ABSTRACT
We report the mutant prevention concentration (MPC) and mutant selection window (MSW) for micafungin and anidulafungin administered to treat Candida glabrata. We also determine the mutation frequency. We studied 20 echinocandin-susceptible, fluconazole-intermediate, and FKS wild-type C. glabrata isolates. Adjusted inocula were stroked directly onto Sabouraud agar plates containing different concentrations of micafungin or anidulafungin and visually inspected daily for up to 5 days of incubation. Individual colonies growing on the plates containing echinocandins at 1 mg/liter were selected for antifungal susceptibility testing. The FKS genes of the resulting individual phenotypically resistant colonies were sequenced, and the MPC, MSW, and mutation frequency were determined. Biofilm was quantified, and the growth kinetics and virulence (Galleria mellonella model) of the resulting individual FKS mutant colonies were studied. For micafungin and anidulafungin, we found similar results for the MPC (0.06 to 2 mg/liter and 0.25 to 2 mg/liter, respectively), MSW (0.015 to 2 mg/liter for both echinocandins), and mutation frequency (3.7 × 10−8 and 2.8 × 10−8, respectively). A total of 12 isolates were able to grow at 1 mg/liter on echinocandin-containing plates, yielding a total of 32 phenotypically resistant colonies; however, FKS2 mutations (ΔF658, S663P, W715L, and E655A) were observed only in 21 colonies. We did not find differences in biofilm formation, the kinetic parameters studied, or the median survival of larvae infected by wild-type isolates and the resulting individual FKS2 mutant colonies. Echinocandin concentrations lower than 2 mg/liter can lead to selection of resistance mutations in C. glabrata isolates in vitro.
KEYWORDS: Candida glabrata, echinocandins, MPC, MSW, FKS mutation, Galleria mellonella
INTRODUCTION
Candida glabrata is one of the most clinically relevant causes of candidemia, and the incidence of candidemia caused by this entity seems to be on the rise (1–3). Echinocandins are currently recommended as the first-line treatment for invasive candidiasis (4–6), and resistance may complicate the management of patients infected by C. glabrata.
Resistance mutations have been identified in patients receiving long-term treatment with echinocandins and in vitro after exposure to increasing or constantly low concentrations of echinocandins (7, 8). Echinocandin resistance is associated with the presence of mutations in hot-spot regions of the genes FKS1 and FKS2 (3, 9, 10).
The cause of increased resistance to echinocandins is unclear. However, 2 recent studies showed that the abdominal compartment and/or colonized mucosa of patients with invasive candidiasis could act as a hidden reservoir of echinocandin-resistant C. glabrata isolates (11, 12). A potential explanation for this finding is the selection of resistance mutations in peritoneal fluid in the presence of low echinocandin concentrations, which are insufficient to inhibit selection of mutations but might promote the selection of resistant mutants (13). In addition, the MSH2 mutator phenotype or specific genotypes may contribute to the development of resistance to echinocandins (14, 15).
The parameters mutant prevention concentration (MPC) and mutant selection window (MSW) are useful when attempting to optimize antibacterial treatment, minimize the emergence of resistant mutants, and understand treatment failure (16). These parameters are mostly unknown for Candida spp., although they might be particularly relevant for C. glabrata.
We report the micafungin and anidulafungin MPC and MSW in C. glabrata and the corresponding mutation frequency. Furthermore, we determined whether the acquisition of FKS mutations entailed a cost for the isolates in terms of fitness and virulence.
(This study was partially presented at the 27th European Congress of Clinical Microbiology and Infectious Diseases in Vienna, Austria, 2017 [ePoster no. EP0626].)
RESULTS
Echinocandin MPC and MSW and mutation frequency.
All of the studied isolates were echinocandin susceptible, fluconazole intermediate, and FKS1 and FKS2 wild type. The number of isolates growing on plates containing micafungin or anidulafungin increased when the incubation period was prolonged to 5 days; overall, the number of isolates growing on anidulafungin-containing plates was higher than the number growing on micafungin-containing plates (Fig. 1a).
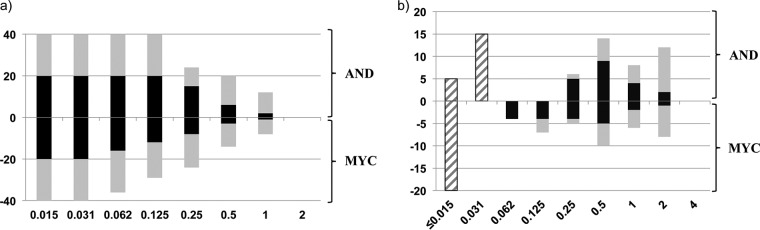
FIG 1.
(a) Number of original isolates growing on anidulafungin-containing plates (positive y axis) and on micafungin-containing plates (negative y axis) after 24 h (black bars) and 5 days (gray bars) of incubation. (b) Distribution of MICs (striped bars) and MPCs (solid bars) of anidulafungin (positive y axis) and micafungin (negative y axis) after 24 h (black bars) and 5 days of incubation (gray bars).
The MIC, MPC, and MSW values for each isolate and overall are shown in Table 1. The observed ranges of micafungin and anidulafungin MPCs were wide (Fig. 1b); none of the isolates was able to grow at echinocandin concentrations above 1 mg/liter. The geometric mean MPCs obtained after 24 h of incubation were significantly lower than the MPCs obtained after 5 days, regardless of the drug studied (P = 0.001). However, the geometric mean MPC of anidulafungin was significantly higher than that of micafungin after 24 h of incubation (0.55 mg/liter versus 0.25 mg/liter; P = 0.046), although the differences did not reach statistical significance after 5 days of incubation (1.15 mg/liter versus 0.73 mg/liter) (Fig. 1b and Table 1). Nevertheless, the MSW of anidulafungin and micafungin was identical after 24 h and 5 days of incubation of the plates (Fig. 1b and Table 1).
TABLE 1.
