ABSTRACT
Isoniazid and rifampin are essential components of first-line antituberculosis (anti-TB) therapy. Understanding the relationship between genetic factors and the pharmacokinetics of these drugs could be useful in optimizing treatment outcomes, but this is understudied in children. We investigated the relationship between N-acetyltransferase type 2 (NAT2) genotypes and isoniazid pharmacokinetics, as well as that between the solute carrier organic anion transporter family member 1B1 (encoded by SLCO1B1) and carboxylesterase 2 (CES2) single nucleotide polymorphisms (SNPs) and rifampin pharmacokinetics in Ghanaian children. Blood samples were collected at times 0, 1, 2, 4, and 8 h postdose in children with tuberculosis on standard first-line therapy for at least 4 weeks. Isoniazid and rifampin concentrations were determined by a validated liquid chromatography-tandem mass spectrometry (LC-MS/MS) method, and pharmacokinetic parameters were calculated using noncompartmental analysis. Genotyping of NAT2, SLCO1B1, and CES2 SNPs were performed using validated TaqMan genotyping assays. The Kruskal-Wallis test was used to compare pharmacokinetic parameters among the three genotypic groups and was followed by the Wilcoxon rank sum test for pairwise group comparisons. Genotype status inferred by the NAT2 4-SNP and 7-SNP genotyping panels identified children with a slow acetylator phenotype but not the rapid genotype. For rifampin, only the rare SLCO1B1*1b homozygous variant was associated with rifampin pharmacokinetics. Our findings suggest that NAT2 and SCLCO1B1*1b genotyping may have minimal clinical utility in dosing decisions at the population level in Ghanaian children, but it could be useful at the individual level or in populations that have a high frequency of implicated genotypes. Further studies in other populations are warranted.
KEYWORDS: NAT2 acetylator genotypes, SLCO1B1 gene, CES2 gene, single nucleotide polymorphisms, rifampin pharmacokinetics, isoniazid pharmacokinetics
INTRODUCTION
Isoniazid and rifampin are essential components of the short-course chemotherapy regimen for first-line treatment of drug-susceptible tuberculosis (TB) (1). The standard first-line regimen consists of isoniazid, rifampin, pyrazinamide, and ethambutol for 2 months, followed by isoniazid and rifampin for an additional 4 months. Resistance to or intolerance of either rifampin or isoniazid extends the treatment duration from 6 months to at least 9 months. Isoniazid alone given for 9 months and rifampin alone for 4 months are also preferred regimens for the treatment of latent TB infection in both adults and children (2). It is expected that the pharmacokinetic profiles of these drugs, associated with efficacy and safety in adults, should apply to children as well (3, 4). Although not validated in randomized or controlled trials, early bactericidal activity (EBA) of isoniazid was related to dose or pharmacokinetics (PK) in adults (5–7). Similarly, the EBAs of rifampin, rifabutin, and rifapentine in adults appear to be associated with drug dose and/or pharmacokinetics (8, 9). Thus, the variability in the pharmacokinetics of isoniazid and/or rifampin due to fixed-weight-band dosing as used in children might influence the clinical outcomes of active or latent TB treatment.
Isoniazid is metabolized primarily by the genetically polymorphic N-acetyltransferase type 2 (NAT2) enzyme (10). In adults, the trimodal distribution of isoniazid elimination and area under the concentration-time curve (AUC) is explained by a number of polymorphisms in the NAT2 gene, which defines the acetylator phenotype (rapid, intermediate, or slow) (11, 12). In a study among South African children who were treated according to previous anti-TB drug dosing guidelines, a trimodal distribution of isoniazid AUC and 2- to 5-hour postdose concentrations was observed (13). The above-mentioned study also found that younger children eliminate isoniazid faster than older children, and as a group, children eliminate isoniazid faster than adults across all acetylator genotype groups (13). Children with rapid and intermediate acetylator genotypes were shown to be at risk of suboptimal isoniazid concentrations when given weight-based dosage similar to that of adults, prompting the investigators to recommend a higher daily dose in milligrams per kilogram of body weight for children (13, 14). In 2010, the World Health Organization (WHO) recommended a higher dosage in milligrams per kilogram of the antituberculosis drugs for all children (15), but it is not yet known if the increased dosage of isoniazid in children resolves the variability in isoniazid pharmacokinetics due to the NAT2 acetylator genotype.
Rifampin undergoes extensive hepatic deacetylation by β-esterase to form the 25-deacetylated metabolite (16). The organic anion transporter polypeptide encoded by the solute carrier organic anion transporter family member 1B1 gene (SLCO1B1) mediates the hepatic uptake and elimination of a range of drugs (17). While some studies in adults found a significant relationship between SLCO1B1 single nucleotide polymorphisms (SNPs) c.463C>A (rs11045819) and SLCO1B1 rs4149032 (intron 2 haplotype tagging SNP; tSNP) and low rifampin plasma exposure (18, 19), others failed to replicate these associations (20, 21). The human carboxylesterase (CES) belongs to the β-esterase family, members of which are thought to be also involved in the metabolism of rifampin. Given the structural similarity between rifampin and substrates of carboxylesterase 2 (CES2), one study explored and demonstrated a relationship between CES2 polymorphisms and rifampin metabolism (22). In particular, the CES2 c.-2263A>G in the promoter region that is closely linked to c.269-965AG and c.1612 + 13GA was associated with rifampin metabolism through expression of the gene (22). To the best of our knowledge, the pharmacogenetic determinants of rifampin pharmacokinetics have not previously been studied in children.
The aim of this study was to examine the relationship between NAT2 acetylator genotypes and isoniazid pharmacokinetics as well as SLCO1B1 and CES2 SNPs and rifampin pharmacokinetics in Ghanaian children with TB who were predominantly given the World Health Organization (WHO)-recommended revised anti-TB drug dosages for children.
RESULTS
Study population.
Of the 113 study participants, 59 (52.2%) were HIV coinfected, 63 (55.8%) were male, and 24 (21.2%) were aged <2 years old as we previously reported (23). None of the HIV-infected patients were receiving antiretroviral therapy at the time of pharmacokinetic sampling. The median (interquartile range [IQR]) isoniazid dose was 11.2 (9.1 to 12.8 mg/kg of body weight) and that for rifampin was 15.8 (13.6 to 18.8 mg/kg).
