ABSTRACT
Fluoroquinolone resistance in Gram-negative bacteria is multifactorial, involving target site mutations, reductions in fluoroquinolone entry due to reduced porin production, increased fluoroquinolone efflux, enzymes that modify fluoroquinolones, and Qnr, a DNA mimic that protects the drug target from fluoroquinolone binding. Here we report a comprehensive analysis, using transformation and in vitro mutant selection, of the relative importance of each of these mechanisms for fluoroquinolone nonsusceptibility using Klebsiella pneumoniae as a model system. Our improved biological understanding was then used to generate 47 rules that can predict fluoroquinolone susceptibility in K. pneumoniae clinical isolates. Key to the success of this predictive process was the use of liquid chromatography-tandem mass spectrometry to measure the abundance of proteins in extracts of cultured bacteria, identifying which sequence variants seen in the whole-genome sequence data were functionally important in the context of fluoroquinolone susceptibility.
KEYWORDS: Klebsiella pneumoniae, antibiotic resistance, susceptibility testing
INTRODUCTION
The use of whole-genome sequencing (WGS) to predict antibacterial drug susceptibility/nonsusceptibility in the clinical setting was recently the subject of a subcommittee report from the European Committee on Antimicrobial Susceptibility Testing (1). Aside from issues around universal WGS data availability and quality, particularly regarding clinical samples with low bacterial titers, the committee considered that a lack of a basic biological understanding of antibacterial resistance (ABR) mechanisms was slowing down progress in this area (1). There have been some successes in predicting susceptibility/nonsusceptibility directly from WGS data for key Gram-negative human pathogens, such as Escherichia coli and Klebsiella pneumoniae, carrying well-known plasmid-encoded ABR mechanisms or commonly encountered antibacterial target site mutations (2, 3). However, when ABR is multifactorial, as typified by fluoroquinolone resistance, prediction of susceptibility/nonsusceptibility is more difficult (1, 4).
Fluoroquinolones have a fluorine atom attached to the central quinolone ring system (5–7). They have a broad spectrum of activity against both Gram-negative and Gram-positive bacteria and play an important role in clinical applications (8–10). Their mode of action involves binding to type II DNA topoisomerases, GyrA/B and ParC/E, resulting in lethal double-strand breaks in DNA (8, 11). A single point mutation in the quinolone-resistance-determining region (QRDR) of GyrA confers nalidixic acid resistance to E. coli, but additional factors are required for fluoroquinolone resistance, for example, QRDR mutations in ParC, a secondary target of fluoroquinolones (12), or mutations in the transcriptional repressor gene ramR, which lead to the overproduction of the resistance-nodulation-cell division (RND) family efflux pump AcrAB-TolC (13–16). The overproduction of OqxAB, another chromosomally encoded RND efflux pump that probably works with TolC, also confers reduced fluoroquinolone susceptibility to K. pneumoniae and results from mutations in the transcriptional repressor gene oqxR (17).
When carried on a plasmid in E. coli, oqxAB also confers fluoroquinolone resistance via drug efflux (18, 19). Other plasmid-mediated quinolone resistance (PMQR) genes include qnrA (20), which encodes a pentapeptide repeat protein that protects GyrA from inhibition by fluoroquinolones, reducing susceptibility. Other qnr gene families, including qnrB (21), qnrC (22), qnrS (23), and qnrD (24), are commonly seen in clinical Enterobacteriaceae isolates, as is the PMQR gene aac(6′)-Ib-cr, encoding an aminoglycoside acetyltransferase capable of acetylating fluoroquinolones at the amino nitrogen if they have a piperazinyl substituent, e.g., ciprofloxacin (25).
Our aim in performing the work set out below was to complete an extensive analysis of the individual and combined contributions of these known mechanisms to fluoroquinolone nonsusceptibility in K. pneumoniae using sequential mutant selection and transformation, with liquid chromatography-tandem mass spectrometry (LC-MS/MS) proteomics being used to monitor protein production and with DNA sequencing being used to identify mutations. Our aim was to generate a series of “rules” that might be applied to WGS data to predict fluoroquinolone susceptibility in K. pneumoniae. We then tested our rules against 40 K. pneumoniae clinical isolates.
RESULTS AND DISCUSSION
Fluoroquinolone nonsusceptibility of K. pneumoniae requires combinations of different mechanisms.
A total of 192 mutants with reduced fluoroquinolone susceptibility were generated from K. pneumoniae Ecl8 and the Ecl8 ΔramR mutant. Among the 29 representative mutants that were extensively characterized, 16 had quinolone target site mutations in gyrA clustered into four different alleles, each encoding a single amino acid substitution within the QRDR: Ser83Phe, Ser83Tyr, Asp87Tyr, or Gly81Cys. Multiple mutations in gyrA or mutations in other topoisomerase genes, e.g., parC, were not identified. Mutations in known regulators of drug efflux were also seen. OqxR mutations were found in 12/29 fully characterized mutants, including frameshift mutations and nonsense and missense mutations at various positions (see Table S3 in the supplemental material). In contrast, only a single (Thr124Pro) RamR mutation was found, and no mutations in AcrR were identified. The roles of two PMQR genes were also assessed, qnr (represented by the qnrA1 variant) and aac(6′)-Ib-cr, with both genes being provided on a vector with expression driven by their native promoters.
