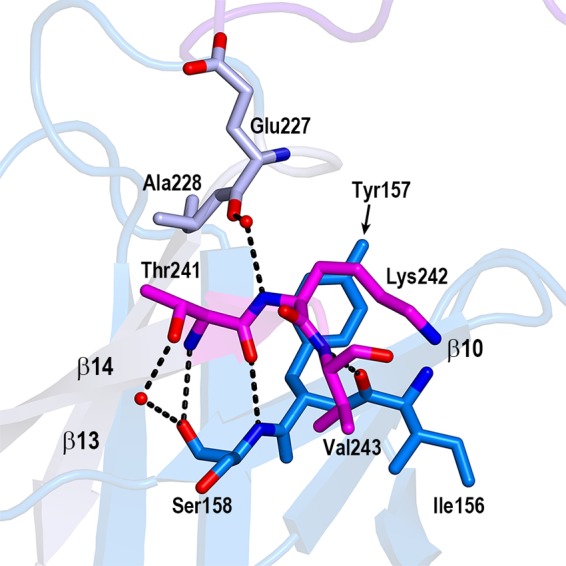
The 5-amino-acid fragment in the β-barrel core of the C-terminal domain. Closeup view of the interactions of the 5-amino-acid fragment with the neighboring residues in the β-barrel structure. Only three residues (Thr241, Lys242, and Val243) are shown. The last two C-terminal residues (Gly244 and Asn245) are invisible due to the poor electron density map. The residues belonging to the 5-amino-acid fragment (magenta), the interacting residues from β13 (gray), and the interacting residues from β10 (blue) are shown in the stick models. The dashed lines and red ovals indicate the hydrogen bonds and water molecules, respectively.
FIG 4.
The cleft and groove of NisI22-C. The distribution of the electrostatic potentials of NisI22-C. The potentials are shown in the range of −80 kT/e (red, negative potential) to + 80 kT/e (blue, positive potential). The views are from the shallow groove (A) and from the deep cleft (B). The residues involved in the interaction with nisin (in blue rectangles) and residues belonging to the C-terminal 21-amino-acid fragment (in gray rectangle) are shown on the negative patch of the C-terminal domain. The orientation of the surface diagram in (B) is almost identical to that of Fig. 2A. The sulfate ion is also shown in the stick model. (C) Zoomed-in view of the deep cleft. The region marked with the rectangle in (B) is magnified. (D) View of cross section after clipping the molecules along the straight black line as in (B). The view is facing the N-terminal domain. The solvent-inaccessible areas are shown in gray. The sulfate ion and the water molecule below the sulfate ion are shown with the stick model and the green sphere, respectively. (E) The distribution of residues critical for the immunity. Residues belonging to the C-terminal 21-amino-acid fragment (light gray) and the C-terminal 5-amino-acid fragment (magenta), which is also a part of the C-terminal 21-amino-acid fragment, are plotted on the surface model of NisI22-C using the same color scheme as in Fig. 2A.
In the studies that identified a region of NisI that was responsible for immunity function using a series of C-terminally truncated mutants, the immune activity of the isolated C-terminal 21-amino-acid fragment alone was also examined (27). Because SpaI is a lipoprotein known to be located on the extracellular side of the plasma membrane and is thought to be structurally unrelated to NisI based on low sequence identity (20, 27, 39, 40), a SpaI′-′NisI fusion mutant, where the C-terminal 21-amino-acid fragment (residues 145 to 165) of SpaI was replaced with the C-terminal 21-amino-acid fragment (residues 225 to 245) of NisI, was used to target the 21-amino-acid fragment to the extracellular side of the cytoplasmic membrane (27). However, our structural studies and the NMR structure of the isolated C-terminal domain show that SpaI and NisI possess structurally similar folds and superpose well on each other (Fig. 1 and 2B). When the C-terminal 21 amino acids of SpaI are replaced with the C-terminal 21 amino acids of NisI, β5 and β6 of SpaI are replaced with β13 and β14 of NisI. As the β5/β6 sheet and the β13/β14 sheet are structurally equivalent in the SpaI fold, the SpaI′-′NisI fusion mutant results in a domain swap between structurally similar proteins rather than a domain fusion to a shuttle protein to target the extracellular side of the bacterial cell. Therefore, it seems that the inherent structural properties of the C-terminal 21-amino-acid fragment of NisI have not changed significantly, except for the environment surrounding the C-terminal 21-amino-acid fragment due to the domain swapping to SpaI and the different domain structure of SpaI, where there is only one SpaI fold. The region swapping of a part important for specificity between the structurally similar domains might explain the fact that the SpaI′-′NisI mutant still has immune activity against nisin, which was measured to be approximately 50% compared with that of wild-type NisI. This was an interesting observation because SpaI was known to have no cross immunity against nisin (22).