Micafungin and anidulafungin MIC, MPC, and MSW after 24 h and 5 days of incubation for each isolate
| Isolate | Value(s) (mg/liter) fora: |
|||||
|---|---|---|---|---|---|---|
| MYC |
AND |
|||||
| MIC | MPC | MSW | MIC | MPC | MSW | |
| 1 | 0.015 | 2/2 | 0.015–2/0.015–2 | 0.03 | 0.5/2 | 0.03–0.5/0.03–2 |
| 2 | 0.015 | 0.25/2 | 0.015–0.25/0.015–2 | 0.015 | 1/2 | 0.015–1/0.015–2 |
| 3 | 0.015 | 0.125/2 | 0.015–0.125/0.015–2 | 0.015 | 0.5/2 | 0.015–0.5/0.015–2 |
| 4 | 0.015 | 0.5/2 | 0.015–0.5/0.015–2 | 0.03 | 0.5/2 | 0.03–0.5/0.03–2 |
| 5 | 0.015 | 0.5/0.5 | 0.015–0.5/0.015–0.5 | 0.03 | 0.25/2 | 0.03–0.25/0.03–2 |
| 6 | 0.015 | 0.25/2 | 0.015–0.25/0.015–2 | 0.03 | 0.5/1 | 0.03–0.5/0.03–1 |
| 7 | 0.015 | 1/2 | 0.015–1/0.015–2 | 0.03 | 0.5/1 | 0.03–0.5/0.03–1 |
| 8 | 0.015 | 0.125/1 | 0.015–0.125/0.015–1 | 0.03 | 0.5/2 | 0.03–0.5/0.03–2 |
| 9 | 0.015 | 0.25/1 | 0.015–0.25/0.015–1 | 0.03 | 0.25/2 | 0.03–0.25/0.03–2 |
| 10 | 0.015 | 0.25/0.5 | 0.015–0.25/0.015–0.5 | 0.03 | 1/2 | 0.03–1/0.03–2 |
| 11 | 0.015 | 0.06/0.5 | 0.015–0.06/0.015–0.5 | 0.03 | 2/2 | 0.03–2/0.03–2 |
| 12 | 0.015 | 1/2 | 0.015–1/0.015–2 | 0.015 | 2/2 | 0.015–2/0.015–2 |
| 13 | 0.015 | 0.125/0.125 | 0.015–0.125/0.015–0.125 | 0.03 | 0.5/0.5 | 0.03–0.5/0.03–0.5 |
| 14 | 0.015 | 0.06/0.125 | 0.015–0.06/0.015–0.125 | 0.015 | 1/1 | 0.015–1/0.015–1 |
| 15 | 0.015 | 0.125/0.125 | 0.015–0.125/0.015–0.125 | 0.03 | 0.5/0.5 | 0.03–0.5/0.03–0.5 |
| 16 | 0.015 | 0.06/0.25 | 0.015–0.06/0.015–0.25 | 0.03 | 0.25/0.25 | 0.03–0.25/0.03–0.25 |
| 17 | 0.015 | 0.5/1 | 0.015–0.5/0.015–1 | 0.015 | 0.25/0.5 | 0.015–0.25/0.015–0.5 |
| 18 | 0.015 | 0.06/1 | 0.015–0.06/0.015–1 | 0.03 | 0.25/0.5 | 0.03–0.25/0.03–0.5 |
| 19 | 0.015 | 0.5/0.5 | 0.015–0.5/0.015–0.5 | 0.03 | 0.5/1 | 0.03–0.5/0.03–1 |
| 20 | 0.015 | 0.5/0.5 | 0.015–0.5/0.015–0.5 | 0.03 | 1/1 | 0.03–1/0.03–1 |
| Overall (GM) | 0.015 | 0.25/0.73 | 0.025 | 0.55/1.16 | ||
| Range | 0.015 | 0.06–2/0.125–2 | 0.015–2/0.015–2 | 0.015–0.03 | 0.25–2/0.25–2 | 0.015–2/0.015–2 |
MYC, micafungin; AND, anidulafungin. Values for MPC and MSW are shown for 24 h/5 days of incubation for each isolate. Boldface characters (isolates codes 1 to 12) represent isolates that were able to grow on plates containing 1 mg/liter of echinocandin.
A total of 12 isolates (numbered 1 to 12; Table 1) grew on agar plates containing 1 mg/liter of echinocandin and yielded 32 colonies, 12 in the micafungin-containing plates and 20 in the anidulafungin-containing plates (Fig. 2). These colonies proved to be phenotypically resistant to both echinocandins according to the EUCAST procedure (subculture on an antifungal-free plate after 24 h of incubation) and despite the fact that only 21 carried FKS2 mutations, whereas the FKS1 sequence was wild type (11 in the micafungin-containing plates and 10 in the anidulafungin-containing plates) (Table 2 and Fig. 2). The mutations found were ΔF658 (n = 14), S663P (n = 4), W715L (n = 2), and E655A (n = 1) (Fig. 2). Significant differences were found in the echinocandin geometric mean MICs against the 11 FKS2 wild-type isolates and the 21 FKS2 mutant colonies (P = 0.02) (Table 2 and Fig. 2). However, the geometric mean MICs of fluconazole against both groups of isolates did not differ significantly (6.73 mg/liter versus 5.75 mg/liter). FKS2 mutations and phenotypic resistance were stable after 5 propagations on echinocandin-free agar plates. Genotyping showed no potential contamination of the isolates during the study (data not shown).
FIG 2.
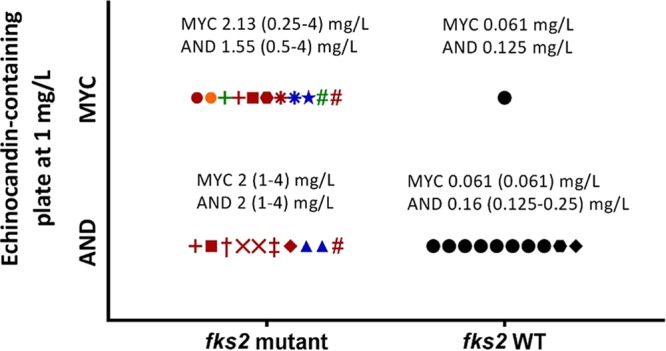
Echinocandin-resistant colonies growing on echinocandin-containing plates (at 1 mg/liter) grouped according to the presence or absence of FKS2 mutations. The range and geometric mean echinocandin MICs for each group are shown. Each symbol represents a colony; colonies with the same symbol shape come from the same isolate. The colors represent different FKS2 mutations (red, ΔF658; Blue, S663P; Green, W715L; orange, E655A). Symbols in black represent echinocandin-resistant but FKS2 wild-type colonies.
TABLE 2.
