Abstract
Incomplete killing of bacterial pathogens by antibiotics is an underlying cause of treatment failure and accompanying complications. Among those avoiding chemotherapy are persisters being individual cells in a population that for extended periods of time survive high antibiotic concentrations proposedly by being in a quiescent state refractory to antibiotic killing. While investigating the human pathogen Staphylococcus aureus and the influence of growth phase on persister formation, we noted that spent supernatants of stationary phase cultures of S. aureus or S. epidermidis, but not of distantly related bacteria, significantly reduced the persister cell frequency upon ciprofloxacin challenge when added to exponentially growing and stationary phase S. aureus cells. Curiously, the persister reducing activity of S. aureus supernatants was also effective against persisters formed by either S. carnosus or Listeria monocytogenes. The persister reducing component, which resisted heat but not proteases and was produced in the late growth phase in an agr quorum-sensing dependent manner, was identified to be the phenol-soluble modulin (PSM) toxins. S. aureus express several PSMs, each with distinct cytolytic and antimicrobial properties; however, the persister reducing activity was specifically linked to synthesis of the PSMα family. Correspondingly, a high-persister phenotype of a PSMα mutant was observed upon fluoroquinolone or aminoglycoside challenge, demonstrating that the persister reducing activity of PSMs can be endogenously synthesized or extrinsically added. Given that PSMs have been associated with lytic activity against bacterial membranes we propose that PSM toxins increase the susceptibility of persister cells to killing by intracellularly acting antibiotics and that chronic and re-occurring infections with quorum sensing, agr negative mutants may be difficult to treat with antibiotics because of persister cells formed in the absence of PSM toxins.
Keywords: S. aureus, persister cells, quorum sensing, agr, phenol-soluble modulins, supernatant
Introduction
Bacterial pathogens that are refractory to antibiotic treatment represent a serious threat to human health. Much attention has been paid to genetically defined antibiotic resistance mechanisms, while more recent awareness has been turned toward non-inherited or epigenetic resistance mechanisms as exemplified by the identification of intrinsic resistance genes (Morris et al., 2005; Vestergaard et al., 2016; El-Halfawy et al., 2017; Ito et al., 2017) or genes that upon induction provide tolerance phenotypes (Geisinger and Isberg, 2015; Haaber et al., 2015). Related to this category is the phenomenon of persisters. Persisters are formed when a fraction of a bacterial population enters a state of dormancy or slow growth that enables them to survive very high concentrations of antibiotics (Balaban et al., 2004; Lewis, 2010; Maisonneuve and Gerdes, 2014; Prax and Bertram, 2014; Schumacher et al., 2015). Clinically, persisters appear to be of critical importance as they contribute to persistent infections that are recurring and difficult to eradicate by antibiotics (Mulcahy et al., 2010; Conlon et al., 2015; Schumacher et al., 2015; Fisher et al., 2017; Van den Bergh et al., 2017). Strikingly, persisters can form a sub-population from which genetically encoded resistant variants emerge (Cohen et al., 2013; Levin-Reisman et al., 2017; Sebastian et al., 2017) and high-persister mutants are selected for by antibiotic exposure (Mechler et al., 2015; Van den Bergh et al., 2016). Consequently the formation of persister cells and methods to eradicate them is of great interest.
Persister cells appear to be formed stochastically during growth (termed type II persisters) (Balaban et al., 2004) and in response to adverse conditions. In Escherichia coli the stochastically induced persisters rely on the stringent response-mediated expression of guanosine tetraphosphate (ppGpp) and the concomitant cease in cell growth results in antibiotic tolerance (Amato et al., 2013; Maisonneuve et al., 2013; Verstraeten et al., 2015; Van den Bergh et al., 2016). Environmental conditions such as acid and oxidative stress, interactions with host environments and various fluctuations in metabolism or respiration also influences persister generation (Hong et al., 2012; Helaine et al., 2014; Orman and Brynildsen, 2015; Harms et al., 2016; Radzikowski et al., 2016). In stationary growth phase persister cells (termed type I persisters) generally form in a much greater fraction of the bacterial population than in exponential growth phase (Balaban et al., 2004; Keren et al., 2004; Lechner et al., 2012), and it has been speculated that biofilms may be largely composed of persister cells (Conlon et al., 2015; Waters et al., 2016). Entry into stationary growth phase is for many bacterial pathogens associated with induction of quorum sensing systems. The timing of quorum sensing activation is reflecting entry into stationary growth phase via sensing of so-called auto-inducers reaching a threshold concentration. Apart from that, quorum sensing systems are fundamentally different in Gram-negative and Gram-positive bacteria with the auto-inducing molecule generally being a peptide in the latter group. Quorum sensing coordinates diverse phenotypes such as expression of virulence factors, genetic competence, and biofilm formation (Miller and Bassler, 2001; Moreno-Gámez et al., 2017). Interestingly, quorum sensing induction itself has also been implicated in induction of antibiotic tolerance and persister cell generation in both Gram-negative and -positive bacteria (Möker et al., 2010; Leung and Lévesque, 2012; Que et al., 2013).
