ABSTRACT
Genomic comparison of the first six Dutch vanD-type vancomycin-resistant Enterococcus faecium (VRE) isolates with four vanD gene clusters from other enterococcal species and anaerobic gut commensals revealed that the vanD gene cluster was located on a genomic island of variable size. Phylogenetic inferences revealed that the Dutch VRE isolates were genetically not closely related and that genetic variation of the vanD-containing genomic island was not species specific, suggesting that this island is transferred horizontally between enterococci and anaerobic gut commensals.
KEYWORDS: Enterococcus faecium, genomic island, vanD type, vancomycin resistance
TEXT
The vanD-type gene cluster encoding vancomycin resistance has been identified among different bacterial species, including enterococci and anaerobic gut commensals (1, 2). Analysis of vanD vancomycin-resistant enterococci revealed some general characteristics. In all of these strains, (i) the d-Ala-d-Ala ligase housekeeping enzyme is not active due to mutations, insertions, and/or deletions in the ddl gene; (ii) vancomycin resistance is constitutively expressed due to mutations, insertions, and/or deletions of the vancomycin sensor and regulator genes vanS and vanR; (iii) the vanD gene cluster is chromosomally located and not shown to be transferable (1, 3, 4).
In the Netherlands, so far, only vanA and vanB types of resistance have been found in Enterococcus faecium isolates, but over the last 4 years, a total of six vanD-positive vancomycin-resistant E. faecium (VRE) isolates were identified in five different patients from four different hospitals in the Netherlands (Table 1). All patients received antibiotics before detection of the vanD-positive VRE isolate, including vancomycin in four of the five patients (Table 1). For patient D, two ampicillin-resistant E. faecium (ARE) (E9352 and E9353) were isolated from blood cultures before the isolation of two vanD-positive VRE isolates (E8429 and E9354). Whole-genome sequencing (WGS) (Illumina NextSeq and, for 3 strains, MinION nanopore [see Table S1 in the supplemental material for assembly statistics]) was performed to determine the genetic relatedness of the six vanD-positive VRE isolates and two ARE strains and the genomic organization of the vanD gene clusters. Based on allelic variation in the 1,423 core genome (cg) MLST loci using Ridom SeqSphere + v3.5.0 (5), a phylogenetic neighbor-joining (NJ) tree was generated that revealed that the vanD-positive VRE isolates from the four different hospitals had different cgMLST profiles, indicating that they were not clonally related (see Fig. S1 in the supplemental material) and belonged to the phylogenetic group clade A1 (data not shown; 6–8). In contrast, the four strains from patient D (hospital 3), two ARE and two VRE, appeared very similar, e.g., the two vanD-positive VRE strains differed in no more than 3 or 4 of 1,423 loci from ARE E9352, indicating that they are genetically very closely related. All six vanD-positive VRE isolates contained a mutated or truncated ddl gene, presumably resulting in a nonfunctional housekeeping ligase (Table 1). Of note, the two VRE isolates from patient D appeared to have two different mutations in ddl. The vanSD vancomycin sensor gene of the six vanD-positive VRE isolates contained several of the previously described nonsynonymous mutations (Table 1). It has been shown that these amino acid substitutions in VanSD may lead to constitutive expression of the vanD gene clusters, thereby rescuing vanD-positive VRE with a nonfunctional housekeeping ddl, which would have otherwise been lethal (1, 9, 10).
TABLE 1.