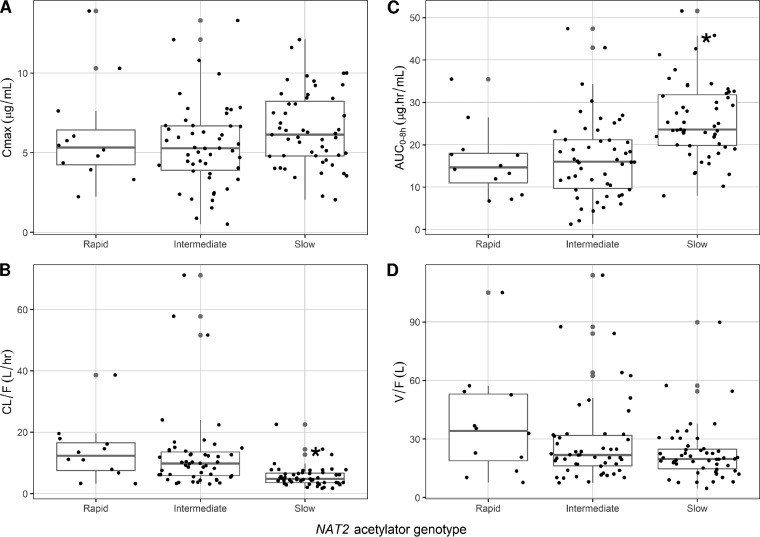
The coefficient of variation of isoniazid maximum concentration in serum (Cmax) and the area under the time-concentration curve from 0 to 8 h (AUC0–8h) were 43.7% and 49.1%, respectively. There was a significant difference in median values of isoniazid AUC0–8h and the predicted apparent oral clearance (CL/F) but no difference in time to Cmax (Tmax), Cmax, or predicted apparent volume of distribution (V/F) among the three NAT2 acetylator phenotype groups (Table 1). As shown in Fig. 1, there was a considerable overlap of isoniazid pharmacokinetic parameters among the three groups, and the differences in median AUC0–8h and CL/F values were significant between slow and rapid as well as slow and intermediate acetylator phenotype groups. There was no significant difference in isoniazid pharmacokinetics in children with a rapid genotype compared to those with an intermediate genotype. Of the 113 study participants, 12 (11%) had an isoniazid Cmax of <3 μg/ml (low Cmax) and 49 (43%) had a Cmax of >6 μg/ml (high) compared to the typical 2-h postdose sample concentration range. Of the 12 participants with low Cmax, only 1 (0.8%) had a rapid and 2 (16.7%) had a slow NAT2 genotype. Of the 49 with high isoniazid, only 26 (53%) had a slow NAT2 genotype. Multivariate analysis suggested that isoniazid dose and slow NAT2 genotype were jointly associated with isoniazid Cmax and AUC0–8h, age and slow NAT2 genotype were associated with CL/F, while isoniazid dose, age, and slow NAT2 genotype were jointly associated with V/F (Table 2).
TABLE 1.
Pharmacokinetic parameters of isoniazid by NAT2 acetylator genotype status in 113 Ghanaian children with TBa
| Genotype (%) | Tmax (h) | Cmax (μg/ml) | AUC0–8h (μg · h/ml) | CL/F (liters/h) | V/F (liters) |
|---|---|---|---|---|---|
| Rapid (10.6) | 1.00 (0.99–1.10) | 5.32 (4.14–6.83) | 14.62 (10.05–18.27) | 12.28 (7.26–17.02) | 34.11 (17.07–53.45) |
| Intermediate (44.3) | 1.05 (1.00–1.17) | 5.27 (3.84–6.70) | 16.01 (9.41–21.23) | 9.79 (5.77–13.83) | 21.75 (16.04–32.03) |
| Slow (45.1) | 1.05 (1.00–1.32) | 6.13 (4.77–8.40) | 23.59 (19.73–32.11) | 4.78 (3.50–6.75) | 19.72 (14.44–24.90) |
| P value | 0.443 | 0.156 | <0.001 | <0.001 | 0.121 |
PK parameter values are medians (IQR). Tmax, time to maximum concentration; Cmax, maximum concentration; AUC0–8h, area under the time-concentration curve from time 0 to 8 h postdose; CL/F, apparent oral clearance; V/F, apparent predicted volume of distribution.
FIG 1.
Relationship between isoniazid Cmax (A), AUC0–8h (B), CL/F (C), and V/F (D) and NAT2 acetylator genotypes in children with tuberculosis. *, significant differences in AUC0–8h and CL/F between slow and rapid or intermediate groups.
TABLE 2.
Multivariate analysis showing coefficient estimates factors that were jointly associated with isoniazid and rifampin pharmacokinetic parametersa
| Drug | PK parameter | Predictor | Estimate | SE | Standardized estimate | P value |
|---|---|---|---|---|---|---|
| Isoniazid | Cmax | Dose | 0.312 | 0.078 | 0.362 | <0.001 |
| Male versus female | −0.750 | 0.476 | −0.144 | 0.118 | ||
| Slow versus nonslow NAT2 genotype | 1.058 | 0.463 | 0.203 | 0.024 | ||
| AUC0–8h | Age | 0.393 | 0.226 | 0.147 | 0.086 | |
| Dose | 1.353 | 0.287 | 0.400 | <0.001 | ||
| Slow versus nonslow NAT2 genotype | 9.642 | 1.602 | 0.472 | <0.001 | ||
| CL/F | Age | 0.593 | 0.234 | 0.222 | 0.013 | |
| Male versus female | 3.118 | 1.800 | 0.152 | 0.086 | ||
| Slow versus nonslow NAT2 genotype | −7.668 | 1.770 | −0.375 | <0.001 | ||
| V/F | Age | 2.313 | 0.477 | 0.447 | <0.001 | |
| Dose | 1.369 | 0.624 | 0.209 | 0.030 | ||
| BMI | 1.325 | 0.787 | 0.148 | 0.095 | ||
| Slow versus nonslow NAT2 genotype | −6.977 | 3.377 | −0.176 | 0.041 | ||
| Rifampin | Cmax | Age | 0.255 | 0.079 | 0.298 | 0.002 |
| Dose | 0.329 | 0.081 | 0.379 | <0.001 | ||
| Male versus female | −0.994 | 0.585 | −0.151 | 0.092 | ||
| TB/HIV coinfection versus TB | −1.132 | 0.566 | −0.173 | 0.048 | ||
| AUC0–8h | Age | 1.201 | 0.330 | 0.326 | <0.001 | |
| Dose | 1.495 | 0.338 | 0.401 | <0.001 | ||
| Male versus female | −5.649 | 2.433 | −0.200 | 0.022 | ||
| TB/HIV coinfection | −6.016 | 2.351 | −0.214 | 0.012 | ||
| CL/F | BMI | 0.639 | 0.393 | 0.151 | 0.106 | |
| SLCO1B1 c.388GG versus AG/AA | −3.430 | 1.850 | −0.172 | 0.066 | ||
| V/F | Age | 1.771 | 0.756 | 0.214 | 0.021 | |
| SLCO1B1 c.388GG versus AG/AA | −12.746 | 6.178 | −0.189 | 0.041 |
Cmax, maximum concentration; AUC0–8h, area under the time concentration curve from time 0 to 8 h postdose; CL/F, apparent oral clearance; V/F, apparent predicted volume of distribution.
