ABSTRACT
The biological signal molecule nitric oxide (NO) was found to induce biofilm dispersal across a range of bacterial species, which led to its consideration for therapeutic strategies to treat biofilms and biofilm-related infections. However, biofilms are often not completely dispersed after exposure to NO. To better understand this phenomenon, we investigated the response of Pseudomonas aeruginosa biofilm cells to successive NO treatments. When biofilms were first pretreated with a low, noneffective dose of NO, a second dose of the signal molecule at a concentration usually capable of inducing dispersal did not have any effect. Amperometric analysis revealed that pretreated P. aeruginosa cells had enhanced NO-scavenging activity, and this effect was associated with the production of the flavohemoglobin Fhp. Further, quantitative real-time reverse transcription-PCR (qRT-PCR) analysis showed that fhp expression increased by over 100-fold in NO-pretreated biofilms compared to untreated biofilms. Biofilms of mutant strains harboring mutations in fhp or fhpR, encoding a NO-responsive regulator of fhp, were not affected in their dispersal response after the initial pretreatment with NO. Overall, these results suggest that FhpR can sense NO to trigger production of the flavohemoglobin Fhp and inhibit subsequent dispersal responses to NO. Finally, the addition of imidazole, which can inhibit the NO dioxygenase activity of flavohemoglobin, attenuated the prevention of dispersal after NO pretreatment and improved the dispersal response in older, starved biofilms. This study clarifies the underlying mechanisms of impaired dispersal induced by repeated NO treatments and offers a new perspective for improving the use of NO in biofilm control strategies.
KEYWORDS: Pseudomonas aeruginosa, biofilms, dispersal, flavohemoglobin, nitric oxide
INTRODUCTION
The biofilm mode of growth, in which microorganisms form aggregates that are encased in a matrix of extracellular macromolecules and can be either suspended or attached to a surface, is essential to the ecology and biology of bacteria (1, 2). Cells in biofilms are generally characterized by a high level of genetic and physiological heterogeneity. This heterogeneity of biofilms, which is caused by microscale chemical gradients, adaptation to environmental conditions, stochastic gene expression, and genotypic variation (3), is central to the function of biofilms and often leads to increased resistance to external stressors, biocides, antibiotics, and immune systems compared to free-floating planktonic cells (4, 5). The increased resistance of biofilms to antimicrobials, which can lead to the failure of treatment strategies to control them, is the cause of many problems in industrial and clinical settings. In the food industry, biofilms limit the run length of manufacturing plants and cause product spoilage and safety problems (6). In drinking water distribution systems, biofilms can cause corrosion or increase the risk of releasing microbial pathogens into the water (7). In clinical settings, formation of a biofilm by the opportunistic pathogen Pseudomonas aeruginosa is responsible for many infections, including chronic lung infections in cystic fibrosis patients and wound infection in severe-burn victims (8). Hence, it is of great interest to study biofilms with the aim of inhibiting biofilm formation.
Bacterial biofilms develop through several stages, including initial reversible attachment, irreversible attachment, maturation, and dispersal (9). The final stage, dispersal, is an active, highly regulated response of biofilm bacteria that is different from the passive, mechanically induced sloughing and erosion of cells from the biofilm outer layers (10, 11). Biofilm dispersal can be triggered by a variety of environmental cues, including changes in nutrient availability and iron level (12–15), changes in temperature (16), and oxygen depletion (17, 18), as well as low levels of nitric oxide (NO) (11, 19). NO, which is a hydrophobic, highly reactive, and short-lived free radical that can diffuse freely through cellular membranes, is a widespread biological messenger. Previous research has found that low, nontoxic concentrations (in the nanomolar to low micromolar range) of NO cause a transition from the sessile biofilm to the motile planktonic phenotype in bacteria. The role of NO in mediating biofilm dispersal was first identified in P. aeruginosa (19) and was later found to be conserved across a broad range of microorganisms (references 20, 21, 22, and 23; reviewed in reference 24), as well as multispecies biofilms from water systems (25–28). Overall, these observations suggested a promising approach to define novel strategies based on NO delivery to disperse and control biofilms and biofilm-associated infections with applications in both clinical and industrial contexts (29).
Before the identification of NO as a signal for biofilm dispersal, the role of NO in bacteria had been mostly studied in the context of nitrosative stress, when NO is produced at higher concentrations from endogenous sources in bacteria or by immune response defenses, such as macrophages (30). These studies have led to the characterization of a number of cellular systems capable of responding to NO to alleviate nitrosative stress. Upon reaction with oxygen or superoxide, NO generates reactive nitrogen species (RNS) that can broadly damage cellular proteins, for instance, by attacking cysteine thiols or iron-sulfur (Fe-S) clusters in respiratory and other metabolic enzymes, as well as nucleic acids and lipids (31). To prevent such damage, bacteria possess a number of NO-sensing proteins, which contain redox sensors, such as heme iron cofactors, non-heme iron or copper centers, Fe-S clusters, or cysteine thiols, and can regulate specific defense responses (32–35). For example, P. aeruginosa can detoxify NO by using either a NO reductase (NOR) (36) or a flavohemoglobin, Fhp (36, 37). Fhp activity requires oxygen, and the system is thus used under aerobic conditions, while NOR is induced under anaerobic conditions. In P. aeruginosa, induction of NOR occurs via the heme NO sensor-containing transcription regulator DNR (dissimilatory nitrate respiration regulator) (38), while Fhp is regulated by FhpR (37), which is homologous to the non-heme iron cofactor protein NorR, which is capable of sensing NO in Escherichia coli (39). The NO-sensing and protective mechanisms appear to be highly diverse in bacteria, and in E. coli, NorR regulates a flavorubredoxin, NorV, rather than the Fhp homologue flavohemoglobin Hmp (40), the expression of which is regulated by the Fe-S cluster regulator NsrR (41). In E. coli and P. aeruginosa, the interplay between NO-producing and NO-scavenging systems was recently shown to generate oscillating cycles of NO levels, thus ensuring that intracellular NO remains present at nontoxic levels (42, 43). In several bacterial species, the molecular mechanisms of NO-mediated dispersal have been shown to involve bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP), a soluble second messenger, which is the key element of a genetic network governing motile-to-sessile transitions and is conserved in Gram-negative bacteria. In P. aeruginosa, although a direct sensor of NO that mediates c-di-GMP degradation has yet to be identified, recent studies have uncovered a novel NO-sensing protein domain, NosP, involved in the regulation of biofilm formation and dispersal via a multicomponent phosphorelay system (44).
While the exogenous addition of a single dose of NO can disperse a significant portion of biofilms, it was recently found that multiple-dose NO treatment does not improve biofilm dispersal (45). Thus, P. aeruginosa biofilms that had been grown for 24 h or 48 h did not show any enhanced dispersal after multiple dosing with the NO donor compound MAHMA NONOate (45). In this study, we hypothesized that after a first exposure to NO, a subpopulation of biofilm cells has an altered signaling pathway and becomes insensitive to NO. We investigated the response of P. aeruginosa biofilm cells to successive NO treatments and show that preexposure of biofilms to very low concentrations of NO inhibits the subsequent dispersal response when biofilms are exposed to higher concentrations of NO. Further, the flavohemoglobin encoded by fhp and regulated by fhpR was found to contribute to the NO pretreatment-induced impairment of dispersal. Finally, we show that this impaired dispersal can be restored by combined treatments with imidazole, a well-known antifungal drug that blocks the NO dioxygenase (NOD) activity of flavohemoglobin. Our findings offer a new perspective on reducing NO compound dosage and improving the use of NO as a biofilm control strategy.
RESULTS
NO-pretreated biofilms show impaired dispersal.