No single acquisition event, whether GyrA, RamR, or OqxR mutations or carriage of a PMQR gene, conferred clinically relevant ciprofloxacin nonsusceptibility to K. pneumoniae Ecl8. Combinations of an oqxR or ramR mutation plus a gyrA mutation or of an oqxR or ramR mutation plus carriage of qnr were the only double combinations that conferred ciprofloxacin nonsusceptibility. Of the 10 triple and 5 quadruple combinations tested, all combinations conferred fluoroquinolone nonsusceptibility except for the ramR oqxR double mutant plus aac(6′)-Ib-cr (Table S4).
Predicting fluoroquinolone susceptibility in K. pneumoniae clinical isolates from WGS data.
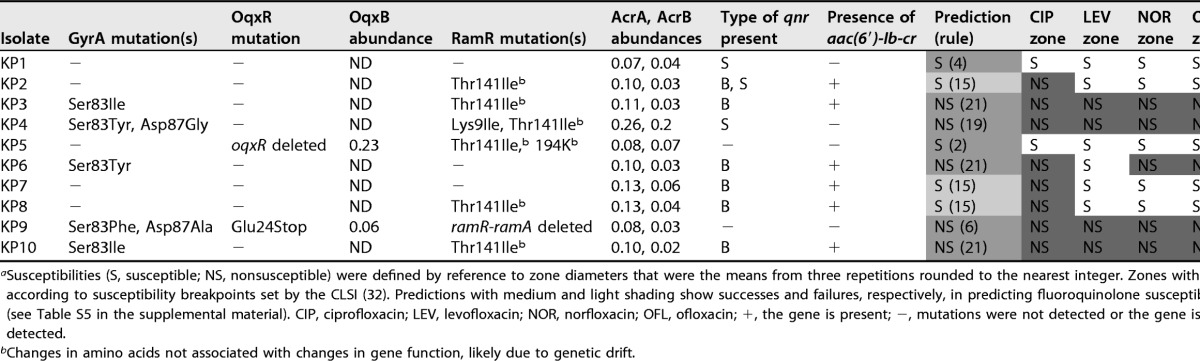
The results from in vitro K. pneumoniae Ecl8 mutant and transformant characterizations (see Table S4 in the supplemental material) were used to generate a set of 31 rules to predict fluoroquinolone susceptibility/nonsusceptibility in K. pneumoniae, as shown in Table S5. The 31 rules were then applied to 10 K. pneumoniae clinical isolates based on WGS data. The presence of PMQR genes was defined as a binary result from the WGS data, given that all genes were fully intact. The presence of QRDR mutations in GyrA and of single nucleotide polymorphisms (SNPs) affecting the amino acid sequences of RamR and OqxR was read directly from the WGS data. To confirm whether the identified regulatory SNPs activated efflux pump production in these clinical isolates, whole-cell LC-MS/MS proteomics was performed. Clinical isolates carrying Thr141Ile and 194K insertion mutations in RamR (6/10 clinical isolates carrying one or both mutations) did not hyperproduce AcrA and AcrB (Table 1), as would be expected from a true RamR loss-of-function mutation (26), so these variants were considered functionally of the wild type and to have arisen by random genetic drift. Two out of 10 isolates carried OqxR mutations, and both isolates were confirmed to hyperproduce OqxB (Table 1).
TABLE 1.
Combination of disc test, proteomics, WGS, and predictions of fluoroquinolone susceptibility for K. pneumoniae clinical isolatesa
Susceptibilities (S, susceptible; NS, nonsusceptible) were defined by reference to zone diameters that were the means from three repetitions rounded to the nearest integer. Zones with dark shading are nonsusceptible according to susceptibility breakpoints set by the CLSI (32). Predictions with medium and light shading show successes and failures, respectively, in predicting fluoroquinolone susceptibility by using the preliminary rules (see Table S5 in the supplemental material). CIP, ciprofloxacin; LEV, levofloxacin; NOR, norfloxacin; OFL, ofloxacin; +, the gene is present; −, mutations were not detected or the gene is absent; ND, protein was not detected.
Changes in amino acids not associated with changes in gene function, likely due to genetic drift.
By applying our predictive rules (Table S5), WGS information on the 10 clinical isolates, with proteomics being used to confirm/deny suspected oqxR or ramR loss-of-function mutations, allowed the correct prediction of ciprofloxacin susceptibility/nonsusceptibility for 7/10 isolates. The three remaining isolates were incorrectly predicted to be ciprofloxacin susceptible, as defined by rule 15, which is the production of Qnr and Aac-6′-Ib-cr in an otherwise wild-type background (Table S5). The rule 15 clinical isolates were in fact ciprofloxacin nonsusceptible, although they were susceptible to all other fluoroquinolones tested (Table 1).