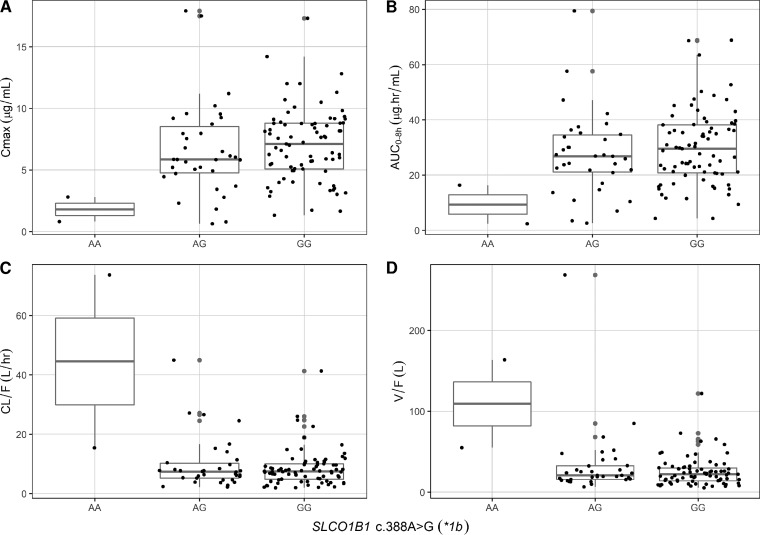
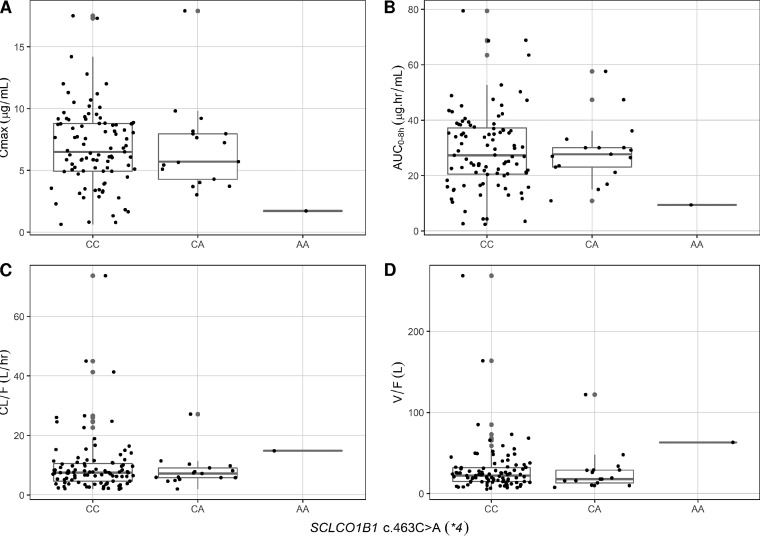
The coefficient of variation of rifampin Cmax and AUC0–8h were 48.0% and 48.9%, respectively. None of the evaluated SLCO1B1 and CES2 SNPs were significantly associated with rifampin pharmacokinetics in the primary analysis (Table 3). The genetic variation in SLCO1B1 c.388A>G (*1b) SNP showed a trend toward a significant relationship with rifampin PK parameters (Table 3). The two patients with the *1b homozygous variant (AA genotype) had significantly lower rifampin Cmax and AUC0–8h and higher CL/F and V/F than did those with the wild type (GG genotype) in pairwise analysis (Fig. 2). Also, the one participant with SLCO1B1 c.463AA had much lower rifampin Cmax and AUC and higher CL/F and V/F than did participants with SLCO1B1 c.463 CC or CA (Table 3), but pairwise comparison was not done because there was just one participant with the homozygous variant (Fig. 3). Of the 113 participants, 73 (65%) had rifampin Cmax of <8 μg/ml (low Cmax), but we found no association between any of the studied SNPs and risk of low Cmax. In a multivariate model, none of the studied SNPs were associated with rifampin Cmax, AUC0–8h, and CL/F (Table 2). The SLCO1B1 c.388GG compared to AA/AG and age were jointly associated with rifampin V/F (Table 2).
TABLE 3.