The cleft and groove of NisI22-C.
One of the major differences between NisI22-C and SpaI is that NisI has two β-barrel domains, yielding a polypeptide chain approximately two times longer than SpaI (Fig. 2) (24, 29). The presence of two structurally stable β-barrel domains in one polypeptide chain might increase the chance of making interdomain associations and creating clefts and grooves. In the case of NisI22-C, the interdomain association produces one deep cleft (Fig. 4B) and one shallow groove (Fig. 4A), which are surrounded by the N- and C-terminal domains. Residues from the link loop region of the N-terminal domain make up the bottom of the deep cleft, and both peripheral sides are contributed by residues from the N- and C-terminal domains (Fig. 4E). The bottom of the deep cleft has a positively charged surface, while the entrance of the cleft is negatively charged (Fig. 4B and C). Inside the deep cleft, there is a deep cavity forming a narrow channel to the opposite side of the cleft, and one sulfate ion lies on the positively charged surface of the cavity (Fig. 4C). Under the sulfate ion, one water molecule resides at the narrowest region of the channel (Fig. 4D). The shallow groove, which is on the opposite side of the deep cleft, is wide open with a positively charged bottom and is flanked by extended negatively charged patches (Fig. 4A). Interestingly, the residues, such as Tyr172, Asp174, Glu232, and Asn234, which were shown to be involved in the nisin interaction (24), are distributed in the negative patch of the C-terminal domain (Fig. 4A).
The sulfate ion in the deep cleft is involved in the interactions between the link loop regions of the N-terminal domain and the C-terminal domain, using extensive hydrogen bonds and hydrophobic interactions. Arg125 from the link loop region of the N-terminal domain and Tyr152 from the C-terminal domain interacts with O2 or O3 oxygen atoms of the sulfate ion using the atoms of the side chains, whereas the interactions of Ala123 from the link loop region and Leu225 and Thr226 from the C-terminal domain with the sulfate ion are mediated through hydrogen bonds involving amide and carbonyl groups of the peptide backbone (Fig. 5). Gly119 and Lys122 from the link loop region and Ser224 from the C-terminal domain also contribute to interactions with the sulfate ion through hydrophobic interactions. Notably, Leu225 and Thr226, which belong to the 21-amino-acid fragment that was shown to influence the immune activity of NisI, make contacts with the sulfate ion (Fig. 5B). It is also worth noting that the deep cavity has an extra space open to the channel, which is capable of holding approximately one more molecule similar in size to a sulfate ion and which is not dominated by negative or positive charges (Fig. 4C).
FIG 5.
