ABSTRACT
G-quadruplexes are DNA or RNA secondary structures that can be formed from guanine-rich nucleic acids. These four-stranded structures, composed of stacked quartets of guanine bases, can be highly stable and have been demonstrated to occur in vivo in the DNA of human cells and other systems, where they play important biological roles, influencing processes such as telomere maintenance, DNA replication and transcription, or, in the case of RNA G-quadruplexes, RNA translation and processing. We report for the first time that DNA G-quadruplexes can be detected in the nuclei of the malaria parasite Plasmodium falciparum, which has one of the most A/T-biased genomes sequenced and therefore possesses few guanine-rich sequences with the potential to form G-quadruplexes. We show that despite this paucity of putative G-quadruplex-forming sequences, P. falciparum parasites are sensitive to several G-quadruplex-stabilizing drugs, including quarfloxin, which previously reached phase 2 clinical trials as an anticancer drug. Quarfloxin has a rapid initial rate of kill and is active against ring stages as well as replicative stages of intraerythrocytic development. We show that several G-quadruplex-stabilizing drugs, including quarfloxin, can suppress the transcription of a G-quadruplex-containing reporter gene in P. falciparum but that quarfloxin does not appear to disrupt the transcription of rRNAs, which was proposed as its mode of action in both human cells and trypanosomes. These data suggest that quarfloxin has potential for repositioning as an antimalarial with a novel mode of action. Furthermore, G-quadruplex biology in P. falciparum may present a target for development of other new antimalarial drugs.
KEYWORDS: malaria, Plasmodium falciparum, G-quadruplex, quarfloxin
INTRODUCTION
Protozoan Plasmodium parasites are the causative agents of human malaria, a disease responsible for widespread morbidity and about half a million deaths each year (1). Most malaria deaths are caused by the species Plasmodium falciparum, although five other Plasmodium species also infect humans. P. falciparum has one of the most A/T-biased genomes ever sequenced, at ∼81% A/T (2). Interestingly, not all Plasmodium species share this feature: the genome of the second major human malaria parasite, P. vivax, is only ∼58% A/T, whereas the A/T content of the genome of the well-studied rodent malaria species, P. berghei, is more similar to that of P. falciparum at ∼78% (3, 4).
The A/T-biased genome in species like P. falciparum results in an extreme paucity of guanine-rich sequences and, hence, of putative G-quadruplex-forming sequences (PQSs) (5). The G-quadruplex is one of many non-double-helical secondary structures that can be formed by DNA, alongside triplexes, hairpins, etc. To form an intramolecular G-quadruplex motif, four tracts of at least three guanines are required in close proximity, in order to form the type of structure shown in Fig. 1A (6). Genome sequences can thus be mined for PQSs by searching for the consensus sequence G3N1–7G3N1–7 G3N1–7G3 (7), although there is increasing evidence that G-quadruplexes may also fold with longer loops between the guanine tracts, with nonguanine bulges and in intermolecular modes that involve both the sense and antisense strands of a DNA sequence (8, 9). Nevertheless, when the P. falciparum genome was analyzed for conventional intramolecular PQSs, only 80 were found in genes or intergenic regions (5, 10). This translates to only one PQS per ∼300 kb of the nontelomeric P. falciparum genome, compared to an average of one PQS per kb in the human genome (7). Several hundred PQSs were also found in the P. falciparum telomeres, which have the repeat sequence GGGTT(T/C)A and are inherently able to form G-quadruplexes (11, 12). Predicting the expected PQS density of a given genome is not straightforward due to variable genome compositions and biased base dyad frequencies, but under the simplest predictive algorithm (a Bernoulli stream of random bases at 81% A/T), a genome with the size and composition of the P. falciparum genome contains no PQSs at all (5); therefore, the maintenance of PQSs in specific genomic regions is likely to be biologically functional.
FIG 1.
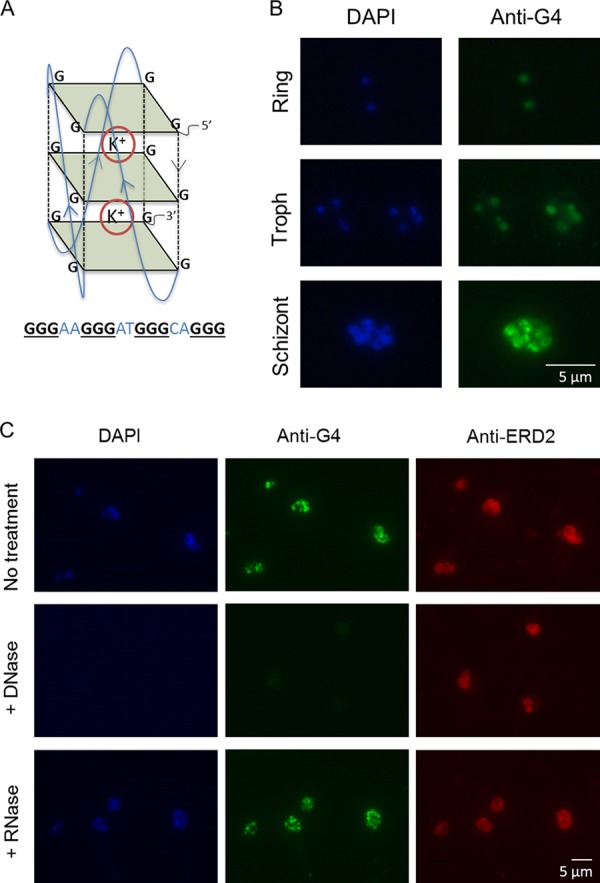
G-quadruplexes can be detected in P. falciparum parasites. (A) Schematic of a G-quadruplex DNA motif. Guanine tetrads are shown as green squares, and guanine backbones are shown as dashed black lines. G-quartets stack on top of one another to form the quadruplex, which is stabilized by cations, such as potassium (K+). An example of a PQS that could fold into such a structure is shown below. (B) Immunofluorescence images showing G-quadruplexes detected with the structure-specific antibody IH6 in P. falciparum intraerythrocytic stages (3D7 parasite strain). Images are representative of those from 3 independent experiments examining mixed-stage cultures. Troph, trophozoite. (C) DNase treatment abolishes G-quadruplex (IH6) staining but has no effect on the distribution of a protein, ERD2, used as a control (middle). RNase treatment has no discernible effect on G-quadruplex (IH6) staining (bottom). Images are representative of those from 3 independent experiments.
G-quadruplexes can have many important biological roles, including regulating the telomere structure (13), inhibiting gene transcription (14), and promoting recombination, probably via stalling of RNA polymerases and replicative polymerases (15, 16). In P. falciparum, we have recently shown that PQSs are primarily clustered within the coding sequence or upstream of the major virulence gene family var and that mitotic recombination events among var genes are strongly spatially associated with PQSs (5), so one role for G-quadruplexes in P. falciparum could be to promote the generation of var gene diversity. Here, we set out to establish experimentally whether G-quadruplexes are more broadly important for the biology of P. falciparum. If so, then interfering with G-quadruplex metabolism might be expected to affect the growth of the parasite, and this would raise the possibility of repositioning G-quadruplex-binding compounds as antimalarial drugs. Many G-quadruplex-binding compounds have been developed in recent years primarily as anticancer agents because several oncogenes, such as c-myc, are controlled by G-quadruplexes (14) and G-quadruplexes are also involved in telomere regulation (13). Thus, drugs that interfere with these processes can affect the viability of cancer cells more severely than that of normal cells. Similarly to rapidly dividing cancer cells, all single-celled eukaryotic pathogens must maintain their telomeres in order to survive as they replicate within their hosts, so telomere disruption would be one possible avenue for the antimalarial activity of G-quadruplex-binding compounds. In fact, some such compounds have already been shown to bind to the P. falciparum telomere repeat (11). Alternatively, antimalarial activity might be achieved by disrupting the transcription or translation of essential G-quadruplex-containing genes or by inducing a DNA damage response to G-quadruplex-stalled replication forks.