Study of virulence in the Galleria mellonella larvaea
| Isolate no. and echinocandin plate exposure | MIC (mg/liter) |
FKS2 HS1 sequence (no. of colonies) | Median survival (days) | |
|---|---|---|---|---|
| MYC | AND | |||
| 1 | ||||
| None | 0.015 | 0.03 | WT | 5 |
| MYC | 4 | 2 | ΔF658 (1) | 6 |
| 0.25 | 0.5 | E655A (1) | 5 | |
| 0.06 | 0.125 | WT (1) | 5.5 | |
| AND | 0.06 | 0.125 | WT (6) | 4.5 |
| 0.06 | 0.25 | WT (2) | 4 | |
| 2 | ||||
| None | 0.015 | 0.015 | WT | 5 |
| MYC | 0.5 | 0.5 | W715L (1) | 5 |
| 2 | 1 | ΔF658 (1) | 5 | |
| AND | 2 | 2 | ΔF658 (1) | 5.5 |
| 3 | ||||
| None | 0.015 | 0.015 | WT | 3 |
| MYC | 4 | 4 | ΔF658 (1) | 3 |
| AND | 2 | 4 | ΔF658 (1) | 3 |
| 4 | ||||
| None | 0.015 | 0.03 | WT | 3.5 |
| MYC | 4 | 2 | ΔF658 (1) | 3.5 |
| AND | 0.06 | 0.25 | WT (1) | 4.5 |
| 5 | ||||
| None | 0.015 | 0.03 | WT | 2 |
| AND | 1 | 1 | ΔF658 (1) | 2 |
| 6 | ||||
| None | 0.015 | 0.03 | WT | 3 |
| MYC | 4 | 2 | ΔF658 (1) | 5.5 |
| 4 | 2 | S663P (1) | 3.5 | |
| 7 | ||||
| None | 0.015 | 0.03 | WT | 2 |
| MYC | 2 | 1 | S663P (1) | 2 |
| 8 | ||||
| None | 0.015 | 0.03 | WT | 6 |
| AND | 4 | 2 | ΔF658 (1) | 6.5 |
| 2 | 2 | ΔF658 (1) | 6 | |
| 9 | ||||
| None | 0.015 | 0.03 | WT | 2.5 |
| AND | 1 | 2 | ΔF658 (1) | 2.5 |
| 10 | ||||
| None | 0.015 | 0.03 | WT | 4 |
| AND | 1 | 2 | ΔF658 (1) | 7 |
| 0.06 | 0.25 | WT (1) | 4 | |
| 11 | ||||
| None | 0.015 | 0.03 | WT | 3 |
| AND | 2 | 2 | S663P (1) | 3 |
| 4 | 2 | S663P (1) | 4 | |
| 12 | ||||
| None | 0.015 | 0.015 | WT | 3 |
| MYC | 4 | 2 | W715L (1) | 3 |
| 2 | 4 | ΔF658 (1) | 3.5 | |
| AND | 4 | 2 | ΔF658 (1) | 5 |
Results are larvae infected by 32 colonies (FKS2 mutant and FKS2 wild type) obtained from the 12 isolates streaked onto plates containing 1 mg/liter echinocandin.
The geometric mean of the mutation frequency and its range in the presence of micafungin (3.7 × 10−8 and 6.4 × 10−8 to 2.3 × 10−8) did not differ from those of anidulafungin (2.8 × 10−8 and 1.5 × 10−7 to 1.7 × 10−8) (P = 0.5). However, if only colonies with FKS2 mutations are selected, the geometric mean of the mutation frequency and its range were lower and significantly different (P = 0.02) in the presence of micafungin (3.4 × 10−8 and 5.3 × 10−8 to 2.2 × 10−8) than that in the presence of anidulafungin (2.2 × 10−8 and 3.9 × 10−8 to 1.7 × 10−8).
Genotyping analysis and MSH2 gene sequencing.
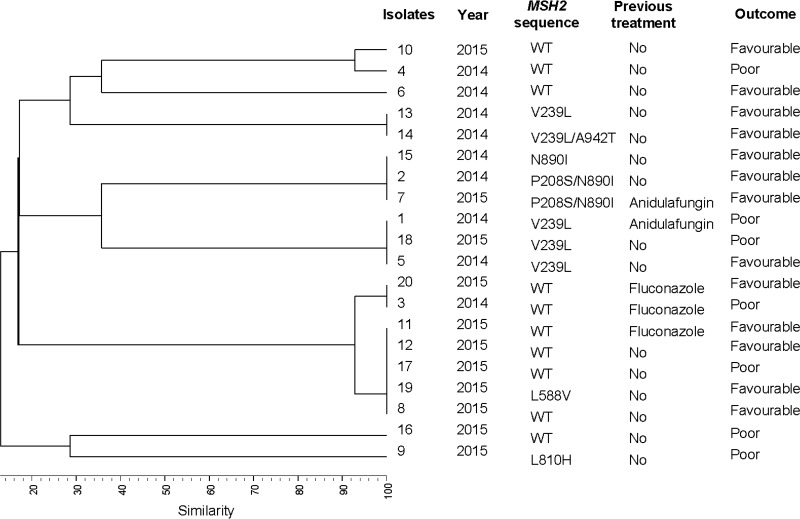
Genotyping of the 20 wild-type isolates revealed the presence of 10 genotypes, 5 of which were clusters involving 75% of isolates (Fig. 3). The MSH2 gene sequence of wild-type isolates revealed the presence of the following mutations in 50% of isolates (Fig. 3): V239L (n = 4), V239L/A942T (n = 1), P208S/N890I (n = 2), N890I (n = 1), L810H (n = 1), and L588V (n = 1). L810H and L588V have not been previously described. Most of the MSH2 mutations were found in isolates involved in clusters, and although the bulk of isolates from each cluster showed the same mutation, some isolates showed an MSH2 sequence that differed from the remaining isolates in the cluster. The presence of MSH2 mutations in wild-type isolates was not associated with the secondary acquisition of resistance to echinocandins, type of FKS2 mutation, patient outcome, or previous antifungal treatment.
FIG 3.
Genetic relationship between the 20 Candida glabrata isolates studied; the MSH2 sequence of each isolate, patient outcome (mortality), and the previous antifungal received are also shown.
Pathogenicity study of wild-type and resulting C. glabrata echinocandin-resistant isolates.
We did not find differences in the average growth/time to maximum rate between the wild-type isolates (4.76 × 10−6 s−1/8.36 × 104 s) and the FKS2 mutant colonies (4.69 × 10−6 s−1/1.16 × 105 s).
Wild-type isolates were classified as low (65%) or moderate (35%) biofilm formers. They exhibited high (80%) and moderate (20%) metabolic activity. The individual FKS2 mutant colonies were low (81%) or moderate (19%) biofilm formers and exhibited high (33.3%) and moderate (66.7%) metabolic activity. The differences did not reach statistical significance.