Strain characteristics and WGS sequence statistics
| Strain | Resistance classificationa | Hospital | Patient | Age (yrs) | Underlying disease | Isolation date (mo-day-yr) | Isolation site | Antibiotic use | Interval vancomycin treatment and VanDb | MICs (mg/liter) |
Variation in Ddlc | Variation in VanSc | Size of genomic island (bp) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vancomycin | Teicoplanin | Ampicillin | |||||||||||||
| E7962 | VRE | 1 | A | 73 | Acute myeloid leukemia | 11-25-2013 | Rectum | Vancomycin cotrimoxazole, ciprofloxacin | Unknown | 16 | <1 | >256 | S185F | S57R, K169E, T170P, V280L | 185,412 |
| E8043 | VRE | 2 | B | 57 | Acute lymphoid leukemia | 3-24-2014 | Rectum | vancomycin | Unknown | 16 | <1 | >256 | Insertion 2 aa at pos. 280 | S57R, A165V, V280L | 168,512 |
| E9242 | VRE | 2 | C | 18 | Acute lymphoid leukemia | 3-31-2016 | Rectum | Cotrimoxazole, ciprofloxacin | Unknown | 256 | 4 | >256 | Δ2 bp at pos. 319 | K308Q | 144,229 |
| E9352d | ARE | 12-9-2012 | Blood | Amoxicillin-clavulanic acid, cefotaxime, ciprofloxacin, metronidazole, vancomycin | NA | <1 | <1 | >256 | Normal | NA | |||||
| E9353d | ARE | 3 | D | 65 | Cardiovascular disease, cholangitis | 12-27-2012 | Blood | NA | <1 | <1 | >256 | Normal | NA | ||
| E8429d | VRE | 1-22-2013 | Rectum | 43 days | 256 | 2 | >256 | Δ1 bp at pos. 165 | S57R, A68V, K308Q, V280L | 120,918 | |||||
| E9354d | VRE | 1-27-2013 | Rectum | 48 days | 256 | 2 | >256 | Δ4 aa at pos. 280 | S57R, A68V, K308Q, V280L | 120,844 | |||||
| E9641 | VRE | 4 | E | 63 | Non-Hodgkin lymphoma | 2-27-2017 | Rectum | Ciprofloxacin, ceftazidime, piperacillin-tazobactam, vancomycin | 4 mo | 8 | <0.5 | >256 | S185F | S57R, V280L | 168,541 |
VRE, vancomycin-resistant E. faecium; ARE, ampicillin-resistant E. faecium.
NA, not available.
Indicated positions are based on amino acid (aa) sequence for Ddl (d-Ala-d-Ala ligase) and VanS (vancomycin sensor protein).
These four strains were isolated from the same patient at different time points.
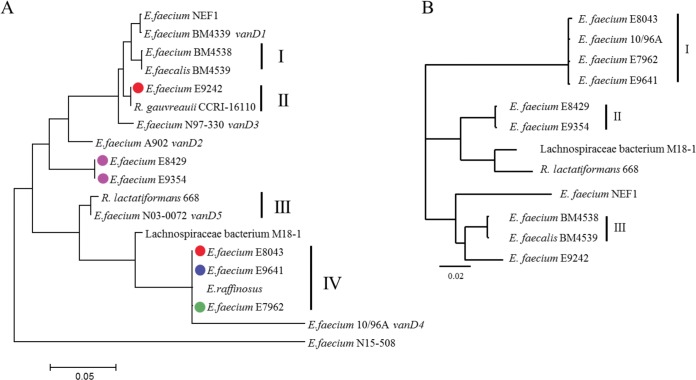
Phylogenetic analysis of the complete vanD gene clusters from the 6 Dutch isolates and 13 vanD gene clusters retrieved from GenBank, including representative sequences of the 5 previously described different types of vanD gene clusters (vanD1 to vanD5) (9, 11–14); 3 vanD gene clusters from other E. faecium strains (1, 15, 16); vanD gene clusters from Enterococcus faecalis (16), Enterococcus raffinosus (17), anaerobic gut commensals Ruminococcus gauvreauii (2), Ruthenibacterium lactatiformans 668 (18), and Lachnospiraceae bacterium M18-1 (see Table S2 in the supplemental material), revealed that the vanD gene clusters of the Dutch VRE isolates did not cluster together in a single branch and that vanD gene clusters did not cluster according to the species in which they were contained (Fig. 1A). In contrast, highly similar clusters, including multiple species, were observed. This may suggest genetic exchange of the vanD gene cluster between species and genera. Recently, Boyd et al. (15) described a vanD gene cluster-containing element, designated IMEEfm15508 (integrative mobilizable element in E. faecium strain N15-508). Based on our analysis, this vanD gene cluster clustered completely separately from all other vanD gene clusters. A phylogenetic NJ tree based on aligned protein sequences of all ligases conferring vancomycin resistance indicated that the E. faecium N15-508 VanD belonged to a separate lineage between the VanB and VanD ligases (see Fig. S2 in the supplemental material).
