ABSTRACT
The presence of the mcr-1 gene in Escherichia coli isolated from retail freshwater fish was investigated. Seven (3.65%) clonally unrelated original E. coli isolates from grass carp were positive for mcr-1. The mcr-1 genes were encoded by either chromosomes (n = 2) or conjugative plasmids (2 IncI2, 2 IncP, and 1 IncX4). The IncP plasmids were similar to other mcr-1-harboring IncP plasmids from China, though the insertion sites varied. Our report warrants further surveillance of resistance genes in aquaculture.
KEYWORDS: colistin resistance, mcr-1, Escherichia coli, fish, IncP, Enterobacteriaceae, plasmid mediated
TEXT
Since the first identification of the plasmid-mediated colistin resistance gene mcr-1, it has been detected worldwide, mainly in Escherichia coli from livestock, food, humans, and the environment (1–3). Recently, the presence of mcr-1 has been reported in scampi, rivers, well water, and sewage from farms and hospitals (4–8), suggesting its possible spread to aquaculture. It has been proposed that aquaculture may promote the origination, mobilization, and selection of mcr genes (9). However, a systematic study on the prevalence of mcr-1 genes in aquaculture is lacking. Therefore, we carried out an investigation of the mcr-1 gene in E. coli from retail grass carp, one of the leading freshwater fishes produced in China.
Between March 2016 and December 2016, a total of 192 nonduplicated E. coli strains were recovered from 700 grass carp collected from 11 fish markets (A to K) located throughout Guangzhou, the largest trading center of aquatic products in southern China. The susceptibilities of these isolates to 19 antimicrobial agents were determined by the agar dilution method or the broth microdilution method (colistin) (10). Eight isolates showed reduced susceptibility to colistin, with MICs of 4 to 8 mg/liter, and exhibited a multidrug resistance phenotype (Table 1).
TABLE 1.
Characterization of mcr-1-carrying E. coli isolates and plasmids
| Isolate | Isolation date | Market | MLSTa | Resistance patternb | Colistin (mg/liter) | Other resistance genes | Location and plasmid size (kb) | Conjugation frequency (cells per recipient) | Genetic context |
|---|---|---|---|---|---|---|---|---|---|
| GDP6F1 | Sep 2016 | A | ST48 (ST10 Cplx) | AMP, STR, NEO, CHL, FLR, SXT, TET, DOX | 4 | aadA2, aadA1, aph(3′)-Ic, blaTEM-176, blaOXA-10, blaCARB-2, mcr-1, qnrS1, oqxB, oqxA, cmlA1, floR, arr-2, sul2, sul3, tet(A), dfrA14, dfrA16 | IncP, 50.4 | 5.0 × 10−4 | IncP-mcr-1-hp-IncP |
| GDT6F13 | Apr 2016 | B | ST4014 | AMP, CTX, FOS, STR, GEN, NEO, CHL, FLR, SXT, TET, DOX | 8 | strA, strB, aph(3′)-Ic, aac(3)-IId, blaCTX-M-55, mcr-1, oqxA, oqxB, fosA3, erm(B), mph(A), floR, sul2, tet(A), dfrA17 | chromosome | ISApl1-mcr-1-hp-ISApl1 | |
| GDT6F36 | May 2016 | C | ST7508 | STR, CHL, FLR, TET | 8 | strA, strB, mcr-1, qnrS1, floR, sul2, tet(A) | IncP, 52.7 | 1.4 × 10−4 | ISApl1-mcr-1-hp-ISApl1 |
| GDT6F38 | May 2016 | C | ST101 (ST101Clpx) | AMP, NEO, CHL, FLR, SXT, TET, DOX | 8 | aadA1, aph(3′)-Ia, aadA2, mcr-1, floR, cmlA1, sul2, sul3, tet(M), tet(A), dfrA12 | chromosome | ISApl1-mcr-1-hp-IS1294- ISApl1 | |
| GDT6F49 | May 2016 | C | ST2040 | AMP, CTX, STR, CHL, FLR, SXT, TET, DOX | 8 | aadA1, blaOXA-10, blaCTX-M-65, mcr-1, qnrS1, cmlA1, floR, arr-2, tet(A), tet(A), dfrA14 | IncX4, 33.3 | 5.5 × 10−4 | IncX4-mcr-1-IncX4 |
| GDT6F93 | July 2016 | D | ST7013 | AMP, CL, STR, CHL, FLR, CIP, SXT, TET, DOX | 4 | strA, strB, mcr-1, aadA5, aadA1, blaOXA-10, blaTEM-1A, qnrS1, cmlA1, floR, arr-2, sul2, tet(A), dfrA17, dfrA14 | IncI2, 60.8 | 1.7 × 10−3 | IncI2-mcr-1-IncI2 |
| GDT6F97 | July 2017 | D | ST156 | AMP, CTX, FOS, STR, GEN, NEO, CHL, FLR, CIP, SXT, TET, DOX | 4 | tet(A), aac(3)-Iid, fosA3, dfrA12, floR, strB, tet(M), aadA22, strA, aph(3′)-Iia, oqxA, oqxB, erm(B), sul2, blaCTX-M-14 | IncI2, 60 | 4.8 × 10−4 | IncI2-mcr-1-IncI2 |
MLST, multilocus sequence type.