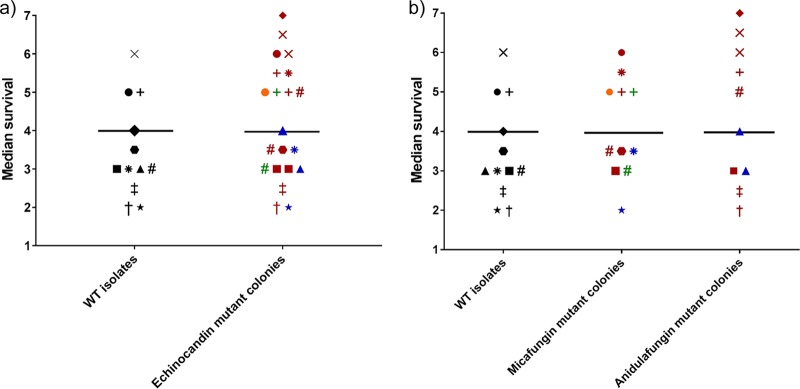
We found no differences in the median survival of larvae infected with FKS2 mutant colonies (4 days) and those infected by the wild-type isolates (4 days), regardless of the type of FKS2 mutation or the echinocandin-containing plate with the mutant (Table 2 and Fig. 4). All FKS2 mutant colonies in the same wild-type isolate showed similar median survival regardless of the type of FKS2 mutations found; however, we observed median differences in survival between isolates with the same FKS2 mutation.
FIG 4.
(a) Median survival of wild-type (WT) isolates and mutant colonies on plates containing 1 mg/liter echinocandin and the median survival of each group. (b) Median survival of WT isolates and mutant colonies on plates containing 1 mg/liter micafungin and anidulafungin and the median of each group. Phenotypic echinocandin-resistant isolates showing the WT FKS2 gene were excluded from the analysis. Each symbol represents a colony; colonies with the same symbol shape come from the same isolate.
DISCUSSION
Our study shows that C. glabrata isolates are able to develop secondary resistance to echinocandins when the drug concentration is below 2 mg/liter. The presence of mutations in the MSH2 gene did not seem to influence the selection of resistance mutations, and FKS2 mutations did not have a significant impact on the mortality of G. mellonella-infected larvae.
The rate of echinocandin resistance in C. glabrata has been increasing over the last few years, particularly in areas of northern Europe and North America (2, 3). However, the rate of resistance to echinocandins in C. glabrata in other regions, such as Spain, is much lower (1, 17). The reasons for these apparently heterogeneous findings are unclear, although they could be the result of variations in policies toward the use of echinocandins. It has been shown that long-term treatment with echinocandins is a risk factor for the presence of C. glabrata-resistant isolates (2, 18, 19).
Even though few patients are primarily infected by echinocandin-resistant isolates, promotion of in vitro resistance is relatively easy to achieve in C. glabrata (7, 8, 20–23). We recently showed that C. glabrata isolates could become echinocandin resistant in vitro after serial propagation onto plates containing low to increasing micafungin concentrations (7, 8). Consequently, the combination of a high inoculum and low concentrations of drug within the MSW are required to obtain resistant isolates. The present study was designed to complement our previous observations by assessing the mutation frequency, the MPC, and the MSW. Ideally, the concentration of an antimicrobial drug in the body should be above the MPC to rule out the selection of resistant mutants (i.e., FKS mutants), particularly at sites where C. glabrata cells are abundant. The parameters MPC and MSW were previously used for antibacterial drugs to optimize the response to antimicrobial treatment and to optimize prevention of emergence of resistant isolates (24–27). However, they have received little attention in the study of Candida species infection.
We found that concentrations of micafungin and anidulafungin of ≥2 mg/liter could prevent the emergence of C. glabrata-resistant mutants. Furthermore, concentrations between the MIC and 1 mg/liter may enable the emergence of these mutants. This observation is consistent with findings from our previous study, in which a concentration of 0.031 mg/liter of micafungin, which is within the MSW, was able to promote the presence of C. glabrata-resistant mutants (8). The echinocandin MIC endpoint is defined as a ≥50% reduction in fungal growth in the presence of the drug compared with the drug-free control well, meaning that a residual number of viable yeast cells may be present. However, it is not surprising that the EUCAST procedure could prove inefficient for selection of mutants, considering the inoculum suspension plated (1 × 105 CFU/ml) and the C. glabrata mutation frequency in the presence of micafungin or anidulafungin reported here (5 × 10−8 to 2 × 10−8 and 4 × 10−8 to 2 × 10−8, respectively). For that reason, and as reported for bacteria, we used very high inocula (109 CFU/ml) to ensure the presence of mutants in order to calculate the MPC and the MSW.
MPCs should be evaluated in line with drug pharmacokinetics and the suspected origin of the infection, because using high doses to overcome the MPC in tissues may induce drug-related toxicity. This limitation can be addressed by combining various drugs, as previously shown for antibacterials (27). However, considering the poor support that antifungal combination has received in recent guidelines (5, 6), this approach may be reserved for scenarios of high echinocandin resistance rates. Pharmacokinetic studies reported the maximum concentration of drug in serum (Cmax) of micafungin, anidulafungin, and caspofungin to be equal to 4.95 mg/liter, 3.5 mg/liter, and 7.64 mg/liter, respectively, after standard single doses (28, 29). However, in patients with peritonitis, the administration of a dose of micafungin yielded consistently low levels of the drug in the peritoneum (below 2 mg/liter), thus enabling the emergence of mutations in C. glabrata over time (13). These concentrations of echinocandins in the bloodstream clearly exceed the MPC and thus may be able to prevent the selection of resistance mutations because of the purportedly low number of circulating cells. However, echinocandins are partially excreted in feces, where the number of C. glabrata cells may be much higher, enabling spontaneously generated mutants to become selected (30). The presence of FKS mutants in C. glabrata isolates from sites with impaired penetration of candins, such as the skin or the peritoneum, has been reported (11–13).
According to EUCAST, C. glabrata isolates with micafungin or anidulafungin MICs above 0.031 mg/liter and 0.062 mg/liter, respectively, are resistant (31, 32). Of the 32 phenotypically resistant colonies found, 11 did not harbor FKS mutations. This phenomenon was more frequently observed in colonies grown on plates containing anidulafungin than on plates containing micafungin, and the MICs tended to be no more than 1-fold or 2-fold concentrations above the breakpoint. To date, the FKS mutation is the only reported mechanism of resistance, and FKS wild-type isolates with slightly high echinocandin MICs might not be considered truly resistant. Other authors have reported the same observations (particularly on plates containing anidulafungin), even in isolates with higher echinocandin MICs, thus warranting future research into this issue (21, 33). In order to calculate the mutation frequency, we chose 1 mg/liter of echinocandins as our minimum, as colony counting was not possible at lower concentrations. We did not find significant differences between micafungin and anidulafungin if the mutation frequency was calculated based on resistant colonies. However, taking into account only the mutants, significant differences were observed, and the mutation frequency reported here is similar to that of other studies (21, 23).