Staphylococcus aureus is a versatile pathogen causing a wide range of infections including abscesses, osteomyelitis, wound infections, endocarditis, and infections related to indwelling devices (Lowy, 1998; DeLeo et al., 2010). Chronic infections are also common and here persister cell formation appears to be central as exemplified by eradication of a chronic infection with an anti-persister compound (Conlon et al., 2013; Conlon, 2014). In contrast to other organisms the stringent response is not involved in S. aureus persister generation; however, the importance of the stationary growth phase for persister formation in S. aureus has been highlighted by the finding that genes normally expressed in stationary phase are induced in persister cells formed during exponential growth, and that these cells contain less ATP compared to growing cells again characterizing cells in the stationary growth phase (Conlon et al., 2016). Inspired by recent reports demonstrating that culture supernatants of S. aureus stimulate resuscitation of otherwise non-culturable S. aureus biofilm cells (Pasquaroli et al., 2013) and artificially generated S. aureus dormant cells (Pascoe et al., 2014), we set out to explore whether such spent medium would affect the frequency of persister cells observed in S. aureus cultures.
Results and Discussion
S. aureus Exponential Phase Persister Cell Frequency Is Reduced by Medium Spent by Staphylococci
Persister cell formation in S. aureus has previously been studied in response to lethal concentrations of ciprofloxacin or oxacillin using a variety of different strains (Keren et al., 2004; Lechner et al., 2012; Johnson and Levin, 2013; Conlon et al., 2016). Since some S. aureus strains are known to aggregate even at low cell densities and such aggregation confers antibiotic tolerance (Haaber et al., 2012), we chose to study formation of persister cells in exponential phase using strain Newman that reportedly is not or low-aggregating (Haaber et al., 2012). We conducted time-kill experiments by adding 20x MIC ciprofloxacin to growing cultures and observed the characteristic bi-phasic killing kinetics reflecting persister cell formation (Supplementary Figure S1) with 0.0001–0.001% surviving after 24 h, which is comparable to what was reported previously (Johnson and Levin, 2013). When repeating the time-kill protocol with persister cells collected at 24 h the killing kinetics and colony forming units (CFU) remained essentially unchanged demonstrating that the survivors are true persisters. In subsequent assays, we recorded persisters 24 h after antibiotic treatment.
When studying the influence of growth phases on persister formation in S. aureus strain Newman we discovered, by serendipity, that supplementing the TSB growth medium with 25% spent supernatant from stationary phase cells strikingly reduced exponential phase persister cells surviving lethal concentrations of ciprofloxacin by 10 to 100-fold when compared to cells just grown in plain TSB. Similar reduction was observed with supernatant obtained from the closely related species, S. epidermidis, while no effect was observed with supernatants from S. carnosus or Escherichia coli (Figure 1A). The supernatant effect is dose-dependent with moderate reduction in ciprofloxacin persisters observed already when supplemented with 1% supernatant while maximum activity, approaching the assay detection limit (1 CFU/ml), required 10–25% supernatant (Figure 1B). Importantly, the ciprofloxacin minimum inhibitory concentration (MIC) for S. aureus Newman (0.5 μg/ml) was unaffected by the presence of spent supernatant. We observed that the same supernatants affected the persister frequency also when selecting with oxacillin while the supernatant effect was negligible or absent when selecting for persisters with vancomycin (Supplementary Figure S2). To probe the generality of our observation we found that persister frequencies of two other S. aureus strains, SA564 and 8325-4, that are characterized as being able to form persister cells at low and relatively high frequencies, respectively, were also greatly affected by their respective supernatants (Supplementary Figure S3). We conclude that a component present in spent supernatants from S. aureus and closely related staphylococcal species significantly reduces persister cell formation and that the effect depends on the antibiotic with which they are isolated. Hence, we focused on persisters selected with ciprofloxacin onwards.
