ABSTRACT
Six imported pigs originating from Guangdong, Henan, and Hunan provinces in China during October 2015 to February 2017 were cultured and found to be positive for meropenem-resistant Escherichia coli. The samples yielded 9 E. coli isolates of diverse sequence types carrying blaNDM-5 on IncX3 (8 isolates from 5 farms) or IncFII (1 isolate from 1 farm) plasmids. The mcr-1 gene was coharbored by 4 isolates. The IncX3 plasmids (∼46 kb) carrying blaNDM-5 were identical or nearly identical to each other.
KEYWORDS: carbapenems, antimicrobial resistance epidemiology, molecular epidemiology, Enterobacteriaceae
TEXT
The presence of carbapenemase-producing Enterobacteriaceae (CPE) in livestock animals is of concern because this may facilitate expansion of the gene pool from which pathogenic bacteria can pick up resistance genes, and consumers may subsequently be exposed through the food chain (1, 2). For this reason, there is a need to enhance the monitoring of carbapenem resistance in the food supply (1). In Hong Kong, 80% of the food animals are imported from mainland China and involve farm suppliers from multiple provinces in the country (3).
From September 2008 to February 2017, rectal swabs were obtained from randomly selected fresh pig carcasses at a centralized slaughterhouse in Hong Kong by trained veterinary staff, as part of an ongoing surveillance (3). Each swab was collected from a single animal and was inoculated into nutrient broth with 10 mg/liter vancomycin and 0.5 mg/liter meropenem (4), followed by subculture on a MacConkey agar plate supplemented with 2 mg/liter meropenem. Five to ten colonies from each selective plate were picked. Matrix-assisted laser desorption ionization–time of flight mass spectrometry was used for bacterial identification. The agar dilution (for colistin) and disc diffusion (for other antibiotics) methods were used to determine antimicrobial susceptibility (5, 6). Isolates from the same animal were considered to be unique if the resistance profiles for meropenem and colistin were different.
In total, 856 pigs were cultured over 263 sampling dates (see Table S1 in the supplemental material). Six pigs originating from six different farms were cultured and found to be positive for meropenem-resistant Escherichia coli (Table 1). According to the susceptibility patterns, a total of nine isolates were considered to be unique and were investigated further. All isolates had positive CarbaNP test results and were resistant to ertapenem, imipenem, and meropenem. The presence of carbapenemase genes and mcr-1 was investigated by PCR and sequencing (7–9). The blaNDM-5 gene was identified in all nine isolates, of which four were resistant to colistin and coharbored mcr-1. The isolates were further investigated by multilocus sequence typing (MLST) and replicon typing (7, 9, 10). Plasmids carrying blaNDM-5 were X3 (n = 8, ∼45 kb) or F36 (n = 1, ∼100 kb). Of the four mcr-1 genes identified, three were harbored on plasmids of different replicon types (X4, FIB, and Y). In conjugation experiments, no cotransfer of carbapenem and colistin coresistance was seen, but the plasmids carrying blaNDM or mcr-1 could be transferred separately at frequencies of 10−4 to 10−5 and 10−1 to 10−6 transconjugants per donor cell, respectively (9, 10).
TABLE 1.
Sources and characteristics for nine NDM-positive E. coli isolates
| Strain | Specimen | Sourceb | Date collected | MLST | blaNDM-5 | mcr-1 | Replicon type of plasmid harboring: |
Resistance patternd | |
|---|---|---|---|---|---|---|---|---|---|
| blaNDM | mcr-1 | ||||||||
| P744A | Pig 1 | Henan (A1) | October 2015 | ST10 | + | − | X3 | None | Chl, Nit |
| P744Ta | Pig 1 | Henan (A1) | October 2015 | ST1602 | + | + | X3 | X4 | Chl, Cip, Nit |
| P748a | Pig 2 | Hunan (B) | January 2016 | ST167 | + | − | F36 | None | Gen, Chl, Cip, Nit |
| P768 | Pig 3 | Henan (A2) | May 2016 | ST117 | + | − | X3 | None | Gen, Chl, Cip |
| P768-11a | Pig 3 | Henan (A2) | May 2016 | ST871 | + | + | X3 | FIB | Cip |
| P785a | Pig 4 | Guangdong (C1) | June 2016 | ST7512 | + | + | X3 | Chromosomalc | Gen, Chl, Cip, Nit |
| P788A | Pig 5 | Guangdong (C2) | June 2016 | ST1286 | + | − | X3 | None | Nit |
| P788A-32a | Pig 5 | Guangdong (C2) | June 2016 | ST7510 | + | + | X3 | Y | Gen, Chl, Nit |
| P855a | Pig 6 | Guangdong (C3) | February 2017 | ST7511 | + | − | X3 | None | Gen, Chl, Cip, Nit |
The six isolates were investigated further by genome sequencing.