Rifampin pharmacokinetic parameter by SLCO1B1 and CES2 single nucleotide polymorphism status in 113 Ghanaian children with TBa
| Genetic factor (%) | Cmax (μg/ml) | AUC0–8h (μg · h/ml) | CL/F (liters/h) | V/F (liters) |
|---|---|---|---|---|
| SLCO1B1c.388A>G (*1b) | ||||
| AA (1.8) | 1.81 (0.81–2.80) | 9.33 (2.35–16.31) | 44.54 (15.38–73.69) | 109.23 (54.86–163.59) |
| AG (30.1) | 5.86 (4.71–8.72) | 26.84 (20.90–34.71) | 7.38 (5.22–10.37) | 20.73 (15.64–32.76) |
| GG (68.1) | 7.11 (5.08–8.79) | 29.50 (20.79–38.71) | 7.43 (4.86–10.06) | 21.98 (14.20–30.12) |
| P value | 0.052 | 0.085 | 0.093 | 0.067 |
| SLCO1B1c.388A>G (*1b) | ||||
| AA/AG (31.9) | 5.84 (4.08–8.34) | 26.35 (16.67–34.27) | 7.58 (5.26–12.73) | 22.07 (15.95–40.54) |
| GG (68.1) | 7.11 (5.08–8.79) | 29.50 (20.79–38.71) | 7.43 (4.86–10.06) | 21.98 (14.20–30.12) |
| P value | 0.147 | 0.181 | 0.583 | 0.265 |
| SLCO1B1 rs4149032 (intron SNP) | ||||
| CC (57.5) | 7.00 (5.10–8.77) | 29.50 (21.23–36.58) | 7.34 (4.99–9.79) | 21.20 (13.46–31.00) |
| CT (31.9) | 6.55 (4.59–8.81) | 26.35 (16.74–37.82) | 7.58 (4.62–11.56) | 21.80 (16.64–30.48) |
| TT (10.6) | 5.43 (3.25–6.90) | 24.07 (13.58–29.58) | 8.37 (5.88–14.74) | 23.59 (13.02–49.79) |
| P value | 0.228 | 0.262 | 0.462 | 0.657 |
| SLCO1B1 c.463C>A (*4) | ||||
| CC (84.1) | 6.50 (4.94–8.79) | 27.25 (20.37–37.46) | 7.52 (4.63–10.67) | 22.13 (14.72–32.10) |
| CA (15.0) | 5.70 (4.28–7.96) | 27.67 (23.05–30.06) | 7.18 (5.78–9.10) | 17.72 (13.16–28.79) |
| AA (0.09) | 1.72 (1.72, 1.72) | 9.35 (9.35, 9.35) | 14.81 (14.81, 14.81) | 63.01 (63.01, 63.01) |
| P value | 0.262 | 0.304 | 0.422 | 0.248 |
| SLCO1B1 c.463C>A (*4) | ||||
| CC (84.1) | 6.50 (4.94–8.79) | 27.25 (20.37–37.46) | 7.52 (4.63–10.67) | 22.13 (14.72–32.10) |
| CA/AA (15.9) | 5.68 (4.02–7.96) | 27.30 (21.11–30.06) | 7.26 (5.78–9.79) | 18.46 (13.16–28.95) |
| P value | 0.417 | 0.621 | 0.919 | 0.701 |
| SLCO1B1 c.521T>C (*5) | ||||
| TT (97.3) | 6.32 (4.92–8.79) | 27.08 (20.62–36.58) | 7.49 (4.86–10.37) | 21.59 (14.72–31.00) |
| TC (2.7) | 8.05 (3.23–8.68) | 29.83 (16.44–35.30) | 5.10 (4.99–12.50) | 49.09 (7.82–58.79) |
| P value | 0.986 | 0.915 | 0.761 | 0.464 |
| CES2 c.–2263A>G | ||||
| AA (49.6) | 7.17 (4.94–8.81) | 29.00 (20.95–38.31) | 7.41 (5.04–9.69) | 19.89 (14.02–30.56) |
| AG (41.6) | 5.85 (3.69–8.17) | 24.48 (16.31–34.71) | 7.71 (5.57–12.50) | 23.36 (15.64–36.76) |
| GG (8.8) | 6.90 (5.90–9.24) | 30.62 (25.12–43.45) | 4.48 (2.25–9.80) | 21.80 (10.17–27.42) |
| P value | 0.134 | 0.179 | 0.207 | 0.487 |
PK parameter values are medians (IQR). Tmax, time to maximum concentration; Cmax, maximum concentration; AUC0–8h, area under the time-concentration curve from time 0 to 8 h postdose; CL/F, apparent oral clearance; V/F, apparent predicted volume of distribution.
FIG 2.
Relationship between rifampin Cmax (A), AUC0–8h (B), CL/F (C), and V/F (D) and SLCO1B1 c.388A>G genotypes in children with tuberculosis. The differences in median values of all pharmacokinetic parameters between genotypes AA and GG were significant in post hoc analysis.
FIG 3.
Relationship between rifampin Cmax (A), AUC0–8h (B), CL/F (C), and V/F (D) and SLCO1B1 c.463C>A genotypes in children with tuberculosis.
Overall, 99 (87.6%) subjects completed therapy, 12 (10.6%) were lost to follow-up, and 2 (1.8%) died. No study participants discontinued therapy or required modification of therapy because of medication side effects. Of 74 children who had liver enzyme test results available at baseline and at week 4 of therapy, there were no significant differences in the median changes in aspartate transferase (AST) or alanine transferase (ALT) levels at week 4 from baseline by NAT2 acetylator genotype status (P > 0.05).
DISCUSSION
In this study, the 4-SNP or 7-SNP NAT2 acetylator genotype identified slow compared to rapid or intermediate metabolizers of isoniazid among children treated according to the revised WHO dosing guidelines. However, we found no difference in isoniazid pharmacokinetics in children with a rapid genotype compared to those with an intermediate genotype. For rifampin, we found no significant relationship between pharmacokinetic parameters and SLCO1B1 and CES2 SNPs, except that the rare SLCO1B1*1b homozygous AA variant (found in only 2% of participants) was associated with low rifampin concentrations. Overall, the modest effect of NAT2 genotypes to fully discriminate between children with low, intermediate, and high isoniazid plasma exposure without significant overlap and the rare occurrence of the SLCO1B1 c.388AA genotype suggest that genotyping for the studied SNPs in Ghanaian children may have minimal clinical utility in making isoniazid and rifampin dosing decisions at the population level.
Inferring the genotype status by the 4-SNP NAT2 genotyping panel is considered more economical than using the recommended 7-SNP panel. The experimentally determined NAT2 4-SNP acetylator genotype inferred NAT2 acetylator phenotype status with 98.4% accuracy (24). The 4-SNP NAT2 genotyping panel also had a 100% agreement with acetylator phenotypes inferred by the recommended 7-SNP panel in a diverse population of non-European ancestry (25). In the current study, we found 100% agreement between acetylator status inferred by the 4-SNP and 7-SNP genotyping panels. However, NAT2 genotypes failed to clearly discriminate between isoniazid phenotype groups, as we observed a considerable overlap of isoniazid pharmacokinetic parameters across the three groups (Fig. 1). Unlike the trimodal distribution of isoniazid pharmacokinetics reported in some studies in adults (11, 12), the bimodal distribution observed in our study is consistent with other studies in children, in which the pharmacokinetic parameters in those rapid and intermediate genotypes were similar (13, 26, 27). The bimodal distribution of NAT2 acetylator phenotype in children may be explained by differences in enzyme maturation with age. NAT2 enzyme maturation based on weight-normalized CL/F was demonstrated to increase with age in children up to 2 years old for rapid and intermediate acetylators, but no significant change was observed in slow acetylators (28).