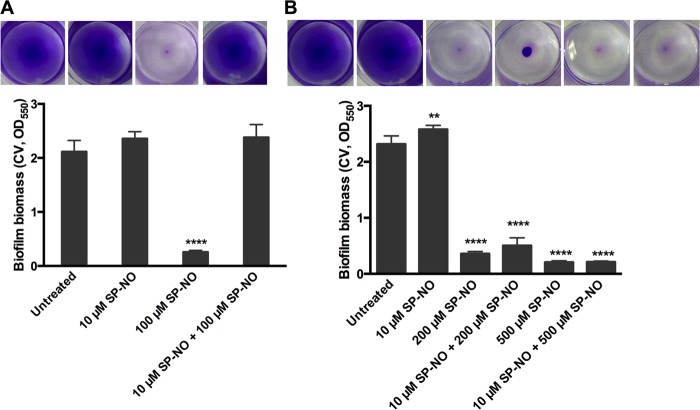
Previously, it was found that multiple doses of NO did not significantly increase biofilm dispersal (45). To better understand this phenomenon, the response of P. aeruginosa biofilm cells to NO was investigated by using a batch culture multiwell plate biofilm dispersal assay (46). Biofilms with no pretreatment, grown for a total of 6 h 15 min, including a dispersal treatment (15 min), were reduced by 88% after exposure to a dispersing dose, 100 μM, of the NO donor (Z)-1-[N-[3-aminopropyl]-N-[4-(3-aminopropylammonio)butyl]-amino]diazen-1-ium-1,2-diolate (spermine NONOate [SP-NO]) compared to control, nondispersed biofilms as seen with crystal violet (CV) staining (Fig. 1A; see Fig. S1A a and c in the supplemental material). According to the confocal image analysis of LIVE/DEAD stained biofilms, 87% of 6-h biofilm live cells were dispersed by the same concentration of NO for 15 min, and there was no significant increase in dead biofilm cells (see Fig. S2 in the supplemental material), showing that 100 μM SP-NO did not affect the viability of the biofilm cells. This is comparable to the results of CV staining, where 6-h biofilms were reduced by 88% (Fig. 1A). In this system, biofilms that were pretreated with 10 μM SP-NO (a nondispersing dose) for 3 h before inducing dispersal with 100 μM SP-NO for 15 min had similar biomass with and without the second NO treatment (Fig. 1A; see Fig. S1A b and d). The biomass of these biofilms was also similar to that of control biofilms that were not treated at all during the entire experiment. These data suggest that the bacterial response to a dispersing concentration of NO (100 μM SP-NO) was altered after preexposure to nondispersing concentrations of NO (10 μM SP-NO). However, pretreated biofilms were dispersed when the concentration of the second dose of NO was increased to 200 or 500 μM (Fig. 1B). This suggests that the factor responsible for the pretreatment effect could be titrated away by increasing the effective concentration of NO. Two strains that overproduce alginate were also tested using the pretreatment assay: PDO300, which is a mucA22 derivative of PAO1 constructed by allelic exchange (47), and an isogenic ΔmucA mutant strain (48). Both strains could be dispersed by a single dose of 100 μM NO, as was observed for the nonmucoid P. aeruginosa PAO1 strain. Biofilms of PDO300 and the ΔmucA mutant that were preexposed to 10 μM SP-NO were also impaired in their dispersal response after a subsequent exposure to a second, effective dose of NO (100 μM SP-NO) (see Fig. S3 in the supplemental material), which is similar to the wild-type (WT) strain PAO1. In the same system, exposure of biofilms formed by the wild-type PAO1 strain to 10 μM SP-NO did not prevent carbon or oxygen starvation-induced dispersal (see Fig. S4 and S5 in the supplemental material), suggesting that the pretreatment induces a NO-specific response.
FIG 1.
Biofilms that were pretreated with a low dose of NO required increased concentrations of NO to induce dispersal. P. aeruginosa biofilms were grown in multiwell plate batch cultures for a total of 6 h 15 min, including or not a pretreatment with 10 μM SP-NO at t = 3 h and a dispersal treatment with 100 μM (A), 200 μM (B), or 500 μM (B) SP-NO at t = 6 h. At the end of the incubation, biofilm biomass was analyzed by CV staining. Experimental details are shown in Fig. S1A in the supplemental material. The error bars indicate standard deviations (n = 3). The asterisks indicate statistically significant differences compared to untreated control samples (**, P < 0.01; ****, P < 0.0001). Each image represents the CV-stained biofilms.
NO-scavenging activity is cell associated and not secreted.
Because the change in dispersal behavior after pretreatment appeared to be specific to NO-mediated dispersal and could be overcome by using higher levels of the signal molecule, we hypothesized that biofilm bacteria may induce a response to scavenge and reduce NO levels. Instantaneous levels of NO in solution liberated from donor compounds were measured amperometrically using NO-specific electrodes in the presence of various bacterial cultures. The NO donor SP-NO was again used, and at a concentration of 100 μM donor compound, the amount of NO in solution was found to reach a steady state of 3.7 μM after 7 min (Fig. 2). While the addition of untreated P. aeruginosa cells had no effect on the release of NO, when NO-pretreated cells were added to the 100 μM SP-NO solution, the amount of NO measured was dramatically reduced (Fig. 2). In contrast, the cell-free supernatant of pretreated cells did not have any impact on NO levels (see Fig. S6 in the supplemental material), suggesting that the NO-scavenging capacity was not due to secreted factors but was associated with intracellular components. This was also supported by the fact that replacing the supernatants of biofilm cultures did not alleviate the dispersal prevention effect of pretreatment with NO (see Fig. S7 in the supplemental material). Taken together, these results indicate that NO pretreatment induced the production of NO-scavenging factors, which were cell associated and not secreted.
FIG 2.
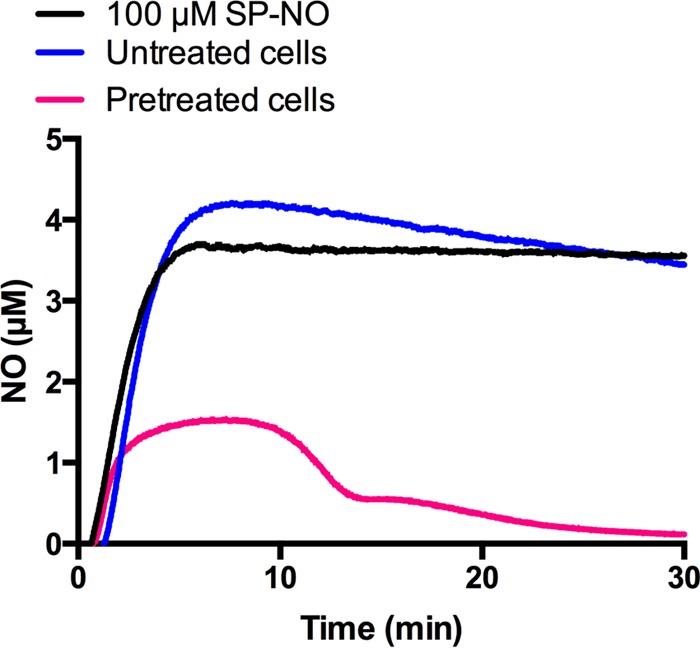
NO-pretreated P. aeruginosa cells scavenge free NO. NO levels in solution liberated from 100 μM SP-NO were measured amperometrically in the absence or presence of P. aeruginosa cells grown in multiwell plates for 3 h and treated or not with 10 μM SP-NO for a further 3 h.
fhp and fhpR are required for inhibiting dispersal.
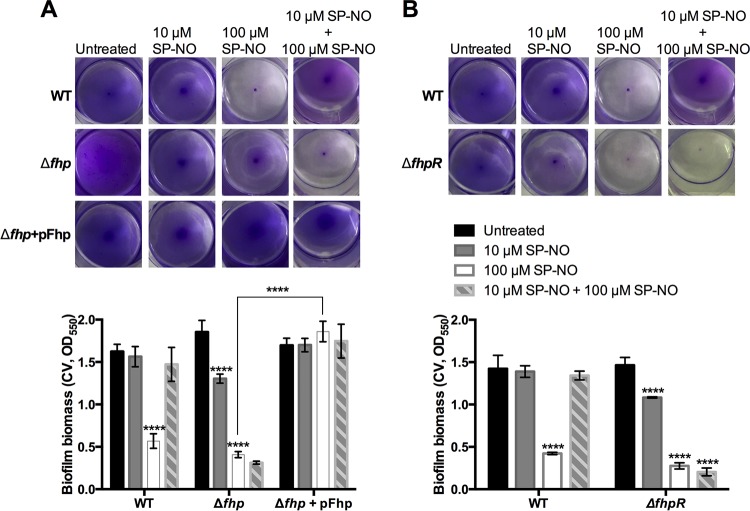
In P. aeruginosa, several proteins have been characterized that have the ability to prevent nitrosative stress by scavenging NO through reduction reactions, such as the NO reductase NOR (encoded by norCB), or oxidation reactions, such as the dioxygenases Fhp, HmgA, and Hpd (49). To determine if these proteins might be involved in scavenging NO and preventing dispersal, transposon mutant strains, including P. aeruginosa Δfhp, ΔnorB, ΔnorC, ΔhmgA, and Δhpd, were tested using the pretreatment assay. In contrast to all of the other mutants tested, the P. aeruginosa Δfhp mutant was readily dispersed in response to 100 μM SP-NO despite pretreatment with 10 μM SP-NO (Fig. 3A; see Fig. S8 in the supplemental material). Complementation of the Δfhp mutant restored the wild-type phenotype with regard to the dispersal defect. Further, even in the absence of pretreatment, the complemented strain showed enhanced resistance to 100 μM SP-NO-induced dispersal compared to the wild type (Fig. 3A). This was probably due to the high copy number of the plasmid pUCP22, leading to high expression of Fhp and, hence, NO scavenging. Taken together, these data suggest that Fhp production is the main contributor to the NO pretreatment-induced impairment of dispersal.
FIG 3.