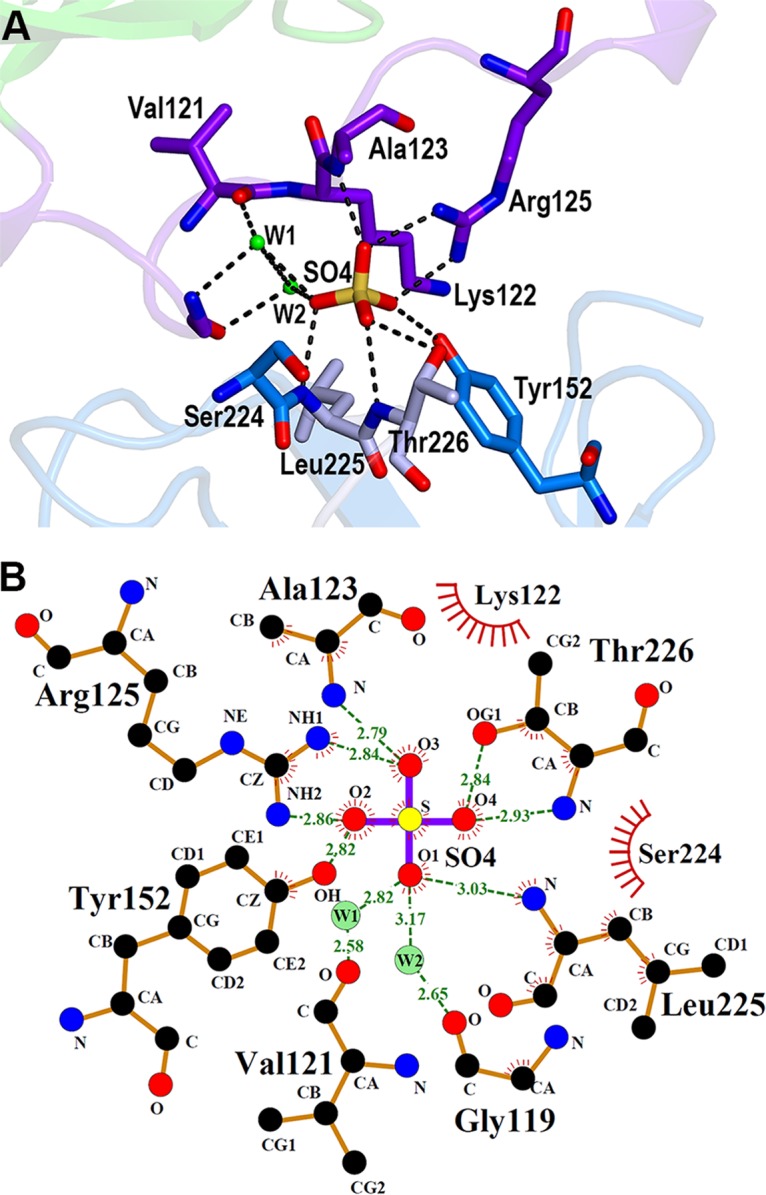
Sulfate ion in the deep cleft of NisI22-C. (A) Detailed view of the interactions of the sulfate ion with NisI22-C. The side chain and backbone atoms making contacts with a sulfate ion are shown with the stick models (carbon, as in Fig. 1B) (nitrogen in blue, oxygen in red, and sulfur in yellow). The hydrogen bonds are represented with dashed lines, and the green ovals indicate water-mediated interactions. (B) Two-dimensional view of the interaction. The diagram was produced using LigPlot (46). The hydrogen bonds are shown using dashed lines, and hydrophobic contacts are shown using an arc with spokes radiating to a binding partner. Water molecules (W1 and W2) are also included.
Lipid II provides the docking sites for nisin on the membrane where pore formation is initiated (14). In this study, the structure of NisI22-C was determined and found to contain a sulfate ion residing in a deep cleft formed by the N- and C-terminal domains. Interestingly, the sulfate ion resembles the phosphate group of pyrophosphate moiety of the lipid II intermediate, which has been shown to be the target of nisin (see Fig. S2 in the supplemental material). Therefore, it may be possible that the deep cleft of NisI where the sulfate ion is found is used for a binding site for lipid II, thereby increasing the chances of encountering nisin. When a farnesyl diphosphate backbone containing 3 isoprene repeats is docked into the cleft using the AutoDock Vina program, the phosphate group of the diphosphate moiety fits well on the sulfate ion in the cleft (Fig. 6). The inner deep side of the cleft interacts with the diphosphate group, while the other side is open, suggesting a space that could accommodate a bulky molecule such as the peptidoglycan subunit of the lipid II, consisting of N-acetylglucosamine and N-acetylmuramic acid with a pentapeptide group (Fig. 6). It is also worthwhile to note that the nisin-interacting residues are prevalent in the negative patch opposite to the deep cleft (Fig. 4A). The subsequent biochemical and structural studies involving both NisI and a derivative containing the properties of lipid II are anticipated to corroborate this suggested idea.