Clinical isolates with low OmpK35/OmpK36 porin ratios have reduced fluoroquinolone susceptibility.
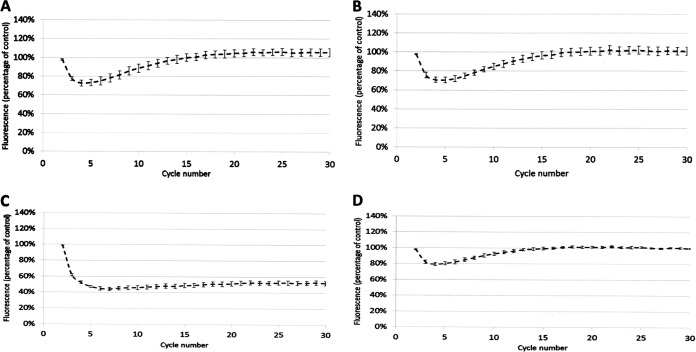
We hypothesized that the three “rule 15” clinical isolates incorrectly predicted to be fluoroquinolone susceptible (Table 1) have reduced envelope permeability compared with Ecl8, the isolate used to define our predictive rules (see Tables S4 and S5 in the supplemental material). Fluorescent dye accumulation assays revealed that the shape of the dye accumulation curve for representative rule 15 clinical isolates KP7 and KP8 (Fig. 1A and B) was like that seen previously upon micF overexpression in Ecl8, i.e., causing reduced OmpF (OmpK35) porin production (26). Porin reduction retards the entry of the dye but does not prevent the accumulation of the dye over time. This phenotype is in stark contrast to efflux-mediated reduced envelope permeability, which gives a persistent level of reduced dye accumulation, as seen in the Ecl8 ΔramR mutant previously (26) and, for example, in clinical isolate KP21, which has a ramR mutation and overproduces AcrAB-TolC (Table 2 and Fig. 1C).
FIG 1.
Accumulation of H33342 dye by K. pneumoniae clinical isolates over a 30-cycle (45-min) incubation period. (A) KP7; (B) KP8; (C) KP21; (D) KP46. In each case, permeability was compared with that of Ecl8 (set to 100%) in cells grown in Mueller-Hinton broth. Each line shows mean data for three biological replicates with 8 technical replicates each, and error bars define the standard errors of the means.
TABLE 2.
Combination of disc test, proteomics, and genome sequencing data and predictions of fluoroquinolone susceptibility for K. pneumoniae clinical isolatesa
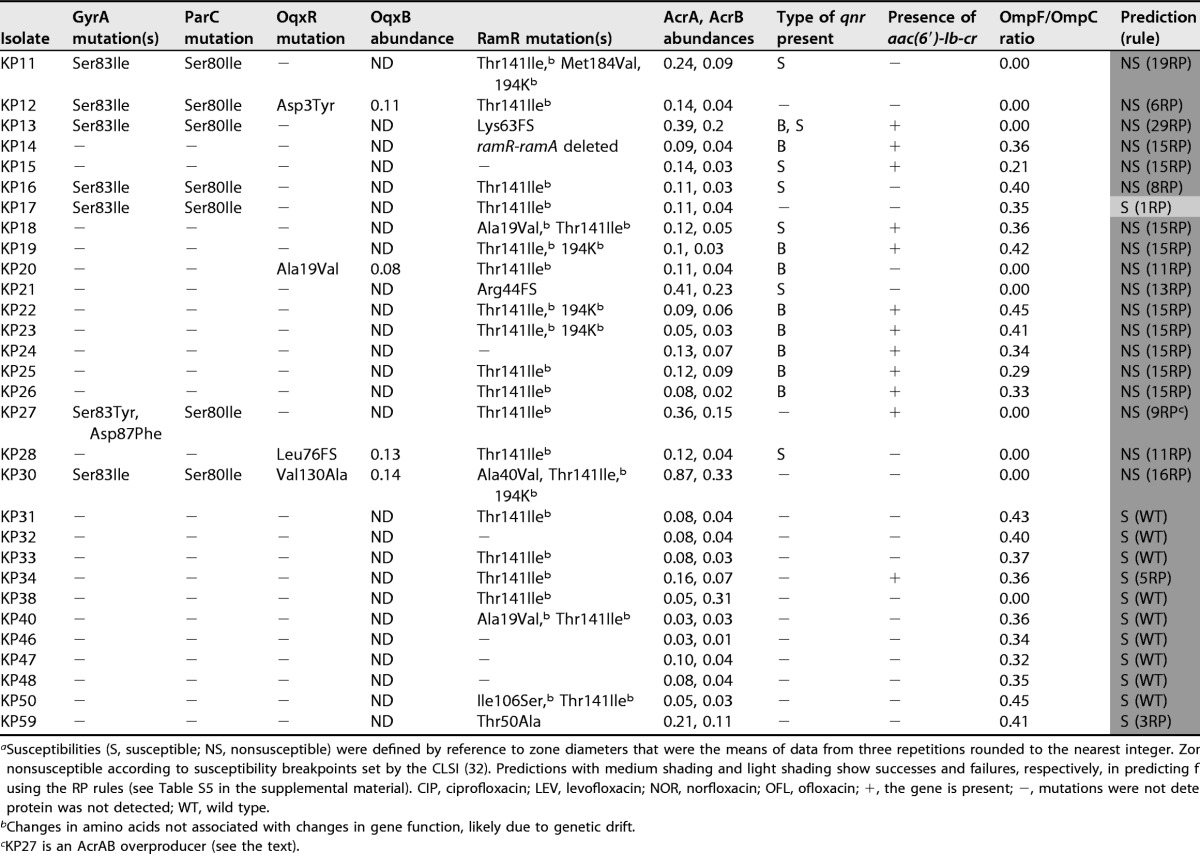
Susceptibilities (S, susceptible; NS, nonsusceptible) were defined by reference to zone diameters that were the means of data from three repetitions rounded to the nearest integer. Zones with dark shading are nonsusceptible according to susceptibility breakpoints set by the CLSI (32). Predictions with medium shading and light shading show successes and failures, respectively, in predicting fluoroquinolone susceptibility by using the RP rules (see Table S5 in the supplemental material). CIP, ciprofloxacin; LEV, levofloxacin; NOR, norfloxacin; OFL, ofloxacin; +, the gene is present; −, mutations were not detected or the gene is absent; ND, protein was not detected; WT, wild type.
Changes in amino acids not associated with changes in gene function, likely due to genetic drift.
KP27 is an AcrAB overproducer (see the text).