P. aeruginosa Δfhp and ΔfhpR mutant strains are not affected by NO pretreatment. Biofilms of P. aeruginosa WT, Δfhp (A) and ΔfhpR (B) transposon mutants, and the Δfhp strain complemented with pFhp plasmid (A) were grown in multiwell plate batch cultures for a total of 6 h 15 min, including or not a pretreatment with 10 μM SP-NO at t = 3 h and a dispersal treatment with 100 μM SP-NO at t = 6 h. At the end of the incubation, the biofilm biomass was analyzed by CV staining. The error bars indicate standard deviations (n = 3). The asterisks indicate statistically significant differences compared to untreated control samples or between samples, as indicated (****, P < 0.0001). Each image represents the CV-stained biofilms.
The transcriptional activity of the fhp promoter is known to be dependent on the regulators FhpR, AsrA, and PA3697 (37, 50). To determine which regulator of fhp may be involved in the dispersal impairment, the ΔfhpR, ΔasrA, and PA3697 transposon mutant strains were tested. The results showed that NO-pretreated biofilms formed by P. aeruginosa ΔfhpR also lost their defect in dispersal (Fig. 3B). In contrast, the ΔasrA and PA3697 transposon mutants were not affected (see Fig. S9 in the supplemental material). This suggests that fhpR may play a role in regulating the production of NO-scavenging proteins after exposure to low levels of NO.
Expression of fhp increases significantly after exogenous NO pretreatment and moderately over the developmental time course of untreated biofilms.
Since previous results indicated that Fhp contributes to the NO pretreatment-induced impairment of dispersal, we hypothesized that fhp might be upregulated after exogenous pretreatment stimulation. Quantitative real-time reverse transcription-PCR (qRT-PCR) was used to measure transcription levels of fhp in biofilms and planktonic cells with and without NO pretreatment. After 3 h, biofilms and planktonic cells showed similar fhp expression levels before exposure to NO (Fig. 4). Expression levels of fhp were also similar in 6-h planktonic cells and 6-h biofilms (data not shown). Treatment with 10 μM SP-NO induced a strong increase in fhp expression in both 3-h biofilms and planktonic cells, by over 100- and 600-fold, respectively, compared to the untreated controls (Fig. 4), suggesting that fhp expression does not happen only in planktonic cells but can also occur in biofilms, which could compromise biofilm control strategies that rely on using NO as a dispersing agent.
FIG 4.
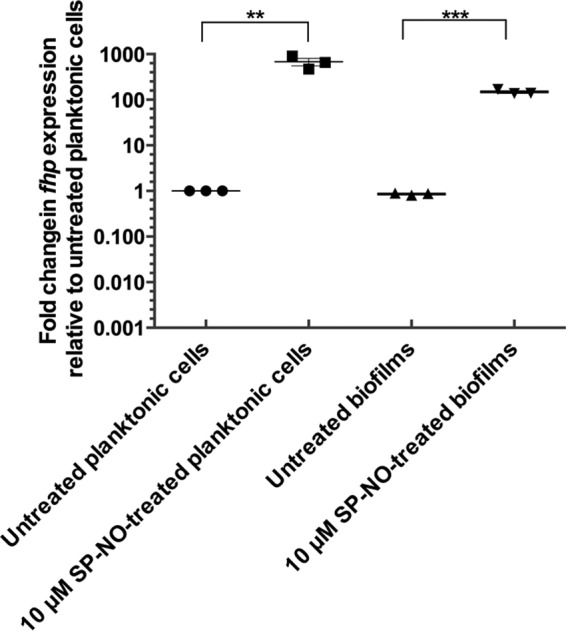
NO pretreatment induces the expression of fhp. P. aeruginosa PAO1 was grown in multiwell plate batch cultures for 3 h before being treated or not with 10 μM SP-NO. After 15 min, mRNAs were extracted from biofilm and planktonic bacteria, and fhp mRNA levels were quantified by qRT-PCR analysis. The data show fold changes in fhp transcript levels relative to untreated planktonic cells. The error bars indicate standard deviations (n = 3). The asterisks indicate statistically significant differences between samples as indicated (**, P < 0.01; ***, P < 0.001).
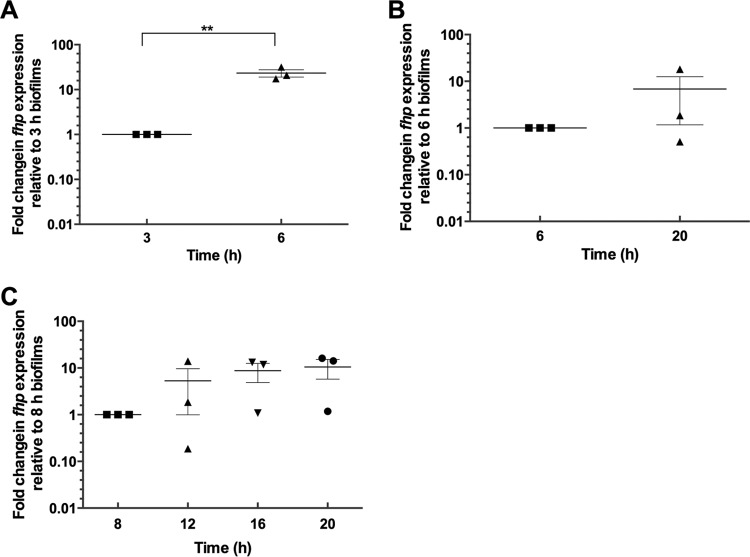
To further investigate the role of fhp in biofilm development, we then sought to determine its expression dynamics during biofilm formation. Expression levels of fhp in biofilms were found to increase by 23.3-fold without any exogenous stimulation in 6-h biofilms compared to 3-h biofilms, suggesting that fhp is induced in older biofilms (Fig. 5A). Under these experimental conditions, which were optimized for the fast formation of attached biofilm biomass, dispersal of biofilm cells is known to occur naturally after 8 to 10 h due to the depletion of the carbon source and the onset of starvation (51). Therefore, in order to study the expression of fhp in biofilms older than 10 h, biofilms were grown at room temperature with shaking at 120 rpm instead of 37°C at 180 rpm, which allowed the biofilm biomass to increase continuously for over 20 h. By using this system, the average of normalized transcript levels of fhp was found to be 6.8-fold higher in 20-h biofilms than in 6-h biofilms (Fig. 5B), although these changes were not statistically significant due to the large standard error. In this system, fhp was also induced in 12-, 16-, and 20-h unstimulated biofilms compared to 8-h biofilms (Fig. 5C). Expression levels of fhp were similar in 20-h biofilms and overnight culture (16-h planktonic cells) (data not shown). Taken together, these results suggest that fhp is endogenously induced, albeit to varying degrees, over time during biofilm development depending on the experimental conditions.
FIG 5.
fhp expression increases over time during biofilm development. fhp mRNA levels were quantified by qRT-PCR analysis and compared at different time points in biofilms grown at 37°C with fast agitation (A) or compared between 6-h biofilms grown at 37°C with fast agitation and 20-h biofilms grown at room temperature with slower agitation, a system that allows continuous increase in biofilm biomass over 20 h (B). (C) Time course analysis of fhp mRNA levels in the 20-h biofilm system. The error bars indicate standard errors (n = 3). The asterisks indicate statistically significant differences between samples (**, P < 0.01).
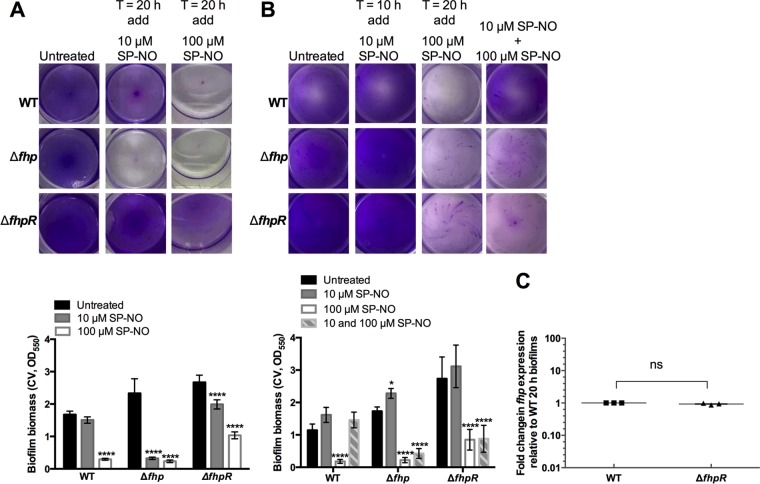
In older biofilms, inactivation of fhp, but not fhpR, leads to enhanced sensitivity to NO-mediated induction of dispersal.