FIGURE 1.
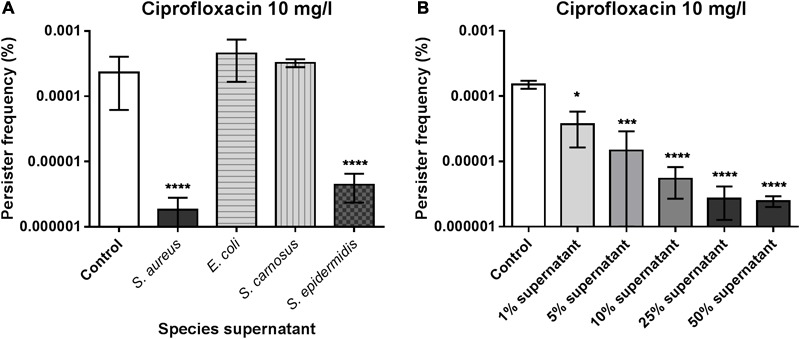
Staphylococcus aureus type II persister cell frequency is reduced in the presence of endogenous or closely related staphylococcal supernatants. S. aureus strain Newman was grown exponentially in the presence of 25% S. aureus (Newman), E. coli, S. carnosus or S. epidermidis stationary phase supernatants and challenged with 20x MIC of ciprofloxacin (A). The persister cell frequency was determined as the fraction of survivors in individual cultures following 24 h antibiotic treatment. The dose-dependence of the S. aureus supernatant was assessed by 1–50% supplementation using 20x MIC of ciprofloxacin (B). All experimental conditions were compared to the un-supplemented control (TSB medium). The data represent the mean persister frequencies ± SD calculated from three biological replicates (∗P < 0.05, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001).
The persister cell frequency increases dramatically when S. aureus enters stationary phase (Keren et al., 2004). Therefore, we examined if persister cells formed in exponential phase are stationary phase cells carried over by dilution and if the effect of the spent supernatant was preserved when added to cells passaged for multiple growth cycles. By employing a successive back-dilution strategy (Figure 2A) we demonstrated that, in our assay, the persister cells formed during exponential growth are not carried over from the inoculum (Figure 2B) and that the spent supernatant remains equally active in reducing persister cells formation when added to cells after two rounds of passaging (Figure 2B). A previous study reported that S. aureus stationary phase supernatants are able to resuscitate dormant cells and that this effect could be mediated by two putative transglycosylases, IsaA and SceD (Pascoe et al., 2014). However, inactivation of either isaA, sceD or both genes did not alter the supernatant activity in our persister assay (Supplementary Figure S4), indicating that other factors are responsible for the observed effect.
FIGURE 2.
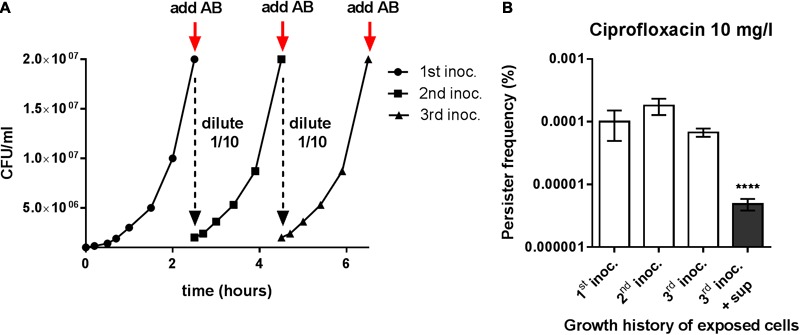
Persister reducing activity is unrelated to the stationary phase inoculum. (A) Setup of the re-inoculation experiment to assay for true type II persisters. The initial culture (1st inoc.) was generated by a 1000-fold dilution of stationary phase cells in fresh TSB followed by growth for 2.5 h to enter exponential growth and reach a cell density of approximately 2 × 107 CFU/ml. The subsequent cultures (2nd and 3rd inoc.) were derived from the initial culture by successive back-dilution (1/10) in fresh TSB and growth until reaching the same cell density. (B) Persister frequencies obtained after challenging the different S. aureus strain Newman cultures with 20x MIC of ciprofloxacin. The third culture was grown in the presence or absence of 25% stationary phase endogenous supernatant. The data represent the mean persister frequencies ± SD calculated from three biological replicates (∗∗∗∗P < 0.0001).