The apparent ability of C. glabrata isolates to acquire resistance to echinocandins in vitro is not consistent with the low number of resistant isolates from clinical invasive samples. A potential explanation is that the fitness of mutant cells is inferior to that of the wild type. We recently reported a positive correlation in mortality between the patients and the G. mellonella model, showing that this model can be suitable for the study of virulence in Candida (34). Overall, we did not find significant differences in median survival between wild-type isolates and their FKS2 mutant colonies. However, the wild-type isolates were more virulent than the resulting FKS2 mutant colonies in 5 of the 12 isolates, whereas median survival was similar to that of the wild-type isolates in the remaining mutant colonies. These observations, which are not consistent with our recently reported results, may be explained in part by the differences between the mean mortality of larvae infected by wild-type isolates versus those infected by FKS mutant isolates reported previously (3 versus 5 days) (8) or in the present study (4 versus 4 days). Similarly, we found no differences in biofilm formation between mutant and wild-type isolates. C. glabrata frequently forms biofilms featuring low biomass and high metabolic activity (35). Therefore, the lack of differences in biofilm formation between mutant and wild-type isolates is not surprising. Biofilm formation is lower in C. glabrata than in C. albicans, and the patients infected by the latter have higher mortality (36). The low early mortality (within 30 days of diagnosis) of our patients also may be associated with the low biofilm formation.
Although some authors showed that the MSH2 mutator phenotype or specific genotypes may contribute to the development of resistance to echinocandins (14, 15), we were unable to confirm these findings, as the modifications in the MSH2 gene were not related to more marked acquisition of echinocandin resistance in vitro.
Alternative mechanisms of echinocandin resistance, such as compensatory mutations or genomic rearrangement, have been reported in C. glabrata (15, 37). However, we cannot rule out the presence of the heteroresistant subpopulations rather than genetic drift in this clonal species (38).
We conclude that C. glabrata isolates are able to develop secondary resistance to echinocandins when the concentrations of the drugs are below 2 mg/liter. The presence of mutations in the MSH2 gene did not seem to influence the promotion of resistance, and FKS2 mutations did not have a significant impact on the mortality of G. mellonella-infected larva.
MATERIALS AND METHODS
Yeast isolates and patients.
We studied 20 Candida glabrata isolates recovered from blood cultures of patients with candidemia (1 isolate per patient) admitted to Gregorio Marañón Hospital (Madrid, Spain) between 2014 and 2015. The isolates were identified using chromogenic agar plates and confirmed by amplification and sequencing of the ITS1-5.8S-ITS2 regions (39). A total of 25% of the patients studied had received fluconazole or anidulafungin during the month before the diagnosis of candidemia. Early mortality (within 30 days of diagnosis) and late mortality (30 days after diagnosis) were 25% and 35%, respectively.
Antifungal susceptibility testing.
All isolates were tested for susceptibility to micafungin (Astellas Pharma, Inc., Tokyo, Japan), anidulafungin, and fluconazole (Pfizer Pharmaceutical Group, New York, NY, USA) according to the EUCAST EDef 7.2 microdilution procedure (31, 32, 40, 41). The echinocandins and fluconazole were tested at concentrations ranging from 0.015 to 8 mg/liter and 0.25 to 128 mg/liter, respectively. Inoculated plates were incubated for 24 h at 35°C. Strains were classified according to the resistance breakpoints proposed by EUCAST: micafungin, MIC of >0.031 mg/liter; anidulafungin, MIC of >0.062 mg/liter; and fluconazole, MIC of >32 mg/liter (31, 32, 40).
Echinocandin mutant prevention concentration and mutant selection window.
Exposure to echinocandins on agar plates was as previously described, with some modifications (7). Briefly, isolate suspensions were adjusted to 3 × 109 to 7 × 109 CFU/ml (mean of 4.8 × 109 ± 0.96 × 109) using a Neubauer chamber and stroked directly (100 μl) onto Sabouraud agar plates containing 8 concentrations (0.015 mg/liter to 2 mg/liter; 2-fold concentrations) of micafungin and anidulafungin. Plates were incubated at 35°C and visually inspected every 24 h for up to 5 days of incubation. MPCs were defined as the lowest echinocandin concentration leading to complete inhibition of fungal growth on echinocandin-containing agar plates at 2 time points (24 h and 5 days) (16).
The MPCs obtained at the 2 incubation time points were compared using the Wilcoxon signed-rank test (IBM SPSS Statistics for Windows, version 21.0; Armonk, NY, USA). A P value of <0.05 was considered statistically significant.
The MSW for each isolate was defined as the range of concentrations between the MIC, obtained by microdilution, and the MPC, considering the 2 incubation time points (24 h and 5 days) (42).
Mutation frequency.
The calculation of mutation frequency was based on an echinocandin concentration of 1 mg/liter; lower concentrations may not have enabled individualization of colonies owing to heavy growth. The individual colonies growing on the plates containing echinocandins at concentrations equal to 1 mg/liter after 5 days of incubation were studied for susceptibility to micafungin, anidulafungin, and fluconazole according to the EUCAST procedure, and the hot spots of FKS1 and FKS2 genes were sequenced (43, 44). Mutation frequency was defined as the ratio between the number of phenotypically resistant colonies on micafungin-containing or anidulafungin-containing plates and the number of cells stroked. The mutation frequencies obtained for both echinocandins were compared (Wilcoxon signed-rank test; IBM SPSS Statistics for Windows, version 21.0).
MSH2 sequencing.
The MSH2 gene of the wild-type and resulting C. glabrata echinocandin-resistant isolates was amplified and sequenced according to Delliere et al. (14), with the following modifications: 1.25 U of AmpliTaq gold (Applied Biosystems), 0.2 mM deoxynucleoside triphosphates, 2 mM MgCl2, and 100 ng of extracted DNA.
Microsatellite typing.
The wild-type and the resulting C. glabrata echinocandin-resistant isolates were genotyped using a panel of 14 microsatellite markers (45–48). Singleton genotypes were defined as those found only once, whereas a cluster was defined as the presence of ≥2 isolates with the same genotype.
Pathogenicity study of wild-type and resulting C. glabrata echinocandin-resistant isolates.