Because fhp expression appeared to be induced with time in biofilms, we then tested if the dispersal response to NO might be altered in older biofilms. The effects of 10 and 100 μM SP-NO were tested on 20-h biofilms of P. aeruginosa wild-type and isogenic Δfhp and ΔfhpR mutant strains. In this biofilm assay, a low dose of NO generated from 10 μM SP-NO dispersed 86% of the biofilm biomass in the P. aeruginosa Δfhp mutant, while only 10% and 26% of biofilms formed by P. aeruginosa wild type and ΔfhpR were dispersed by 10 μM SP-NO (Fig. 6A). In the presence of 100 μM SP-NO, 90% of both wild-type and Δfhp biofilms were removed, and ΔfhpR biofilms were dispersed by about 61%. This suggests that 20-h biofilms of Δfhp were more sensitive to NO signals than those of the wild-type and ΔfhpR mutant strains. qRT-PCR analysis of 20-h wild-type and ΔfhpR biofilms did not show any significant differences in fhp mRNA levels between the strains (Fig. 6C), suggesting that fhpR was not necessary for the nonstimulated, endogenous induction of fhp in 20-h biofilms. However, in contrast to the wild-type strain, the Δfhp and ΔfhpR mutant strains still retained their dispersal capacity after pretreatment stimulation with an initial low dose of NO in the 20-h biofilm system (Fig. 6B), which correlates with the results previously obtained with the 6-h biofilm system (Fig. 1A).
FIG 6.
(A) The Δfhp mutant strain is more sensitive to NO-mediated induction of dispersal than the wild-type strain and the ΔfhpR mutant in 20-h biofilms. Biofilms of P. aeruginosa WT and the Δfhp and ΔfhpR transposon mutant strains grown in multiwell plate batch cultures at room temperature for 20 h were treated with 10 μM or 100 μM SP-NO for 15 min before CV staining. Experimental details are shown in Fig. S1B in the supplemental material. The error bars indicate standard deviations (n = 3). The asterisks indicate statistically significant differences compared to untreated control samples (****, P < 0.0001). Each image represents the stained biofilms. (B) Δfhp and ΔfhpR mutant strains still retain their dispersal capacity after pretreatment with an initial low dose of NO in the 20-h biofilm system. Biofilms of P. aeruginosa WT and Δfhp and ΔfhpR transposon mutant strains were grown in multiwell plate batch cultures for a total of 20 h 15 min, including or not a pretreatment with 10 μM SP-NO at t = 10 h and a dispersal treatment with 100 μM SP-NO at t = 20 h. At the end of the incubation, the biofilm biomass was analyzed by CV staining. Experimental details are shown in Fig. S1C in the supplemental material. The error bars indicate standard deviations (n = 3). The asterisks indicate statistically significant differences compared to untreated control samples (*, P < 0.1; ****, P < 0.0001). Each image represents the stained biofilms. (C) No significant differences were found in fhp mRNA levels between 20-h biofilms formed by P. aeruginosa wild type and the ΔfhpR mutant. The error bars indicate standard errors (n = 3). ns, no significant difference.
Imidazole blocks the NO pretreatment effect and enhances dispersal by inhibiting flavohemoglobin.
Because the production of Fhp impaired NO-mediated dispersal, we hypothesized that inhibiting the NOD activity of Fhp could restore dispersal in pretreated biofilms. One such candidate inhibitor compound is imidazole, which has been previously reported to hinder the NOD activity of bacterial and fungal flavohemoglobins by coordinating and constraining their heme (52). In the current study, imidazole was tested for its ability to inhibit purified flavohemoglobin and flavohemoglobin produced in P. aeruginosa biofilms. The NO consumption activity of purified histidine-tagged Fhp from P. aeruginosa with and without imidazole treatment was investigated by using NO-specific electrodes and an appropriate buffer system to study NOD kinetics (52). As expected, the addition of 5 nM Fhp purified from P. aeruginosa dramatically reduced the amount of NO released from 100 μM SP-NO (Fig. 7A). In contrast, Fhp that had been treated with imidazole did not have any effect on the release of NO. Furthermore, truncated Fhp proteins that lacked the flavin-binding or heme-binding domain both lost the ability to scavenge NO (see Fig. S10 in the supplemental material).
FIG 7.
Imidazole can inhibit the NOD activity of Fhp and restore the dispersal response of P. aeruginosa biofilms pretreated with NO. (A) NO released from 100 μM SP-NO in the absence and presence of Fhp and imidazole was measured amperometrically in 100 mM sodium phosphate buffer, pH 7.0, containing 0.3 mM EDTA, 100 μM NADPH, and 1 μM FAD at room temperature. Fhp (5 nM), either untreated or treated with imidazole (5 mM), was added 15 min after adding SP-NO. (B) P. aeruginosa biofilms were grown in multiwell plate batch cultures for 6 h. Then, the biofilms were treated or not with 5 mM imidazole (Im) for 15 min, after which time dispersal was induced with 100 μM SP-NO for 15 min before quantifying the attached biomass by CV staining. For the pretreatment experiments, biofilms were grown for 3 h, exposed to 10 μM SP-NO, and incubated for a further 3 h (6 h total) before being exposed to imidazole and NO, followed by assessment of the total biofilm biomass by CV staining. The error bars indicate standard deviations (n = 3). The asterisks indicate statistically significant differences compared to untreated control samples (***, P < 0.001; ****, P < 0.0001). The images show the CV-stained biofilms.
In the absence of imidazole, 6-h biofilms pretreated with a low dose of NO (10 μM SP-NO) were impaired in their dispersal response to 100 μM SP-NO, as expected. However, the addition of 5 mM imidazole for 15 min prior to the final treatment with 100 μM SP-NO restored the dispersal response, resulting in the detachment of 64% of biofilms (Fig. 7B). Treatment with imidazole alone showed only a 12% reduction in biofilm biomass. Overall, these data strongly suggest that imidazole can inhibit the flavohemoglobin function in NO-pretreated biofilms and enhance biofilm dispersal.
We further investigated whether the addition of imidazole could improve the dispersal response to NO in P. aeruginosa biofilms by inhibiting flavohemoglobin produced naturally during biofilm development rather than after induction with exogenous NO. In this experiment, biofilms were not preexposed to NO but were treated with imidazole and a low dose of NO (10 μM SP-NO) successively. It was found that combined treatments of imidazole (5 or 10 mM) and 10 μM SP-NO failed to disperse 6-h P. aeruginosa biofilms (Fig. 8A). In contrast, in 20-h biofilms, although 10 μM SP-NO alone also had no impact on the biofilms, combined treatments of imidazole (5 and 10 mM) and 10 μM SP-NO dispersed about 88% of the biofilms (Fig. 8B).
FIG 8.
Imidazole enhances the sensitivity of the dispersal response to NO in 20-h biofilms. Biofilms were grown in multiwell plate batch cultures for 5 h (A) or 19 h (B) and subsequently treated with 5 and 10 mM imidazole for 1 h prior to addition of 10 μM SP-NO for 15 min. The error bars indicate standard deviations (n = 3). At the end of the incubation, the biofilm biomass was analyzed by CV staining. The asterisks indicate statistically significant differences compared to untreated control samples (****, P < 0.0001). The images show the CV-stained biofilms.
DISCUSSION
An endogenous mechanism to limit biofilm dispersal.
In this work, an experimental system was designed to test the effect of successive exposures to NO signals on biofilm dispersal. We were able to replicate the previously observed attenuation of dispersal responses after multiple treatments with NO by first exposing biofilms to a low, nondispersing concentration, followed by NO concentrations that would normally disperse biofilms (Fig. 1). By using this system, we found that the pretreatment effect was due to strong induction of fhp, encoding the NO-scavenging flavohemoglobin, probably after direct sensing of NO by FhpR. Thus, this work has uncovered a novel role for the flavohemoglobin Fhp and the regulator FhpR, which, in addition to protecting bacteria from nitrosative stress, are also involved in mediating dispersal responses to NO signals. Interestingly, the pretreatment-induced repression of dispersal appeared to be specific for the NO-mediated signaling pathway, while dispersal responses induced by oxygen or nutrient limitation were not affected by NO pretreatment (see Fig. S4 and S5 in the supplemental material). This clarifies the fact that, despite the observation that all three dispersal responses to oxygen depletion, starvation, and NO operate through modification of intracellular c-di-GMP concentrations and require the periplasmic protease LapG (53), the oxygen- and nutrient-controlled dispersal pathways are independent of NO signaling and, for instance, do not involve production of Fhp.
The results presented here show that the NO dispersal pathway can be modulated by Fhp activity, the production of which is induced in response to NO exposure, through the regulator FhpR. Fhp has a dioxygenase function that converts NO to nitrate (NO3−) (37, 54), and this reaction is dependent on NADPH, flavin adenine dinucleotide (FAD), and oxygen (55). Our results showed that the expression level of fhp was highly induced by a nondispersing dose of NO (10 μM SP-NO) in both biofilms and planktonic cells. The induction of fhp after exposure to low levels of NO was quite surprising, given that a 1.8-fold downregulation of fhp in response to NO has been previously observed in P. aeruginosa biofilms (11). However, the experimental conditions in the previous studies were different, as biofilms were grown for 5 days and exposed to the NO donor sodium nitroprusside for 1 h compared to biofilms treated with NO after 3 h, that is, almost immediately after cell attachment in the current work. Thus, it is possible that in more mature biofilms, fhp exhibits different expression and regulation profiles. This is supported by our observation that fhp expression was higher in 20-h biofilms than in 6-h biofilms.