In addition to the established potential of G-quadruplex-binding drugs as anticancer agents, a precedent for their potential as antiparasitic agents was recently published. Rudenko and coworkers, in the course of investigating RNA polymerase I inhibitors for their potency against Trypanosoma brucei, showed that the G-quadruplex-binding fluoroquinolone compound quarfloxin is trypanocidal, with a 50% inhibitory concentration of only 155 nM (17). Quarfloxin disrupts RNA polymerase I activity by binding to G-quadruplexes in repetitive rRNA genes in the nucleolus, disrupting their physiological interaction with nucleolin, and, ultimately, preventing rRNA transcription (18, 19). Cancer cells are hypersensitive to RNA polymerase I disruption (20), so quarfloxin was originally tested against neuroendocrine tumors and leukemias, but it was found to have poor bioavailability and to accumulate particularly in blood cells (21). The latter property may have reduced its effectiveness against solid tumors, but it raised the possibility of targeting intraerythrocytic malaria parasites instead. We have tested this concept, showing that quarfloxin is indeed a potent and fast-acting antiplasmodial compound in vitro but that its mode of action is probably different from that demonstrated on human cells and trypanosomes.
RESULTS
G-quadruplexes can be detected in P. falciparum nuclei.
The existence of G-quadruplexes in the P. falciparum genome has been predicted bioinformatically (5, 10), and PQSs from this genome have been shown to fold into G-quadruplexes in vitro (10, 11). To visualize G-quadruplex structures in cultured cells, structure-specific antibodies can be used, as demonstrated in mammalian cells (22–24) and ciliate macronuclei (25). Using immunofluorescence microscopy, we tested whether the G-quadruplex-specific antibody IH6 (24) could similarly recognize G-quadruplexes in P. falciparum. Clear nuclear staining was seen in all intraerythrocytic parasite stages (Fig. 1B), and DNase treatment abolished this staining (but not the staining of a parasite protein, ERD2, used as a control). RNase treatment, in contrast, had no observable effect (Fig. 1C). A second independent G-quadruplex-specific antibody, BG4 (23), also gave comparable nuclear staining (see Fig. S1 in the supplemental material). G-quadruplex structures in DNA therefore appear to be consistently present during P. falciparum intraerythrocytic development.
G-quadruplex-stabilizing drugs inhibit the growth of blood-stage malaria parasites.
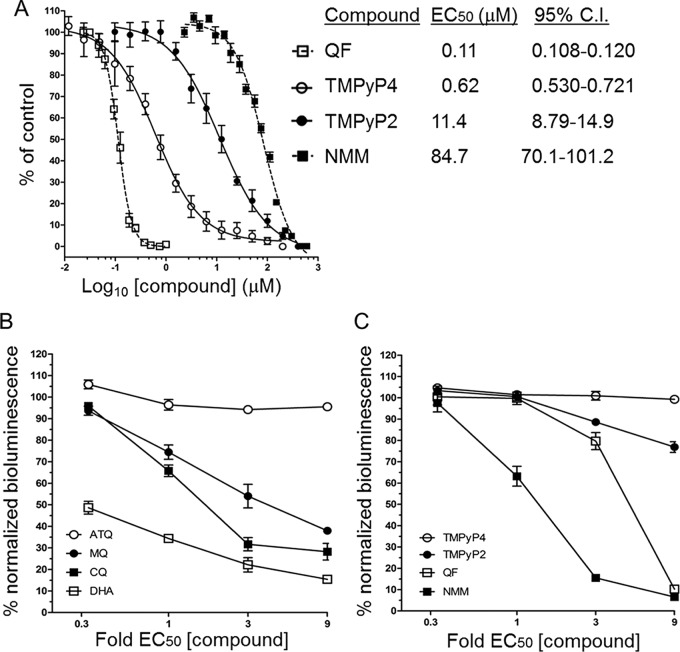
If the G-quadruplexes detected in P. falciparum are important for parasite biology, G-quadruplex stabilization might be expected to affect the growth of the parasite. We therefore tested the effect of several G-quadruplex-stabilizing compounds on the in vitro growth of P. falciparum in a standard 48-h assay encompassing one complete parasite growth cycle (26). The antiplasmodial potency of the compounds N-methyl-mesoporphyrin IX (NMM) (27, 28), 5,10,15,20-tetra-(N-methyl-4-pyridyl)porphine (TMPyP4) (29), and quarfloxin (18) varied widely, with 50% effective concentrations (EC50s) ranging from 114 nM for quarfloxin to 84 μM for NMM (Fig. 2A). This variation could be due to the compounds having different G-quadruplex-stabilizing capacities or various affinities for different G-quadruplex isoforms, but it could also be due to off-target effects (particularly given the high EC50s determined for some of the compounds). Therefore, to focus on G-quadruplex-binding properties alone, we directly compared the antimalarial potency of one of the compounds, TMPyP4, with that of a closely matched analogue, 5,10,15,20-tetra-(N-methyl-2-pyridyl)porphine (TMPyP2), which is structurally very similar to TMPyP4 but which has a markedly lower G-quadruplex affinity via an alternative binding mode (30). Parasite growth was affected ∼18-fold more severely by TMPyP4 than by TMPyP2, suggesting that unimpeded G-quadruplex metabolism does have an important role in the healthy growth of parasites. Furthermore, the drug with the most potent antiplasmodial activity in our panel of drugs was quarfloxin, a drug that has already reached phase 2 clinical trials.
FIG 2.
G-quadruplex-binding drugs inhibit parasite growth. (A) The G-quadruplex-binding drugs NMM, quarfloxin, TMPyP2, and TMPyP4 inhibit the in vitro growth of P. falciparum (3D7 parasite strain) to various extents. Percent parasite growth is plotted as a function of the compound concentration, and error bars are standard errors of the means. The mean EC50s from 3 independent malaria SYBR green I-based fluorescence (MSF) assays are tabulated, and 95% confidence intervals (C.I.) are shown. (B, C) EC50-dependent loss of normalized bioluminescence signal in BRRoK assays against the concentrations of benchmark antimalarials (B) and G-quadruplex-stabilizing compounds (C). The mean bioluminescence signal (normalized against that for an untreated control) remaining in P. falciparum Dd2 luc after a 6-h exposure to the indicated fold EC50 of each drug or compound is plotted. Error bars represent ±SDs from three biological replicates. ATQ, atovaquone; CQ, chloroquine; DHA, dihydroartemisinin; MQ, mefloquine; NMM, N-methyl-mesoporphyrin IX; QF, quarfloxin; TMPyP2, 5,10,15,20-tetra-(N-methyl-2-pyridyl)porphine; TMPyP4, 5,10,15,20-tetra-(N-methyl-4-pyridyl)porphine.