FIG 1.
(A) Phylogenetic neighbor-joining (NJ) tree of the vanD gene cluster. The phylogenetic NJ tree was generated from a ClustalW multiple alignment of DNA sequences using MegAlign, DNAStar Lasergene v.14 software. The six Dutch isolates are indicated with green, red, purple, and blue dots (hospitals 1 through 4, respectively). Vertical lines and Roman numerals indicate clusters of highly related (>90% identity) vanD gene clusters, including multiple species. (B) Phylogeny of the 120- 190-kb genomic island. The phylogenetic NJ tree was generated from a ClustalW multiple alignment of the vanD-containing genomic island using Geneious 8.1.2 software. Vertical lines and Roman numerals indicate clusters of highly related genomic islands (>90% identity).
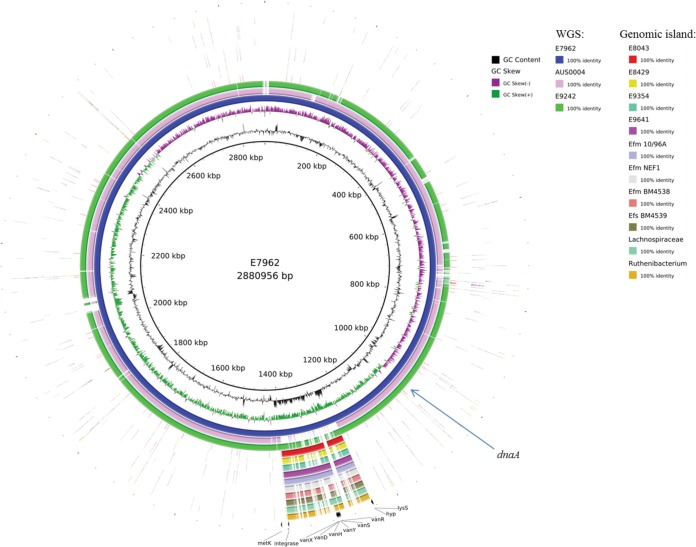
To investigate whether the vanD gene cluster in the Dutch VRE isolates was located on a plasmid or other mobile genetic element, we performed long-read nanopore sequencing for strains E7962, E8429, and E9242 and, in combination with short-read Illumina reads, generated a hybrid assembly using SPAdes 3.8.0. This resulted in completely assembled chromosomes for strains E7962 and E9242 of 2,880,956 and 2,831,933 nucleotides, respectively. A comparative circular alignment, using E7962 as reference and including the vanD-negative complete genome sequence of E. faecium AUS0004 (accession no. NC_017022) revealed that the vanD gene cluster was part of a large genomic island of 185 kb with a divergent GC content compared to the rest of the genome of strain E7962 and that this island in E9242 is slightly smaller (144 kb) (Fig. 2 and Table 1). In the four other Dutch VRE isolates and the six vanD-positive strains from GenBank for which WGSs were available, the vanD gene cluster was also part of a similar large genomic island with a variable size of 120 to 168 kb (Fig. 2 and Table 1; see also Table S2). The previously mentioned IMEEfm15508 element, however, is different in gene content and size (66.7 kb) from the genomic island we described in this study and was therefore excluded from further analysis (15).
FIG 2.
Circular representation (BLAST Ring Image Generator [24]) of complete genomes and genomic islands using E. faecium E7962 as reference.