The in vitro growth kinetics, biofilm quantification, and virulence on final-instar larvae of Galleria mellonella of the wild-type isolates and the resulting individual FKS mutant colonies were studied as previously described (8).
The average growth rate and time to maximum rate were calculated (49). Briefly, 100 μl of adjusted inocula (3 × 105 CFU/ml) of each isolate was added to 100 μl of double-concentrated RPMI 1640 medium supplemented with 2% glucose (Merck KGaA, Darmstadt, Germany) and morpholinepropanesulfonic acid (MOPS; Sigma-Aldrich, Co., St. Louis, MO, USA) in flat-bottomed 96-well microdilution trays. Each study was performed in triplicate. Trays were incubated at 35°C with moderate shaking in a spectrophotometer (Thermo Fisher Scientific, Madrid, Spain) for 35 h. The optical density in each well was measured every 15 min at 490 nm to calculate the kinetic parameters (average growth rate and time to maximum rate). Differences between kinetic parameters were studied using the Kruskal-Wallis test.
The biofilm was formed according to the method proposed by Marcos-Zambrano and colleagues (35). Briefly, the cells were grown at 30°C with shaking overnight on 10 ml of yeast-peptone-dextrose broth (Difco, Becton Dickinson, Madrid, Spain) before being washed and resuspended in 5 ml of RPMI 1640 broth medium adjusted to approximately 1 × 106 cells/ml. A total of 100 μl of the suspension was inoculated in 96-well trays and incubated for 24 h at 37°C. Each strain was tested in triplicate. Biofilm production was quantified using crystal violet staining; the metabolic activity of biofilm was measured using the XTT [2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide salt] reduction assay. We followed the cutoff points proposed in a previous paper to classify the isolates as low, moderate, and high biofilm forming and as having low, moderate, and high metabolic activity (35).
We compared the mortality caused by wild-type isolates and the resulting individual FKS2 mutant colonies in final-instar larvae of G. mellonella (Bichosa, Salceda de Caselas, Spain). Briefly, infecting inocula were prepared using a Neubauer chamber and checked in Sabouraud dextrose agar. Inocula ranging from 3 × 106 to 7 × 106 CFU per larva were accepted because the inoculum ranges did not affect the mortality of the larvae. Ten G. mellonella larvae per isolate were infected with 10 μl of a suspension of C. glabrata, and 2 control groups were established, one comprising 10 larvae inoculated with 10 μl of phosphate-buffered saline to monitor trauma and another comprising 10 noninjected larvae. Larvae were incubated at 37°C for up to 7 days after infection, and the number of dead larvae was recorded daily (50). Survival curves were obtained for wild-type isolates and the resulting individual echinocandin-resistant colonies using the Kaplan-Meier method (Graph Pad Prism 5.02 statistical software; GraphPad, La Jolla, CA, USA), and the differences were evaluated (log-rank test).
This study was approved by the Ethics Committee of Hospital Gregorio Marañón (CEIC-A1; study no. 208/16).
ACKNOWLEDGMENTS
We thank Thomas O'Boyle for editing the article.
The study was supported by grants PI14/00740 and MSI15/00115 from the Fondo de Investigación Sanitaria (FIS; Instituto de Salud Carlos III; Plan Nacional de I+D+I 2013-2016). It was also supported by grant CM-SANTANDER (GR3/2014; group 920200) and a grant from the Spanish Network for Research in Infectious Diseases (REIPI RD12/0015). The study was cofunded by the European Regional Development Fund (FEDER), “A way of making Europe.” The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
P.E. (CPI15/00115) and J.G. (CPII15/00006) are recipients of a Miguel Servet contract supported by the FIS; L.J.M.-Z. (PI14/00740) is supported by FIS; M.A.B.-C. received a predoctoral grant from the Instituto de Investigación Sanitaria Gregorio Marañón (II-Predoc-2016-IISGM).
J.G. has received funds for speaking at symposia organized on behalf of Astellas, Gilead, MSD, Scynexis, and United Medical; he has also received funds for research from Fondo de Investigación Sanitaria, Gilead, Scynexis, and Cidara. R.C. has received funds for speaking at symposia organized on behalf of Gilead and MSD. All other authors have no conflicts to declare.
REFERENCES
- 1.Guinea J, Zaragoza O, Escribano P, Martin-Mazuelos E, Peman J, Sanchez-Reus F, Cuenca-Estrella M. 2014. Molecular identification and antifungal susceptibility of yeast isolates causing fungemia collected in a population-based study in Spain in 2010 and 2011. Antimicrob Agents Chemother 58:1529–1537. doi: 10.1128/AAC.02155-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander BD, Johnson MD, Pfeiffer CD, Jimenez-Ortigosa C, Catania J, Booker R, Castanheira M, Messer SA, Perlin DS, Pfaller MA. 2013. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin Infect Dis 56:1724–1732. doi: 10.1093/cid/cit136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arendrup MC, Perlin DS. 2014. Echinocandin resistance: an emerging clinical problem? Curr Opin Infect Dis 27:484–492. doi: 10.1097/QCO.0000000000000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pappas PG, Kauffman CA, Andes D, Benjamin DK Jr, Calandra TF, Edwards JE Jr, Filler SG, Fisher JF, Kullberg BJ, Ostrosky-Zeichner L, Reboli AC, Rex JH, Walsh TJ, Sobel JD. 2009. Clinical practice guidelines for the management of candidiasis. Clin Infect Dis 48:503–535. doi: 10.1086/596757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, Zaoutis TE, Sobel JD. 2016. Clinical practice guideline for the management of candidiasis: 2016 update. Clin Infect Dis 62:e1–e50. doi: 10.1093/cid/civ1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornely OA, Bassetti M, Calandra T, Garbino J, Kullberg BJ, Lortholary O, Meersseman W, Akova M, Arendrup MC, Arikan-Akdagli S, Bille J, Castagnola E, Cuenca-Estrella M, Donnelly JP, Groll AH, Herbrecht R, Hope WW, Jensen HE, Lass-Florl C, Petrikkos G, Richardson MD, Roilides E, Verweij PE, Viscoli C, Ullmann AJ. 2012. ESCMID* guideline for the diagnosis and management of Candida diseases 2012: non-neutropenic adult patients. Clin Microbiol Infect 18(Suppl 7):S19–S37. doi: 10.1111/1469-0691.12039. [DOI] [PubMed] [Google Scholar]
- 7.Bordallo-Cardona MA, Escribano P, de la Pedrosa EG, Marcos-Zambrano LJ, Canton R, Bouza E, Guinea J. 2017. In vitro exposure to increasing micafungin concentrations easily promotes echinocandin resistance in Candida glabrata isolates. Antimicrob Agents Chemother 61:e01542–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bordallo-Cardona MÁ, Escribano P, Marcos-Zambrano LJ, Díaz-García J, de la Pedrosa EG, Cantón R, Bouza E, Guinea J. 8 December 2017. Low and constant micafungin concentrations may be sufficient to lead to resistance mutations in FKS2 gene of Candida glabrata. Med Mycol doi: 10.1093/mmy/myx124. [DOI] [PubMed] [Google Scholar]