During growth in the same medium used to perform proteomic analyses of our clinical isolates (nutrient broth), the OmpF/OmpC ratio seen for Ecl8 was approximately 1:1 (26). For these 10 test clinical isolates, the OmpF/OmpC ratio was in a range of 0.25:1 to 0.40:1, or it was zero due to the apparent loss of OmpF (Table 1). This observed reduced OmpF/OmpC ratio in the clinical isolates relative to Ecl8 rationalized the observed envelope permeability assay data (Fig. 1) and supported our hypothesis that clinical rule 15 isolates have reduced OmpF porin levels compared with those of Ecl8, creating a ciprofloxacin-nonsusceptible phenotype in the presence of Qnr and Aac-6′-Ib-cr.
Following analysis of 30 additional K. pneumoniae clinical isolates, proteomics analysis revealed that all the clinical isolates in our collection have an OmpF/OmpC ratio of <0.5 (Table 2), so we predicted that these clinical isolates also have reduced envelope permeability compared with Ecl8. This proved to be correct, e.g., for representative isolate KP46, where the dye accumulation curve was like that for the rule 15 clinical isolates (Fig. 1A, B, and D).
Based on this finding of reduced OmpF/OmpC ratios in the clinical isolates relative to Ecl8, we set out to refine our predictive rules (Table S5), defined by using Ecl8, to take this reduced permeability (RP) into consideration. Comparisons between the MICs of ciprofloxacin against Ecl8 variants and those against equivalent clinical isolates (having reduced OmpF/OmpC ratios) were possible for rule 4 (qnr) (0.063 mg · liter−1 for Ecl8 versus 0.25 mg · liter−1 for KP1), rule 5 (aac-6′-Ib-cr) (0.016 mg · liter−1 for Ecl8 versus for 0.063 mg · liter−1 for KP34), rule 15 (qnr plus aac-6′-Ib-cr) (0.5 mg · liter−1 for Ecl8 versus 2 mg · liter−1 for KP8), and rule 21 (gyrA plus qnr plus aac-6′-Ib-cr) (2 mg · liter−1 for Ecl8 versus 8 mg · liter−1 for KP6) isolates. Based on the finding that, in each case, the reduced OmpF/OmpC ratio increases the MIC of ciprofloxacin by 4-fold, we modified our predictive rules. To do this, we first determined the ciprofloxacin MIC against the 12/31 ciprofloxacin-susceptible Ecl8 derivatives (Table S5) and applied a 4-fold MIC increase correction to account for RP caused by a reduced background OmpF/OmpC ratio. In this way, 5/31 of the originally defined predictive rules were altered to predict ciprofloxacin nonsusceptibility based on MIC breakpoints after applying this RP correction (Table S5).
Impact of multiple QRDR mutations on fluoroquinolone susceptibility.