FIG 6.
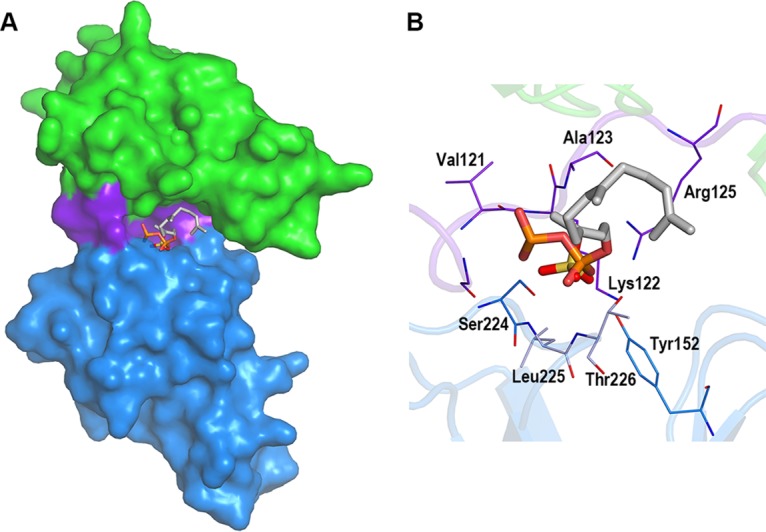
Simulated docking of a farnesyl diphosphate backbone variant of lipid II. The diphosphate group and 3 isoprene repeats were docked on the sulfate ion in the cleft. (A) Modeled farnesyl diphosphate variant and sulfate ion in the cleft of NisI22-C shown with the surface model (the N-terminal domain, green; the link region, purple; the C-terminal domain, dark blue). The farnesyl diphosphate variant and sulfate ion are shown with stick models (P in orange, S in yellow, O in red, and C in cyan). The view is from the deep cleft. (B) Zoomed-in view in the cleft. Residues making contacts with a sulfate ion are shown in stick models as in Fig. 5B.
MATERIALS AND METHODS
Protein production.
Selenomethionine (SeMet)-labeled NisI22-C was produced using PASM autoinduction medium (41) and purified as described previously (35). The construct for encoding NisI22-C (residues 22 to the C terminus) was designed to express the Gln22 residue as the start residue, which is the second residue after the Cys20 residue to which a diacylglycerol moiety is attached. Briefly, SeMet-labeled NisI22-C was expressed with a N-terminal tobacco etch virus (TEV) protease-cleavable His6-tag in Escherichia coli BL21 Star (DE3) cells (Invitrogen). The protein was purified with the sequential steps of His affinity chromatography, His6-tag cleavage by TEV protease, a second His affinity chromatography to remove cleaved His6 tags, and size exclusion chromatography. The reductive methylation of SeMet-labeled NisI22-C was performed with the same procedures as that for the native protein as described previously (35).
Crystallization and data collection.
The crystals of methylated SeMet-labeled NisI22-C were optimized with the hanging-drop vapor-diffusion method with protein samples at a concentration of 20 mg · ml−1 and VDX plates (Hampton Research) using the optimized crystallization condition of native NisI22-c reported previously (35). The best crystals were grown similarly under a condition consisting of 30 to 40 mM ammonium sulfate, 0.1 M Na-acetate pH 4.6, and 16 to 18% polyethylene glycol (PEG) 4000. A single crystal detached from a cluster of orthorhombic crystals using a nylon loop was soaked briefly in the reservoir solution supplemented with 30% glycerol and then flash-frozen directly in a 100-K nitrogen stream. A single anomalous dispersion (SAD) data set consisting of 360 frames of X-ray diffraction data at selenium peak energy (0.97917 Å) was collected on an ADSC Q270 charge-coupled-device (CCD) detector at beamline 7A of the Pohang Light Source II (PLS II) at the Pohang Accelerator Laboratory (PAL, Pohang, South Korea) using 1° oscillations. The data were indexed and scaled with the HKL-2000 software package (42). The crystallographic data statistics are summarized in Table 1.