Factors affecting the viability of a compound as a potential antimalarial drug include not only the EC50 but also the rate of kill. Therefore, to further define the antiplasmodial activities of the G-quadruplex-stabilizing compounds, their initial rates of kill were determined in trophozoite intraerythrocytic stages using the bioluminescence relative rate of kill (BRRoK) assay (31). This assay uses a P. falciparum clone genetically engineered to express a luciferase reporter gene under the control of a trophozoite-specific promoter. Thus, it can determine the concentration-dependent effect of a drug upon cell viability by measuring the bioluminescence signal from the rapidly turned over luciferase protein. The loss of bioluminescence is benchmarked against the loss caused by a range of antimalarial compounds with well-defined rate of kill dynamics, shown here as dihydroartemisinin > chloroquine > mefloquine > atovaquone (Fig. 2B). Comparison of the loss of the bioluminescence signal after 6 h of exposure to the G-quadruplex-stabilizing compounds indicated a range of initial rates of kill, with the rank order being NMM > quarfloxin > TMPyP2 = TMPyP4 (Fig. 2C). Relative to the antimalarial benchmarks, NMM exhibited a rapid cytocidal activity comparable to that of chloroquine, while the cytocidal activity of quarfloxin was comparable to that of mefloquine at low concentrations and to that of chloroquine and dihydroartemisinin at high concentrations. Neither TMPyP2 nor TMPyP4 showed cytocidal activity during this 6-h assay, potentially reflecting a lag phase in their antiplasmodial activity (31). These data were consistent with the BRRoK data generated with a second strain of P. falciparum carrying the same luciferase reporter construct (Fig. S2A and B), and the EC50s were likewise consistent across the two different strains (Fig. S2C).
Quarfloxin targets all stages of the erythrocytic life cycle, including the ring stage.
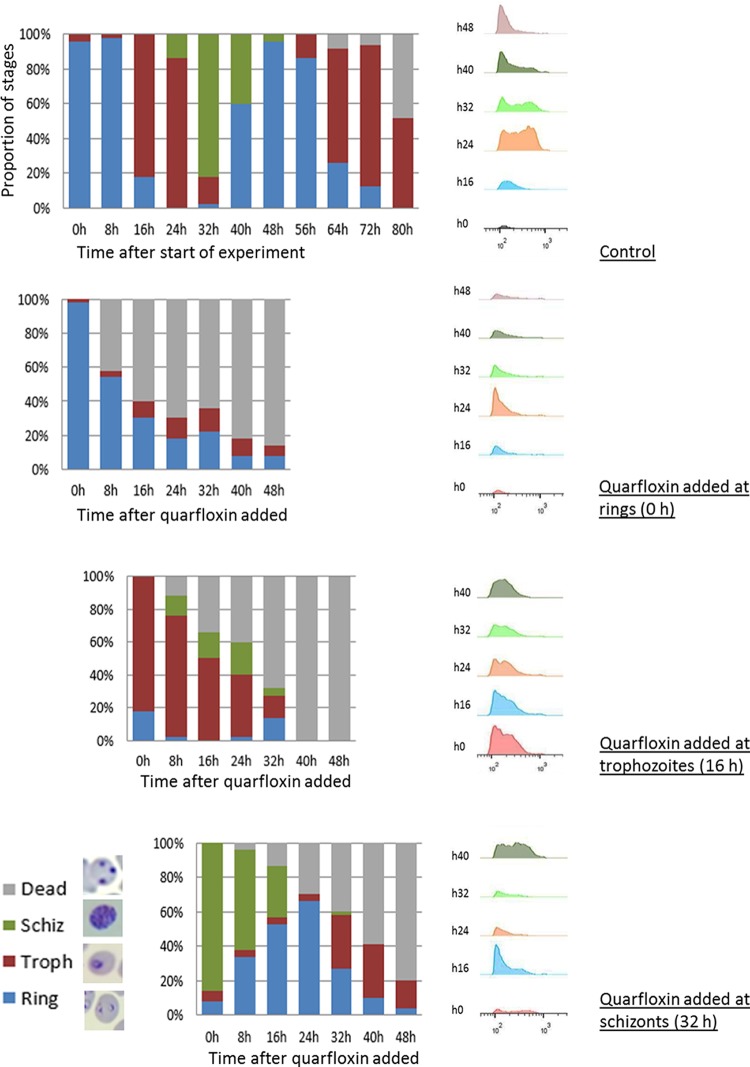
While G-quadruplexes can be detected at all stages of the intraerythrocytic cycle (Fig. 1B), the BRRoK assay effectively measures the rate of kill only in trophozoites because the luciferase reporter gene used in this assay is expressed from a trophozoite-specific promoter (31). We therefore investigated the cytocidal effect of quarfloxin and NMM at other stages of the erythrocytic cycle by visual assessment of synchronized parasites together with flow cytometric measurement of their DNA content. Dead parasites do not synthesize new DNA, they tend to become visibly pyknotic, and they do not progress to the next morphological stage at the expected time point. Figure 3 shows that quarfloxin was highly active against ring stages, preventing the great majority of them from ever progressing to trophozoites. It was almost equally active against trophozoites but less active against schizonts, a substantial proportion of which reinvaded to rings before dying in the next cycle. In contrast, although NMM appeared to be more potent than quarfloxin in the BRRoK assay, it showed little activity against rings, which were largely able to progress to trophozoites (Fig. S3). Corroborating the findings of the BRRoK assay, however, NMM was potent against trophozoites and prevented them from progressing to schizonts. These data point to different modes of antiplasmodial action for these two G-quadruplex-binding compounds.
FIG 3.
Quarfloxin kills ring-stage parasites as well as replicative stages. The developmental stages of 3D7 parasites were assessed visually at 8-h intervals over a single growth cycle, starting at the mid-ring stage, either in the absence of drug or with quarfloxin added at 210 nM (approximately 2 times the EC50) at the ring, trophozoite (Troph), or schizont (Schiz) stage. Parasites were also analyzed at 8-h intervals for DNA content by flow cytometry under each of these conditions. For morphology assessment, 50 parasites were counted at each time point, and the photographs show the representative parasite morphology for each stage; dead parasites stained as a dense intracellular dot without clear morphological features. For flow cytometry, 200,000 parasites were counted in each sample.
Quarfloxin does not affect telomere maintenance.
G-quadruplex-binding compounds may kill cells via a variety of molecular mechanisms, including inducing telomere dysfunction, disrupting DNA replication, or disrupting the transcription of important genes (such as rRNAs, as discussed in the introduction). The disruption of DNA replication seemed an unlikely mode of action for quarfloxin because it was highly potent against prereplicative rings, but both the disruption of gene transcription and an acute response to telomere dysfunction were possibilities. We therefore investigated these possible mechanisms, starting with the possibility of telomere dysfunction because the great majority of the PQSs in the P. falciparum genome are found in telomere repeats (10). Parasites were grown in relatively low levels of quarfloxin (approximate EC50, 120 nM), which could be tolerated for up to 15 growth cycles, but no evidence of telomere shortening or lengthening was detected by telomere restriction fragment Southern blotting after long-term exposure to quarfloxin (Fig. S4).
The mode of action of quarfloxin may involve deregulated expression of PQS-containing genes.
To investigate the possibility that the transcription of essential G-quadruplex-containing genes was deregulated by quarfloxin, we assessed the transcription of a validated G-quadruplex-containing reporter gene and also the transcription of rRNA genes. First, reporter genes were designed and constructed using PQS-encoding genes that appear naturally in the P. falciparum genome. The predominant gene group containing PQSs is the var virulence genes, but these are prohibitively large for cloning and many of the var-associated PQSs actually occur in the var upstream region rather than the coding sequence. Therefore, two genes from a second family of variantly expressed virulence genes were chosen: the rifin genes PF3D7_0700200 and PF3D7_1254400. Only 3 out of ∼200 rifin genes encode PQSs, whereas a larger proportion (30 out of 63) of the var genes encode PQSs (5), but crucially, the rifin genes have small sequences of ∼1 kb that are tractable for cloning. The complete open reading frames of these genes were therefore cloned into expression vectors with C-terminal tags which allowed their transcripts to be distinguished from those of the many other highly homologous rifin genes in the family. PF3D7_0700200 encodes a PQS in the sense orientation, and PF3D7_1254400 encodes a PQS in the antisense orientation. The latter gene proved not to be expressed when transfected into parasites (a common issue with variantly expressed gene families; data not shown), but the other gene, PF3D7_0700200, was successfully transcribed in transfected parasites.