Province (farm) origin of the pig.
Chromosomal location of mcr-1 in the isolate was confirmed by genome sequencing.
Resistance patterns for amikacin (Ak), chloramphenicol (Chl), ciprofloxacin (Cip), fosfomycin (Fos), gentamicin (Gen), and nitrofurantoin (Nit).
Six isolates (one from each animal) were sequenced by an Illumina MiSeq platform at >150-fold coverage (Table 1). The plasmids were assembled de novo using a CLC Genomics Workbench (Qiagen, Redwood City, CA), and gaps were closed by additional PCR and Sanger sequencing (7, 9, 10). ISfinder (https://www-is.biotoul.fr/about.php) was used to identify and annotate insertion sequences. In strain P748, blaNDM-5 was found in a contig (∼32 kb) with 100% coverage and 98% identity to p28078-NDM (GenBank accession no. MF156713).
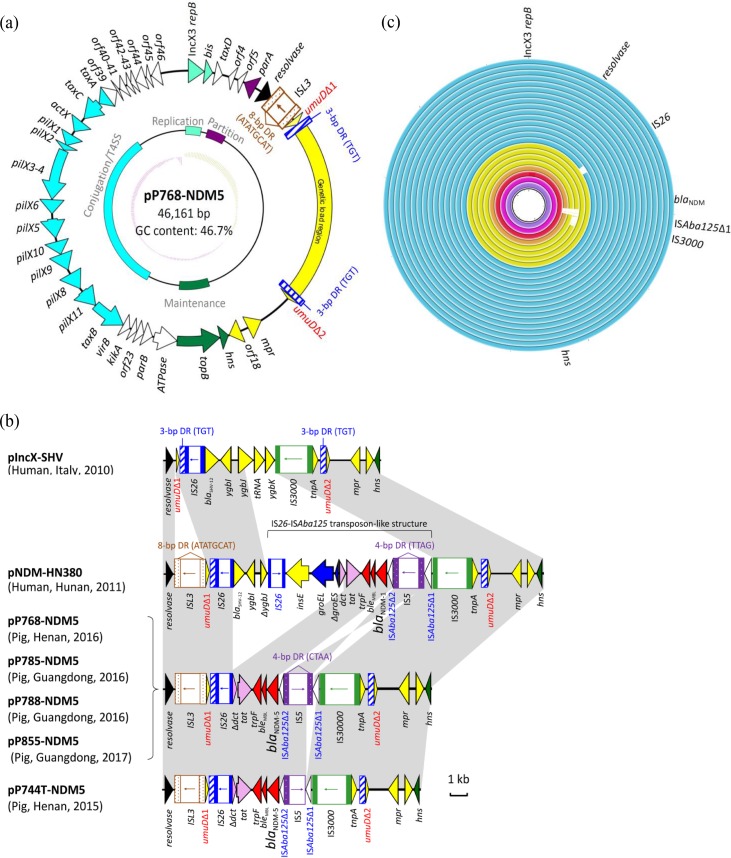
Complete sequences of the five ∼46-kb IncX3 plasmids were obtained (see Table S2 in the supplemental material). They were found to have plasmid scaffolds typical of IncX3 plasmids (Fig. 1a). The genetic load regions in the five plasmids were compared with two reference IncX3 plasmids (pNDM-HN380 and pIncX-SHV) (Fig. 1b). In the blaNDM-carrying plasmids, an ISL3 with 8-bp flanking direct repeats (ATATGCAT) was found downstream of the resolvase gene. The umuD gene was split into two fragments (umuDΔ1 and umuDΔ2) at the same position as in pIncX-SHV, resulting in a pair of 3-bp direct repeats (TGT). In pNDM-HN380, blaNDM was inserted as an IS26-ISAba125 transposon-like structure (Fig. 1b). Subsequently, the upstream ISAba125 was disrupted by IS5 (10). In four plasmids with links to the Guangdong and Henan provinces, the sequences inserted between the two umuD fragments were 100% identical (10,117 bp in length). This inserted sequence differed from that in pNDM-HN380 by a deletion of 7,874 bp (Fig. 1b). The remaining plasmid with a link to Henan had an additional deletion (616 bp) at the junction between the IS5 and ISAba125Δ1 remnants. In the five NDM-5 plasmids, IS5 was inserted at the same position, leading to the flanking 4-bp direct repeats (CTAA). In pNDM-HN380, IS5 was inserted at a different position in the opposite orientation.
FIG 1.