Antimicrobial susceptibility was determined and evaluated according to CLSI document no. M100-S27 (https://clsi.org/standards/products/microbiology/documents/m100/). Resistance to florfenicol (>16 mg/L) and neomycin (>8 mg/L) was interpreted according to EUCAST clinical breakpoints and epidemiological cutoff values (http://mic.eucast.org/Eucast2/). AMP, ampicillin; CTX, cefotaxime; FOS, fosfomycin; STR, streptomycin; GEN, gentamicin; NEO, neomycin; CHL, chloramphenicol; FFC, florfenicol; CIP, ciprofloxacin; SXT, sulfamethoxazole/trimethoprim; TET, tetracycline; DOX, doxycycline.
The presence of mcr-1 was confirmed by PCR screening with specific primers described by Liu et al. (1) and by sequencing. Colistin-resistant isolates were also screened for mcr-2, mcr-3, mcr-4, and mcr-5 genes as detailed previously (11–14). Seven E. coli isolates (3.65%) were positive for mcr-1. One E. coli isolate exhibited reduced susceptibility to colistin, but was negative for all mcr genes. Although the origin of mcr-1 in retail freshwater fish is unclear, improper disposal of human or animal sewage to the aquatic environment might be one source, as Cabello and Godfrey reported that aquacultural water is likely to be contaminated by human and animal pathogens (9). Additionally, mcr-1 may occur in integrated agriculture where aquacultured fish was fed animal manure containing colistin. Furthermore, the possibility of mcr-1 originating from aquatic bacteria could not be ruled out (15). It has been reported that enzymes encoded by mcr-1, such as ethanolamine phosphotransferase (EptA and PmrC), were detected in Shewanella algae, a member of a genus that contains opportunistic fish and human pathogens (16). Furthermore, the amino acid sequence of the mcr-1-encoding protein is significantly similar to that of the phosphoethanolamine transferase of Enhydrobacter aerosaccus, an aquatic bacterium (1). The emergence of mcr-1 in aquatic products should raise concerns, as mcr-1 may spread globally via international trade, as evidenced by reports in Norway (4). In addition, considering high consumption of aquatic products and the habit of eating sashimi (raw fish meat), it is highly possible that mcr-1 could be transferred directly to humans through the food chain.
Molecular typing results demonstrated that mcr-1-positive isolates belonged to different sequence types (STs), including ST48 (ST10 Cplx), ST4014, ST101 (ST101 Cplx), ST2040, ST7013, ST156, and a novel ST, ST7508 (Table 1). Interestingly, ST48 and ST156 E. coli isolates were also found as carriers of mcr-1 in scampi and Muscovy duck, respectively (4, 17).
S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) and Southern blotting indicated that the mcr-1 genes were located on chromosomes in two isolates (GDT6F13 and GDT6F38) and on plasmids in the remaining five isolates (GDP6F1, GDT6F36, GDT6F49, GDT6F93, and GDT6F97). Streptomycin-resistant E. coli strain C600 was used as a recipient in the conjugation experiment to study the transferability of the mcr-1 genes. All five mcr-1-bearing plasmids were successfully transferred to the recipient strain at frequencies varied from 1.4 × 10−4 to 5.5 × 10−4 cells per donor cell (Table 1).