Following the application of the revised rules allowing for an OmpF/OmpC ratio of <0.5 (see Table S5 in the supplemental material), we were successful in predicting ciprofloxacin susceptibility/nonsusceptibility for 29/30 additional clinical isolates not previously tested (Table 2). To understand the incorrect prediction of ciprofloxacin susceptibly in isolate KP17 (predictive rule 1RP), we considered additional mechanisms that might be involved. We noted from the WGS data that isolate KP17 carries both a GyrA mutation and a ParC topoisomerase mutation, which was never seen in our Ecl8 mutants and thus was not included in our 31 predictive rules but was relatively common in our clinical isolates (Tables 1 and 2). To assess the impact of a second QRDR mutation, we compared ciprofloxacin MICs against clinical isolates having a single or double QRDR mutation but otherwise defined by the same predictive rule. For example, the ciprofloxacin MIC against KP6 (rule 21, one QRDR mutation) was 8 mg · liter−1, and the MIC against KP3 (rule 21, two QRDR mutations) was 4-fold higher, at 32 mg · liter−1. The application of this 4-fold MIC correction to the predicted MIC of ciprofloxacin against a single QRDR mutant rule 1RP isolate (1 mg · liter−1, susceptible) (Table S5) led to a predicted ciprofloxacin MIC against a double QRDR mutant of 4 mg · liter−1 (nonsusceptible, now correctly predicting the observed phenotype of KP17) (Table 2). We therefore added additional predictive rules to consider the existence of double GyrA or GyrA/ParC QRDR mutations, making 47 rules in total (Table S6).
Mutations in repressor binding sites upregulate efflux pump production and influence fluoroquinolone susceptibility.
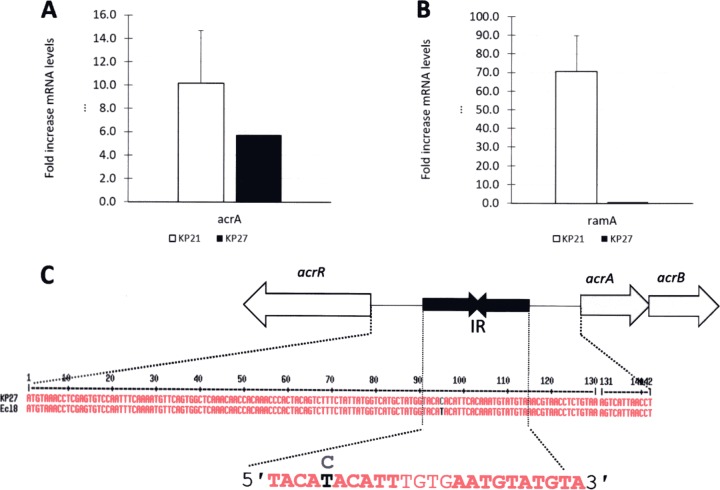
During our analysis, we noticed that while clinical isolate KP27 has a wild-type ramR sequence, the proteomics data showed that it hyperproduces AcrAB (Table 2). This was confirmed by using quantitative reverse transcription-PCR (qRT-PCR) for acrA relative to a wild-type control isolate, KP47, but ramA was not overexpressed in KP27, validating the observation that RamR is of the wild type (Fig. 2A and B and Table 2) (26). WGS identified a mutation in the acrR-acrA intergenic region in KP27, causing a T-to-C transition at the fifth position (in boldface type) of the first AcrR binding motif (5′-TACATACATT-3′) (Fig. 2C), predicted to derepress acrAB expression (27). While it does not affect our designation of KP27 as a ciprofloxacin-nonsusceptible isolate, this example shows how LC-MS/MS can allow the identification of previously unknown regulatory mutations involved in phenotypically relevant efflux pump overproduction.
FIG 2.
(A and B) Expression levels of acrA (A) and ramA (B) in K. pneumoniae clinical isolates KP21 (ramR mutant) and KP27 (ramR wild type), both of which were normalized to expression levels in a control isolate, KP17, using qRT-PCR. Data are presented as means ± standard errors of the means (n = 3 preparations of RNA). (C) A single nucleotide polymorphism is present in the putative AcrR binding site (inverted repeat [IR]) upstream of acrAB in KP27.
Conclusions.
We have demonstrated stepwise combinations of mechanisms leading to fluoroquinolone nonsusceptibility in K. pneumoniae and characterized the importance of each mechanism and the interplay between them. Using this information, we have successfully applied WGS to predict ciprofloxacin susceptibility/nonsusceptibility of K. pneumoniae clinical isolates. However, LC-MS/MS proteomics was essential for this process, particularly for determining the relative importance of different SNPs seen in the efflux pump regulators OqxR and RamR. The more phenotypically relevant SNPs that we can identify (e.g., see Table S3 in the supplemental material for a list of OqxR loss-of-function mutations uncovered here), the more we will be able to rely on WGS alone to identify phenotypically relevant mutations.
LC-MS/MS also allowed us to uncover new biology in this study. We incorrectly predicted ciprofloxacin susceptibility in genotypically rule 15 clinical isolates, a particularly common group, because they have reduced envelope permeability compared with Ecl8. LC-MS/MS revealed that this is due to the downregulation of OmpF and a consequent reduction in the OmpF/OmpC porin ratio. In fact, it would seem likely that this difference is due to a genetic change in Ecl8 rather than an equivalent genetic change in all the clinical isolates, so we propose that the predictive rules listed in Table S6, which allow for this reduced OmpF/OmpC ratio, are appropriate to use for all clinical isolates. Allowing for GyrA/ParC double mutations expanded the non-wild-type rules to a total number of 47, only 7 of which predict ciprofloxacin susceptibility.