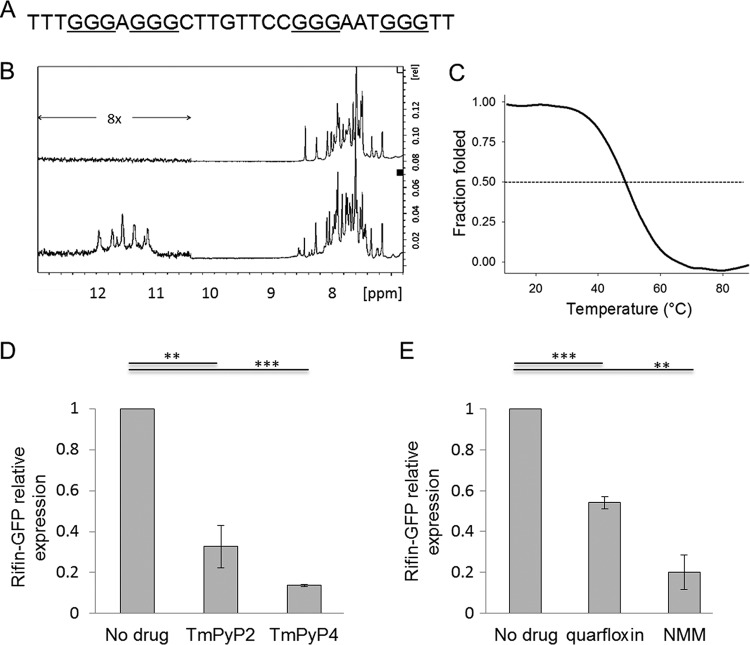
To confirm that the PQS within this reporter gene could indeed form a G-quadruplex structure, 1H nuclear magnetic resonance (NMR) experiments were conducted with a synthetic oligonucleotide from this gene (Fig. 4A), revealing the presence of characteristic imino proton signals (chemical shift, between 10 and 12 ppm) when the oligonucleotide was folded in the presence of potassium (Fig. 4B). The oligonucleotide also showed a 295-nm thermal denaturation curve typical of G-quadruplexes, with half denaturation occurring at 49°C (melting temperature) (Fig. 4C), indicating that the G-quadruplex formed by this sequence is likely to be stable at physiological temperature.
FIG 4.
G-quadruplex-binding drugs affect the expression of a G-quadruplex reporter gene. (A) Oligonucleotide sequence from the gene PF3D7_0700200, used in biophysical assays to confirm the folding of a G-quadruplex structure. (B) 1H NMR spectra, acquired at 37°C in 150 mM Li+ (top) and 150 mM K+ (bottom). Imino proton peaks are present only in K+, in agreement with the inability of Li+ to stabilize G-quadruplex structures. The relatively low intensity of imino peaks under the K+ condition seems to indicate the presence of a floppy structure, potentially the consequence of the second long loop. (C) Stability of the G-quadruplex as evaluated by thermal denaturation, followed at 295 nm. (D) Expression of the GFP-tagged G-quadruplex reporter gene PF3D7_0700200 in transgenic parasite cultures treated with 0.75 μM TMPyP4 or TMPyP2 for 6 h. Expression was determined by quantitative real-time PCR using primer pair GFP_F/R and compared to the average expression of three housekeeping genes encoding actin, seryl-tRNA synthetase, and fructose bisphosphate aldolase. The experiment was carried out in both biological and technical duplicates, error bars are standard errors of the means, and statistical significance was assessed by a 1-tailed t test (**, P < 0.01; ***, P < 0.001). (E) Expression of the GFP-tagged G-quadruplex reporter gene PF3D7_0700200 in transgenic parasite cultures treated with 120 nM quarfloxin or 80 μM NMM for 1 h, determined as described in the legend to panel D.
To confirm that this G-quadruplex structure could be targeted by G-quadruplex-binding compounds, parasites carrying the construct were treated for 6 h with 0.75 μM TMPyP4 or TMPyP2 (0.75 μM is approximately the 48-h EC50 dose of TMPyP4). Because these compounds do not have any immediate cytocidal effect, the 6-h exposure did not cause any visible change in parasite morphology, but it clearly suppressed reporter gene expression relative to the levels of expression of housekeeping genes. TMPyP4, which is the more potent G-quadruplex-binding analog, had a much stronger effect on transcription than TMPyP2 (Fig. 4D). The same assay was then carried out with ∼48-h EC50 doses of quarfloxin and NMM, but owing to the rapid cytocidal effect previously established for these compounds, only 1 h of exposure was used in an attempt to avoid nonspecific results associated with parasite death. Reporter gene expression was markedly suppressed by both quarfloxin and NMM (Fig. 4E).
Having shown that quarfloxin, in common with other G-quadruplex-binding compounds, can indeed affect the expression of a PQS-encoding gene, we turned to the expression of rRNA genes, which was previously shown to be suppressed by quarfloxin in both human cells and trypanosomes (17, 18). Plasmodium does not encode its rRNA in conventional tandem arrays and possesses only a few rRNA genes scattered on various chromosomes (2) (Fig. 5A); these are expressed stage specifically, with A-type genes being used in the blood stages and S-type genes being used in the mosquito stages (32). Therefore, reverse transcription-PCR was conducted for the A-type genes after 1 h of quarfloxin exposure, and the results are shown in Fig. 4E. In contrast to the effect seen in T. brucei, in which rRNA precursor transcripts were dramatically suppressed after just 15 min of quarfloxin exposure (17), there was no loss of these transcripts in P. falciparum; in fact, the transcript level rose somewhat (Fig. 5B), and this effect was broadly mirrored in the level of mature 18S rRNA (Fig. 5C). Thus, the mode of action of quarfloxin in P. falciparum is evidently different from that previously characterized in both T. brucei and human cells.
FIG 5.
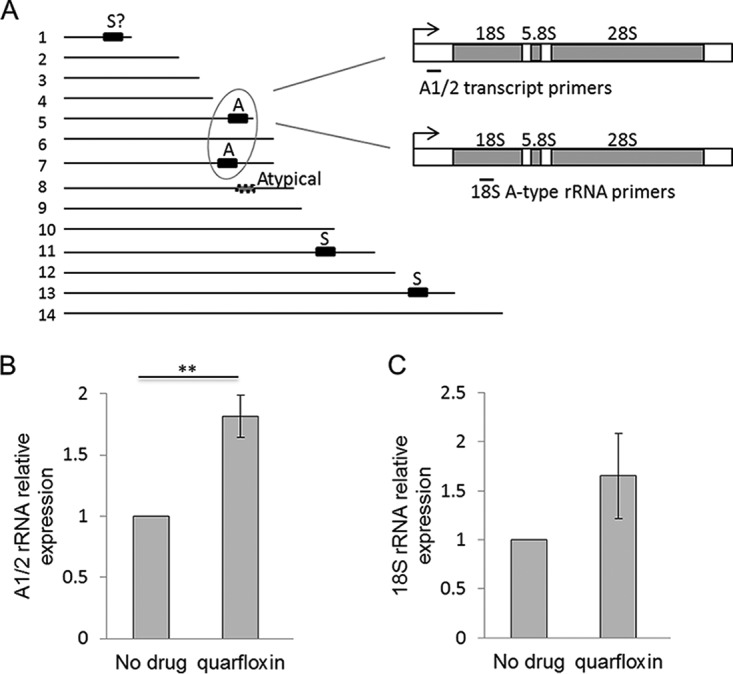
Quarfloxin does not inhibit the expression of rRNA genes. (A) Schematic showing the locations of rRNA-encoding units on the 14 chromosomes of the P. falciparum genome, together with their type (A, blood-stage expressed; S, mosquito-stage expressed). The locations of the primers used to measure rRNA transcription are also shown. (B) Expression of rRNA A1 and A2 transcripts in the same parasites used in the assay whose results are shown in Fig. 4E after treatment with 120 nM quarfloxin for 1 h. Target gene expression relative to that of three control genes was determined as in the assay whose results are shown in Fig. 4; error bars are standard errors of the means, and statistical significance was assessed by a 1-tailed t test (**, P < 0.01). (C) Expression of mature 18S rRNA in the same parasites used in the assay whose results are shown in Fig. 4E after treatment with 120 nM quarfloxin for 1 h.