To investigate the genetic backgrounds of the mcr-1-positive isolates, whole-genome sequencing was conducted on an Illumina HiSeq 2500-PE125 platform (Beijing Novogene Bioinformatics Cp., Ltd.). The sequences were assembled using SOAPdenovo (http://soap.genomics.org.cn/soapdenovo.html) and analyzed using the online tools MLST (MultiLocus Sequence Typing), PlasmidFinder, and ResFinder (http://cge.cbs.dtu.dk/services). Based on reference plasmids, mcr-1-bearing contigs were comparatively analyzed, and gaps between contigs were filled by PCR and Sanger sequencing. Four complete sequences of mcr-1-bearing plasmids were obtained, namely, IncP plasmids pHNGDF1-1 and pHNGDF36-1, IncX4 plasmid pHNGDF49, and IncI2 plasmid pHNGDF93. Previous studies revealed that the IncX4 and IncI2 plasmids were dominant carriers of mcr-1 (18, 19). In this study, pHNGDF49 and pHNGDF93 were highly similar to those reported mcr-1-bearing IncX4 and IncI2 plasmids from various origins, including from animals, humans, and the environment (see supplemental material), suggesting the dissemination of these epidemic mcr-1-carrying plasmids to aquaculture.
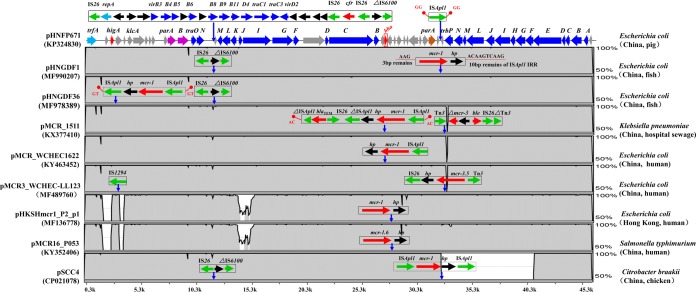
Additionally, broad-host-range IncP plasmids were also the vectors for mcr-1 in this study. mcr-1-positive IncP plasmids have been described in various species of bacteria in China, including Klebsiella pneumoniae (pMCR_1511, GenBank accession number KX377410), E. coli (pMCR_WCHEC1622, KY463452; pMCR3_WCHEC-LL123, MF489760; and pHKSHmcr_P2_p1, MF136778), Citrobacter braakii (pSCC4, NZ_CP021078), and Salmonella enterica serovar Typhimurium (pMCR16_P053, KY352406), originating from diverse sources such as hospital sewage, pig feces, chickens, and humans (Fig. 1) (20, 21), highlighting the significance of IncP in the transmission of mcr-1. The backbones of pHNGDF1-1 and pHNGDF36-1 were almost identical to that of pHNFP671 (KP324830), which belonged to a new IncP-1 plasmid clade and did not harbor mcr-1. pHNFP671 was found in an E. coli isolate obtained from swine feces in China. Comparative analysis of pHNGDF1-1, pHNGDF36-1, other IncP plasmids encoding mcr-1, and pHNFP671 revealed a high level of conservation across the plasmid backbones, except for pSCC4, which has an ∼8,400 bp deletion of the transfer region (Fig. 1).
FIG 1.
Levels of identity between the pHNGDF1-1, pHNGDF36-1, pMCR_1511, pMCR_WCHEC1622, pMCR3_WCHEC-LL123, pHKSHmcr_P2_p1, pMCR16_P053, and pSCC4 backbones and the pHNFP671 backbone. The scale of identity is displayed on the right. With tra genes shown by appropriate capital letters, the extents and directions of specific genes are shown by labeled arrows in different color. Labeled vertical arrows are used to annotate the insertion loci of mobile elements that were removed before alignment of the backbones. Sequences of other plasmids (accession numbers shown in the figure) were obtained from GenBank.
Although mcr-1-carrying IncP plasmids shared conserved backbones with pHNFP671, the variable regions were different (Fig. 1). In pHNFP671, ISApl1 was present ∼630 bp upstream of trbP. Interestingly, mcr-1-hp and ISApl1-mcr-1-hp-ISApl1 were also inserted within the same locus in pHNGDF1-1 and pSCC4, respectively (Fig. 1). This suggests that this locus is a hot spot for the insertion of ISApl1. As for pHNGDF36-1, it represented a common mechanism mediating the spread of mcr-1. A composite transposon, ISApl1-mcr-1-hp-ISApl1, flanked by 2-bp (GT) direct repeats (DRs), was inserted downstream of higA, which was in agreement with the report that ISApl1 could generate 2-bp DRs (22).