Overall, this work enhanced our biological understanding of fluoroquinolone resistance in K. pneumoniae and will improve our ability to predict ciprofloxacin susceptibility in clinical isolates based on WGS data. Particularly, it shows the value of using proteomics to clarify genotype-to-phenotype relationships and to bridge the gap between WGS and antimicrobial susceptibility testing, where mechanisms of resistance are multifactorial and complex.
MATERIALS AND METHODS
Bacterial strains and antibacterial drug susceptibility testing.
The isogenic pair K. pneumoniae Ecl8 (28) and Ecl8 ΔramR (29) was used throughout. Forty human K. pneumoniae clinical isolates were studied. Details of these isolates are given in Table S1 in the supplemental material. Disc susceptibility testing was performed and MICs were determined according to CLSI methodologies (30, 31). Susceptibility/nonsusceptibility was interpreted by using CLSI performance standards (32).
Selection of in vitro K. pneumoniae mutants conferring reduced fluoroquinolone susceptibility and transformation with PMQR genes.
Mutants were generated by plating 100 μl of a culture grown overnight onto LB agar containing ciprofloxacin or nalidixic acid at various doubling concentrations below the CLSI-defined nonsusceptibility breakpoint. First-step mutants were then reselected by using increasing concentrations of ciprofloxacin to generate fluoroquinolone-nonsusceptible mutants. PCR-sequencing to identify mutations was performed by using primers listed in Table S2 in the supplemental material. The PMQR genes qnrA1 and aac(6′)-Ib-cr were synthesized (Eurofins Genomics), as defined in Table S2, to include native promoters and ligated into the pEX-K4 cloning vector (Eurofins). Recombinant plasmids were used to transform K. pneumoniae to kanamycin (30 mg/liter) resistance by using electroporation, as is standard for E. coli laboratory strains.
Quantitative analysis of the whole-cell proteome via Orbitrap LC-MS/MS and qRT-PCR.
Cells were cultured in 50 ml nutrient broth (Oxoid) with shaking (160 rpm) at 37°C until the optical density at 600 nm (OD600) reached 0.5 to 0.7. RNA was purified, and qRT-PCR was performed as previously described (26, 33). For proteomics, cells in cultures were pelleted by centrifugation (10 min at 4,000 × g at 4°C), resuspended in 20 ml of 30 mM Tris-HCl (pH 8), and broken by sonication with a cycle of 1 s on and 0.5 s off for 3 min at an amplitude of 63% by using a Sonics Vibracell VC-505 instrument (Sonics and Materials Inc., Newton, CT, USA). Following centrifugation (15 min at 4°C at 8,000 rpm) (Sorvall RC5B with an SS34 rotor), the protein concentration in the supernatant was quantified by using Bio-Rad Protein Assay Dye Reagent Concentrate according to the manufacturer's instructions. One microgram of total protein was separated by SDS-PAGE using 11% acrylamide–0.5% bis-acrylamide (Bio-Rad) gels and a Bio-Rad model 3000X1 Min-Protein Tetracell chamber. Gels were run at 200 V until the dye front had moved approximately 1 cm into the separating gel. Proteins in gels were stained with Instant Blue (Expedeon) for 20 min and destained in water.
The 1 cm of the gel lane containing protein was cut out, and proteins were subjected to in-gel tryptic digestion using a DigestPro automated digestion unit (Intavis Ltd.). The resulting peptides were fractionated by using an Ultimate 3000 nanoHPLC system in line with an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific) according to our previously reported protocol (26, 33). The raw data files were processed and quantified by using Proteome Discoverer software v1.4 (Thermo Scientific) and searched against the UniProt K. pneumoniae strain ATCC 700721/MGH 78578 database (5,126 protein entries; UniProt accession number 272620) by using the SEQUEST (ver. 28, rev. 13) algorithm. Protein area measurements were calculated from peptide peak areas by using the Top 3 method (34) and were then used to calculate the abundance of each protein. Proteins with fewer than three peptide hits were excluded from the analysis. The raw abundance of each protein was divided by the average abundance of a ribosomal protein to normalize for sample-to-sample loading variability.
Fluorescent Hoechst 33342 dye accumulation assay.
Envelope permeability in bacteria grown in cation-adjusted Muller-Hinton broth (Sigma) was estimated as described previously (26, 33), using an established fluorescent dye accumulation assay (35). Hoechst 33342 (H33342) dye (Sigma) was used at a final concentration of 2.5 μM.
Whole-genome sequencing and data analysis.