In our multivariate analysis, isoniazid dose and NAT2 acetylator genotype status were joint predictors of isoniazid Cmax and AUC0–8h, while dose, age, and acetylator NAT2 genotype jointly predicted CL/F. Thus, the dose of isoniazid likely influences the relationship between NAT2 acetylator genotypes and isoniazid pharmacokinetics in children. In a study that used the previous recommended isoniazid dose of 5 mg/kg for children in South Africa, 35% of children with homozygous rapid genotype had a low 2-hour concentration with a faster elimination of isoniazid in younger than older children, prompting the investigators to recommend an isoniazid dose of at least 10 mg/kg for children younger than 5 years old (13). In our study, in which the median isoniazid dose was 11.2 mg/kg, a low isoniazid Cmax was uncommon and only one child with rapid acetylator genotype had a low Cmax.
Adult studies that examined the relationship between rifampin pharmacokinetics and SLCO1B1 SNPs reported 36% lower rifampin AUC0–24 in participants with the SLCO1B1 c.463CA genotype than in those with the c.463CC genotype (19) and 18% and 28% lower AUC in patients heterozygous and homozygous, respectively, for the rs4149032 allele (18). However, two recent studies in Indian and Malawian adult patients failed to replicate or confirm the above-mentioned associations (20, 21). We found no significant association between the studied SCLCO1B1 and CES2 SNPs in the primary analysis. In post hoc analysis, the rare SLCO1B1 c.388AA genotype (found in 2 children) was associated with low rifampin concentrations compared to those with c.388GG. Also, one patient with the SLCO1B1 c.463AA genotype appeared to have low rifampin Cmax and AUC, but this could be due to chance. The relationship between SLCO1B1 c.388A>G SNP and rifampin pharmacokinetics requires further evaluation, especially in populations where the homozygous variant is frequent, as our study was highly limited by sample size and the rare occurrence of the variant. For the CES2 genetic variation and rifampin pharmacokinetics, our study was limited since we examined only one SNP with the strongest effect in vitro (22). It is possible that other CES2 SNPs that we did not include in our study may influence rifampin metabolism.
In summary, our results suggest that genotyping for the NAT2 acetylator genotype status or the selected SCLCO1B1 and CES2 SNPs may have only minimal clinical utility in isoniazid and rifampin dosing decisions at the population level in Ghanaian children given the modest effect of the NAT2 acetylator genotype and the rarity of the implicated SLCO1B1 SNP. However, at the individual level or in other populations with different allele frequencies of the implicated SNPs, host genetics may have a role in individualizing therapy. Further study with a larger sample size or in other populations in which the distribution of the studied SNPs may be different from that of our population is warranted.
MATERIALS AND METHODS
Study population and design.
Children aged 3 months to 14 years old with clinical diagnosis of TB were enrolled in a study at Komfo Anokye Teaching Hospital (KATH), Kumasi, Ghana. Briefly, enrolled children were treated with a regimen consisting of 7 to 15 mg/kg isoniazid, 10 to 20 mg/kg rifampin, 30 to 40 mg/kg pyrazinamide, and 15 to 25 mg/kg ethambutol daily for 2 months and then 7 to 15 mg/kg isoniazid and 10 to 20 mg/kg rifampin daily for 4 months. Pharmacokinetic sampling was performed after 4 weeks of anti-TB treatment as previously described (23, 29). The details of the study population and study procedures were previously reported (23). The Institutional Review Board (IRB) of KATH, Ghana, and Lifespan Hospitals, Providence, RI, USA, reviewed and approved the study. All parents or guardians of study participants provided signed informed consent.
Blood samples were collected at times 0 (predose), 1, 2, 4, and 8 hours postdose. This sampling scheme, when conducted at steady state, is considered sufficient to estimate key pharmacokinetic parameters such as Cmax and area under the concentration-time curve (AUC) (30). The blood samples collected in EDTA-coated tubes were placed immediately on ice and centrifuged within 30 min at 3,000 × g for 10 min. Plasma was stored at −80°C until shipment on dry ice to University of Cape Town, Cape Town, South Africa, for drug concentration assays. Drug concentrations were determined using validated liquid chromatography-tandem mass spectrometry (LC-MS/MS) methods as we previously described (29). The observed Cmax and time to Cmax (Tmax) were determined by inspection of the concentration of drug in serum-time graphs for each drug. The calculation of AUC from time 0 to 8 h (AUC0–8h), predicted apparent oral clearance (CL/F), and predicted apparent volume of distribution (V/F) was performed using noncompartmental analysis (Phoenix Software; Pharsight Corporation, Mountain View, CA).
Genotyping.
The isolated genomic DNA samples from blood leukocytes were genotyped by the TaqMan genotyping method on ViiA 7 real-time PCR system according to the manufacturer's recommendations (Applied Biosystems, Thermo Fisher Scientific, Foster City, CA). For NAT2, four SNPs were genotyped, rs1801279 (191G>A), rs1801280 (341T>C), rs1799930 (590G>A), and rs1799931 (857G>A). Those homozygous wild-type samples for all SNPs were classified as rapid acetylator phenotype, those heterozygous for any one of the SNPs were classified as intermediate acetylator phenotype, and those homozygous variants for one or more SNPs or heterozygous for two or more SNPs were classified as slow acetylator phenotype according to the established criteria for the 4-SNP genotype panel (24). We further genotyped the samples for NAT2 gene SNPs, rs1041983 (282C>T), rs1799929 (481C>T), and rs1208 (803A>G) to infer 7-SNP genotypes. In our study participants, the 4-SNP- and 7-SNP panel-inferred acetylator genotypes were similar. Thus, in this paper, genotype-phenotype associations described for 4-SNP are the same as for 7-SNP. For rifampin pharmacogenetics, we genotyped for SLCO1B1c.388A>G (*1b, rs2306283), c.463C>A (*4, rs11045819), c.521T>C (*5, rs4149056), and rs4149032 (intron 2 haplotype tagging SNP; tSNP) and CES2 c.-2263A>G. These SNPs were selected based on reported effects on rifampin pharmacokinetics in previous studies in adults (18, 19, 22). The departure from Hardy Weinberg equilibrium (HWE) was tested for all the SNPs from NAT2, CES2, and SLCO1B1 using the chi-square test with 1 degree of freedom. The genotype distribution of all SNPs (except NAT2) 282C>T (rs1041983) in the 7-SNP NAT2 panel (Table 4) were in HWE by χ2 analysis (31).