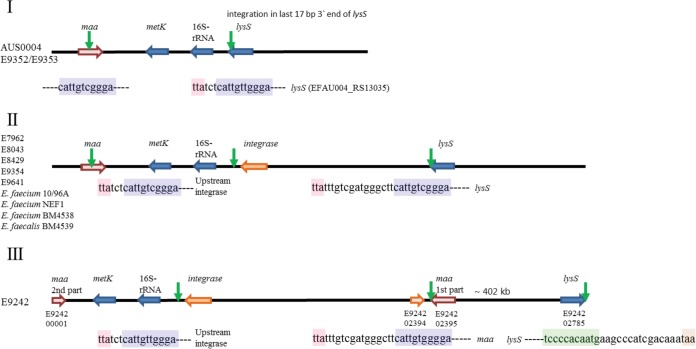
Based on comparison with the E. faecium AUS0004 genome sequence, the insertion site of the vanD-containing genomic island was located between the 16S ribosomal rRNA gene (EFAU004_RS13030) and lysS (EFAU004_RS13035) (Fig. 3, I). In both E. faecium and E. faecalis strains, insertion had occurred in the last 11 bp of the 3′ end of lysS (Fig. 3, I), resulting in a duplication of this 11-bp insertion site at both ends of the island, thereby generating an alternative stop codon for lysS (Fig. 3, II; Table S2). In strain E9242, the lysS gene (E9242_02785) was at the end of the contig in the forward orientation (Fig. 3, III). PCR analysis confirmed a large genomic rearrangement of ∼402 kb (data not shown), which should have occurred after integration of the island via a 29-bp inversed repeat region in the maa and lysS genes (Fig. 3, III). The presence of an integrase gene and duplication of the insertion site suggest exchange via a circular intermediate, which is a general characteristic for an integrative and conjugative element (19–21).
FIG 3.
Genomic organization (not drawn to scale) indicating the integration of the genomic island. (I) vanD-negative isolates AUS0004, E9352, and E9353. Green arrows indicate the presence of an 11-bp repeat unit (blue box) in the 3′ end of lysS (red box, stop codon) and maa genes. (II) vanD-positive isolates E. faecium E7962, E8043, E8429, E9354, E9641, 10/96A, NEF1, and BM4538 and E. faecalis BM4539. In these strains, integration always occurred by the 11-bp repeat sequence present in lysS, resulting in a duplication at the 11-bp repeat unit upstream to the integrase and an alternative stop codon for lysS. (III) Genomic rearrangement in vanD-positive strain E9242. In this strain, after integration of the genomic island, a large genomic rearrangement had occurred between the 11-bp repeat present in the lysS and maa genes, resulting in a split of the maa gene and an inverted repeat (green box) in the lysS gene.
A phylogenetic tree based on a multiple alignment of the vanD-containing genomic islands revealed three clusters of closely related islands (Fig. 1B). Cluster I in Fig. 1B contained Dutch isolates E7962, E8043, and E9641 and strain E. faecium 10/96A from Brazil; cluster II contained the two vanD-positive VRE isolates from patient D; and cluster III contained E. faecium BM4538 and E. faecalis BM4539. The genomic island of vanD-positive VRE E9242 clustered separately from the other Dutch isolates. A multiple alignment of the vanD-containing genomic islands revealed five conserved blocks of genes (see regions A through E in Fig. S3A and B in the supplemental material), including the vanD gene cluster (region D, Fig. S3B and C) interspersed with variable regions (Fig. S3A and B). A nonsupervised orthologous group (eggNOG v4.5 [22]) analysis revealed that the overall distribution of Clusters of Orthologous Groups (COG) categories was very similar for all the strains (see Fig. S4 and Tables S3 to S14 in the supplemental material), including a high number of transcriptional regulators and putative two-component systems (category K, T and depicted as blue and purple genes, respectively, in Fig. S3A and B) and genes involved in replication, recombination, and repair (category L, green genes in Fig. S3A and B). In addition to the vancomycin resistance genes (category M, orange genes in Fig. S3A and B), at least 12 other putative antibiotic resistance genes were identified (yellow genes in Fig. S3A and B).