Structure determination.
Determination of the location of the selenium sites, phasing, electron density modification of the initial map, and initial model building were carried out with the AutoSol wizard in the PHENIX package (43) using the SAD data set. The missing parts of the autotraced model were built manually using Coot (44). The model then was subjected to iterative manual model building and refinement with Coot and the PHENIX package, respectively. The final model was completed with the addition of water molecules, which was carried out by automatic addition using the PHENIX package and further manual inspection using Coot with the aid of the Fo-Fc map. The model of NisI22-C converged to an Rwork of 17.6% and an Rfree of 20.9%, which was calculated with 9.1% of the reflections. The statistics of refinement are presented in Table 1. All figures of the molecular model were prepared with PyMOL (http://www.pymol.org).
Docking simulation.
Molecular docking of a farnesyl diphosphate backbone variant of lipid II (the diphosphate group and 3 isoprene repeats) was performed using the Autodock Vina program (45). The variant of lipid II was derived from the nisin-lipid II complex (PDB accession number 1WCO) (16). The center of the grid box (x: 28 Å, y: 62 Å, z: 51 Å) was set between the Ca atoms of Thr36 and Thr226 and the size of the grid was 20 × 20 × 20 Å.
Accession number(s).
The atomic coordinates and structure factors were deposited in the Protein Data Bank under accession number 5XHB.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (grant 2016R1D1A1B03932947). Experiments at beamline 7A (SB I) of PLS were supported in part by MSIP and POSTECH.
We declare that we have no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01966-17.
REFERENCES
- 1.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins—viable alternative to antibiotics? Nat Rev Microbiol 11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 2.Severina E, Severin A, Tomasz A. 1998. Antibacterial efficacy of nisin against multidrug-resistant Gram-positive pathogens. J Antimicrob Chemother 41:341–347. doi: 10.1093/jac/41.3.341. [DOI] [PubMed] [Google Scholar]
- 3.Cotter PD, Hill C, Ross RP. 2005. Bacterial lantibiotics: strategies to improve therapeutic potential. Curr Protein Pept Sci 6:61–75. doi: 10.2174/1389203053027584. [DOI] [PubMed] [Google Scholar]
- 4.Piper C, Cotter PD, Ross RP, Hill C. 2009. Discovery of medically significant lantibiotics. Curr Drug Discov Technol 6:1–18. doi: 10.2174/157016309787581075. [DOI] [PubMed] [Google Scholar]
- 5.Willey JM, van der Donk WA. 2007. Lantibiotics: peptides of diverse structure and function. Annu Rev Microbiol 61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- 6.Rogers LA, Whittier EO. 1928. Limiting factors in the lactic fermentation. J Bacteriol 16:211–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wirawan RE, Klesse NA, Jack RW, Tagg JR. 2006. Molecular and genetic characterization of a novel nisin variant produced by Streptococcus uberis. Appl Environ Microbiol 72:1148–1156. doi: 10.1128/AEM.72.2.1148-1156.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaletta C, Entian KD. 1989. Nisin, a peptide antibiotic: cloning and sequencing of the nisA gene and posttranslational processing of its peptide product. J Bacteriol 171:1597–1601. doi: 10.1128/jb.171.3.1597-1601.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karakas Sen A, Narbad A, Horn N, Dodd HM, Parr AJ, Colquhoun I, Gasson MJ. 1999. Post-translational modification of nisin. The involvement of NisB in the dehydration process. Eur J Biochem 261:524–532. [DOI] [PubMed] [Google Scholar]
- 10.Koponen O, Tolonen M, Qiao M, Wahlström G, Helin J, Saris PE. 2002. NisB is required for the dehydration and NisC for the lanthionine formation in the post-translational modification of nisin. Microbiology 148:3561–3568. doi: 10.1099/00221287-148-11-3561. [DOI] [PubMed] [Google Scholar]