In P. aeruginosa, NO could also be reduced to N2O by NOR (56, 57), although this pathway was not necessary for the NO scavenging observed in the 6-h biofilms. The expression of the denitrification genes norB and norC occurs under anaerobic or low-oxygen conditions (37). This suggests that the biofilms studied here were not experiencing oxygen limitation based on the incubation conditions.
We further demonstrated that the regulator fhpR was required for the pretreatment-induced impaired dispersal. fhp and fhpR are adjacent and share a bidirectional promoter region, which is independent of DNR or ANR (37). FhpR is a homologue of NorR in E. coli that regulates the production of flavorubredoxin (39, 40). It was previously reported that the fhp promoter is dependent on two additional regulators, AsrA and PA3697 (50), although neither appeared to be required in our assays for pretreatment-induced impaired dispersal (see Fig. S9 in the supplemental material). Interestingly, fhpR was not required for these effects in the 20-h biofilms, which suggests that an additional regulatory mechanism controls fhp expression in the older, 20-h biofilms.
Flavohemoglobin is distributed broadly among prokaryotic and eukaryotic microorganisms (58). In E. coli and Staphylococcus aureus, the expression of genes encoding flavohemoglobins has been shown to be stimulated by NO and nitrosative stresses (59, 60). Therefore, it may protect mixed-community biofilms found in natural and engineered systems from NO-induced dispersal.
Flavohemoglobin may also play a role across a biofilm-host interface. NO, which at high concentrations can cause nitrosative damage to bacteria, is known to be produced by NO synthases in infected host tissues in both mammals and plants (61, 62). In the context of a symbiotic relationship between bacteria and a host, flavohemoglobin could potentially confer resistance to the host immune defenses. Thus, in the squid-Vibrio symbiosis, Hmp has been shown to alleviate nitrosative stress in the Vibrio fischeri symbiont and to allow its successful colonization of the host while other invading bacterial species are excluded (63, 64).
Overall, our data have uncovered an intrinsic mechanism by which bacteria can modulate their response to NO-induced dispersal. In the context of biofilm development, NO production from the deep anaerobic zones of mature microcolonies has been linked to the death of a subpopulation of cells and the formation of hollow voids within microcolonies (19). Induction of fhp expression could be a potential strategy by which biofilms may limit the extent of biofilm dispersal induced by endogenously produced NO in mature biofilms, e.g., in microcolonies undergoing seeding dispersal. Future studies in our laboratory will investigate the dynamics and spatial localization of fhp expression during biofilm development and dispersal.
Combined treatments with NO and Fhp inhibitors to improve biofilm control.
Previous studies have shown that imidazole compounds can inhibit NOD function by coordinating heme iron and “fitting” within the heme pocket, since they have bulky aromatic substituents (65–68). Here, imidazole was found to block the NO pretreatment-induced impairment of dispersal and to work synergistically with NO to disperse biofilms at very low concentrations of NO donors. Thus, it is possible that imidazole or related compounds could improve the efficacy of NO as a dispersing agent for biofilms formed by a broad range of microorganisms. It was particularly surprising that in the presence of imidazole, 20-h biofilms were able to disperse in response to 10 μM SP-NO while 6-h biofilms did not disperse in response to such a low dose of NO (Fig. 8). There are several possible explanations for this observation. First, cells in younger biofilms (6-h biofilms) have additional mechanisms of NO defense, in addition to Fhp, that are capable of scavenging NO to some level and that could be absent in 20-h biofilms, while older biofilms (20-h biofilms) may rely mostly on Fhp. Alternatively, older biofilms may have an altered pathway for inducing dispersal in response to lower levels of NO rather than a pathway for scavenging NO. Further experiments will need to focus on these possibilities to better define the mechanisms of NO response for younger and older biofilms.
Naturally occurring and synthetic derivatives of imidazole have been well documented (69–71). For example, sponges of the genera Leucetta and Clathrina are rich sources of imidazole alkaloids (72–74). In the future, it will be interesting to investigate the combined effects of low doses of NO and different imidazole derivatives with low toxicity and high bioavailability on biofilms formed by single and multiple species.
In conclusion, this study has identified a mechanism to explain the previous observations that multiple doses of NO do not improve biofilm dispersal. A novel role for the NO-scavenging flavohemoglobin Fhp has been identified, where Fhp is induced in an NO-dependent fashion by FhpR in younger biofilms and subsequently impairs the dispersal responses to NO. Further, this work highlights an innovative strategy for potentiating or synergizing the activity of NO as a biofilm control agent by inhibiting natural scavenging mechanisms. Our study offers a new perspective on improving the use of NO for biofilm control in both clinical and industrial settings.
MATERIALS AND METHODS
Bacteria and growth conditions.
The P. aeruginosa WT strain PAO1 (75); strain PDO300, which is a mucA22 derivative of PAO1 constructed by allelic exchange (47); and an isogenic ΔmucA mutant strain (48) were used in this study. P. aeruginosa mutant strains containing a transposon Tn5-derived insertion element (Tcr) in key genes involved in NO detoxification, which were obtained from the University of Washington P. aeruginosa PAO1 transposon mutant library (supported by grant number NIH P30 DK089507), were also used: Δfhp (PA2664), strain PW5458 fhp-F06::ISlacZ/hah (an influenza hemagglutinin [HA] epitope and a hexahistidine motif); ΔfhpR (PA2665), strain PW5460 fhpR-H11::ISphoA/hah; ΔnorB (PA0524), strains PW1961 norB-A04::ISphoA/hah and PW1962 norB-A11::ISphoA/hah; ΔnorC (PA0523), strains PW1959 norC-B02::ISlacZ/hah and PW1960 norC-H04::ISphoA/hah; ΔhmgA (PA2009), strain PW4489 hmgA-C03::ISphoA/hah; Δhpd (PA0865), strains PW2577 hpd-H01::ISlacZ/hah and PW2578 hpd-H02::ISlacZ/hah; ΔasrA (PA0779), strains PW2411 asrA-A07::ISlacZ/hah, PW2413 asrA-G11::ISlacZ/hah, and PW2414 asrA-C03::ISlacZ/hah; and ΔPA3697, strains PW7256 PA3697-H04::ISlacZ/hah, PW7257 PA3697-C03::ISlacZ/hah, and PW7258 PA3697-D09::ISphoA/hah (76). For experiments involving the transposon mutants, the isogenic wild-type strain MPAO1 was used (77); no difference in dispersal behavior or response to NO between strains PAO1 and MPAO1 has been observed. Bacteria were routinely grown in Luria-Bertani (LB)-Miller broth (BD Difco) overnight at 37°C, with agitation at 200 rpm for 16 h to prepare the cells for experiments.
NO donor compounds.
The NO donor compound SP-NO was obtained from Cayman Chemical. SP-NO spontaneously dissociates to liberate 2 moles of NO per mole of the parent compound, with half-lives of 39 min and 230 min at 37°C and 22 to 25°C, respectively (pH 7.4) (78, 79). Aliquots of 100 mM stock solutions of SP-NO dissolved in 10 mM NaOH (Merck) were kept at −20°C for up to 3 months and thawed only once for each experiment. Further dilutions were freshly made in 10 mM NaOH and used immediately.
Biofilm dispersal assays.
Biofilms were grown as previously described (46) with some modifications. Briefly, overnight cultures of P. aeruginosa PAO1 wild type and mutants were diluted 200-fold to an optical density at 600 nm (OD600) of 0.003 in M9 minimal medium (containing 9 mM NaCl, 22 mM KH2PO4, 48 mM Na2HPO4, 19 mM NH4Cl, 2 mM MgSO4, 100 µM CaCl2, and 0.4% glucose, pH 7.0), which was freshly prepared for every experiment. One milliliter of the diluted culture was added to 24-well microtiter plates (Nunc, Thermo Fisher Scientific), and the plates were incubated on an orbital shaker at 180 rpm for 6-h biofilms, which were cultivated at 37°C, or at 120 rpm for 20-h biofilms, which were cultivated at room temperature. After 6 h or 20 h growth, SP-NO was added to the cultures at a final concentration of 100 μM to induce biofilm dispersal, and the plates were incubated for a further 15 min. For the untreated control, an equal amount of 10 mM NaOH was added. After the final incubation, the biofilm biomass was analyzed by CV staining. Briefly, biofilms grown on the interior surfaces of the multiwell plate wells were washed with phosphate-buffered saline (PBS) containing 137 mM NaCl, 2.7 mM KCl, 2 mM KH2PO4, and 10 mM Na2HPO4 (pH 7.4) to remove loosely attached bacteria before staining for 20 min with 1 ml of 0.1% CV solution made by diluting a 1% CV aqueous solution (Sigma) 1:10 in Milli-Q water. The stained biofilms were washed twice with 1 ml PBS, and the remaining CV was dissolved in 1 ml of absolute ethanol. The biofilm biomass was quantified by measurement of the absorbance at 550 nm (OD550) in an Infinite Pro2000 microplate reader (Tecan). All biofilm assays were performed in independent triplicate experiments.