- 9.Perlin DS. 2007. Resistance to echinocandin-class antifungal drugs. Drug Resistance Updates 10:121–130. doi: 10.1016/j.drup.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perlin DS. 2015. Mechanisms of echinocandin antifungal drug resistance. Ann N Y Acad Sci 1354:1–11. doi: 10.1111/nyas.12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shields RK, Nguyen MH, Press EG, Clancy CJ. 2014. Abdominal candidiasis is a hidden reservoir of echinocandin resistance. Antimicrob Agents Chemother 58:7601–7605. doi: 10.1128/AAC.04134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen RH, Johansen HK, Soes LM, Lemming LE, Rosenvinge FS, Nielsen L, Olesen B, Kristensen L, Dzajic E, Astvad KM, Arendrup MC. 2015. Posttreatment antifungal resistance among colonizing Candida isolates in candidemia patients: results from a systematic multicenter study. Antimicrob Agents Chemother 60:1500–1508. doi: 10.1128/AAC.01763-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grau S, Luque S, Campillo N, Samso E, Rodriguez U, Garcia-Bernedo CA, Salas E, Sharma R, Hope WW, Roberts JA. 2015. Plasma and peritoneal fluid population pharmacokinetics of micafungin in post-surgical patients with severe peritonitis. J Antimicrob Chemother 70:2854–2861. doi: 10.1093/jac/dkv173. [DOI] [PubMed] [Google Scholar]
- 14.Delliere S, Healey K, Gits-Muselli M, Carrara B, Barbaro A, Guigue N, Lecefel C, Touratier S, Desnos-Ollivier M, Perlin DS, Bretagne S, Alanio A. 2016. Fluconazole and echinocandin resistance of Candida glabrata correlates better with antifungal drug exposure rather than with MSH2 mutator genotype in a french cohort of patients harboring low rates of resistance. Front Microbiol 7:2038. doi: 10.3389/fmicb.2016.02038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Healey KR, Zhao Y, Perez WB, Lockhart SR, Sobel JD, Farmakiotis D, Kontoyiannis DP, Sanglard D, Taj-Aldeen SJ, Alexander BD, Jimenez-Ortigosa C, Shor E, Perlin DS. 2016. Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat Commun 7:11128. doi: 10.1038/ncomms11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drlica K. 2003. The mutant selection window and antimicrobial resistance. J Antimicrob Chemother 52:11–17. doi: 10.1093/jac/dkg269. [DOI] [PubMed] [Google Scholar]
- 17.Marcos-Zambrano LJ, Escribano P, Sanchez C, Muñoz P, Bouza E, Guinea J. 2014. Antifungal resistance to fluconazole and echinocandins is not emerging in yeast isolates causing fungemia in a Spanish tertiary care center. Antimicrob Agents Chemother 58:4565–4572. doi: 10.1128/AAC.02670-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cleary JD, Garcia-Effron G, Chapman SW, Perlin DS. 2008. Reduced Candida glabrata susceptibility secondary to an FKS1 mutation developed during candidemia treatment. Antimicrob Agents Chemother 52:2263–2265. doi: 10.1128/AAC.01568-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lortholary O, Desnos-Ollivier M, Sitbon K, Fontanet A, Bretagne S, Dromer F. 2011. Recent exposure to caspofungin or fluconazole influences the epidemiology of candidemia: a prospective multicenter study involving 2,441 patients. Antimicrob Agents Chemother 55:532–538. doi: 10.1128/AAC.01128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartizal K, Gill CJ, Abruzzo GK, Flattery AM, Kong L, Scott PM, Smith JG, Leighton CE, Bouffard A, Dropinski JF, Balkovec J. 1997. In vitro preclinical evaluation studies with the echinocandin antifungal MK-0991 (L-743,872). Antimicrob Agents Chemother 41:2326–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Locke JB, Almaguer AL, Zuill DE, Bartizal K. 2016. Characterization of in vitro resistance development to the novel echinocandin CD101 in Candida species. Antimicrob Agents Chemother 60:6100–6107. doi: 10.1128/AAC.00620-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balashov SV, Park S, Perlin DS. 2006. Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob Agents Chemother 50:2058–2063. doi: 10.1128/AAC.01653-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan KSR, Ellen Press G, Hong Nguyen M, Clancy CJ. 2015. Mutational frequency and FKS mutations rates of Candida glabrata vary by echinocandin agent. Abstr 55th Intersci Conf Antimicrob Agents Chemother. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong Y, Zhao X, Domagala J, Drlica K. 1999. Effect of fluoroquinolone concentration on selection of resistant mutants of Mycobacterium bovis BCG and Staphylococcus aureus. Antimicrob Agents Chemother 43:1756–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong Y, Zhao X, Kreiswirth BN, Drlica K. 2000. Mutant prevention concentration as a measure of antibiotic potency: studies with clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother 44:2581–2584. doi: 10.1128/AAC.44.9.2581-2584.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blondeau JM, Zhao X, Hansen G, Drlica K. 2001. Mutant prevention concentrations of fluoroquinolones for clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother 45:433–438. doi: 10.1128/AAC.45.2.433-438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canton R, Morosini MI. 2011. Emergence and spread of antibiotic resistance following exposure to antibiotics. FEMS Microbiol Rev 35:977–991. doi: 10.1111/j.1574-6976.2011.00295.x. [DOI] [PubMed] [Google Scholar]
- 28.Nicasio AM, Tessier PR, Nicolau DP, Knauft RF, Russomanno J, Shore E, Kuti JL. 2009. Bronchopulmonary disposition of micafungin in healthy adult volunteers. Antimicrob Agents Chemother 53:1218–1220. doi: 10.1128/AAC.01386-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen SC, Slavin MA, Sorrell TC. 2011. Echinocandin antifungal drugs in fungal infections: a comparison. Drugs 71:11–41. doi: 10.2165/11585270-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 30.Maraki S, Hamilos G, Dimopoulou D, Andrianaki AM, Karageorgiadis AS, Kyvernitakis A, Lionakis S, Kofteridis DP, Samonis G. 2015. Study on the comparative activity of echinocandins on murine gut colonization by Candida albicans. Med Mycol 53:597–602. doi: 10.1093/mmy/myv028. [DOI] [PubMed] [Google Scholar]
- 31.European Committee on Antimicrobial Susceptibility Testing. 2013. Anidulafungin: rationale for the clinical breakpoints, version 2.0. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Rationale_documents/Anidulafungin_rationale_2_0_2013.pdf.