A BLAST search against the GenBank WGS database showed that the pMCR_1511-like IncP plasmid carrying mcr-1 was also present in the E. coli strain CT37C A1 isolated from Gallus gallus feces in Netherlands (GenBank accession number FLZF01000032; 99% coverage and 99% nucleotide identity), a Salmonella enterica subsp. enterica serovar Typhimurium ST34 strain GMR-S-1257 isolated from human feces in Colombia (MVPR01000083; 99% coverage and 99% nucleotide identity) (23), and three E. coli strains isolated from human vaginal secretions in Colombia (MVPN01000072; 99% coverage and 99% nucleotide identity). It is likely that the pMCR_1511-like IncP plasmid is widely distributed.
pHNGDF1-1 and pHNGDF36-1 were stably maintained after passage (data not shown). Additionally, pairwise competition assays were carried out using transformants, which were obtained by chemical transformation with E. coli DH5α as the recipient and competed with E. coli DH5α. It was found that that carriage of pHNGDF1-1 and pHNGDF36-1 enhanced biological fitness in the host (Fig. 2). Recently, IncP plasmids were described as mediating the dissemination of the mcr-3 gene between Enterobacteriaceae and Aeromonas spp., which are pathogens of aquacultured fish (24). Moreover, Zhao et al. proposed that the IncP plasmid could mediate the transmission of mcr-1 from Enterobacteriaceae to other Gram-negative bacteria, such as Pseudomonas aeruginosa (20). Thus, considering its high conjugation frequency and broad host range, the IncP plasmid may facilitate the dissemination of mcr-1 across various hosts, and it might potentially become as dominant a carrier of mcr-1 as the IncI2 and IncX4 plasmids.
FIG 2.
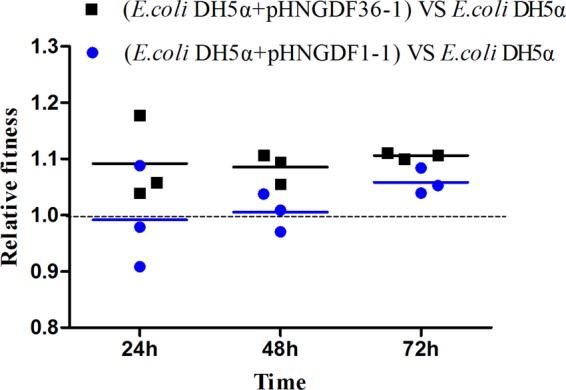
Fitness cost of pHNGDF1-1 and pHNGDF36-1 in E. coli DH5α in vitro. Growth competition between recipient E. coli DH5α and transformants containing pHNGDF1-1 and pHNGDF36-1. Each transformant was cultured in the presence of DH5α but without any antibiotics. The results were calculated and expressed as relative fitness against DH5α. The initial ratio was 1:1.
In summary, to the best of our knowledge, ours is the first report of the mcr-1 gene in fish products. The presence of mcr-1 in retail freshwater fish is of great concern, given that this gene has the possibility to spread globally via international trade of aquatic products and to threaten human health through the food chain. This study warrants further investigation in aquaculture to prevent the spread of antimicrobial resistance.
Accession number(s).
Plasmids pHNGDF1-1, pHNGDF36-1, pHNGDF49, and pHNGDF93 have been deposited in GenBank under the accession numbers MF990207, MF978389, MF978387, and MF978388, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work, including the efforts of Jian-Hua Liu, was supported in part by grants from the National Natural Science Foundation of China (grans 31625026 and 81661138002), and the National Training Program of Innovation and Entrepreneurship for Undergraduates (grant 201610564120).
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02378-17.
REFERENCES
- 1.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 2.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo J, Yao X, Lv L, Doi Y, Huang X, Huang S, Liu JH. 2017. Emergence of mcr-1 in Raoultella ornithinolytica and Escherichia coli isolates from retail vegetables in China. Antimicrob Agents Chemother 61:e01139-. doi: 10.1128/AAC.01139-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slettemeås JS, Jannice S, Urdahl AM, Mo SS, Johannessen GS, Grave K, Norström M, Steinbakk M, Sunde M. 2017. Imported food and feed as contributors to the introduction of plasmid-mediated colistin-resistant Enterobacteriaceae to a ‘low prevalence’ country. J Antimicrob Chemother 72:2675–2677. doi: 10.1093/jac/dkx161. [DOI] [PubMed] [Google Scholar]
- 5.Lekunberri I, Balcázar JL, Borrego CM. 2017. Detection and quantification of the plasmid-mediated mcr-1 gene conferring colistin resistance in wastewater. Int J Antimicrob Agents 50:734–736. doi: 10.1016/j.ijantimicag.2017.08.018. [DOI] [PubMed] [Google Scholar]
