ABSTRACT
Sulbactam is a plausible option for treating Acinetobacter infections because of its intrinsic antibacterial activity against the members of the Acinetobacter genus, but the mechanisms of sulbactam resistance have not been fully studied in Acinetobacter baumannii. In this study, a total of 2,197 clinical A. baumannii isolates were collected from 27 provinces in China. Eighty-eight isolates with various MICs for sulbactam were selected on the basis of their diverse clonality and underwent multilocus sequence typing (MLST), antimicrobial susceptibility testing, and resistance gene screening. The copy number and relative expression of blaTEM-1D and ampC were measured via quantitative PCR and quantitative reverse transcription-PCR, respectively. The genetic structure of multicopy blaTEM-1D was determined using the whole-genome sequencing technology. The cefoperazone-sulbactam resistance rate of the 2,197 isolates was 39.7%. The rate of positivity for blaTEM-1D or ISAba1-ampC in the sulbactam-nonsusceptible group (64.91% and 78.95%, respectively) was significantly higher than that in the sulbactam-susceptible group (0% and 0%, respectively; P < 0.001). The MIC of sulbactam (P < 0.001) varied considerably between the groups expressing ampC with or without upstream ISAba1. Notably, the genetic structure of the multicopy blaTEM-1D gene in strain ZS3 revealed that blaTEM-1D was embedded within four tandem copies of the cassette IS26-blaTEM-1D-Tn3-IS26. Therefore, blaTEM-1D and ISAba1-ampC represent the prevalent mechanism underlying sulbactam resistance in clinical A. baumannii isolates in China. The structure of the four tandem copies of blaTEM-1D first identified may increase sulbactam resistance.
KEYWORDS: Acinetobacter baumannii, sulbactam, blaTEM-1, ISAba1-ampC, multicopy
INTRODUCTION
Acinetobacter baumannii is a notorious pathogen that causes severe nosocomial infections and exhibits a remarkable ability to develop multidrug or even pandrug resistance (1). In recent decades, the rate of multidrug-resistant Acinetobacter baumannii (MDRAB) strains has dramatically increased worldwide; therefore, treatment options are limited (2). Sulbactam is a plausible option for treating carbapenem-resistant Acinetobacter baumannii (CRAB) and MDRAB infections because of its intrinsic antibacterial activity against the members of the Acinetobacter genus (3, 4).
Sulbactam is a common β-lactamase inhibitor that is typically administered in combination with ampicillin or cefoperazone. Recent experimental and clinical studies have demonstrated that sulbactam has a promising effect against CRAB or MDRAB compared with that of colistin and imipenem-cilastatin (5, 6). However, a survey in the United States reported that the rate of resistance to ampicillin-sulbactam increased from 35.2% during the period from 2003 to 2005 to 41.2% during the period from 2009 to 2012 (7). Data from the Chinese surveillance system (CHINET) showed that the rate of Acinetobacter sp. resistance to cefoperazone-sulbactam increased from 25% to approximately 40% from 2005 to 2014 in China (8).
Limited studies have shown that the β-lactamase TEM-1 and an Acinetobacter-derived cephalosporinase (ADC-30) could confer sulbactam resistance to A. baumannii (9, 10). However, their partial contributions and other underlying mechanisms of sulbactam resistance have not been fully studied, especially in strains with a background different from that of A. baumannii as well as strains with different sulbactam MICs.
This study aimed to explore the epidemiological characteristics and contributions of known mechanisms of sulbactam resistance in A. baumannii strains with diverse genetic backgrounds in China, and we discovered the first clinical strain of A. baumannii with four tandem chromosomal copies of blaTEM-1D originating from China.
RESULTS AND DISCUSSION
Epidemiology of sulbactam-resistant isolates in Chinese hospitals.
A total of 2,197 A. baumannii clinical isolates were collected from 27 provinces and areas representing a broad regional distribution in China. The resistance rate of all A. baumannii isolates was 39.7% for cefoperazone-sulbactam, 76.6% for imipenem, 78.5% for meropenem, 86.8% for amikacin, and 93.3% for cefepime. Compared with the rate of resistance to sulbactam in combination with other compounds in other countries during the same period, the rate of resistance in China was similar to that in the United States (41.2%) from 2009 to 2012 (7). However, the rate in China was slightly lower than that in Vietnam (66.7%) from 2009 to 2011 (11).