TABLE 4.
Distribution of studied NAT2, SLCO1B1, and CES2 single nucleotide polymorphisms in Ghanaian children with TBa
| Single nucleotide polymorphism | Mutation | dbSNP ID | MAF | Genotype | No. of times observed | Frequency (%) | HWE P value |
|---|---|---|---|---|---|---|---|
| NAT2 191G>A | Arg64Gln | rs1801279 | 0.12 | GG | 87 | 77 | 0.38 |
| GA | 26 | 23 | |||||
| AA | 0 | 0 | |||||
| NAT2 282C>T | Tyr94Tyr | rs1041983 | 0.42 | CC | 45 | 39.8 | 0.044 |
| CT | 42 | 37.2 | |||||
| TT | 26 | 23 | |||||
| NAT2 341C>T | Ile114Thr | rs1801280 | 0.30 | CC | 10 | 8.5 | 0.99 |
| CT | 48 | 42.5 | |||||
| TT | 55 | 49 | |||||
| NAT2 481C>T | Leu161Leu | rs1799929 | 0.25 | CC | 61 | 54 | 0.55 |
| CT | 47 | 42 | |||||
| TT | 5 | 4 | |||||
| NAT2 590G>A | Arg197Gln | rs1799930 | 0.26 | GG | 64 | 57 | 0.74 |
| GA | 40 | 35 | |||||
| AA | 9 | 8 | |||||
| NAT2 803A>G | Arg268Lys | rs1208 | 0.41 | AA | 42 | 37 | 0.68 |
| AG | 50 | 44 | |||||
| GG | 21 | 19 | |||||
| NAT2 191G>A | Gly286Glu | rs1799931 | 0.01 | GG | 111 | 98.2 | 0.99 |
| GA | 2 | 1.8 | |||||
| AA | 0 | 0.0 | |||||
| NAT2 genotypeb | |||||||
| Rapid | 12 | 10.6 | |||||
| Intermediate | 50 | 44.3 | |||||
| Slow | 51 | 45.1 | |||||
| SLCO1B1c.388A>G (*1b) | Asn130Asp | rs2306283 | 0.17 | AA | 2 | 1.8 | 0.72 |
| AG | 34 | 30.1 | |||||
| GG | 77 | 68.1 | |||||
| SLCO1B1 (tSNP) C>T | rs4149032 | 0.27 | CC | 65 | 57.5 | 0.15 | |
| CT | 36 | 31.9 | |||||
| TT | 12 | 10.6 | |||||
| SLCO1B1 c.463C>A (*4) | Pro155Thr | rs11045819 | 0.08 | CC | 95 | 84.1 | 0.97 |
| CA | 17 | 15.0 | |||||
| AA | 1 | 0.9 | |||||
| SLCO1B1 c.521T>C (*5) | Val174Ala | rs4149056 | 0.01 | TT | 110 | 97.3 | 0.99 |
| TC | 3 | 2.7 | |||||
| CC | 0 | 0.0 | |||||
| CES2 c.-2263A>G | 2263A>G | 0.30 | AA | 56 | 49.6 | 0.99 | |
| AG | 47 | 41.6 | |||||
| GG | 10 | 10.8 |
HWE, Hardy Weinberg equilibrium; MAF, minor allele frequency; SLCO1B1 tSNP, SLCO1B1 intron 2 haplotype tagging SNP.
NAT2 genotypes defined by the 4-SNP panel (191G>A, 341T>C, 590G>A, and 857G>A) and the 7-SNP panel (191G>A, 341T>C, 590G>A, and 857G>A plus 282C>T, 481C>T, and 803A>G) were similar.
Statistical analysis.
Multiple-group comparisons of pharmacokinetic parameters were examined by both one-way analysis of variance (ANOVA) and the Kruskal-Wallis test, followed by pairwise group comparisons by two-sample t test and its nonparametric version of the Wilcoxon rank sum test. Multivariate analysis with best subset variable selection based on corrected Akaike's information criterion (AICC) was used to find joint predictors of PK parameters. In addition to genotypes, HIV coinfection status (positive versus negative), sex (male versus female), age, body mass index (BMI), and drug dose were included in the multivariate model fitting. Statistical analyses were performed using software SAS 9.4 (SAS Institute Inc., Cary, NC). For all analyses, a P value of <0.05 was considered significant.
ACKNOWLEDGMENTS
We thank the study participants, the supportive staff of the TB and HIV clinics, and the malnutrition ward at KATH who helped with patient enrollment.