For microscopy analysis, P. aeruginosa PAO1 wild-type biofilms grown in multiwell plate batch cultures for 6 h and subsequently left untreated or treated with 100 μM SP-NO for 15 min were rinsed once with PBS before being stained with the LIVE/DEAD BacLight bacterial viability kit reagents (Molecular Probes, Inc.) according to the manufacturer’s procedure. One microliter of each of the two components was mixed thoroughly in 1 ml of PBS, and then 0.5 ml of this solution was added into each well and allowed to incubate at room temperature in the dark for 15 min. Images of untreated and NO-treated biofilms were acquired using confocal laser scanning microscopy (CLSM) (Carl Zeiss Microscopy; LSM 780). Biofilm quantification was performed using the IMARIS software package (Bitplane AG).
To test successive exposures to NO treatments, P. aeruginosa biofilms that had been grown in multiwell plates as described above for 3 h received a pretreatment with 10 μM SP-NO before incubating the plates for another 3 h. After a total of 6 h, the biofilms were treated or not with 100 μM SP-NO for 15 min to induce dispersal before quantifying the biofilm biomass by CV staining.
For experiments involving the alginate-overproducing strains, biofilms of all three strains, including PAO1 wild type, PDO300, and ΔmucA mutant were grown in M9 medium supplemented with 2% Casamino acids, where the higher amount of Casamino acids was necessary for the mucoid strains to form biofilms in our experimental conditions. Exposure to NO donors and biofilm biomass analysis by CV staining were carried out as described above.
The biofilm assay was also used to test the effect on biofilm dispersal of the flavohemoglobin inhibitor imidazole (Sigma-Aldrich), which was prepared in Milli-Q water at different concentrations. Biofilms that had been pretreated with 10 μM SP-NO after 3 h growth (6-h biofilms) were then treated with 5 mM imidazole for 15 min prior to addition of 100 μM SP-NO to induce dispersal. Biofilms that had been grown for 5 h or 19 h without pretreatment were treated with 5 or 10 mM imidazole for 1 h prior to addition of 10 μM SP-NO. The biofilm biomass was quantified by CV staining as described above.
Amperometric measurements of NO.
The concentration of NO liberated from SP-NO was measured amperometrically by using a TBR1025 free radical analyzer (World Precision Instruments) equipped with a NO-specific ISO-NOP 2-mm electrode with a detection range from 1 nM to 100 μM and calibrated by using MAHMA NONOate (Cayman Chemical) as the NO donor.
NOD activity of purified Fhp was also assessed amperometrically by following a protocol previously described by Gardner and Gardner (80). NO was released from 100 μM SP-NO in a NOD activity buffer solution consisting of 100 mM Na3PO4, pH 7.0, 0.3 mM EDTA, 100 μM NADPH (Cayman Chemical), and 1 μM FAD (Sigma-Aldrich) at room temperature; according to the method, NADPH and FAD were added before adding SP-NO. Fhp, imidazole-treated Fhp, and truncated Fhp were added 15 min after adding SP-NO. Imidazole-treated Fhp was obtained by incubating 500 nM Fhp with 500 mM imidazole for 1 h at room temperature. The mixture was added to NOD activity buffer in the presence of FAD, NADPH, and NO. The final concentrations of Fhp and imidazole were 5 nM and 5 mM, respectively.
Generation of the complemented strain.
An fhp complementation plasmid was constructed by amplifying the fhp gene, including 156 bp of the promoter-containing upstream region, with the primers fhp_for/fhp_rev (see Table S1 in the supplemental material) using Pfu polymerase (Thermo Fisher Scientific) and ligating it into EcoRI and BamHI (New England BioLabs) double-cut plasmid pUCP22. The resultant plasmids were transformed into the E. coli donor strain DH5α and transformed into the transposon mutant Δfhp (PW5458 fhp-F06::ISlacZ/hah).
qRT-PCR.
cDNA was reverse transcribed from 0.5 μg of total RNA using the SuperScript III first-strand synthesis system (Invitrogen) according to the manufacturer's protocols. The cDNA was then used as the template for qRT-PCR in a real-time PCR system using SYBR green with the Rox detection system (Kapa Biosystems) and primers, including fhp_RT_for/fhp_RT_rev for fhp and rpoD_RT_for/rpoD_RT_rev for the housekeeping gene rpoD. The primers are listed in Table S1 in the supplemental material. Each sample was independently tested three times and assayed in triplicate during each run. Relative gene expression with respect to rpoD mRNA transcript levels was calculated using the 2−ΔΔCT method (81).
Fhp protein purification.
Cloning and protein expression were carried out at the Protein Production Platform (PPP) at Nanyang Technological University (NTU). Targets of fhp (1,175 bp), truncated fhp without a flavin-containing domain (422 bp), and truncated fhp without a heme-binding domain (716 bp) were amplified using primers Fhp-f7852/Fhp-r7876, Fhp-f7852/Fhp-r7880, and Fhp-f7854/Fhp-r7877 (see Table S1 in the supplemental material). All cloning was performed in 96-well systems using ligation-independent cloning (LIC) technology in E. coli (82, 83). Histidine-tagged-protein production was induced with IPTG (isopropyl-β-d-thiogalactopyranoside) and analyzed on SDS-PAGE gels. Clones with acceptable levels of expression were selected for large-scale expression and purification. During the purification steps, cell lysates were loaded on immobilized-metal affinity chromatography (IMAC) columns (1 ml Ni-nitrilotriacetic acid [NTA] Superflow [Qiagen]) and were washed. His-tagged proteins were eluted with 500 mM imidazole, collected, and stored in sample loops on the system and then injected into gel filtration (GF) columns (Superdex 200 HR 26/60 [GE Healthcare]), which separated imidazole from the proteins. Elution peaks were collected in 2-ml fractions and analyzed on SDS-PAGE gels. Relevant peaks were pooled, and tris(2-carboxyethyl)phosphine (TCEP) was added to a total concentration of 2 mM. Purified proteins were kept in buffer containing 20 mM HEPES (pH 7.5), 300 mM NaCl, 10% (vol/vol) glycerol, and 2 mM TCEP. The final protein batch was then aliquoted into smaller fractions, frozen in liquid nitrogen, and stored at −80°C.
Statistical analysis.
Multivariate analyses were performed using t tests, one-way analysis of variance (ANOVA), and two-way ANOVA, followed by the Sidak posttest for individual comparisons.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge financial support from the Singapore Centre for Environmental Life Sciences Engineering, whose research is supported by the National Research Foundation Singapore, Ministry of Education, Nanyang Technological University, and National University of Singapore under its Research Centre of Excellence Programme. N.B. is supported by the French Government's Investissement d'Avenir program, Laboratoire d'Excellence “Integrative Biology of Emerging Infectious Diseases” (grant no. ANR-10-LABX-62-IBEID). This research was partly supported by a research grant (1301-IRIS-57) from the Environment and Water Industry Program Office of Singapore.
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01832-17.