According to the temporary susceptibility breakpoint for sulbactam in A. baumannii (≤4 mg/liter), 57 and 31 isolates were obtained from the sulbactam-nonsusceptible and sulbactam-susceptible groups, respectively. The clinical information and sequence types (STs) for these isolates are provided in Table S2 in the supplemental material. These isolates were distributed in 11 geographically separated provinces in China (Fig. 1). The STs based on the Oxford and Pasteur schemes are listed in the Table S2. The MIC of most of these strains was 32 mg/liter, and the MICs of sulbactam ranged from 0.25 mg/liter to 256 mg/liter.
FIG 1.
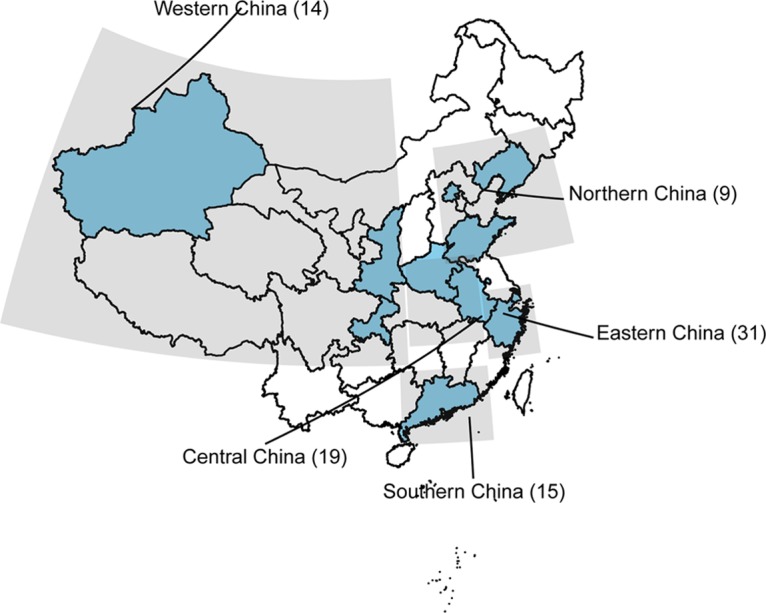
Geographical distribution of the strains collected from 11 different provinces (shaded blue). The numbers in the parentheses represent the numbers of strains.
Prevalence and contribution of blaTEM-1D to sulbactam resistance.
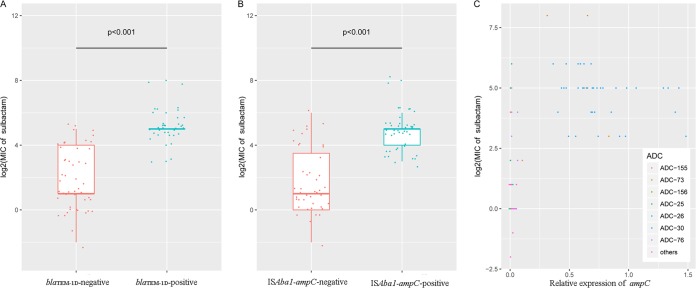
Of the 88 clinical strains in the study, 37 (42.05%) isolates tested positive for blaTEM-1D. All blaTEM-1D-positive strains were identified in the group without susceptibility to sulbactam. A statistically significant difference in the rate of positivity for blaTEM-1D was noted between the sulbactam-nonsusceptible group and the sulbactam-susceptible group (64.91% versus 0%, P < 0.001). Furthermore, when all strains were divided into two groups (strains with blaTEM-1D and strains without blaTEM-1D), the MIC of sulbactam for strains with blaTEM-1D was significantly increased compared with that for strains without blaTEM-1D (P < 0.001) (Fig. 2A). These results indicate that the presence of blaTEM-1D could play an important role in the development of sulbactam resistance in A. baumannii, which is consistent with the findings of a previous study (9).
FIG 2.
Roles of blaTEM-1D and ISAba1-ampC in sulbactam resistance. (A) Comparison of the MICs of sulbactam (in milligrams per liter) between the groups harboring blaTEM-1D or not; (B) comparison of the MICs of sulbactam between the groups with or without ISAba1-ampC; (C) comparison of sulbactam resistance and the relative expression of ampC between the different ADC protein variants.