As a fluoroquinolone, quarfloxin shares limited structural similarity with the well-established 4-aminoquinoline drugs (chloroquine, mefloquine, etc.), which kill Plasmodium parasites by inhibiting hemozoin crystallization in the food vacuole (33). This therefore presented another possible mode of action for quarfloxin, but the drug's high potency against prereplicative ring stages, the fact that its BRRoK profile is distinct from that of chloroquine, and the fact that the drug does not prevent heme crystallization in vitro (Fig. S5) all suggest that quarfloxin does not actually act in the same mode as 4-aminoquinolines. The true target of quarfloxin in Plasmodium parasites remains unknown, but it is apparently neither the heme detoxification pathway (Fig. S5) nor the production of rRNA (Fig. 5).
DISCUSSION
This is, to our knowledge, the first experimental investigation of the biological roles of G-quadruplex motifs in the malaria parasite P. falciparum. We demonstrate that G-quadruplex DNA can be detected in this parasite and that disrupting the normal processing of G-quadruplex motifs impairs parasite growth in vitro. It is important to note that sequences with the potential to form G-quadruplexes are very scarce in the P. falciparum genome and that the G-quadruplexes detected in P. falciparum nuclei using structure-specific antibodies are likely to be primarily in telomeres, since the majority of all PQSs in P. falciparum are tandemly arrayed in telomere repeats (10) and such antibodies probably cannot detect G-quadruplexes at a single-motif resolution (34). (Indeed, the G-quadruplex signal seen in late-stage parasites appears to be strongest in nuclear peripheral foci, as expected of telomeric DNA.) However, it is also important to note that the G-quadruplex-forming potential of the P. falciparum genome may be underestimated by standard in silico algorithms, as was recently demonstrated by sequencing the human genome with a G-quadruplex-sensitive method (8): this empirical approach detected many more G-quadruplexes than the in silico method of scanning the genome for repetitions of G3N1–7 (7). For the P. falciparum genome, a relatively relaxed predictive algorithm was used in our previous study (G3N0–11), and this still found just 80 nontelomeric PQSs (5); however, the algorithm overlooks many of the noncanonical G-quadruplexes detected in the human genome, such as bulged G-quadruplexes (8) and interstrand rather than intrastrand G-quadruplexes (9). Finally, the structure-specific antibody used here detects only DNA and not RNA G-quadruplexes, so the existence of RNA G-quadruplexes in P. falciparum remains to be confirmed.
Several G-quadruplex-stabilizing compounds were shown to inhibit malaria parasite growth, including the phase 2 anticancer drug quarfloxin, which was potent and relatively fast acting against both ring and trophozoite parasites. An ideal antimalarial should kill parasites at all stages of the intraerythrocytic cycle (and, indeed, ideally at nonerythrocytic phases of the life cycle as well). For example, the current first-line antimalarial drug, artemisinin, kills prereplicative ring stages as well as trophozoites and schizonts, whereas older antimalarials, such as chloroquine and antifolates, are highly active only against replicative stages. While it is not essential to elucidate the molecular mechanism(s) of an antiplasmodial compound prior to clinical development—indeed, the mechanism of action of artemisinin remains poorly understood—this information can help in predicting the development of drug resistance and cross-resistance. Therefore, we investigated the mode of action for quarfloxin. We found no evidence that quarfloxin kills parasites by inducing telomere dysfunction, despite the predominance of PQSs in telomere repeats, since telomere maintenance appeared to be normal after up to 15 cycles of growth in quarfloxin. This does not conclusively exclude the possibility of an acutely lethal telomere dysfunction being induced at higher drug exposures, but the tools to assess this in P. falciparum are lacking: the parasite lacks the variant histone γH2AX (35), which is commonly used to detect DNA damage foci, and also lacks all components of the conventional telomere shelterin complex (36).
In the absence of any clear effect on telomeres, we assessed the possibility that quarfloxin might kill parasites by preventing the proper expression of G-quadruplex-containing genes. Forty-one PQSs are predicted in non-var genes (5), many of which may be essential (in contrast, var gene expression is dispensable in vitro [37], albeit it is vital in vivo for virulence phenotypes, such as intravascular adhesion, antigenic variation, and immune evasion). As a proof of concept, a G-quadruplex-encoding rifin reporter gene was cloned, validated, and transfected into parasites, and its expression was shown to be suppressed by all of the G-quadruplex-binding compounds tested. This is the first demonstration of G-quadruplex-dependent gene expression in P. falciparum, and it conforms to a model in which sense-strand G-quadruplexes, when stably formed in DNA, can inhibit transcription by RNA polymerase II.
We then assessed the model for quarfloxin action that was previously reported in trypanosomes and human cells: suppression of rRNA transcription by RNA polymerase I (17, 18). We focused only upon the nuclear rRNA genes and not those encoded in the apicoplast organelle, because interfering with protein synthesis in the apicoplast of P. falciparum gives a characteristic delayed death phenotype, with death occurring only in the second growth cycle after drug addition (38), whereas the parasite death observed here with quarfloxin was very rapid, making apicoplast ribosomes highly unlikely to be a key target.
Although P. falciparum expresses its rRNA genes via RNA polymerase I, it does not maintain these genes in conventional tandem arrays (2), and furthermore, most of the parasite's rRNA genes are not predicted to contain PQSs (5). Only one atypical locus on chromosome 8, PF3D7_0830000, which contains 5.8S and 28S rRNA genes but no gene for 18S rRNA, contains a G3N0–11 motif. It is therefore arguably not surprising that we did not find any evidence for the acute suppression of rRNA transcription by quarfloxin. Instead, another essential gene(s) that encodes PQSs may be targeted by quarfloxin, and the possibility of other G-quadruplex-related modes of action, such as acute induction of DNA damage or telomere dysfunction, cannot yet be ruled out. (It also remains possible that G-quadruplexes are simply not involved; we are currently conducting further work to establish quarfloxin's actual mode of action in P. falciparum, having established that at least one other predictable possibility—the inhibition of hemozoin crystallization—is not consistent with our data.) More generally, the very diverse rates of kill for the different compounds tested here suggest that they may vary not only in their absolute affinity for G-quadruplexes but also in the subset of G-quadruplexes that each compound targets within the genome.
In conclusion, this work shows that G-quadruplex-binding compounds may have the potential to be repositioned as antimalarials. Quarfloxin is potent and fast acting (at least in vitro); it has well-characterized pharmacodynamics in humans, as established from previous cancer trials; and the selective window on P. falciparum is at least 40 times that on human cells (111 nM compared to 4.44 μM on human breast epithelial cells, as reported by Kerry et al. [17]). G-quadruplex biology in malaria parasites thus constitutes an exciting area for future study, both to improve our understanding of basic biology and virulence gene control in this parasite and also to support the discovery of novel targets for antimalarial drug development.
MATERIALS AND METHODS
Parasite culture and drugs.