Comparisons of IncX plasmids in this study. (a) Circular map of plasmid pP768-NDM-5. This plasmid was used to illustrate the backbone shared by all the analyzed plasmids and the location of the genetic load region. (b) Comparison of the genetic load region in 5 plasmids harboring blaNDM-5 with 2 reference plasmids (pIncX-SHV and pNDM-HN380). (c) Alignment of pP768-NDM-5 with 22 plasmids identified in GenBank (last accessed 27 October 2017). The circular maps were generated with the BLAST Ring Image Generator, and each plasmid was colored by the geographic origin (China, blue; Myanmar, yellow; Oman, orange; India, red; Canada, pink; and Kuwait, purple) in the following order (outer to inner circles): pP768-NDM-5, pCREC-A6-NDM, pSCE516-2, pNDM-5_IncX3, pEc1929, pECNDM101, pAD-19R, pNDM-5_WCHEC0215, pK518_NDM-5, pK516_NDM-5, NUHL24835, pNDM-QD28, pNDM-QD29, pEC50-NDM-7, pZHDC40, pJEG027, pM216_X3, pM213_X3 and pM110_X3, pOM26-1, pNDM-MGR194 and pKpN01-NDM7, and pKW53T-NDM (see full details of plasmids in Table S3).
To explore the geographic distribution of potentially related blaNDM-carrying IncX3 plasmids, the complete sequence of pP768-NDM-5 (chosen as a representative) was used to query the GenBank database. Twenty-two plasmids related to pP768-NDM-5 were identified (Fig. 1c), including 14 plasmids from China, 4 from Myanmar, and 1 each from Canada, India, Kuwait, and Oman (see Table S3 in the supplemental material). The plasmids did not carry resistance genes other than blaNDM. Multiple NDM variants were carried by the plasmids. These include NDM-1 and variants that differed by one to three amino acids, including NDM-4 (M154L), NDM-5 (V88L, M154L), NDM-7 (D130N, M154L), and NDM-17 (V88L, M154L, E170K).
We identified the occurrence of similar IncX3 plasmids carrying blaNDM-5 in pigs originating from multiple farms across three different Chinese provinces. The involvement of IncX3 plasmids (represented by pNDM-HN380) in the dissemination of NDM in multiple geographic areas in China was initially reported by our group in 2012 (7). Subsequently, sporadic reports of pNDM-HN380-like plasmids carrying NDM variants have been reported in India, the Arabian Peninsula, Europe, and Australia (11–14). Recently, a Chinese national survey and several provincial studies revealed that IncX3 plasmids harboring different blaNDM variants were frequently found among clinical isolates of different MLSTs and species, suggesting that they represent an important vector responsible for the wide dissemination of NDM in China (15–17). IncX3 plasmids have a narrow host range and have been found mainly in Enterobacteriaceae (18). Our finding from analysis of complete plasmid sequences indicates that the five IncX3 plasmids originating from pigs are related to pNDM-HN380 and plasmids originating from many other geographic areas, thus confirming that this mobile NDM vector is widespread in the ecosystem.
As carbapenems have never been licensed for use in food animals in China, the NDM-producing pig isolates detected in the present study may have been introduced to the farms via human activity or contaminated feeds. It is worrisome that some of the NDM-producing isolates in the present study were found to coharbor mcr-1 in another plasmid or the chromosome. Nonetheless, our isolates that coharbored mcr-1 were recovered before the ban of colistin in animal feeds was implemented in China in November 2016.
In conclusion, this study identified an epidemic IncX3 plasmid carrying blaNDM-5 disseminated among E. coli originating from pigs with epidemiological links to geographically segregated areas in China.
Accession number(s).
The complete sequences of the five IncX3 plasmids were deposited in the GenBank database under accession numbers MF547511 (pP744-NDM-5), MF547510 (pP768-NDM-5), MF547509 (pP785-NDM-5), MF547507 (pP788A-NDM-5), and MF547508 (pP855-NDM-5).
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff at the Food and Environmental Hygiene Department (FEHD) of the Hong Kong Special Administrative Region (HKSAR) for assistance with specimen collection.
This work was supported by grants from the Health and Medical Research Fund of the Food and Health Bureau of the HKSAR and the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Disease for the HKSAR Department of Health.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02295-17.