To evaluate the correlation between the sulbactam MIC and blaTEM-1D expression, we performed quantitative reverse transcription-PCR (qRT-PCR) to assess blaTEM-1D expression. The results are presented in Table S2. In general, a moderate association between the expression of blaTEM-1D and the MIC of sulbactam (log2) was observed in the A. baumannii strains (r = 0.541; P < 0.001), although this correlation was smaller than that reported in a previous study (r = 0.92) (9). Additionally, the levels of blaTEM-1D expression were similar for all blaTEM-1D-positive strains (P > 0.05) except strain ZS3, which exhibited an increased relative expression of blaTEM-1D (11.38 ± 4.76) compared with that of the other strains (P < 0.001). Consistent with this finding, the sulbactam MIC of strain ZS3 was the highest (256 mg/liter) among all the clinical isolates tested. However, for most of the isolates, the MICs of sulbactam for the blaTEM-1D-positive strains, except for strain ZS3, ranged from 8 to 256 mg/liter, and these strains presented the same level of blaTEM-1D expression, suggesting that additional mechanisms contribute to sulbactam resistance in A. baumannii. For example, overexpression of other β-lactamases, mutations of penicillin-binding proteins, inactivation or downregulation of porins, and overexpression of the efflux pump are common factors that lead to β-lactam resistance in A. baumannii (1, 10, 12).
The promoter and the copy number are two common parameters associated with the mRNA expression of resistance genes. Therefore, the promoter region was also amplified and sequenced. Promoter P4 was the only promoter type in all blaTEM-1D-positive strains. P4, which is upstream of the blaTEM genes, contributes to increases in the MICs of β-lactams for Escherichia coli according to the findings of a previous study (13). Regarding the blaTEM-1D copy number, all blaTEM-1D-positive strains except strain ZS3 possessed one copy of blaTEM-1D; strain ZS3 carried at least two copies of blaTEM-1D. Therefore, the multicopy blaTEM-1D gene is a likely factor contributing to the high level of sulbactam resistance in strain ZS3 because it increases the production of the β-lactamase TEM-1D.
Structure and mechanism of multicopy blaTEM-1D.
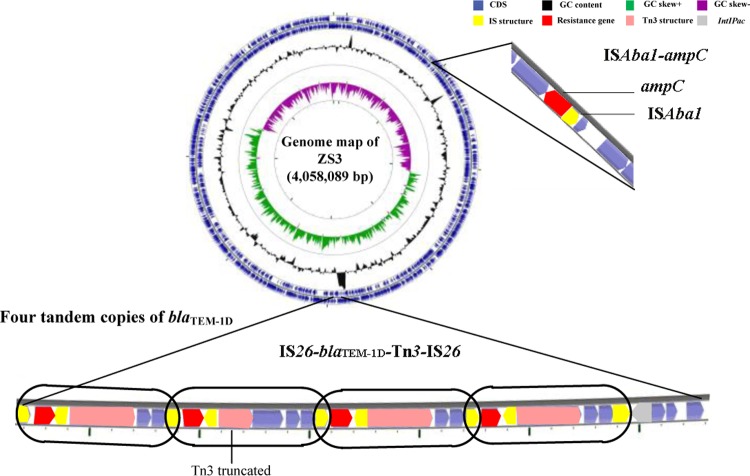
To investigate the structure and mechanism of multicopy blaTEM-1D, the whole genome of strain ZS3 (which carries multiple copies of blaTEM-1D) was sequenced using the single-molecule real-time (SMRT) sequencing technology. Interestingly, the blaTEM-1D gene was identified in the chromosome of strain ZS3 and embedded within a long quartic-duplicated fragment (7.6 kb) (Fig. 3). The duplicated fragment consisted of a cluster of genes (IS26-blaTEM-1D-Tn3-IS26), which is equivalent to that noted in other strains with single copies of blaTEM-1D. Four contiguous duplicated units were arranged together in the same direction and shared the same internal element, namely, IS26.
FIG 3.
Circular map of the A. baumannii ZS3 genome and the gene environment of multiple copies of blaTEM-1D and ISAba1-ampC. The genome of A. baumannii ZS3 is represented by the outer blue circle. The GC content is represented by the black circle, and the GC skewed circle is the inner circle represented in green and purple. Blue arrows, coding sequences (CDS); red arrows, resistance genes; yellow rectangles, insertion sequence (IS) structures; pink arrows, Tn3 structures; gray arrow, IntIPac. The blaTEM-1D repeat fragment is highlighted with a black frame.