Based on the diversity in cgMLST allelic profiles, sequence variation in the vanD gene clusters and in the genomic islands carrying the vanD gene clusters, we conclude that the six Dutch vanD-positive VRE isolates are not epidemiologically linked and thus have not emerged through either clonal spread or horizontal transmission of the vancomycin resistance genes. In contrast, our results point toward independent acquisition of a large genomic island containing the vanD gene cluster, possibly from the patient's own anaerobic microbiota, which might have occurred in patient D. Domingo et al. (23) described high prevalences of vanB-, vanG-, and vanD-type resistance genes not associated with enterococci present in the human fecal flora. The level of similarity among the genomic islands containing the vanD gene clusters between the anaerobic bacteria and E. faecium described in this study support the hypothesis that anaerobic gut commensals may represent a reservoir for the vanD type of vancomycin resistance; however, so far, there is no experimental evidence for genetic exchange between gut commensals and enterococci.
The fact that we did not find indications for clonal spread of vanD-positive VRE suggests that these VREs do not transmit easily between patients, in contrast to vanA- or vanB-positive VRE. However, because the genomic island described in this study contains a high number of additional antibiotic resistance genes, acquisition of the island and subsequent infection with E. faecium strains containing the island may lead to particularly difficult or even nontreatable infections.
Accession number(s).
The raw reads obtained for the eight E. faecium strains used in this study have been deposited at the European Nucleotide Archive under the following project accession numbers: PRJEB21556 and PRJEB21647.
Supplementary Material
ACKNOWLEDGMENTS
J.C. was supported by the Finnish COIN Center of Excellence and ERC grant no. 742158. The work of the German National Reference Centre for Staphylococci and Enterococci (G.W.) is supported by a grant from the German Federal Ministry of Health.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01793-17.
REFERENCES
- 1.Depardieu F, Foucault ML, Bell J, Dubouix A, Guibert M, Lavigne JP, Levast M, Courvalin P. 2009. New combinations of mutations in VanD-type vancomycin-resistant Enterococcus faecium, Enterococcus faecalis, and Enterococcus avium strains. Antimicrob Agents Chemother 53:1952–1963. doi: 10.1128/AAC.01348-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Domingo MC, Huletsky A, Giroux R, Picard FJ, Bergeron MG. 2007. vanD and vanG-like gene clusters in a Ruminococcus species isolated from human bowel flora. Antimicrob Agents Chemother 51:4111–4117. doi: 10.1128/AAC.00584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Courvalin P. 2006. Vancomycin resistance in Gram-positive cocci. Clin Infect Dis 42(Suppl 1):S25–S34. [DOI] [PubMed] [Google Scholar]
- 4.Miller WR, Munita JM, Arias CA. 2014. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti Infect Ther 12:1221–1236. doi: 10.1586/14787210.2014.956092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jünemann S, Sedlazeck FJ, Prior K, Albersmeier A, John U, Kalinowski J, Mellmann A, Goesmann A, von Haeseler A, Stoye J, Harmsen D. 2013. Updating benchtop sequencing performance comparison. Nat Biotechnol 31:294–296. doi: 10.1038/nbt.2522. [DOI] [PubMed] [Google Scholar]
- 6.de Been M, Pinholt M, Top J, Bletz S, Mellmann A, van Schaik W, Brouwer E, Rogers M, Kraat Y, Bonten M, Corander J, Westh H, Harmsen D, Willems RJL. 2015. Core genome multilocus sequence typing scheme for high-resolution typing of Enterococcus faecium. J Clin Microbiol 53:3788–3797. doi: 10.1128/JCM.01946-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJL, Earl AM, Gilmore MS. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4:pii=e00534-13. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer KL, Godfrey P, Griggs A, Kos VN, Zucker J, Desjardins C, Cerqueira G, Gevers D, Walker S, Wortman J, Feldgarden M, Haas B, Birren B, Gilmore MS. 2012. Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio 3:e00318-00311. doi: 10.1128/mBio.00318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Depardieu F, Reynolds PE, Courvalin P. 2003. VanD-type vancomycin-resistant Enterococcus faecium 10/96A. Antimicrob Agents Chemother 47:7–18. doi: 10.1128/AAC.47.1.7-18.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parkinson JS, Kofoid EC. 1992. Communication modules in bacterial signaling proteins. Annu Rev Genet 26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 11.Casadewall B, Courvalin P. 1999. Characterization of the vanD glycopeptide resistance gene cluster from Enterococcus faecium BM4339. J Bacteriol 181:3644–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyd DA, Kibsey P, Roscoe D, Mulvey MR. 2004. Enterococcus faecium N03-0072 carries a new VanD-type vancomycin resistance determinant: characterization of the VanD5 operon. J Antimicrob Chemother 54:680–683. doi: 10.1093/jac/dkh391. [DOI] [PubMed] [Google Scholar]
- 13.Boyd DA, Conly J, Dedier H, Peters G, Robertson L, Slater E, Mulvey MR. 2000. Molecular characterization of the vanD gene cluster and a novel insertion element in a vancomycin-resistant enterococcus isolated in Canada. J Clin Microbiol 38:2392–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostrowsky BE, Clark NC, Thauvin-Eliopoulos C, Venkataraman L, Samore MH, Tenover FC, Eliopoulos GM, Moellering RC, Gold HS. 1999. A cluster of VanD vancomycin-resistant Enterococcus faecium: molecular characterization and clinical epidemiology. J Infect Dis 180:1177–1185. doi: 10.1086/315030. [DOI] [PubMed] [Google Scholar]
- 15.Boyd DA, Lalancette C, Lévesque S, Golding GR. 2016. Characterization of a genomic island harbouring a new vanD allele from Enterococcus faecium N15-508 isolated in Canada. J Antimicrob Chemother 71:2052–2054. doi: 10.1093/jac/dkw063. [DOI] [PubMed] [Google Scholar]
- 16.Depardieu F, Kolbert M, Pruul H, Bell J, Courvalin P. 2004. VanD-type vancomycin-resistant Enterococcus faecium and Enterococcus faecalis. Antimicrob Agents Chemother 48:3892–3904. doi: 10.1128/AAC.48.10.3892-3904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanimoto K, Nomura T, Maruyama H, Tomita H, Shibata N, Arakawa Y, Ike Y. 2006. First VanD-type vancomycin-resistant Enterococcus raffinosus isolate. Antimicrob Agents Chemother 50:3966–3967. doi: 10.1128/AAC.00607-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shkoporov AN, Chaplin AV, Shcherbakova VA, Suzina NE, Kafarskaia LI, Bozhenko VK, Efimov BA. 2016. Ruthenibacterium lactatiformans gen. nov., sp. nov., an anaerobic, lactate-producing member of the family Ruminococcaceae isolated from human faeces. Int J Syst Evol Microbiol 66:3041–3049. doi: 10.1099/ijsem.0.001143. [DOI] [PubMed] [Google Scholar]
- 19.Johnson CM, Grossman AD. 2015. Integrative and conjugative elements (ICEs): what they do and how they work. Annu Rev Genet 49:577–601. doi: 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Top J, Sinnige JC, Majoor EAM, Bonten MJM, Willems RJL, van Schaik W. 2011. The recombinase IntA is required for excision of esp-containing ICEEfm1 in Enterococcus faecium. J Bacteriol 193:1003–1006. doi: 10.1128/JB.00952-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.León-Sampedro R, Novais C, Peixe L, Baquero F, Coque TM. 2016. Diversity and evolution of the Tn5801-tet(M)-like integrative and conjugative elements among Enterococcus, Streptococcus, and Staphylococcus. Antimicrob Agents Chemother 60:1736–1746. doi: 10.1128/AAC.01864-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, Jensen LJ, von Mering C, Bork P. 2016. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Domingo MC, Huletsky A, Giroux R, Boissinot K, Picard FJ, Lebel P, Ferraro MJ, Bergeron MG. 2005. High prevalence of glycopeptide resistance genes vanB, vanD, and vanG not associated with enterococci in human fecal flora. Antimicrob Agents Chemother 49:4784–4786. doi: 10.1128/AAC.49.11.4784-4786.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.