REFERENCES
- 1.Woodford N, Wareham DW, Guerra B, Teale C. 2014. Carbapenemase-producing Enterobacteriaceae and non-Enterobacteriaceae from animals and the environment: an emerging public health risk of our own making? J Antimicrob Chemother 69:287–291. doi: 10.1093/jac/dkt392. [DOI] [PubMed] [Google Scholar]
- 2.Mollenkopf DF, Stull JW, Mathys DA, Bowman AS, Feicht SM, Grooters SV, Daniels JB, Wittum TE. 2017. Carbapenemase-producing Enterobacteriaceae recovered from the environment of a swine farrow-to-finish operation in the United States. Antimicrob Agents Chemother 61:e01298-. doi: 10.1128/AAC.01298-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho PL, Chow KH, Lai EL, Lo WU, Yeung MK, Chan J, Chan PY, Yuen KY. 2011. Extensive dissemination of CTX-M-producing Escherichia coli with multidrug resistance to ‘critically important’ antibiotics among food animals in Hong Kong, 2008-10. J Antimicrob Chemother 66:765–768. doi: 10.1093/jac/dkq539. [DOI] [PubMed] [Google Scholar]
- 4.Darling LA, Evans AM, Stellrecht KA, Nattanmai SM, Montero CI. 2017. A Triple-disk enrichment method for carbapenem-resistant Enterobacteriaceae (CRE) screening. J Clin Microbiol 55:3557–3559. doi: 10.1128/JCM.01185-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing—27th ed. CLSI supplement M100S. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 6.Wong SC, Tse H, Chen JH, Cheng VC, Ho PL, Yuen KY. 2016. Colistin-resistant Enterobacteriaceae carrying the mcr-1 gene among patients in Hong Kong. Emerg Infect Dis 22:1667–1669. doi: 10.3201/eid2209.160091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho PL, Li Z, Lo WU, Cheung YY, Lin CH, Sham PC, Cheng VC, Ng TK, Que TL, Chow KH. 2012. Identification and characterization of a novel incompatibility group X3 plasmid carrying blaNDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg Microbes Infect 1:e39. doi: 10.1038/emi.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho PL, Cheung YY, Wang Y, Lo WU, Lai EL, Chow KH, Cheng VC. 2016. Characterization of carbapenem-resistant Escherichia coli and Klebsiella pneumoniae from a healthcare region in Hong Kong. Eur J Clin Microbiol Infect Dis 35:379–385. doi: 10.1007/s10096-015-2550-3. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Lo WU, Lai RW, Tse CW, Lee RA, Luk WK, Cheng VC, Que TL, Chow KH, Ho PL. 2017. IncN ST7 epidemic plasmid carrying blaIMP-4 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. J Antimicrob Chemother 72:99–103. doi: 10.1093/jac/dkw353. [DOI] [PubMed] [Google Scholar]
- 10.Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, Ang I, Tong AH, Bao JY, Lok S, Lo JY. 2011. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One 6:e17989. doi: 10.1371/journal.pone.0017989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wailan AM, Paterson DL, Caffery M, Sowden D, Sidjabat HE. 2015. Draft genome sequence of NDM-5-producing Escherichia coli sequence type 648 and genetic context of blaNDM-5 in Australia. Genome Announc 3(2):e00194-15. doi: 10.1128/genomeA.00194-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petrosillo N, Vranic-Ladavac M, Feudi C, Villa L, Fortini D, Barisic N, Bedenic B, Ladavac R, D'Arezzo S, Andrasevic AT, Capone A. 2016. Spread of Enterobacter cloacae carrying blaNDM-1, blaCTX-M-15, blaSHV-12 and plasmid-mediated quinolone resistance genes in a surgical intensive care unit in Croatia. J Glob Antimicrob Resist 4:44–48. doi: 10.1016/j.jgar.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Choudhury NA, Paul D, Chakravarty A, Bhattacharjee A, Dhar Chanda D. 2018. IncX3 plasmid mediated occurrence of blaNDM-4 within Escherichia coli ST448 from India. J Infect Public Health 1:111–114. doi: 10.1016/j.jiph.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Riazzo C, Lopez-Cerero L, Rojo-Martin MD, Hoyos-Mallecot Y, Fernandez-Cuenca F, Martin-Ruiz JL, Pascual-Hernandez A, Naas T, Navarro-Mari JM. 2017. First report of NDM-1-producing clinical isolate of Leclercia adecarboxylata in Spain. Diagn Microbiol Infect Dis 88:268–270. doi: 10.1016/j.diagmicrobio.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 15.Zhang R, Liu L, Zhou H, Chan EW, Li J, Fang Y, Li Y, Liao K, Chen S. 2017. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine 19:98–106. doi: 10.1016/j.ebiom.2017.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Q, Fang L, Fu Y, Du X, Shen Y, Yu Y. 2015. Dissemination of NDM-1-producing Enterobacteriaceae mediated by the IncX3-type plasmid. PLoS One 10:e0129454. doi: 10.1371/journal.pone.0129454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang F, Xie L, Wang X, Han L, Guo X, Ni Y, Qu H, Sun J. 2016. Further spread of blaNDM-5 in Enterobacteriaceae via IncX3 plasmids in Shanghai, China. Front Microbiol 7:424. doi: 10.3389/fmicb.2016.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. 2012. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68:43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.