REFERENCES
- 1.Webb JS, Givskov M, Kjelleberg S. 2003. Bacterial biofilms: prokaryotic adventures in multicellularity. Curr Opin Microbiol 6:578–585. doi: 10.1016/j.mib.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 2.Wong GC, O'Toole GA. 2011. All together now: integrating biofilm research across disciplines. MRS Bulletin 36:339–342. doi: 10.1557/mrs.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat Rev Microbiol 6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 4.Buckingham-Meyer K, Goeres DM, Hamilton MA. 2007. Comparative evaluation of biofilm disinfectant efficacy tests. J Microbiol Methods 70:236–244. doi: 10.1016/j.mimet.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Costerton JW, Stewart PS, Greenberg E. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 6.Brooks JD, Flint SH. 2008. Biofilms in the food industry: problems and potential solutions. Int J Food Sci Technol 43:2163–2176. doi: 10.1111/j.1365-2621.2008.01839.x. [DOI] [Google Scholar]
- 7.Mattila-Sandholm T, Wirtanen G. 1992. Biofilm formation in the industry: a review. Food Rev Int 8:573–603. doi: 10.1080/87559129209540953. [DOI] [Google Scholar]
- 8.Lyczak JB, Cannon CL, Pier GB. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect 2:1051–1060. doi: 10.1016/S1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 9.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. 2011. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol 10:39–50. doi: 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- 11.Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. 2009. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J Bacteriol 191:7333–7342. doi: 10.1128/JB.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gjermansen M, Ragas P, Sternberg C, Molin S, Tolker-Nielsen T. 2005. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ Microbiol 7:894–904. doi: 10.1111/j.1462-2920.2005.00775.x. [DOI] [PubMed] [Google Scholar]
- 13.Hunt SM, Werner EM, Huang B, Hamilton MA, Stewart PS. 2004. Hypothesis for the role of nutrient starvation in biofilm detachment. Appl Environ Microbiol 70:7418–7425. doi: 10.1128/AEM.70.12.7418-7425.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glick R, Gilmour C, Tremblay J, Satanower S, Avidan O, Déziel E, Greenberg EP, Poole K, Banin E. 2010. Increase in rhamnolipid synthesis under iron-limiting conditions influences surface motility and biofilm formation in Pseudomonas aeruginosa. J Bacteriol 192:2973–2980. doi: 10.1128/JB.01601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreau-Marquis S, Bomberger JM, Anderson GG, Swiatecka-Urban A, Ye S, O'Toole GA, Stanton BA. 2008. The ΔF508-CFTR mutation results in increased biofilm formation by Pseudomonas aeruginosa by increasing iron availability. Am J Physiol Lung Cell Mol Physiol 295:L25–L37. doi: 10.1152/ajplung.00391.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan JB, Fine DH. 2002. Biofilm dispersal of Neisseria subflava and other phylogenetically diverse oral bacteria. Appl Environ Microbiol 68:4943–4950. doi: 10.1128/AEM.68.10.4943-4950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.An S, Wu J, Zhang L-H. 2010. Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl Environ Microbiol 76:8160–8173. doi: 10.1128/AEM.01233-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thormann KM, Saville RM, Shukla S, Spormann AM. 2005. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J Bacteriol 187:1014–1021. doi: 10.1128/JB.187.3.1014-1021.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barraud N, Hassett DJ, Hwang S-H, Rice SA, Kjelleberg S, Webb JS. 2006. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol 188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt I, Steenbakkers PJ, op den Camp HJ, Schmidt K, Jetten MS. 2004. Physiologic and proteomic evidence for a role of nitric oxide in biofilm formation by Nitrosomonas europaea and other ammonia oxidizers. J Bacteriol 186:2781–2788. doi: 10.1128/JB.186.9.2781-2788.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Potter AJ, Kidd SP, Edwards JL, Falsetta ML, Apicella MA, Jennings MP, McEwan AG. 2009. Thioredoxin reductase is essential for protection of Neisseria gonorrhoeae against killing by nitric oxide and for bacterial growth during interaction with cervical epithelial cells. J Infect Dis 199:227–235. doi: 10.1086/595737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlson HK, Vance RE, Marletta MA. 2010. H-NOX regulation of c-di-GMP metabolism and biofilm formation in Legionella pneumophila. Mol Microbiol 77:930–942. doi: 10.1111/j.1365-2958.2010.07259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu N, Xu Y, Hossain S, Huang N, Coursolle D, Gralnick JA, Boon EM. 2012. Nitric oxide regulation of cyclic di-GMP synthesis and hydrolysis in Shewanella woodyi. Biochemistry 51:2087–2099. doi: 10.1021/bi201753f. [DOI] [PubMed] [Google Scholar]
- 24.Arora DP, Hossain S, Xu Y, Boon EM. 2015. Nitric oxide regulation of bacterial biofilms. Biochemistry 54:3717–3728. doi: 10.1021/bi501476n. [DOI] [PubMed] [Google Scholar]
- 25.Barraud N, Storey MV, Moore ZP, Webb JS, Rice SA, Kjelleberg S. 2009. Nitric oxide-mediated dispersal in single-and multi-species biofilms of clinically and industrially relevant microorganisms. Microb Biotechnol 2:370–378. doi: 10.1111/j.1751-7915.2009.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abbas A, Adams C, Scully N, Glennon J, O'Gara F. 2007. A role for TonB1 in biofilm formation and quorum sensing in Pseudomonas aeruginosa. FEMS Microbiol Lett 274:269–278. doi: 10.1111/j.1574-6968.2007.00845.x. [DOI] [PubMed] [Google Scholar]
- 27.Barnes RJ, Low JH, Bandi RR, Tay M, Chua F, Aung T, Fane AG, Kjelleberg S, Rice SA. 2015. Nitric oxide treatment for the control of reverse osmosis membrane biofouling. Appl Environ Microbiol 81:2515–2524. doi: 10.1128/AEM.03404-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo J, Zhang J, Barnes RJ, Tan X, McDougald D, Fane AG, Zhuang G, Kjelleberg S, Cohen Y, Rice SA. 2015. The application of nitric oxide to control biofouling of membrane bioreactors. Microb Biotechnol 8:549–560. doi: 10.1111/1751-7915.12261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barraud N, Kelso JM, Rice AS, Kjelleberg S. 2015. Nitric oxide: a key mediator of biofilm dispersal with applications in infectious diseases. Curr Pharm Des 21:31–42. doi: 10.2174/1381612820666140905112822. [DOI] [PubMed] [Google Scholar]
- 30.MacMicking J, Xie Q-W, Nathan C. 1997. Nitric oxide and macrophage function. Annu Rev Immunol 15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 31.Fang FC. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat Rev Microbiol 2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 32.Plate L, Marletta MA. 2012. Nitric oxide modulates bacterial biofilm formation through a multicomponent cyclic-di-GMP signaling network. Mol Cell 46:449–460. doi: 10.1016/j.molcel.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poole R. 2005. Nitric oxide and nitrosative stress tolerance in bacteria. Biochem Soc Trans 33:176–180. doi: 10.1042/BST0330176. [DOI] [PubMed] [Google Scholar]
- 34.Spiro S. 2007. Regulators of bacterial responses to nitric oxide. FEMS Microbiol Rev 31:193–211. doi: 10.1111/j.1574-6976.2006.00061.x. [DOI] [PubMed] [Google Scholar]
- 35.Green J, Paget MS. 2004. Bacterial redox sensors. Nat Rev Microbiol 2:954–966. doi: 10.1038/nrmicro1022. [DOI] [PubMed] [Google Scholar]
- 36.Arai H, Igarashi Y, Kodama T. 1995. The structural genes for nitric oxide reductase from Pseudomonas aeruginosa. Biochim Biophys Acta 1261:279–284. doi: 10.1016/0167-4781(95)00018-C. [DOI] [PubMed] [Google Scholar]
- 37.Arai H, Hayashi M, Kuroi A, Ishii M, Igarashi Y. 2005. Transcriptional regulation of the flavohemoglobin gene for aerobic nitric oxide detoxification by the second nitric oxide-responsive regulator of Pseudomonas aeruginosa. J Bacteriol 187:3960–3968. doi: 10.1128/JB.187.12.3960-3968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giardina G, Rinaldo S, Castiglione N, Caruso M, Cutruzzola F. 2009. A dramatic conformational rearrangement is necessary for the activation of DNR from Pseudomonas aeruginosa. Crystal structure of wild-type DNR. Proteins 77:174–180. doi: 10.1002/prot.22428. [DOI] [PubMed] [Google Scholar]
- 39.Tucker NP, D'Autréaux B, Yousafzai FK, Fairhurst SA, Spiro S, Dixon R. 2008. Analysis of the nitric oxide-sensing non-heme iron center in the NorR regulatory protein. J Biol Chem 283:908–918. doi: 10.1074/jbc.M705850200. [DOI] [PubMed] [Google Scholar]
- 40.Hutchings MI, Mandhana N, Spiro S. 2002. The NorR protein of Escherichia coli activates expression of the flavorubredoxin gene norV in response to reactive nitrogen species. J Bacteriol 184:4640–4643. doi: 10.1128/JB.184.16.4640-4643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bodenmiller DM, Spiro S. 2006. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J Bacteriol 188:874–881. doi: 10.1128/JB.188.3.874-881.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson JL, Brynildsen MP. 2016. Discovery and dissection of metabolic oscillations in the microaerobic nitric oxide response network of Escherichia coli. Proc Natl Acad Sci U S A 113:E1757–E1766. doi: 10.1073/pnas.1521354113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson JL, Jaslove JM, Murawski AM, Fazen CH, Brynildsen MP. 2017. An integrated network analysis reveals that nitric oxide reductase prevents metabolic cycling of nitric oxide by Pseudomonas aeruginosa. Metab Eng 41:67–81. doi: 10.1016/j.ymben.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 44.Hossain S, Boon EM. 2017. Discovery of a novel nitric oxide binding protein and nitric oxide-responsive signaling pathway in Pseudomonas aeruginosa. ACS Infect Dis 3:454–461. doi: 10.1021/acsinfecdis.7b00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barnes RJ, Bandi RR, Wong WS, Barraud N, McDougald D, Fane A, Kjelleberg S, Rice SA. 2013. Optimal dosing regimen of nitric oxide donor compounds for the reduction of Pseudomonas aeruginosa biofilm and isolates from wastewater membranes. Biofouling 29:203–212. doi: 10.1080/08927014.2012.760069. [DOI] [PubMed] [Google Scholar]