The work was supported primarily by the Eunice Kennedy Shriver National Institute of Child Health and Human Development at the National Institutes of Health (grant number HD071779). X. Tang was supported in part by Lifespan/Tufts/Brown Center for AIDS Research [P30 AI042853] and A. Topletz by T32 grant award DA013911. The pharmacokinetic laboratory at University of Cape Town is supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health Awards UM1 AI068634, UM1 AI068636, and UM1 AI106701, under the auspices of the Adult Clinical Trial Group. H. Yang utilized core services and support from the University of Rochester Center for AIDS Research (CFAR), an NIH-funded program (P30 AI078498). M. H. Court was supported by the National Institute of General Medical Sciences at the National Institutes of Health (grant number R01 GM102130). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Nahid P, Dorman SE, Alipanah N, Barry PM, Brozek JL, Cattamanchi A, Chaisson LH, Chaisson RE, Daley CL, Grzemska M, Higashi JM, Ho CS, Hopewell PC, Keshavjee SA, Lienhardt C, Menzies R, Merrifield C, Narita M, O'Brien R, Peloquin CA, Raftery A, Saukkonen J, Schaaf HS, Sotgiu G, Starke JR, Migliori GB, Vernon A. 2016. Official American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America clinical practice guidelines: treatment of drug-susceptible tuberculosis. Clin Infect Dis 63:e147–e195. doi: 10.1093/cid/ciw376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Getahun H, Matteelli A, Abubakar I, Aziz MA, Baddeley A, Barreira D, Den Boon S, Borroto Gutierrez SM, Bruchfeld J, Burhan E, Cavalcante S, Cedillos R, Chaisson R, Chee CB, Chesire L, Corbett E, Dara M, Denholm J, de Vries G, Falzon D, Ford N, Gale-Rowe M, Gilpin C, Girardi E, Go UY, Govindasamy D, D Grant A, Grzemska M, Harris R, Horsburgh CR Jr, Ismayilov A, Jaramillo E, Kik S, Kranzer K, Lienhardt C, LoBue P, Lonnroth K, Marks G, Menzies D, Migliori GB, Mosca D, Mukadi YD, Mwinga A, Nelson L, Nishikiori N, Oordt-Speets A, Rangaka MX, Reis A, Rotz L, Sandgren A, Sañé Schepisi M, Schünemann HJ, Sharma SK, Sotgiu G, Stagg HR, Sterling TR, Tayeb T, Uplekar M, van der Werf MJ, Vandevelde W, van Kessel F, van't Hoog A, Varma JK, Vezhnina N, Voniatis C, Vonk Noordegraaf-Schouten M, Weil D, Weyer K, Wilkinson RJ, Yoshiyama T, Zellweger JP, Raviglione M. 2015. Management of latent Mycobacterium tuberculosis infection: WHO guidelines for low tuberculosis burden countries. Eur Respir J 46:1563–1576. doi: 10.1183/13993003.01245-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donald PR, Ahmed A, Burman WJ, Cotton MF, Graham SM, Mendel C, McIlleron H, Mac Kenzie WR, Nachman S, Schaaf HS, Starke JR, Wingfield C, Hesseling AC. 2013. Requirements for the clinical evaluation of new anti-tuberculosis agents in children. Int J Tuberc Lung Dis 17:794–799. doi: 10.5588/ijtld.12.0567. [DOI] [PubMed] [Google Scholar]
- 4.Schaaf HS, Garcia-Prats AJ, Donald PR. 2015. Antituberculosis drugs in children. Clin Pharmacol Ther 98:252–265. doi: 10.1002/cpt.164. [DOI] [PubMed] [Google Scholar]
- 5.Donald PR, Parkin DP, Seifart HI, Schaaf HS, van Helden PD, Werely CJ, Sirgel FA, Venter A, Maritz JS. 2007. The influence of dose and N-acetyltransferase-2 (NAT2) genotype and phenotype on the pharmacokinetics and pharmacodynamics of isoniazid. Eur J Clin Pharmacol 63:633–639. doi: 10.1007/s00228-007-0305-5. [DOI] [PubMed] [Google Scholar]
- 6.Donald PR, Sirgel FA, Botha FJ, Seifart HI, Parkin DP, Vandenplas ML, Van de Wal BW, Maritz JS, Mitchison DA. 1997. The early bactericidal activity of isoniazid related to its dose size in pulmonary tuberculosis. Am J Respir Crit Care Med 156:895–900. doi: 10.1164/ajrccm.156.3.9609132. [DOI] [PubMed] [Google Scholar]
- 7.Jindani A, Aber VR, Edwards EA, Mitchison DA. 1980. The early bactericidal activity of drugs in patients with pulmonary tuberculosis. Am Rev Respir Dis 121:939–949. [DOI] [PubMed] [Google Scholar]
- 8.Sirgel FA, Botha FJ, Parkin DP, Van De Wal BW, Donald PR, Clark PK, Mitchison DA. 1993. The early bactericidal activity of rifabutin in patients with pulmonary tuberculosis measured by sputum viable counts: a new method of drug assessment. J Antimicrob Chemother 32:867–875. doi: 10.1093/jac/32.6.867. [DOI] [PubMed] [Google Scholar]
- 9.Sirgel FA, Fourie PB, Donald PR, Padayatchi N, Rustomjee R, Levin J, Roscigno G, Norman J, McIlleron H, Mitchison DA. 2005. The early bactericidal activities of rifampin and rifapentine in pulmonary tuberculosis. Am J Respir Crit Care Med 172:128–135. doi: 10.1164/rccm.200411-1557OC. [DOI] [PubMed] [Google Scholar]
- 10.Preziosi P. 2007. Isoniazid: metabolic aspects and toxicological correlates. Curr Drug Metab 8:839–851. doi: 10.2174/138920007782798216. [DOI] [PubMed] [Google Scholar]
- 11.Kinzig-Schippers M, Tomalik-Scharte D, Jetter A, Scheidel B, Jakob V, Rodamer M, Cascorbi I, Doroshyenko O, Sorgel F, Fuhr U. 2005. Should we use N-acetyltransferase type 2 genotyping to personalize isoniazid doses? Antimicrob Agents Chemother 49:1733–1738. doi: 10.1128/AAC.49.5.1733-1738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkin DP, Vandenplas S, Botha FJ, Vandenplas ML, Seifart HI, van Helden PD, van der Walt BJ, Donald PR, van Jaarsveld PP. 1997. Trimodality of isoniazid elimination: phenotype and genotype in patients with tuberculosis. Am J Respir Crit Care Med 155:1717–1722. doi: 10.1164/ajrccm.155.5.9154882. [DOI] [PubMed] [Google Scholar]