The Active Component Is Proteinaceous and Present within the >30 kDa Fraction
To assess the nature of the component(s) responsible for the observed reduction in isolated persister cells, we subjected the supernatant to heat or protease treatment (Figure 3A) and spin-column fractionation (Figure 3B) prior to testing in the persister assay. By this, we defined the supernatant entity as being a heat-stable, proteinaceous substance that is enriched in the high molecular weight fraction; both of these characteristics further confirming that the active component is distinct from the resuscitation factor described by Pascoe et al. (2014) that was reported to be heat-sensitive and present in the <30 kDa fraction.
FIGURE 3.
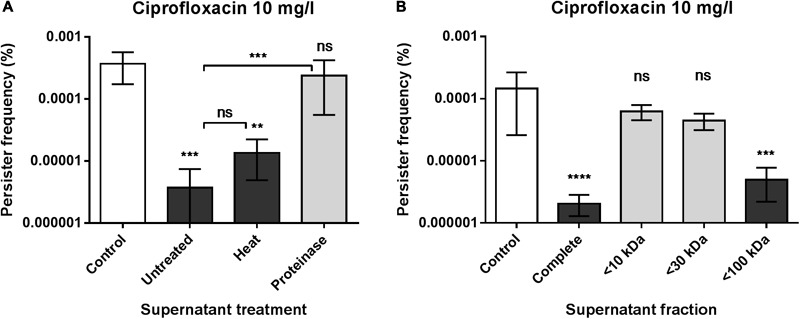
The active components is proteinaceous and present within the >30 kDa fraction. (A) Persister frequencies obtained from S. aureus strain Newman when grown in the presence of 25% untreated, heat- and proteinase-treated endogenous supernatant compared to the un-supplemented control. (B) Effect of supernatant fractions passed through 10, 30, and 100 kDa cut-off membranes on persister levels of S. aureus strain Newman. Comparison was made to the un-supplemented control and cultures added 25% of the complete endogenous supernatant. The data represent the mean persister frequencies ± SD calculated from three biological replicates treated with 20x MIC of ciprofloxacin (∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001).
The Active Supernatant Component Accumulates in Stationary Phase in an agr-Dependent Manner
The central quorum sensing system of S. aureus, agr, is composed of a two component system with the membrane-associated AgrC histidine kinase and the response regulator AgrA that in response to auto-inducing peptides expressed by agrB and agrD induce expression of the effector molecule RNAIII (Novick, 2003). S. carnosus is considered non-pathogenic and strain TM300 is characterized by relatively low exoprotein production, likely a partial consequence of a mutation rendering the agr system non-functional (Rosenstein et al., 2009). This, together with the finding that the spent supernatants displayed persister repressing activity only if taken from post-exponentially growing cells (Figure 4A), suggested that the sought component(s) is under quorum sensing control. Indeed, we find that a supernatant taken from an agr quorum sensing deficient strain showed no modulatory activity on exponential phase persister cells selected with ciprofloxacin (Figure 4B), which we confirmed when obtaining the supernatant from another strain background (Supplementary Figure S5).
FIGURE 4.