- 11.Qiao M, Saris PE. 1996. Evidence for a role of NisT in transport of the lantibiotic nisin produced by Lactococcus lactis N8. FEMS Microbiol Lett 144:89–93. doi: 10.1111/j.1574-6968.1996.tb08513.x. [DOI] [PubMed] [Google Scholar]
- 12.van der Meer JR, Polman J, Beerthuyzen MM, Siezen RJ, Kuipers OP, De Vos WM. 1993. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J Bacteriol 175:2578–2588. doi: 10.1128/jb.175.9.2578-2588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruhr E, Sahl HG. 1985. Mode of action of the peptide antibiotic nisin and influence on the membrane potential of whole cells and on cytoplasmic and artificial membrane vesicles. Antimicrob Agents Chemother 27:841–845. doi: 10.1128/AAC.27.5.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brötz H, Josten M, Wiedemann I, Schneider U, Götz F, Bierbaum G, Sahl HG. 1998. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol Microbiol 30:317–327. doi: 10.1046/j.1365-2958.1998.01065.x. [DOI] [PubMed] [Google Scholar]
- 15.van Kraaij C, Breukink E, Noordermeer MA, Demel RA, Siezen RJ, Kuipers OP, de Kruijff B. 1998. Pore formation by nisin involves translocation of its C-terminal part across the membrane. Biochemistry 37:16033–16040. doi: 10.1021/bi980931b. [DOI] [PubMed] [Google Scholar]
- 16.Hsu ST, Breukink E, Tischenko E, Lutters MA, de Kruijff B, Kaptein R, Bonvin AM, van Nuland NA. 2004. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat Struct Mol Biol 11:963–967. doi: 10.1038/nsmb830. [DOI] [PubMed] [Google Scholar]
- 17.Hasper HE, Kramer NE, Smith JL, Hillman JD, Zachariah C, Kuipers OP, de Kruijff B, Breukink E. 2006. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313:1636–1637. doi: 10.1126/science.1129818. [DOI] [PubMed] [Google Scholar]
- 18.Kuipers OP, Beerthuyzen MM, Siezen RJ, de Vos WM. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur J Biochem 216:281–291. [DOI] [PubMed] [Google Scholar]
- 19.Engelke G, Gutowski-Eckel Z, Kiesau P, Siegers K, Hammelmann M, Entian KD. 1994. Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Appl Environ Microbiol 60:814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein C, Entian KD. 1994. Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC. 6633. Appl Environ Microbiol 60:2793–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegers K, Entian KD. 1995. Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl Environ Microbiol 61:1082–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stein T, Heinzmann S, Solovieva I, Entian KD. 2003. Function of Lactococcus lactis nisin immunity genes nisI and nisFEG after coordinated expression in the surrogate host Bacillus subtilis. J Biol Chem 278:89–94. doi: 10.1074/jbc.M207237200. [DOI] [PubMed] [Google Scholar]
- 23.Takala TM, Koponen O, Qiao M, Saris PEJ. 2004. Lipid-free NisI: interaction with nisin and contribution to nisin immunity via secretion. FEMS Microbiol Lett 237:171–177. doi: 10.1111/j.1574-6968.2004.tb09693.x. [DOI] [Google Scholar]
- 24.Hacker C, Christ NA, Duchardt-Ferner E, Korn S, Göbl C, Berninger L, Düsterhus S, Hellmich UA, Madl T, Kötter P, Entian KD, Wöhnert J. 2015. The solution structure of the lantibiotic immunity protein NisI and its interactions with nisin. J Biol Chem 290:28869–28886. doi: 10.1074/jbc.M115.679969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao M, Immonen T, Koponen O, Saris PE. 1995. The cellular location and effect on nisin immunity of the NisI protein from Lactococcus lactis N8 expressed in Escherichia coli and L. lactis. FEMS Microbiol Lett 131:75–80. doi: 10.1111/j.1574-6968.1995.tb07757.x. [DOI] [PubMed] [Google Scholar]