The 3D7 strain of P. falciparum was obtained from the Malaria Research and Reference Reagent Resource Center (MR4), and the NF54attB strain was obtained from David Fidock. The luciferase-expressing Dd2 strain was previously described (39). Parasites were cultured as previously described (40). Parasite growth and morphology were assessed on blood smears stained with Hemacolor (Merck). Synchronized parasite cultures were obtained by performing two treatments with 5% d-sorbitol (41) 42 h apart, to yield an approximately 4-h window of ring-stage parasites. 3D7 parasites were used in all experiments except the gene expression studies, which were performed in NF54attB, and the BRRoK assays, which were performed in both the NF54 and Dd2 backgrounds. The compounds NMM, TMPyP4, and TMPyP2 were obtained from Frontier Scientific, and quarfloxin was obtained from Tetragene LLC, solubilized in water, and stored without exposure to light at −20°C.
Immunofluorescence assay.
For G-quadruplex detection using the IH6 antibody, air-dried blood smears were fixed with 2% paraformaldehyde–phosphate-buffered saline (PBS) for 2 min and then 90% acetone–10% methanol for 2 min at 4°C. The smears were air dried, blocked with 1% bovine serum albumin–PBS, and sequentially treated with primary and secondary antibodies. The antibodies used were the anti-G-quadruplex monoclonal antibody IH6 (Biorbyt), rabbit polyclonal antibodies to the P. falciparum cis-Golgi marker ERD2 (MRA-1; obtained from MR4 [42]), Cy3-conjugated anti-rabbit IgG (Jackson ImmunoResearch), and Alexa Fluor 488-conjugated anti-mouse IgG (Thermo Fisher Scientific). For enzymatic treatments, slides were incubated after fixation with 0.12 U μl−1 Turbo DNase (Albion) or with 100 μg ml−1 RNase A (Thermo Fisher Scientific) for 1 h at 37°C. Slides were mounted with ProLong Diamond antifade mountant containing DAPI (4′,6-diamidino-2-phenylindole; Thermo Fisher Scientific) and were imaged with an Evos cell imaging system (Thermo Fisher Scientific). Parasites were visualized and images were captured using the same exposure for each fluorochrome across all treatments.
For G-quadruplex detection using the BG4 antibody, parasites were first removed from host erythrocytes using 0.4% saponin and then fixed in 4% paraformaldehyde–PBS. Fixed parasites were dispensed onto a 12-multispot-well slide (Thermo Fisher Scientific) and air dried. All subsequent steps were carried out at room temperature in a 10-μl volume. The parasites were permeabilized with 0.1% Triton X-100 in PBS, washed with PBS, and then blocked in 3% bovine serum albumin in PBS for 1.5 h. Antibody staining steps were done with Flag-tagged BG4 antibody (EMD Millipore) for 1.5 h, rabbit anti-Flag antibody (Cell Signaling Technology) for 1.5 h, and Cy3-conjugated anti-rabbit immunoglobulin secondary antibody (Jackson ImmunoResearch) for 1 h. The slides were washed three times with PBS after each step. Mounting and visualization were as described above.
MSF assay.
Malaria SYBR green I-based fluorescence (MSF) assays were carried out as previously described (26). Trophozoite-stage cultures (75 μl, 1% parasitemia, 4% hematocrit) were added to 96-multiwell plates containing triplicate wells with 75 μl complete medium predosed with compounds. On each assay plate, 3 wells containing 150 μl trophozoite-stage culture (0.5% parasitemia, 2% hematocrit) in the absence of drugs served as a positive control (100% growth). The outer wells of each 96-well plate were filled with 200 μl of medium to prevent evaporation. The plates were incubated for 48 h in a gassed (1% O2, 3% CO2, 96% N2) chamber at 37°C. Following this, 100 μl of the sample from each well was mixed with 100 μl MSF lysis buffer (20 mM Tris, pH 7.5, 5 mM EDTA, 0.008% saponin, 0.8% Triton X-100) supplemented with 0.2 μl ml−1 of SYBR green I (Sigma). After a 1-h incubation in the dark at room temperature, SYBR green I fluorescence was measured using the blue fluorescent module (excitation, 490 nm; emission, 510 to 570 nm) of a GloMax multidetection system (Promega). Percent parasite growth was calculated as follows: 100 × {[μ(s) − μ(−)]/[μ(+) − μ(−)]}, where μ(s), μ(−), and μ(+) are the means of the fluorescent readouts from sample wells, from control wells without drug (100% growth), and from wells with the maximum drug concentration (0% growth), respectively. EC50s were determined by plotting the log concentration-normalized response curves using GraphPad Prism (v5.0) software (GraphPad Software, Inc., San Diego, CA) from the mean from three independent biological repeats.
BRRoK assay.
Bioluminescence relative rate of kill (BRRoK) assays were carried out as described previously (31). Trophozoite cultures of strain Dd2 luc or NF54 luc (100 μl, 2% parasitemia, 4% hematocrit) were added to 96-multiwell plates containing 100 μl of predosed complete culture medium (3-fold EC50 dilution series, 9× EC50 to 0.33× EC50 for each compound tested), mixed by pipetting, and incubated for 6 h at 37°C. Samples of 40 μl were removed from each well, and the bioluminescence signal was measured using the luciferase single-step lysis protocol (43). Controls in each biological replicate consisted of a trophozoite-stage culture with no drug added (100% bioluminescence). The normalized bioluminescence data from three biological repeats were plotted as a proportion of that for the untreated control, with reference data for benchmark antimalarial compounds being sourced from a previous study (31).
Flow cytometry.
Parasite cultures were washed in PBS and then stained for flow cytometry using SYBR green I diluted 1:2,000 in PBS and incubated at room temperature without light exposure for 20 min. Parasites were then fixed in 4% formalin (Sigma) in PBS and incubated at 4°C for 15 min, before thorough washing in PBS and analysis using forward scatter and the green fluorescence channel of a Guava easyCyte system (Merck Millipore).
Southern blotting.
Genomic DNA was extracted from parasites using a QIAamp DNA blood minikit (Qiagen) and digested with the restriction enzymes AluI, DdeI, MboII, and RsaI as previously described (44). Digested genomic DNA was resolved on a 1% agarose gel, transferred to a GeneScreen Plus hybridization transfer membrane (PerkinElmer), hybridized with a probe specific for telomeres (45), labeled with alkaline phosphatase (AlkPhos direct labeling and detection system; GE Healthcare), and then imaged according to the kit manufacturer's directions.
Plasmid construction and parasite transfection to generate Rifin-GFP reporter line.
To generate the G-quadruplex reporter line, the rifin gene PF3D7_0700200 was amplified using the primers TGCCTAGGATGAAAATCCATTATATTAAT and ACCGTACGTTCTTTTAATAGTTTTATGTATG and then cloned upstream of the C-terminal green fluorescent protein (GFP) tag in the pLN-ENR-GFP plasmid (46) to generate the plasmid pLN-Rif0700200-GFP. NF54attB parasites were transfected with pLN-Rif0700200-GFP and maintained on BSD selection as previously described (46, 47).
Biophysical characterization of G-quadruplex formation.
Oligonucleotide TTTGGGAGGGCTTGTTCCGGGAATGGGTT was purchased from Eurofins Genomics (Ebersberg, Germany) and was resuspended in Milli-Q water to generate a stock solution at 400 μM. Before the experiments, the oligonucleotide was annealed by heating at 90°C for 3 min. Cation solution (KCl or LiCl) and lithium cacodylate were then added to reach final concentrations of 150 and 10 mM, respectively, and a pH of 7.4. For thermal denaturation, the oligonucleotide final concentration was 2.5 μM and UV absorption was recorded at 295 nm on a Uvikon XS spectrophotometer (Seconam), using a 1-cm-path-length quartz cell (type no. 115B-QS; Hellma, France). For NMR, samples were prepared by dissolving the oligonucleotides in H2O-D2O (95:5) and 10 mM lithium cacodylate, pH 7.4, for a final oligonucleotide concentration of 200 μM. NMR data were collected at 500 MHz on a Bruker Avance spectrometer (fitted with a TCI triple-resonance cryo-probe with a z-axis gradient). One-dimensional 1H spectra were recorded at a temperature of 25°C. Water suppression was achieved using excitation sculpting with gradients (48).