This is the first study to discover that four tandem copies of blaTEM-1D are detected in the chromosome of A. baumannii. The mechanism of tandem gene amplification was revealed in a previous review (14). Gene duplication and amplification (GDA) were achieved by nonequal homologous recombination, in which the two flanking IS26 elements provided homology for recombination. The IS26 element was formed at the joint between the amplified units.
Chromosomal gene amplification can lead to increased antibiotic resistance by increasing the expression of a modifying or degrading enzyme, which explains why strain ZS3 exhibited a higher level of blaTEM-1D gene expression (11.38 ± 4.76) and the highest sulbactam MIC (256 mg/liter). However, the correlation between the copy number and the MIC of sulbactam could not be verified because of a lack of a sufficient number of strains harboring multiple copies of the blaTEM-1D gene.
Prevalence of ISAba1-blaADC and contribution of ISAba1-blaADC to sulbactam resistance.
All 88 clinical strains tested possessed the ADC β-lactamase, and 41 (46.59%) isolates harbored ADC-30 (ampC allele 2). In addition to ADC-30, other types of ADC protein variants were found, including ADC-76 (ampC allele 10 [ampC-10], 12.50%), ADC-155 (ampC-36, 5.68%), ADC-25 (ampC-19, 5.68%), ADC-26 (ampC-56, 5.68%), ADC-73 (ampC-20, 4.55%), ADC-156 (ampC-52, 3.41%), and others. Of these, some novel ampC alleles and ADC protein variants (such as ADC-154, -155, -156, and -157) were identified and submitted to the GenBank database (accession and ID numbers are listed in Table S2 in the supplemental material). Notably, ADC-30 and ADC-73 were found only in the sulbactam-nonsusceptible group, whereas ADC-76, ADC-155, and ADC-25 were detected in the sulbactam-nonsusceptible and sulbactam-susceptible groups. The other ADC protein variants were present only in the sulbactam-susceptible group.
The correlation between the ampC alleles and STs based on the Pasteur multilocus sequence typing scheme is listed in Table S3, and the single ampC allele type was not included. The predominant allele, ampC-2, was present in the isolates of clonal complex 2 (CC2) and ST215. Additionally, isolates from CC2 had other ampC alleles, alleles 2, 19, 20, and 28, which were similar to those found in a previous study (15). In addition, ampC allele 10 was detected in all CC10 isolates (ST10 and ST23), but also in two isolates of other STs. The linkages between the ampC alleles and STs could provide the foundation for searching for clinically important clones of A. baumannii. ADCs, which are also known as AmpC cephalosporinases, are inherent to A. baumannii strains and are chromosomally encoded (1, 16, 17). Furthermore, the basal level of ampC expression does not reduce the efficacy of expanded-spectrum cephalosporins (18), and the relationship between ampC expression and the efficacy of sulbactam remains unknown.
To assess the correlation between the sulbactam MIC and ampC expression, we measured ampC expression via qRT-PCR. The results illustrated that the relative expression of ampC was significantly lower in the sulbactam-susceptible group than in the sulbactam-nonsusceptible group (P < 0.001). The level of ampC expression was moderately correlated with the MIC of sulbactam (log2) in the A. baumannii isolates (r = 0.553; P < 0.001). In Fig. 2C, the dots which represent ADC-30 and ADC-73 are located in the top center and are clearly separated from the dots for the other ADC protein variants. The MIC of sulbactam (log2) and the level of ampC expression in the ADC-30 and ADC-73 groups were increased compared with those in the groups of the remaining ADC protein variant types (P < 0.001). In addition, previous studies have shown that the AmpC β-lactamase, which is located on the chromosome of A. baumannii, is noninducible, in contrast to other Gram-negative organisms, because of the absence of the ampR gene (1, 17, 19); therefore, induction experiments with sulbactam were not performed.
The data from the copy number assay demonstrated that all isolates possessed only one copy of ampC. Interestingly, the insertion element ISAba1, associated with ADC-30 (ampC-2) in a previous study (10), was identified only upstream of ampC-2 and ampC-20 and was not identified to be present in combination with the other ampC alleles in this study. The surrounding region of ampC with ISAba1 is presented in Fig. 3. The rate of ISAba1-ampC-positive strains in the sulbactam-nonsusceptible group was significantly increased compared with that in the sulbactam-susceptible group (78.95% versus 0%, P < 0.001). To explore the role of ISAba1, we determined the differences between the relative expression of ampC and the MIC of sulbactam in groups with and groups without ISAba1. There was a notable difference in the expression of ampC (P < 0.001) and the MIC of sulbactam (log2) (P < 0.001) (Fig. 2B) between the two groups.