- 13.Schaaf HS, Parkin DP, Seifart HI, Werely CJ, Hesseling PB, van Helden PD, Maritz JS, Donald PR. 2005. Isoniazid pharmacokinetics in children treated for respiratory tuberculosis. Arch Dis Child 90:614–618. doi: 10.1136/adc.2004.052175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McIlleron H, Willemse M, Werely CJ, Hussey GD, Schaaf HS, Smith PJ, Donald PR. 2009. Isoniazid plasma concentrations in a cohort of South African children with tuberculosis: implications for international pediatric dosing guidelines. Clin Infect Dis 48:1547–1553. doi: 10.1086/598192. [DOI] [PubMed] [Google Scholar]
- 15.World Health Organization. 2014. Guidance for national tuberculosis programmes on the management of tuberculosis in children, 2nd ed WHO/HTM/TB/2014.03.1-146. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 16.Jamis-Dow CA, Katki AG, Collins JM, Klecker RW. 1997. Rifampin and rifabutin and their metabolism by human liver esterases. Xenobiotica 27:1015–1024. doi: 10.1080/004982597239994. [DOI] [PubMed] [Google Scholar]
- 17.Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KL, Chu X, Dahlin A, Evers R, Fischer V, Hillgren KM, Hoffmaster KA, Ishikawa T, Keppler D, Kim RB, Lee CA, Niemi M, Polli JW, Sugiyama Y, Swaan PW, Ware JA, Wright SH, Yee SW, Zamek-Gliszczynski MJ, Zhang L. 2010. Membrane transporters in drug development. Nat Rev Drug Discov 9:215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chigutsa E, Visser ME, Swart EC, Denti P, Pushpakom S, Egan D, Holford NH, Smith PJ, Maartens G, Owen A, McIlleron H. 2011. The SLCO1B1 rs4149032 polymorphism is highly prevalent in South Africans and is associated with reduced rifampin concentrations: dosing implications. Antimicrob Agents Chemother 55:4122–4127. doi: 10.1128/AAC.01833-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiner M, Peloquin C, Burman W, Luo CC, Engle M, Prihoda TJ, Mac Kenzie WR, Bliven-Sizemore E, Johnson JL, Vernon A. 2010. Effects of tuberculosis, race, and human gene SLCO1B1 polymorphisms on rifampin concentrations. Antimicrob Agents Chemother 54:4192–4200. doi: 10.1128/AAC.00353-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramesh K, Hemanth Kumar AK, Kannan T, Vijayalakshmi R, Sudha V, Manohar Nesakumar S, Bharathiraja T, Lavanya J, Swaminathan S, Ramachandran G. 2016. SLCO1B1 gene polymorphisms do not influence plasma rifampicin concentrations in a South Indian population. Int J Tuberc Lung Dis 20:1231–1235. doi: 10.5588/ijtld.15.1007. [DOI] [PubMed] [Google Scholar]
- 21.Sloan DJ, McCallum AD, Schipani A, Egan D, Mwandumba HC, Ward SA, Waterhouse D, Banda G, Allain TJ, Owen A, Khoo SH, Davies GR. 2017. Genetic determinants of the pharmacokinetic variability of rifampin in Malawian adults with pulmonary tuberculosis. Antimicrob Agents Chemother 61:e00210-17. doi: 10.1128/AAC.00210-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song SH, Chang HE, Jun SH, Park KU, Lee JH, Lee EM, Song YH, Song J. 2013. Relationship between CES2 genetic variations and rifampicin metabolism. J Antimicrob Chemother 68:1281–1284. doi: 10.1093/jac/dkt036. [DOI] [PubMed] [Google Scholar]
- 23.Antwi S, Yang H, Enimil A, Sarfo AM, Gillani FS, Ansong D, Dompreh A, Orstin A, Opoku T, Bosomtwe D, Wiesner L, Norman J, Peloquin CA, Kwara A. 2017. Pharmacokinetics of the first-line antituberculosis drugs in Ghanaian children with tuberculosis with or without HIV coinfection. Antimicrob Agents Chemother 61:e01701-16. doi: 10.1128/AAC.01701-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hein DW, Doll MA. 2012. Accuracy of various human NAT2 SNP genotyping panels to infer rapid, intermediate and slow acetylator phenotypes. Pharmacogenomics 13:31–41. doi: 10.2217/pgs.11.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suarez-Kurtz G, Vargens DD, Sortica VA, Hutz MH. 2012. Accuracy of NAT2 SNP genotyping panels to infer acetylator phenotypes in African, Asian, Amerindian and admixed populations. Pharmacogenomics 13:851–854, 855. doi: 10.2217/pgs.12.48. [DOI] [PubMed] [Google Scholar]
- 26.Cranswick N, Mulholland K. 2005. Isoniazid treatment of children: can genetics help guide treatment? Arch Dis Child 90:551–553. doi: 10.1136/adc.2004.063610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verhagen LM, Coenen MJ, Lopez D, Garcia JF, de Waard JH, Schijvenaars MM, Hermans PW, Aarnoutse RE. 2014. Full-gene sequencing analysis of NAT2 and its relationship with isoniazid pharmacokinetics in Venezuelan children with tuberculosis. Pharmacogenomics 15:285–296. doi: 10.2217/pgs.13.230. [DOI] [PubMed] [Google Scholar]
- 28.Zhu R, Kiser JJ, Seifart HI, Werely CJ, Mitchell CD, D'Argenio DZ, Fletcher CV. 2012. The pharmacogenetics of NAT2 enzyme maturation in perinatally HIV exposed infants receiving isoniazid. J Clin Pharmacol 52:511–519. doi: 10.1177/0091270011402826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwara A, Enimil A, Gillani FS, Yang H, Sarfo AM, Dompreh A, Ortsin A, Osei-Tutu L, Kwarteng Owusu S, Wiesner L, Norman J, Kurpewski J, Peloquin CA, Ansong D, Antwi S. 2016. Pharmacokinetics of first-line antituberculosis drugs using WHO revised dosage in children with tuberculosis with and without HIV coinfection. J Pediatr Infect Dis Soc 5:356–365. doi: 10.1093/jpids/piv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thee S, Seddon JA, Donald PR, Seifar HI, Werely CJ, Hesseling AC, Rosenkranz B, Roll S, Magdorf K, Schaaf HS. 2011. Pharmacokinetics of isoniazid, rifampin, and pyrazinamide in children younger than two years of age with tuberculosis; evidence for implementation of revised World Health Organization recommendations. Antimicrob Agents Chemother 55:5560–5567. doi: 10.1128/AAC.05429-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaid DJ, Batzler AJ, Jenkins GD, Hildebrandt MA. 2006. Exact tests of Hardy-Weinberg equilibrium and homogeneity of disequilibrium across strata. Am J Hum Genet 79:1071–1080. doi: 10.1086/510257. [DOI] [PMC free article] [PubMed] [Google Scholar]