- 46.Barraud N, Moscoso JA, Ghigo J-M, Filloux A. 2014. Methods for studying biofilm dispersal in Pseudomonas aeruginosa. Methods Mol Biol 1149:643–651. doi: 10.1007/978-1-4939-0473-0_49. [DOI] [PubMed] [Google Scholar]
- 47.Mathee K, Ciofu O, Sternberg C, Lindum PW, Campbell JI, Jensen P, Johnsen AH, Givskov M, Ohman DE, Søren M. 1999. Mucoid conversion of Pseudomonas aeruginos by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145:1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- 48.Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MR. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol 183:5395–5401. doi: 10.1128/JB.183.18.5395-5401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoon SS, Karabulut AC, Lipscomb JD, Hennigan RF, Lymar SV, Groce SL, Herr AB, Howell ML, Kiley PJ, Schurr MJ. 2007. Two-pronged survival strategy for the major cystic fibrosis pathogen, Pseudomonas aeruginosa, lacking the capacity to degrade nitric oxide during anaerobic respiration. EMBO J 26:3662–3672. doi: 10.1038/sj.emboj.7601787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koskenkorva T, Aro-Kärkkäinen N, Bachmann D, Arai H, Frey AD, Kallio PT. 2008. Transcriptional activity of Pseudomonas aeruginosa fhp promoter is dependent on two regulators in addition to FhpR. Arch Microbiol 189:385–396. doi: 10.1007/s00203-007-0329-3. [DOI] [PubMed] [Google Scholar]
- 51.Schleheck D, Barraud N, Klebensberger J, Webb JS, McDougald D, Rice SA, Kjelleberg S. 2009. Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. PLoS One 4:e5513. doi: 10.1371/journal.pone.0005513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Helmick RA, Fletcher AE, Gardner AM, Gessner CR, Hvitved AN, Gustin MC, Gardner PR. 2005. Imidazole antibiotics inhibit the nitric oxide dioxygenase function of microbial flavohemoglobin. Antimicrob Agents Chemother 49:1837–1843. doi: 10.1128/AAC.49.5.1837-1843.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen T-K, Duong HT, Selvanayagam R, Boyer C, Barraud N. 2015. Iron oxide nanoparticle-mediated hyperthermia stimulates dispersal in bacterial biofilms and enhances antibiotic efficacy. Sci Rep 5:18385. doi: 10.1038/srep18385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hausladen A, Gow AJ, Stamler JS. 1998. Nitrosative stress: metabolic pathway involving the flavohemoglobin. Proc Natl Acad Sci U S A 95:14100–14105. doi: 10.1073/pnas.95.24.14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gardner PR, Gardner AM, Martin LA, Salzman AL. 1998. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc Natl Acad Sci U S A 95:10378–10383. doi: 10.1073/pnas.95.18.10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zumft WG. 2005. Nitric oxide reductases of prokaryotes with emphasis on the respiratory, heme-copper oxidase type. J Inorg Biochem 99:194–215. doi: 10.1016/j.jinorgbio.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 57.Hendriks J, Oubrie A, Castresana J, Urbani A, Gemeinhardt S, Saraste M. 2000. Nitric oxide reductases in bacteria. Biochim Biophys Acta 1459:266–273. doi: 10.1016/S0005-2728(00)00161-4. [DOI] [PubMed] [Google Scholar]
- 58.Bonamore A, Boffi A. 2008. Flavohemoglobin: structure and reactivity. IUBMB Life 60:19–28. doi: 10.1002/iub.9. [DOI] [PubMed] [Google Scholar]
- 59.Gonçalves VL, Nobre LS, Vicente JB, Teixeira M, Saraiva LM. 2006. Flavohemoglobin requires microaerophilic conditions for nitrosative protection of Staphylococcus aureus. FEBS Lett 580:1817–1821. doi: 10.1016/j.febslet.2006.02.039. [DOI] [PubMed] [Google Scholar]
- 60.Poole RK, Anjum MF, Membrillo-Hernández J, Kim SO, Hughes MN, Stewart V. 1996. Nitric oxide, nitrite, and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J Bacteriol 178:5487–5492. doi: 10.1128/jb.178.18.5487-5492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fang FC. 1997. Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J Clin Invest 99:2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cueto M, Hernández-Perera O, Martín R, Bentura ML, Rodrigo J, Lamas S, Golvano MP. 1996. Presence of nitric oxide synthase activity in roots and nodules of Lupinus albus. FEBS Lett 398:159–164. doi: 10.1016/S0014-5793(96)01232-X. [DOI] [PubMed] [Google Scholar]
- 63.Wang Y, Dunn AK, Wilneff J, McFall-Ngai MJ, Spiro S, Ruby EG. 2010. Vibrio fischeri flavohaemoglobin protects against nitric oxide during initiation of the squid-Vibrio symbiosis. Mol Microbiol 78:903–915. doi: 10.1111/j.1365-2958.2010.07376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y, Ruby EG. 2011. The roles of NO in microbial symbioses. Cell Microbiol 13:518–526. doi: 10.1111/j.1462-5822.2011.01576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ollesch G, Kaunzinger A, Juchelka D, Schubert-Zsilavecz M, Ermler U. 1999. Phospholipid bound to the flavohemoprotein from Alcaligenes eutrophus. Eur J Biochem 262:396–405. doi: 10.1046/j.1432-1327.1999.00381.x. [DOI] [PubMed] [Google Scholar]
- 66.Ilari A, Bonamore A, Farina A, Johnson KA, Boffi A. 2002. The X-ray structure of ferric Escherichia coli flavohemoglobin reveals an unexpected geometry of the distal heme pocket. J Biol Chem 277:23725–23732. doi: 10.1074/jbc.M202228200. [DOI] [PubMed] [Google Scholar]
- 67.Gardner PR. 2005. Nitric oxide dioxygenase function and mechanism of flavohemoglobin, hemoglobin, myoglobin and their associated reductases. J Inorg Biochem 99:247–266. doi: 10.1016/j.jinorgbio.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 68.Ermler U, Siddiqui RA, Cramm R, Friedrich B. 1995. Crystal structure of the flavohemoglobin from Alcaligenes eutrophus at 1.75 A resolution. EMBO J 14:6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rani N, Sharma A, Singh R. 2013. Imidazoles as promising scaffolds for antibacterial activity: a review. Mini Rev Med Chem 13:1812–1835. doi: 10.2174/13895575113136660091. [DOI] [PubMed] [Google Scholar]
- 70.De Luca L. 2006. Naturally occurring and synthetic imidazoles: their chemistry and their biological activities. Curr Med Chem 13:1–23. [PubMed] [Google Scholar]
- 71.Gupta GK, Kumar V, Kaur K. 2014. Imidazole containing natural products as antimicrobial agents: a review. Nat Prod J 4:73–81. [Google Scholar]
- 72.Carmely S, Kashman Y. 1987. Naamines and naamidines, novel imidazole alkaloids from the calcareous sponge Leucetta chagosensis. Tetrahedron Lett 28:3003–3006. doi: 10.1016/S0040-4039(00)96268-3. [DOI] [Google Scholar]
- 73.Fu X, Barnes JR, Do T, Schmitz FJ. 1997. New imidazole alkaloids from the sponge Leucetta chagosensis. J Nat Prod 60:497–498. doi: 10.1021/np960694i. [DOI] [Google Scholar]
- 74.He HY, Faulkner DJ, Lee AY, Clardy J. 1992. A new imidazole alkaloid from the marine sponge Leucetta microrhaphis. J Org Chem 57:2176–2178. doi: 10.1021/jo00033a051. [DOI] [Google Scholar]
- 75.Holloway B, Krishnapillai V, Morgan A. 1979. Chromosomal genetics of Pseudomonas. Microbiol Rev 43:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Held K, Ramage E, Jacobs M, Gallagher L, Manoil C. 2012. Sequence-verified two-allele transposon mutant library for Pseudomonas aeruginosa PAO1. J Bacteriol 194:6387–6389. doi: 10.1128/JB.01479-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maragos CM, Morley D, Wink DA, Dunams TM, Saavedra JE, Hoffman A, Bove AA, Isaac L, Hrabie JA, Keefer LK. 1991. Complexes of NO with nucleophiles as agents for the controlled biological release of nitric oxide. Vasorelaxant effects. J Med Chem 34:3242–3247. doi: 10.1021/jm00115a013. [DOI] [PubMed] [Google Scholar]
- 79.Keefer LK, Nims RW, Davies KM, Wink DA. 1996. “NONOates” (1-substituted diazen-1-ium-1, 2-diolates) as nitric oxide donors: convenient nitric oxide dosage forms. Methods Enzymol 268:281–293. doi: 10.1016/S0076-6879(96)68030-6. [DOI] [PubMed] [Google Scholar]
- 80.Gardner AM, Gardner PR. 2002. Flavohemoglobin detoxifies nitric oxide in aerobic, but not anaerobic, Escherichia coli: evidence for a novel inducible anaerobic nitric oxide-scavenging activity. J Biol Chem 277:8166–8171. doi: 10.1074/jbc.M110470200. [DOI] [PubMed] [Google Scholar]
- 81.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 82.Gräslund S, Nordlund P, Weigelt J, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, Ming J. 2008. Protein production and purification. Nat Methods 5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Savitsky P, Bray J, Cooper CD, Marsden BD, Mahajan P, Burgess-Brown NA, Gileadi O. 2010. High-throughput production of human proteins for crystallization: the SGC experience. J Struct Biol 172:3–13. doi: 10.1016/j.jsb.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.