Gene expression analysis.
Total RNA was extracted from mixed-stage cultures of parasites transfected with pLN-Rif0700200-GFP using an RNeasy kit (Qiagen). Extracted RNA was treated with DNase I, and cDNA was subsequently synthesized using an iScript cDNA synthesis kit (Bio-Rad). cDNA was checked for genomic DNA contamination by PCR across the intron of the gene PF3D7_0424300, as previously described (49). Relative gene expression was determined by real-time PCR using a StepOnePlus real-time PCR machine (Thermo Fisher Scientific) and a SensiFAST SYBR Hi-ROX kit (Bioline) on synthesized cDNAs. Cycling conditions were 95°C for 3 min and 40 cycles of 95°C for 15 s, 45°C for 40 s, and 60°C for 1 min. The three housekeeping genes used as controls were PF3D7_0717700 (seryl-tRNA synthetase), PF3D7_1444800 (fructose bisphosphate aldolase), and PF3D7_1246200 (actin); primers specific for these genes have been previously described (50). The levels of the A1 and A2 rRNA transcripts and mature 18S rRNA were also measured using primers previously described (51). The GFP-tagged reporter gene was measured using the GFP-specific primers GATGGAAGCGTTCAACTAGCAGACC and AGCTGTTACAAACTCAAGAAGGACC. ΔΔCT analysis (where CT represents the threshold cycle) was used to calculate the relative copy number of each target gene relative to the average CT for the three control genes. All experiments were conducted in both biological and technical duplicate.
Heme crystallization assay.
The heme crystallization assay was carried out as previously described (52), with the slight modification of using NP-40 rather than Tween 20 to initiate crystallization (as described previously [53]). Quarfloxin (500 μM) was added because previous work showed that the equivalent level of chloroquine could produce the maximal (75%) inhibition of crystallization (52). The conventional readout of this assay, heme absorbance at 415 nm, was precluded by interference from the strong intrinsic color of quarfloxin, and the assay could therefore be assessed only by nonquantitative visual observation of the crystals formed.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Tetragene LLC for supplying quarfloxin, to Pascal De Tullio (ULg) for help with NMR experiments, and to Adriana Adolfi and Rinal Sahputra for conducting preliminary work in the lab on G-quadruplex-stabilizing drugs in P. falciparum.
This study was funded by UK Medical Research Council (grants MR/K000535/1 and MR/L008823/1 to C.J.M.), a Jean Shanks Foundation grant to K.R.M., and a Ph.D. studentship from the Nigerian Tertiary Education Trust Fund to M.T.F.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
L.M.H., K.R.M., and F.N. conducted and designed experiments, analyzed data, and produced figures; M.T.F. conducted BRRoK assays; N.S. and C.D. performed biophysical characterization of G-quadruplex DNA; P.H. designed the BRRoK assays, analyzed data, and edited the manuscript; and C.J.M. designed the study, conducted experiments, analyzed data, and wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01828-17.
REFERENCES
- 1.WHO. 2016. World malaria report 2016. WHO, Geneva, Switzerland: http://www.who.int/malaria/publications/world_malaria_report/en/. [Google Scholar]
- 2.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, Cheng Q, Coulson RM, Crabb BS, Del Portillo HA, Essien K, Feldblyum TV, Fernandez-Becerra C, Gilson PR, Gueye AH, Guo X, Kang'a S, Kooij TW, Korsinczky M, Meyer EV, Nene V, Paulsen I, White O, Ralph SA, Ren Q, Sargeant TJ, Salzberg SL, Stoeckert CJ, Sullivan SA, Yamamoto MM, Hoffman SL, Wortman JR, Gardner MJ, Galinski MR, Barnwell JW, Fraser-Liggett CM. 2008. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otto TD, Bohme U, Jackson AP, Hunt M, Franke-Fayard B, Hoeijmakers WA, Religa AA, Robertson L, Sanders M, Ogun SA, Cunningham D, Erhart A, Billker O, Khan SM, Stunnenberg HG, Langhorne J, Holder AA, Waters AP, Newbold CI, Pain A, Berriman M, Janse CJ. 2014. A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biol 12:86. doi: 10.1186/s12915-014-0086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stanton A, Harris LM, Graham G, Merrick CJ. 2016. Recombination events among virulence genes in malaria parasites are associated with G-quadruplex-forming DNA motifs. BMC Genomics 17:859. doi: 10.1186/s12864-016-3183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert DE, Feigon J. 1999. Multistranded DNA structures. Curr Opin Struct Biol 9:305–314. doi: 10.1016/S0959-440X(99)80041-4. [DOI] [PubMed] [Google Scholar]
- 7.Huppert JL, Balasubramanian S. 2005. Prevalence of quadruplexes in the human genome. Nucleic Acids Res 33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers VS, Marsico G, Boutell JM, Di Antonio M, Smith GP, Balasubramanian S. 2015. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol 33:877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- 9.Kudlicki AS. 2016. G-quadruplexes involving both strands of genomic DNA are highly abundant and colocalize with functional sites in the human genome. PLoS One 11:e0146174. doi: 10.1371/journal.pone.0146174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smargiasso N, Gabelica V, Damblon C, Rosu F, De Pauw E, Teulade-Fichou MP, Rowe JA, Claessens A. 2009. Putative DNA G-quadruplex formation within the promoters of Plasmodium falciparum var genes. BMC Genomics 10:362. doi: 10.1186/1471-2164-10-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Cian A, Grellier P, Mouray E, Depoix D, Bertrand H, Monchaud D, Teulade-Fichou MP, Mergny JL, Alberti P. 2008. Plasmodium telomeric sequences: structure, stability and quadruplex targeting by small compounds. Chembiochem 9:2730–2739. doi: 10.1002/cbic.200800330. [DOI] [PubMed] [Google Scholar]
- 12.Calvo EP, Wasserman M. 2016. G-quadruplex ligands: potent inhibitors of telomerase activity and cell proliferation in Plasmodium falciparum. Mol Biochem Parasitol 207:33–38. doi: 10.1016/j.molbiopara.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. 2005. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol 12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 14.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. 2002. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci U S A 99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kruisselbrink E, Guryev V, Brouwer K, Pontier DB, Cuppen E, Tijsterman M. 2008. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr Biol 18:900–905. doi: 10.1016/j.cub.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 16.Koole W, van Schendel R, Karambelas AE, van Heteren JT, Okihara KL, Tijsterman M. 2014. A polymerase theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat Commun 5:3216. doi: 10.1038/ncomms4216. [DOI] [PubMed] [Google Scholar]
- 17.Kerry LE, Pegg EE, Cameron DP, Budzak J, Poortinga G, Hannan KM, Hannan RD, Rudenko G. 2017. Selective inhibition of RNA polymerase I transcription as a potential approach to treat African trypanosomiasis. PLoS Negl Trop Dis 11:e0005432. doi: 10.1371/journal.pntd.0005432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drygin D, Siddiqui-Jain A, O'Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP, Lim JK, Von Hoff D, Anderes K, Rice WG. 2009. Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res 69:7653–7661. doi: 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]
- 19.Balasubramanian S, Hurley LH, Neidle S. 2011. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov 10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drygin D, Rice WG, Grummt I. 2010. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu Rev Pharmacol Toxicol 50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 21.Papadopoulos K, Mita A, Ricart A, Hufnagel D, Northfelt D, Von Hoff D, Darjania L, Lim J, Padgett C, Marschke R. 2007. Pharmacokinetic findings from the phase I study of quarfloxin (CX-3543): a protein-rDNA quadruplex inhibitor, in patients with advanced solid tumors. Mol Cancer Ther 6(Suppl):B93 http://mct.aacrjournals.org/content/6/11_Supplement/B93. [Google Scholar]