A previous study illustrated that ISAba1 provides strong promoter sequences (20), which results in ampC overexpression and could explain why in our study the expression of ampC-2 and ampC-20 with ISAba1 was significantly increased compared with that of the other allele types of ampC without ISAba1. Accordingly, ampC-2 overexpression contributed to sulbactam resistance in A. baumannii, as described in a previous study (10). In addition, other ampC alleles, such as ampC-20, could also exert this effect with the ISAba1 upstream sequence. The role of ampC in sulbactam resistance is related to the presence of ISAba1, which can regulate ampC overexpression. Therefore, exploring a method for detecting ISAba1-ampC rather than detecting ADC-30 alone is suitable for surveillance of sulbactam resistance in clinical A. baumannii isolates.
In summary, both blaTEM-1D and ISAba1-ampC are prevalent factors involved in the sulbactam resistance mechanism of A. baumannii isolates, based on their polyclonal background in China. This report is the first to identify four tandem copies of blaTEM-1D located on the A. baumannii chromosome, and the multiplication of blaTEM-1D may enhance the sulbactam resistance level by increasing the expression of the β-lactamase TEM-1D. ISAba1, which is associated with ampC, plays a key role in sulbactam resistance by upregulating ampC expression.
MATERIALS AND METHODS
Strains.
A total of 2,197 nonduplicate clinical A. baumannii isolates were collected from 64 central Chinese hospitals in 27 provinces from January 2009 to September 2010 (21). Eighty-eight isolates were selected from the nationwide survey program based on their different types according to multilocus sequence typing (MLST), which was performed using the Oxford scheme and the Pasteur scheme, as previously described (22, 23). Several novel alleles and profiles were assigned using the PubMLST database (http://pubmlst.org/abaumannii/). All isolates were identified to the species level using a PCR targeting the 16S-23S rRNA gene intergenic spacer region and the partial RNA polymerase β-subunit (rpoB) gene (24–26).
Antimicrobial susceptibility testing.
The MICs of sulbactam were determined using the disk diffusion method and the broth microdilution method according to the guidelines provided by the Clinical and Laboratory Standards Institute (CLSI) (27). On the basis of the CLSI susceptibility breakpoint for ampicillin-sulbactam (≤8/4 mg/liter) in A. baumannii, we adopted ≤4 mg/liter as the temporary susceptibility breakpoint because of the lack of a clinical breakpoint for sulbactam alone (27). The interpretive criterion of susceptibility for ampicillin-sulbactam (zone diameter ≥ 15 mm) in A. baumannii was adopted as the breakpoint for cefoperazone-sulbactam in the disk diffusion method.
PCR and sequencing.
Isolates were screened for the presence of blaTEM and ampC by PCR using the blaTEM-specific-C1/C2 and blaADC-F/R primers (see Table S1 in the supplemental material) (28). For the positive strains, a second PCR was performed using the primers TEM Promo F/R and ISAba1-blaADC-F/blaADC-R to amplify the full-length sequence containing the promoter region (29). The alleles of ampC and the ADC protein variants were identified in the database for ampC alleles in A. baumannii, which is available at the PubMLST platform for A. baumannii (http://pubmlst.org/abaumannii/) (15). The novel alleles and variants have been submitted to the GenBank database.
Quantitative PCR (qPCR) and quantitative reverse transcription-PCR (qRT-PCR).
To detect the blaTEM and ampC copy numbers, we performed qPCR using a ViiA7 real-time PCR system (Life Technologies) and a SYBR Premix Ex Taq PCR kit (TaKaRa Bio, Japan). Primers targeting the blaTEM (primers RT TEM-F/R), ampC (primers qPCR for ADC-F/R), and rpoB (primers qPCR for rpoB-F/R) genes were used as previously described and are listed in Table S1 (9, 10, 30). Each reaction mixture contained a total volume of 10 μl with 2 ng of genomic DNA as a template, 100 nmol/liter of each primer, and 1× SYBR green PCR master mix (Applied Biosystems, Carlsbad, CA, USA). Based on its complete genome sequence (GenBank accession number CP014541), strain XH856 possesses a single copy of blaTEM and ampC and was used as the control strain. The relative copy number was calculated using the comparative threshold cycle (CT) method (2−ΔΔCT method) (31, 32). Triplicate samples were included in each run, and all qPCRs were performed three times. The definition of multiple copies relies on the one-sample t test, as noted in a previous study (33).