- 6.Yang D, Qiu ZG, Shen ZQ, Zhao H, Jin M, Li HY, Liu WL, Li JW. 2017. The occurrence of the colistin resistance gene mcr-1 in the Haihe river (China). Int J Environ Res Public Health 14:E576. doi: 10.3390/ijerph14060576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun P, Bi ZW, Nilsson M, Zheng BW, Berglund B, Stalsby LC, Borjesson S, Li XW, Chen BL, Yin H, Nilsson LE. 2017. Occurrence of blaKPC-2, blaCTX-M, and mcr-1 in Enterobacteriaceae from well water in rural China. Antimicrob Agents Chemother 61:e02569-. doi: 10.1128/AAC.02569-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Zhang RM, Li JY, Wu ZW, Yin WJ, Schwarz S, Tyrrell JM, Zheng YJ, Wang SL, Shen ZQ, Liu ZH, Liu JY, Lei L, Li M, Zhang QD, Wu CM, Zhang QJ, Wu YN, Walsh TR, Shen JZ. 2017. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol 2:16260. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- 9.Cabello FC, Godfrey HP. 2017. Comment on: Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother 72:636–637. doi: 10.1093/jac/dkw432. [DOI] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing: 27th informational supplement. CLSI document M100-S27. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, Pezzotti G, Magistrali CF. 2017. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill 22(31):pii=30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xavier BB, Lammens C, Ruhal R, Malhotra-Kumar S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21(27):pii=30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 13.Yin WJ, Li H, Shen YB, Liu ZH, Wang SL, Shen ZQ, Zhang R, Walsh TR, Shen JZ, Wang Y. 2017. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e00543-. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. 2017. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother 72:3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 15.Cabello FC, Tomova A, Ivanova L, Godfrey HP. 2017. Aquaculture and mcr colistin resistance determinants. mBio 8:e01229-17. doi: 10.1128/mBio.01229-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Telke AA, Rolain JM. 2015. Functional genomics to discover antibiotic resistance genes: The paradigm of resistance to colistin mediated by ethanolamine phosphotransferase in Shewanella algae MARS 14. Int J Antimicrob Agents 46:648–652. doi: 10.1016/j.ijantimicag.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Yang RS, Feng YJ, Lv XY, Duan JH, Chen J, Fang LX, Xia J, Liao XP, Sun J, Liu YH. 2016. Emergence of NDM-5- and MCR-1-producing Escherichia coli clones ST648 and ST156 from a single Muscovy duck (Cairina moschata). Antimicrob Agents Chemother 60:6899–6902. doi: 10.1128/AAC.01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li R, Xie M, Zhang J, Yang Z, Liu L, Liu X, Zheng Z, Chan EW, Chen S. 2017. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother 72:393–401. doi: 10.1093/jac/dkw411. [DOI] [PubMed] [Google Scholar]
- 19.Zurfluh K, Nüesch-Inderbinen M, Klumpp J, Poirel L, Nordmann P, Stephan R. 2017. Key features of mcr-1-bearing plasmids from Escherichia coli isolated from humans and food. Antimicrob Resist Infect Control 6:91. doi: 10.1186/s13756-017-0250-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao FF, Feng Y, Lü XJ, McNally A, Zong ZY. 2017. IncP plasmid carrying colistin resistance gene mcr-1 in Klebsiella pneumoniae from hospital sewage. Antimicrob Agents Chemother 61:e02229-. doi: 10.1128/AAC.02229-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Webb HE, Granier SA, Marault M, Millemann Y, Bakker HC, Nightingale KK, Bugarel M, Ison SA, Scott HM, Loneragan GH. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:144–145. doi: 10.1016/S1473-3099(15)00538-1. [DOI] [PubMed] [Google Scholar]
- 22.Tegetmeyer HE, Jones SCP, Langford PR, Baltes N. 2008. ISApl1, a novel insertion element of Actinobacillus pleuropneumoniae, prevents ApxIV-based serological detection of serotype 7 strain AP76. Vet Microbiol 128:342–353. doi: 10.1016/j.vetmic.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 23.Saavedra SY, Diaz L, Wiesner M, Correa A, Arévalo SA, Reyes J, Hidalgo AM, Cadena E, Perenguez M, Montaño LA, Ardila J, Ríos R, Ovalle MV, Díaz1 Porras PP, Villegas MV, Arias CA, Beltrán M, Duarte C. 2017. Genomic and molecular characterization of clinical isolates of Enterobacteriaceae harboring mcr-1 in Colombia, 2002–2016. Antimicrob Agents Chemother 61:e00841-17. doi: 10.1128/AAC.00841-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L, Feng Y, Zhang X, Mcnally A, Zong Z. 2017. New variant of mcr-3 in an extensively drug-resistant Escherichia coli clinical isolate carrying mcr-1 and blaNDM-5. Antimicrob Agents Chemother 61:e01757-17. doi: 10.1128/AAC.01757-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.