Genomes were sequenced by MicrobesNG (Birmingham, UK) on a HiSeq 2500 instrument (Illumina, San Diego, CA, USA). Reads were trimmed by using Trimmomatic (36) and assembled into contigs by using SPAdes 3.10.1 (http://cab.spbu.ru/software/spades/). The presence of resistance genes was determined by using MLST 1.8, PlasmidFinder (37), ResFinder 2.1 (3), and the Center for Genomic Research platform (https://cge.cbs.dtu.dk/services/). Assembled contigs were mapped to the K. pneumoniae Ecl8 reference genome (GenBank accession number GCF_000315385.1), obtained from GenBank by using progressive Mauve alignment software (38).
Supplementary Material
ACKNOWLEDGMENTS
Genome sequencing was provided by MicrobesNG (http://www.microbesng.uk/), which is supported by the BBSRC (grant number BB/L024209/1). This work was funded by grant MR/N013646/1 to M.B.A., A.P.M., O.M.W. and K.J.H. and grant NE/N01961X/1 to M.B.A. and A.P.M. from the Antimicrobial Resistance Cross Council Initiative supported by the seven research councils. W.A.K.W.N.I. was funded by a postgraduate scholarship from the Malaysian Ministry of Education. J.-C.J.-C. was funded by a postgraduate scholarship from CONACyT, Mexico.
We have no transparency declarations.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01814-17.
REFERENCES
- 1.Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, Giske C, Grundman H, Hasman H, Holden MTG, Hopkins KL, Iredell J, Kahlmeter G, Köser CU, MacGowan A, Mevius D, Mulvey M, Naas T, Peto T, Rolain J-M, Samuelsen Ø Woodford N. 2017. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect 23:2–22. doi: 10.1016/j.cmi.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Stoesser N, Batty EM, Eyre DW, Morgan M, Wyllie DH, Del Ojo Elias C, Johnson JR, Walker AS, Peto TEA, Crook DW. 2013. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J Antimicrob Chemother 68:2234–2244. doi: 10.1093/jac/dkt180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redgrave LS, Sutton SB, Webber MA, Piddock LJV. 2014. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol 22:438–445. doi: 10.1016/j.tim.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Lesher GY, Froelich EJ, Gruett MD, Bailey JH, Brundage RP. 1962. 1,8-Naphthyridine derivatives. A new class of chemotherapeutic agents. J Med Pharm Chem 5:1063–1065. doi: 10.1021/jm01240a021. [DOI] [PubMed] [Google Scholar]
- 6.Andriole VT. 2005. The quinolones: past, present, and future. Clin Infect Dis 41(Suppl 2):S113–S119. doi: 10.1086/428051. [DOI] [PubMed] [Google Scholar]
- 7.Emmerson AM, Jones AM. 2003. The quinolones: decades of development and use. J Antimicrob Chemother 51:13–20. doi: 10.1093/jac/dkg208. [DOI] [PubMed] [Google Scholar]
- 8.Levine C, Hiasa H, Marians KJ. 1998. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim Biophys Acta 1400:29–43. doi: 10.1016/S0167-4781(98)00126-2. [DOI] [PubMed] [Google Scholar]
- 9.Ruiz J. 2003. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother 51:1109–1117. doi: 10.1093/jac/dkg222. [DOI] [PubMed] [Google Scholar]
- 10.Hooper DC. 1999. Mechanisms of fluoroquinolone resistance. Drug Resist Updat 2:38–55. doi: 10.1054/drup.1998.0068. [DOI] [PubMed] [Google Scholar]
- 11.Shen LL, Mitscher LA, Sharma PN, O'Donnel TJ, Chu DWT, Cooper CS, Rosen T, Pernett AG. 1989. Mechanism of inhibition of DNA gyrase by quinolone antibacterials: a cooperative drug-DNA binding model. Biochemistry 28:3886–3894. doi: 10.1021/bi00435a039. [DOI] [PubMed] [Google Scholar]
- 12.Hooper DC, Jacoby GA. 2015. Mechanisms of drug resistance: quinolone resistance. Ann N Y Acad Sci 1354:12–31. doi: 10.1111/nyas.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martínez-Martínez L, García I, Ballesta S, Benedí VJ, Hernández-Allés S, Pascual A. 1998. Energy-dependent accumulation of fluoroquinolones in quinolone-resistant Klebsiella pneumoniae strains. Antimicrob Agents Chemother 42:1850–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martínez-Martínez L, Pascual A, Conejo MDC, García I, Joyanes P, Doménech-Sánchez A, Benedí VJ. 2002. Energy-dependent accumulation of norfloxacin and porin expression in clinical isolates of Klebsiella pneumoniae and relationship to extended-spectrum beta-lactamase production. Antimicrob Agents Chemother 46:3926–3932. doi: 10.1128/AAC.46.12.3926-3932.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazzariol A, Zuliani J, Cornaglia G, Rossolini GM, Fontana R. 2002. AcrAB efflux system: expression and contribution to fluoroquinolone resistance in Klebsiella spp. Antimicrob Agents Chemother 46:3984–3986. doi: 10.1128/AAC.46.12.3984-3986.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneiders T, Amyes SGB, Levy SB. 2003. Role of AcrR and RamA in fluoroquinolone resistance in clinical Klebsiella pneumoniae isolates from Singapore. Antimicrob Agents Chemother 47:2831–2837. doi: 10.1128/AAC.47.9.2831-2837.