The expression of blaTEM and ampC was quantified using the same primers used for the qPCR to detect the copy numbers of blaTEM and ampC. RNA was extracted with the RNAprotect Bacteria reagent and an RNeasy minikit (Qiagen, Valencia, CA, USA) and was reverse transcribed into single-stranded cDNA with random hexamer transcriptase (TaKaRa, Japan). Each isolate was tested three times. The level of expression of blaTEM and ampC relative to that of the rpoB gene was calculated as previously described (31).
Whole-genome sequencing and annotation.
A. baumannii strain ZS3 was cultured to the mid-logarithmic phase in 50 ml of LB broth at 37°C. DNA was extracted using a QIAamp DNA minikit (Qiagen, Valencia, CA) and was further purified using a PowerClean DNA cleanup kit (Mo Bio Laboratories, Carlsbad, CA) according to the manufacturer's recommendations. The genome of the strain was sequenced by the Tianke Company (Zhejiang, China). Single-molecule real-time (SMRT) sequencing reads were generated using a PacBio RS II platform (Pacific Biosciences, Menlo Park, CA) and subsequently de novo assembled and resequenced using the PacBio Hierarchical Genome Assembly Process workflow available in the SMRT Analysis (version 2.3.0)/Quiver software package.
The assembled genome was annotated using the NCBI Prokaryotic Genome Automatic Annotation Pipeline (PGAAP) and manually assessed by use of the BLASTX (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and InterProScan (34) programs. Antimicrobial resistance genes were detected by use of the ResFinder tool (https://cge.cbs.dtu.dk/services/ResFinder/). Whole genomes were compared using the Artemis comparison tool (version 11.1.1). In silico MLST analyses of the whole-genome sequencing data were performed using the BacWGSTdb server (35). Graphical maps, sequence features, base composition plots, analysis results, and sequence similarity plots were generated using the CGView server (http://stothard.afns.ualberta.ca/cgview_server/) (36).
Statistical analysis.
Categorical variables were compared using chi-square analysis or Fisher's exact test as appropriate. The normal distribution of continuous variables was determined using the Kolmogorov-Smirnov test and histograms. Continuous variables with normally distributed data were compared by analysis of variance (ANOVA) or a t test, whereas Wilcoxon rank-sum tests or Kruskal-Wallis tests were used for nonnormally distributed data. Correlations between the variables were determined using Spearman's correlation test.
Accession number(s).
The genome of strain A. baumannii ZS3 had been submitted to GenBank under accession number CP021496, and the accession numbers of the ampC alleles and the ID numbers of the ADC protein variants are listed in Table S2.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (grant no. 81230039, 81501777, 31670135, 81371858, 81401698, and 81501778), the Medical Scientific Research Foundation of Zhejiang Province, China (2017KY404), the Natural Science Foundation of Zhejiang Province, China (LY15H190004), and the Zhejiang Province Medical Platform Backbone Talent Plan (2016DTA003).
We have no competing interests to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01947-17.