- 26.Koponen O, Takala TM, Saarela U, Qiao M, Saris PE. 2004. Distribution of the NisI immunity protein and enhancement of nisin activity by the lipid-free NisI. FEMS Microbiol Lett 231:85–90. doi: 10.1016/S0378-1097(03)00934-0. [DOI] [PubMed] [Google Scholar]
- 27.Takala TM, Saris PE. 2006. C terminus of NisI provides specificity to nisin. Microbiology 152:3543–3549. doi: 10.1099/mic.0.29083-0. [DOI] [PubMed] [Google Scholar]
- 28.AlKhatib Z, Lagedroste M, Fey I, Kleinschrodt D, Abts A, Smits SH. 2014. Lantibiotic immunity: inhibition of nisin mediated pore formation by NisI. PLoS One 9:e102246. doi: 10.1371/journal.pone.0102246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christ NA, Bochmann S, Gottstein D, Duchardt-Ferner E, Hellmich UA, Düsterhus S, Kötter P, Güntert P, Entian KD, Wöhnert J. 2012. The First structure of a lantibiotic immunity protein, SpaI from Bacillus subtilis, reveals a novel fold. J Biol Chem 287:35286–35298. doi: 10.1074/jbc.M112.401620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walter TS, Meier C, Assenberg R, Au KF, Ren J, Verma A, Nettleship JE, Owens RJ, Stuart DI, Grimes JM. 2006. Lysine methylation as a routine rescue strategy for protein crystallization. Structure 14:1617–1622. doi: 10.1016/j.str.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Means GE. 1977. Reductive alkylation of amino groups. Methods Enzymol 47:469–478. doi: 10.1016/0076-6879(77)47047-2. [DOI] [PubMed] [Google Scholar]
- 32.Rayment I, Rypniewski WR, Schmidt-Bäse K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. 1993. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science 261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 33.Rypniewski WR, Holden HM, Rayment I. 1993. Structural consequences of reductive methylation of lysine residues in hen egg white lysozyme: an X-ray analysis at 1.8-A resolution. Biochemistry 32:9851–9858. doi: 10.1021/bi00088a041. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi M, Kubota M, Matsuura Y. 1999. Crystallization and improvement of crystal quality for X-ray diffraction of maltooligosyl trehalose synthase by reductive methylation of lysine residues. Acta Crystallogr D Biol Crystallogr 55:931–933. doi: 10.1107/S0907444999002115. [DOI] [PubMed] [Google Scholar]
- 35.Jeong JH, Ha SC. 2017. Full-length nisin immunity protein NisI from Lactococcus lactis in a lipid-free form: crystallization and X-ray analysis. Acta Crystallogr F Struct Biol Commun 73:404–408. doi: 10.1107/S2053230X17008214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hacker C, Christ NA, Duchardt-Ferner E, Korn S, Berninger L, Kötter P, Entian KD, Wöhnert J. 2015. NMR resonance assignments of the lantibiotic immunity protein NisI from Lactococcus lactis. Biomol NMR Assign 9:293–297. doi: 10.1007/s12104-015-9595-1. [DOI] [PubMed] [Google Scholar]
- 37.Holm L, Rosenstrom P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res 38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stein T, Heinzmann S, Düsterhus S, Borchert S, Entian KD. 2005. Expression and functional analysis of the subtilin immunity genes spaIFEG in the subtilin-sensitive host Bacillus subtilis MO1099. J Bacteriol 187:822–828. doi: 10.1128/JB.187.3.822-828.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halami PM, Stein T, Chandrashekar A, Entian KD. 2010. Maturation and processing of SpaI, the lipoprotein involved in subtilin immunity in Bacillus subtilis ATCC. 6633. Microbiol Res 165:183–189. doi: 10.1016/j.micres.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 40.Gross E, Morell JL. 1971. The structure of nisin. J Am Chem Soc 93:4634–4635. doi: 10.1021/ja00747a073. [DOI] [PubMed] [Google Scholar]
- 41.Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in the oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laskowski RA, Swindells MB. 2011. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Infect Model 51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.