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veleba M, Higgins PG, Gonzalez G, Seifert H, Schneiders T. 2012. Characterization of RarA, a novel AraC family multidrug resistance regulator in Klebsiella pneumoniae. Antimicrob Agents Chemother 56:4450–4458. doi: 10.1128/AAC.00456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sørensen AH, Hansen LH, Johannesen E, Sørensen SJ. 2003. Conjugative plasmid conferring resistance to olaquindox. Antimicrob Agents Chemother 47:798–799. doi: 10.1128/AAC.47.2.798-799.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen LH, Johannesen E, Burmolle M, Sorensen AH, Sorensen SJ. 2004. Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli. Antimicrob Agents Chemother 48:3332–3337. doi: 10.1128/AAC.48.9.3332-3337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martínez-Martínez L, Pascual A, Jacoby GA. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 21.Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, Hooper DC. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob Agents Chemother 50:1178–1182. doi: 10.1128/AAC.50.4.1178-1182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang M, Guo Q, Xu X, Wang X, Ye X, Wu S, Hooper DC, Wang M. 2009. New plasmid-mediated quinolone resistance gene, qnrC, found in a clinical isolate of Proteus mirabilis. Antimicrob Agents Chemother 53:1892–1897. doi: 10.1128/AAC.01400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hata M, Suzuki M, Matsumoto M, Takahashi M, Sato K, Ibe S, Sakae K. 2005. Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob Agents Chemother 49:801–803. doi: 10.1128/AAC.49.2.801-803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cavaco LM, Hasman H, Xia S, Aarestrup FM. 2009. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob Agents Chemother 53:603–608. doi: 10.1128/AAC.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH, Bush K, Hooper DC. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med 12:83–88. doi: 10.1038/nm1347. [DOI] [PubMed] [Google Scholar]
- 26.Jiménez-Castellanos J-C, Wan Nur Ismah WAK, Takebayashi Y, Findlay J, Schneiders T, Heesom KJ, Avison MB. 2018. Envelope proteome changes driven by RamA overproduction in Klebsiella pneumoniae that enhance acquired β-lactam resistance. J Antimicrob Chemother 73:88–94. doi: 10.1093/jac/dkx345. [DOI] [PubMed] [Google Scholar]
- 27.Su C-C, Rutherford DJ, Yu EW. 2007. Characterization of the multidrug efflux regulator AcrR from Escherichia coli. Biochem Biophys Res Commun 361:85–90. doi: 10.1016/j.bbrc.2007.06.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.George AM, Hall RM, Stokes HW. 1995. Multidrug resistance in Klebsiella pneumoniae: a novel gene, ramA, confers a multidrug resistance phenotype in Escherichia coli. Microbiology 141(Part 8):1909–1920. doi: 10.1099/13500872-141-8-1909. [DOI] [PubMed] [Google Scholar]
- 29.De Majumdar S, Yu J, Fookes M, McAteer SP, Llobet E, Finn S, Spence S, Monahan A, Monaghan A, Kissenpfennig A, Ingram RJ, Bengoechea J, Gally DL, Fanning S, Elborn JS, Schneiders T. 2015. Elucidation of the RamA regulon in Klebsiella pneumoniae reveals a role in LPS regulation. PLoS Pathog 11:e1004627. doi: 10.1371/journal.ppat.1004627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clinical and Laboratory Standards Institute. 2006. M2-A9. Performance standards for antimicrobial disc susceptibility tests; approved standard, 9th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 31.Clinical and Laboratory Standards Institute. 2015. M07-A10. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 10th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 32.Clinical and Laboratory Standards Institute. 2015. M100-S25. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. An informational supplement for global application developed through the Clinical and Laboratory Standards Institute consensus process. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 33.Jiménez-Castellanos J-C, Wan Ahmad Kamil WNI, Cheung CHP, Tobin MS, Brown J, Isaac SG, Heesom KJ, Schneiders T, Avison MB. 2016. Comparative effects of overproducing the AraC-type transcriptional regulators MarA, SoxS, RarA and RamA on antimicrobial drug susceptibility in Klebsiella pneumoniae. J Antimicrob Chemother 71:1820–1825. doi: 10.1093/jac/dkw088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silva JC, Gorenstein MV, Li G-Z, Vissers JPC, Geromanos SJ. 2006. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics 5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Coldham NG, Webber M, Woodward MJ, Piddock LJV. 2010. A 96-well plate fluorescence assay for assessment of cellular permeability and active efflux in Salmonella enterica serovar Typhimurium and Escherichia coli. J Antimicrob Chemother 65:1655–1663. doi: 10.1093/jac/dkq169. [DOI] [PubMed] [Google Scholar]
- 36.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darling AE, Mau B, Perna NT, Batzoglou S, Zhong Y. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.