REFERENCES
- 1.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vila J, Pachon J. 2008. Therapeutic options for Acinetobacter baumannii infections. Expert Opin Pharmacother 9:587–599. doi: 10.1517/14656566.9.4.587. [DOI] [PubMed] [Google Scholar]
- 3.Oliveira MS, Prado GV, Costa SF, Grinbaum RS, Levin AS. 2008. Ampicillin/sulbactam compared with polymyxins for the treatment of infections caused by carbapenem-resistant Acinetobacter spp. J Antimicrob Chemother 61:1369–1375. doi: 10.1093/jac/dkn128. [DOI] [PubMed] [Google Scholar]
- 4.Levin AS. 2002. Multiresistant Acinetobacter infections: a role for sulbactam combinations in overcoming an emerging worldwide problem. Clin Microbiol Infect 8:144–153. doi: 10.1046/j.1469-0691.2002.00415.x. [DOI] [PubMed] [Google Scholar]
- 5.Tripodi MF, Durante-Mangoni E, Fortunato R, Utili R, Zarrilli R. 2007. Comparative activities of colistin, rifampicin, imipenem and sulbactam/ampicillin alone or in combination against epidemic multidrug-resistant Acinetobacter baumannii isolates producing OXA-58 carbapenemases. Int J Antimicrob Agents 30:537–540. doi: 10.1016/j.ijantimicag.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Wood GC, Hanes SD, Croce MA, Fabian TC, Boucher BA. 2002. Comparison of ampicillin-sulbactam and imipenem-cilastatin for the treatment of Acinetobacter ventilator-associated pneumonia. Clin Infect Dis 34:1425–1430. doi: 10.1086/340055. [DOI] [PubMed] [Google Scholar]
- 7.Zilberberg MD, Kollef MH, Shorr AF. 2016. Secular trends in Acinetobacter baumannii resistance in respiratory and blood stream specimens in the United States, 2003 to 2012: a survey study. J Hosp Med 11:21–26. doi: 10.1002/jhm.2477. [DOI] [PubMed] [Google Scholar]
- 8.Hu FP, Guo Y, Zhu DM, Wang F, Jiang XF, Xu YC, Zhang XJ, Zhang CX, Ji P, Xie Y, Kang M, Wang CQ, Wang AM, Xu YH, Shen JL, Sun ZY, Chen ZJ, Ni YX, Sun JY, Chu YZ, Tian SF, Hu ZD, Li J, Yu YS, Lin J, Shan B, Du Y, Han Y, Guo S, Wei LH, Wu L, Zhang H, Kong J, Hu YJ, Ai XM, Zhuo C, Su DH, Yang Q, Jia B, Huang W. 2016. Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005–2014. Clin Microbiol Infect 22(Suppl 1):S9–S14. doi: 10.1016/j.cmi.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Krizova L, Poirel L, Nordmann P, Nemec A. 2013. TEM-1 beta-lactamase as a source of resistance to sulbactam in clinical strains of Acinetobacter baumannii. J Antimicrob Chemother 68:2786–2791. doi: 10.1093/jac/dkt275. [DOI] [PubMed] [Google Scholar]
- 10.Kuo SC, Lee YT, Yang Lauderdale TL, Huang WC, Chuang MF, Chen CP, Su SC, Lee KR, Chen TL. 2015. Contribution of Acinetobacter-derived cephalosporinase-30 to sulbactam resistance in Acinetobacter baumannii. Front Microbiol 6:231. doi: 10.3389/fmicb.2015.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biedenbach DJ, Bouchillon SK, Hoban DJ, Hackel M, Phuong DM, Nga TT, Phuong NT, Phuong TT, Badal RE. 2014. Antimicrobial susceptibility and extended-spectrum beta-lactamase rates in aerobic gram-negative bacteria causing intra-abdominal infections in Vietnam: report from the Study for Monitoring Antimicrobial Resistance Trends (SMART 2009-2011). Diagn Microbiol Infect Dis 79:463–467. doi: 10.1016/j.diagmicrobio.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Chiu CH, Lee HY, Tseng LY, Chen CL, Chia JH, Su LH, Liu SY. 2010. Mechanisms of resistance to ciprofloxacin, ampicillin/sulbactam and imipenem in Acinetobacter baumannii clinical isolates in Taiwan. Int J Antimicrob Agents 35:382–386. doi: 10.1016/j.ijantimicag.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 13.Lartigue MF, Leflon-Guibout V, Poirel L, Nordmann P, Nicolas-Chanoine MH. 2002. Promoters P3, Pa/Pb, P4, and P5 upstream from bla(TEM) genes and their relationship to beta-lactam resistance. Antimicrob Agents Chemother 46:4035–4037. doi: 10.1128/AAC.46.12.4035-4037.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandegren L, Andersson DI. 2009. Bacterial gene amplification: implications for the evolution of antibiotic resistance. Nat Rev Microbiol 7:578–588. doi: 10.1038/nrmicro2174. [DOI] [PubMed] [Google Scholar]