- 22.Biffi G, Di Antonio M, Tannahill D, Balasubramanian S. 2014. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat Chem 6:75–80. doi: 10.1038/nchem.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. 2013. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem 5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson A, Wu Y, Huang YC, Chavez EA, Platt J, Johnson FB, Brosh RM Jr, Sen D, Lansdorp PM. 2014. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res 42:860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. 2001. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci U S A 98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smilkstein M, Sriwilaijaroen N, Kelly JX, Wilairat P, Riscoe M. 2004. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob Agents Chemother 48:1803–1806. doi: 10.1128/AAC.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arthanari H, Basu S, Kawano TL, Bolton PH. 1998. Fluorescent dyes specific for quadruplex DNA. Nucleic Acids Res 26:3724–3728. doi: 10.1093/nar/26.16.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Matteis F, Gibbs AH, Smith AG. 1980. Inhibition of protohaem ferro-lyase by N-substituted porphyrins. Structural requirements for the inhibitory effect. Biochem J 189:645–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izbicka E, Wheelhouse RT, Raymond E, Davidson KK, Lawrence RA, Sun D, Windle BE, Hurley LH, Von Hoff DD. 1999. Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res 59:639–644. [PubMed] [Google Scholar]
- 30.Han FX, Wheelhouse RT, Hurley LH. 1999. Interactions of TMPyP4 and TMPyP2 with quadruplex DNA. Structural basis for the differential effects on telomerase inhibition. J Am Chem Soc 121:3561–3570. doi: 10.1021/ja984153m. [DOI] [Google Scholar]
- 31.Ullah I, Sharma R, Biagini GA, Horrocks P. 2017. A validated bioluminescence-based assay for the rapid determination of the initial rate of kill for discovery antimalarials. J Antimicrob Chemother 72:717–726. doi: 10.1093/jac/dkw449. [DOI] [PubMed] [Google Scholar]
- 32.Waters AP, Syin C, McCutchan TF. 1989. Developmental regulation of stage-specific ribosome populations in Plasmodium. Nature 342:438–440. doi: 10.1038/342438a0. [DOI] [PubMed] [Google Scholar]
- 33.Slater AF. 1993. Chloroquine: mechanism of drug action and resistance in Plasmodium falciparum. Pharmacol Ther 57:203–235. doi: 10.1016/0163-7258(93)90056-J. [DOI] [PubMed] [Google Scholar]
- 34.Kwok CK, Merrick CJ. 2017. G-quadruplexes: prediction, characterization, and biological application. Trends Biotechnol 35:997–1013. doi: 10.1016/j.tibtech.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 35.Miao J, Fan Q, Cui L, Li J, Li J, Cui L. 2006. The malaria parasite Plasmodium falciparum histones: organization, expression, and acetylation. Gene 369:53–65. doi: 10.1016/j.gene.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 36.Bertschi NL, Toenhake CG, Zou A, Niederwieser I, Henderson R, Moes S, Jenoe P, Parkinson J, Bartfai R, Voss TS. 2017. Malaria parasites possess a telomere repeat-binding protein that shares ancestry with transcription factor IIIA. Nat Microbiol 2:17033. doi: 10.1038/nmicrobiol.2017.33. [DOI] [PubMed] [Google Scholar]
- 37.Dzikowski R, Deitsch KW. 2008. Active transcription is required for maintenance of epigenetic memory in the malaria parasite Plasmodium falciparum. J Mol Biol 382:288–297. doi: 10.1016/j.jmb.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dahl EL, Rosenthal PJ. 2007. Multiple antibiotics exert delayed effects against the Plasmodium falciparum apicoplast. Antimicrob Agents Chemother 51:3485–3490. doi: 10.1128/AAC.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong EH, Hasenkamp S, Horrocks P. 2011. Analysis of the molecular mechanisms governing the stage-specific expression of a prototypical housekeeping gene during intraerythrocytic development of P. falciparum. J Mol Biol 408:205–221. doi: 10.1016/j.jmb.2011.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trager W, Jensen JB. 1976. Human malaria parasites in continuous culture. Science 193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 41.Lambros C, Vanderberg JP. 1979. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol 65:418–420. doi: 10.2307/3280287. [DOI] [PubMed] [Google Scholar]
- 42.Elmendorf HG, Haldar K. 1993. Identification and localization of ERD2 in the malaria parasite Plasmodium falciparum: separation from sites of sphingomyelin synthesis and implications for organization of the Golgi. EMBO J 12:4763–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hasenkamp S, Wong EH, Horrocks P. 2012. An improved single-step lysis protocol to measure luciferase bioluminescence in Plasmodium falciparum. Malar J 11:42. doi: 10.1186/1475-2875-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Figueiredo LM, Freitas-Junior LH, Bottius E, Olivo-Marin JC, Scherf A. 2002. A central role for Plasmodium falciparum subtelomeric regions in spatial positioning and telomere length regulation. EMBO J 21:815–824. doi: 10.1093/emboj/21.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bottius E, Bakhsis N, Scherf A. 1998. Plasmodium falciparum telomerase: de novo telomere addition to telomeric and nontelomeric sequences and role in chromosome healing. Mol Cell Biol 18:919–925. doi: 10.1128/MCB.18.2.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adjalley SH, Lee MC, Fidock DA. 2010. A method for rapid genetic integration into Plasmodium falciparum utilizing mycobacteriophage Bxb1 integrase. Methods Mol Biol 634:87–100. doi: 10.1007/978-1-60761-652-8_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adjalley SH, Johnston GL, Li T, Eastman RT, Ekland EH, Eappen AG, Richman A, Sim BK, Lee MC, Hoffman SL, Fidock DA. 2011. Quantitative assessment of Plasmodium falciparum sexual development reveals potent transmission-blocking activity by methylene blue. Proc Natl Acad Sci U S A 108:E1214–E1223. doi: 10.1073/pnas.1112037108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hwang TL, Shaka AJ. 1995. Water suppression that works. Excitation sculpting using arbitrary waveforms and pulsed field gradients. J Magn Reson Ser A 112:275–279. [Google Scholar]
- 49.Frank M, Dzikowski R, Amulic B, Deitsch K. 2007. Variable switching rates of malaria virulence genes are associated with chromosomal position. Mol Microbiol 64:1486–1498. doi: 10.1111/j.1365-2958.2007.05736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dzikowski R, Frank M, Deitsch K. 2006. Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS Pathog 2:e22. doi: 10.1371/journal.ppat.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mancio-Silva L, Lopez-Rubio JJ, Claes A, Scherf A. 2013. Sir2a regulates rDNA transcription and multiplication rate in the human malaria parasite Plasmodium falciparum. Nat Commun 4:1530. doi: 10.1038/ncomms2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huy NT, Uyen DT, Maeda A, Trang DT, Oida T, Harada S, Kamei K. 2007. Simple colorimetric inhibition assay of heme crystallization for high-throughput screening of antimalarial compounds. Antimicrob Agents Chemother 51:350–353. doi: 10.1128/AAC.00985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sandlin RD, Carter MD, Lee PJ, Auschwitz JM, Leed SE, Johnson JD, Wright DW. 2011. Use of the NP-40 detergent-mediated assay in discovery of inhibitors of beta-hematin crystallization. Antimicrob Agents Chemother 55:3363–3369. doi: 10.1128/AAC.00121-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.