- 32.European Committee on Antimicrobial Susceptibility Testing. 2013. Micafungin and Candida spp.: rationale for the clinical breakpoints, version 1.0. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Rationale_documents/Micafungin_rationale_document_1_0_final.pdf.
- 33.Pham CD, Iqbal N, Bolden CB, Kuykendall RJ, Harrison LH, Farley MM, Schaffner W, Beldavs ZG, Chiller TM, Park BJ, Cleveland AA, Lockhart SR. 2014. Role of FKS Mutations in Candida glabrata: MIC values, echinocandin resistance, and multidrug resistance. Antimicrob Agents Chemother 58:4690–4696. doi: 10.1128/AAC.03255-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcos-Zambrano LJ, Puig-Asensio M, Perez-Garcia F, Escribano P, Sanchez-Carrillo C, Zaragoza O, Padilla B, Cuenca-Estrella M, Almirante B, Martin-Gomez MT, Munoz P, Bouza E, Guinea J. 2017. Candida guilliermondii complex is characterized by high antifungal resistance but low mortality in 22 cases of candidemia. Antimicrob Agents Chemother 61:e00099–17. doi: 10.1128/AAC.00099-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marcos-Zambrano LJ, Escribano P, Bouza E, Guinea J. 2014. Production of biofilm by Candida and non-Candida spp. isolates causing fungemia: comparison of biomass production and metabolic activity and development of cut-off points. Int J Med Microbiol 304:1192–1198. doi: 10.1016/j.ijmm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 36.Rajendran R, Sherry L, Nile CJ, Sherriff A, Johnson EM, Hanson MF, Williams C, Munro CA, Jones BJ, Ramage G. 2016. Biofilm formation is a risk factor for mortality in patients with Candida albicans bloodstream infection-Scotland, 2012-2013. Clin Microbiol Infect 22:87–93. doi: 10.1016/j.cmi.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh-Babak SD, Babak T, Diezmann S, Hill JA, Xie JL, Chen YL, Poutanen SM, Rennie RP, Heitman J, Cowen LE. 2012. Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata. PLoS Pathog 8:e1002718. doi: 10.1371/journal.ppat.1002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hiemenz J, Cagnoni P, Simpson D, Devine S, Chao N, Keirns J, Lau W, Facklam D, Buell D. 2005. Pharmacokinetic and maximum tolerated dose study of micafungin in combination with fluconazole versus fluconazole alone for prophylaxis of fungal infections in adult patients undergoing a bone marrow or peripheral stem cell transplant. Antimicrob Agents Chemother 49:1331–1336. doi: 10.1128/AAC.49.4.1331-1336.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White T, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 322 In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA. [Google Scholar]
- 40.European Committee on Antimicrobial Susceptibility Testing. 2013. Fluconazole: rationale for the clinical breakpoints, version 2.0. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Rationale_documents/Fluconazole_rationale_2_0_20130223.pdf.
- 41.Arendrup MC, Cuenca-Estrella M, Lass-Florl C, Hope W. 2012. EUCAST technical note on the EUCAST definitive document EDef 7.2: method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts EDef 7.2 (EUCAST-AFST). Clin Microbiol Infect 18:E246-7. doi: 10.1111/j.1469-0691.2012.03880.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhao X, Drlica K. 2001. Restricting the selection of antibiotic-resistant mutants: a general strategy derived from fluoroquinolone studies. Clin Infect Dis 33(Suppl 3):S147–S156. doi: 10.1086/321841. [DOI] [PubMed] [Google Scholar]
- 43.Thompson GR III, Wiederhold NP, Vallor AC, Villareal NC, Lewis JS II, Patterson TF. 2008. Development of caspofungin resistance following prolonged therapy for invasive candidiasis secondary to Candida glabrata infection. Antimicrob Agents Chemother 52:3783–3785. doi: 10.1128/AAC.00473-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zimbeck AJ, Iqbal N, Ahlquist AM, Farley MM, Harrison LH, Chiller T, Lockhart SR. 2010. FKS mutations and elevated echinocandin MIC values among Candida glabrata isolates from U.S. population-based surveillance. Antimicrob Agents Chemother 54:5042–5047. doi: 10.1128/AAC.00836-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foulet F, Nicolas N, Eloy O, Botterel F, Gantier JC, Costa JM, Bretagne S. 2005. Microsatellite marker analysis as a typing system for Candida glabrata. J Clin Microbiol 43:4574–4579. doi: 10.1128/JCM.43.9.4574-4579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grenouillet F, Millon L, Bart JM, Roussel S, Biot I, Didier E, Ong AS, Piarroux R. 2007. Multiple-locus variable-number tandem-repeat analysis for rapid typing of Candida glabrata. J Clin Microbiol 45:3781–3784. doi: 10.1128/JCM.01603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abbes S, Sellami H, Sellami A, Hadrich I, Amouri I, Mahfoudh N, Neji S, Makni F, Makni H, Ayadi A. 2012. Candida glabrata strain relatedness by new microsatellite markers. Eur J Clin Microbiol Infect Dis 31:83–91. doi: 10.1007/s10096-011-1280-4. [DOI] [PubMed] [Google Scholar]
- 48.Brisse S, Pannier C, Angoulvant A, de Meeus T, Diancourt L, Faure O, Muller H, Peman J, Viviani MA, Grillot R, Dujon B, Fairhead C, Hennequin C. 2009. Uneven distribution of mating types among genotypes of Candida glabrata isolates from clinical samples. Eukaryot Cell 8:287–295. doi: 10.1128/EC.00215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arendrup MC, Perlin DS, Jensen RH, Howard SJ, Goodwin J, Hope W. 2012. Differential in vivo activities of anidulafungin, caspofungin, and micafungin against Candida glabrata isolates with and without FKS resistance mutations. Antimicrob Agents Chemother 56:2435–2442. doi: 10.1128/AAC.06369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borghi E, Andreoni S, Cirasola D, Ricucci V, Sciota R, Morace G. 2014. Antifungal resistance does not necessarily affect Candida glabrata fitness. J Chemother 26:32–36. doi: 10.1179/1973947813Y.0000000100. [DOI] [PubMed] [Google Scholar]