- 15.Karah N, Jolley KA, Hall RM, Uhlin BE. 2017. Database for the ampC alleles in Acinetobacter baumannii. PLoS One 12:e0176695. doi: 10.1371/journal.pone.0176695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hujer KM, Hamza NS, Hujer AM, Perez F, Helfand MS, Bethel CR, Thomson JM, Anderson VE, Barlow M, Rice LB, Tenover FC, Bonomo RA. 2005. Identification of a new allelic variant of the Acinetobacter baumannii cephalosporinase, ADC-7 beta-lactamase: defining a unique family of class C enzymes. Antimicrob Agents Chemother 49:2941–2948. doi: 10.1128/AAC.49.7.2941-2948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bou G, Martinez-Beltran J. 2000. Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC beta-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother 44:428–432. doi: 10.1128/AAC.44.2.428-432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poirel L, Nordmann P. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect 12:826–836. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 19.Jacoby GA. 2009. AmpC beta-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heritier C, Poirel L, Nordmann P. 2006. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin Microbiol Infect 12:123–130. doi: 10.1111/j.1469-0691.2005.01320.x. [DOI] [PubMed] [Google Scholar]
- 21.Ruan Z, Chen Y, Jiang Y, Zhou H, Zhou Z, Fu Y, Wang H, Wang Y, Yu Y. 2013. Wide distribution of CC92 carbapenem-resistant and OXA-23-producing Acinetobacter baumannii in multiple provinces of China. Int J Antimicrob Agents 42:322–328. doi: 10.1016/j.ijantimicag.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 22.Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodriguez-Valera F. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol 43:4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamidian M, Nigro SJ. 2017. Problems with the Oxford multilocus sequence typing scheme for Acinetobacter baumannii: do sequence type 92 (ST92) and ST109 exist? J Clin Microbiol 55:2287–2289. doi: 10.1128/JCM.00533-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen TL, Siu LK, Wu RC, Shaio MF, Huang LY, Fung CP, Lee CM, Cho WL. 2007. Comparison of one-tube multiplex PCR, automated ribotyping and intergenic spacer (ITS) sequencing for rapid identification of Acinetobacter baumannii. Clin Microbiol Infect 13:801–806. doi: 10.1111/j.1469-0691.2007.01744.x. [DOI] [PubMed] [Google Scholar]
- 25.Chang HC, Wei YF, Dijkshoorn L, Vaneechoutte M, Tang CT, Chang TC. 2005. Species-level identification of isolates of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J Clin Microbiol 43:1632–1639. doi: 10.1128/JCM.43.4.1632-1639.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemec A, Krizova L, Maixnerova M, van der Reijden TJ, Deschaght P, Passet V, Vaneechoutte M, Brisse S, Dijkshoorn L. 2011. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res Microbiol 162:393–404. doi: 10.1016/j.resmic.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing:26th ed Informational supplement M100-S26. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 28.Nemec A, Krizova L, Maixnerova M, Diancourt L, van der Reijden TJ, Brisse S, van den Broek P, Dijkshoorn L. 2008. Emergence of carbapenem resistance in Acinetobacter baumannii in the Czech Republic is associated with the spread of multidrug-resistant strains of European clone II. J Antimicrob Chemother 62:484–489. doi: 10.1093/jac/dkn205. [DOI] [PubMed] [Google Scholar]
- 29.Waltner-Toews RI, Paterson DL, Qureshi ZA, Sidjabat HE, Adams-Haduch JM, Shutt KA, Jones M, Tian GB, Pasculle AW, Doi Y. 2011. Clinical characteristics of bloodstream infections due to ampicillin-sulbactam-resistant, non-extended-spectrum-beta-lactamase-producing Escherichia coli and the role of TEM-1 hyperproduction. Antimicrob Agents Chemother 55:495–501. doi: 10.1128/AAC.00797-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hornsey M, Ellington MJ, Doumith M, Thomas CP, Gordon NC, Wareham DW, Quinn J, Lolans K, Livermore DM, Woodford N. 2010. AdeABC-mediated efflux and tigecycline MICs for epidemic clones of Acinetobacter baumannii. J Antimicrob Chemother 65:1589–1593. doi: 10.1093/jac/dkq218. [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Lee C, Kim J, Shin SG, Hwang S. 2006. Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J Biotechnol 123:273–280. doi: 10.1016/j.jbiotec.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 33.Hua X, Shu J, Ruan Z, Yu Y, Feng Y. 2016. Multiplication of blaOXA-23 is common in clinical Acinetobacter baumannii, but does not enhance carbapenem resistance. J Antimicrob Chemother 71:3381–3385. doi: 10.1093/jac/dkw310. [DOI] [PubMed] [Google Scholar]
- 34.Mulder N, Apweiler R. 2007. InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol Biol 396:59–70. doi: 10.1007/978-1-59745-515-2_5. [DOI] [PubMed] [Google Scholar]
- 35.Ruan Z, Feng Y. 2016. BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res 44:D682–D687. doi: 10.1093/nar/gkv1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grant JR, Stothard P. 2008. The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.