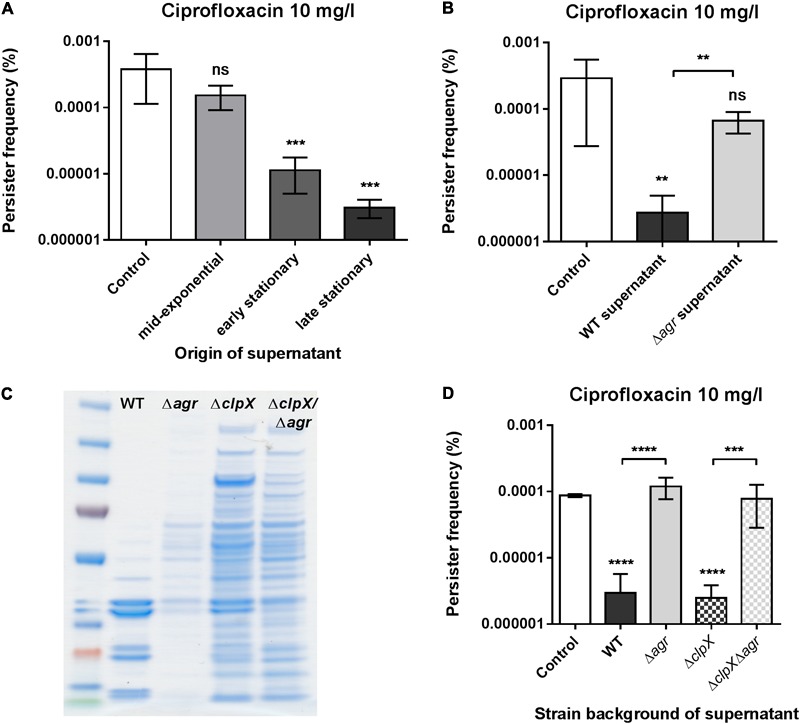
The supernatant activity is growth phase and agr quorum sensing dependent. (A) Persister frequencies obtained from S. aureus strain Newman supplemented with 25% endogenous supernatants obtained from different growth phases. (B) S. aureus strain Newman persister levels obtained when supplemented with 25% stationary phase endogenous supernatant from either wild type or NewmanΔagr cells. (C) SDS-PAGE of total extracellular proteins present in stationary phase supernatants from S. aureus 8325-4 (WT) and its single (Δagr and ΔclpX) mutant and double (ΔagrΔclpX) mutant derivatives. Amount loaded corresponds to 20 μl of protein precipitate obtained from cultures of an equivalent of OD600 of 50. (D) S. aureus strain Newman persister levels obtained when supplemented with 25% stationary phase supernatants from either S. aureus 8325-4 (WT), its single (Δagr and ΔclpX) mutants, or the double (ΔagrΔclpX) mutant. All persister cells were selected using 20x MIC of ciprofloxacin and all supernatant treatments were compared to the un-supplemented control (TSB). The data represent the mean persister frequencies ± SD calculated from three biological replicates (∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001).
agr functionality has been associated with autolytic activity in S. aureus (Fujimoto and Bayles, 1998; Sakoulas et al., 2003) and has also been shown to facilitate release of cytoplasmic proteins, and cellular lipids, nucleic acids, etc. due to membrane damage (Ebner et al., 2017). To rule out that unspecific, spontaneous release of cytosolic proteins is a cause of activity we examined a S. aureus clpX mutant that has growth defects and is characterized by substantial release of cellular proteins into the supernatant (Frees et al., 2003; Bæk et al., 2016). This extraordinary release of cellular proteins was also evident in a clpX/agr double mutant (Figure 4C) and yet, loss of persister-reducing activity of the supernatant by agr dysfunction was equally evident in this background (Figure 4D) suggesting that the sought entity is directly regulated by agr.
Induction of quorum sensing has previously been implicated in persister cell formation in other bacterial species (Möker et al., 2010; Leung and Lévesque, 2012). Although our fractionations had indicated the active factor to be of higher molecular weight, we examined if the small auto-inducing peptides secreted by S. aureus and present in the supernatants might prematurely activate the agr response and hereby influence the observed persister frequency. To test this, we employed the knowledge that a strain belonging to one agr specificity group does not activate the system of another group (Ji et al., 1997). The supernatant from strain Newman reduced the persister frequency of strains belonging to agr specificity groups II and III as did their endogenous supernatants (Figures 5A,B). As a final confirmation that our phenomenon does not rely on quorum sensing induction we found that the wild type supernatant significantly lowers the persister frequency also of an agr mutant (Figure 5C). Interestingly, we noted that the supernatant from S. aureus also negatively affects ciprofloxacin persister levels of S. carnosus and even Listeria monocytogenes, still in an agr-dependent manner (Supplementary Figure S6). This observation suggests that the supernatant affects persister cells via physiological means rather than some specific regulatory mechanism.
FIGURE 5.
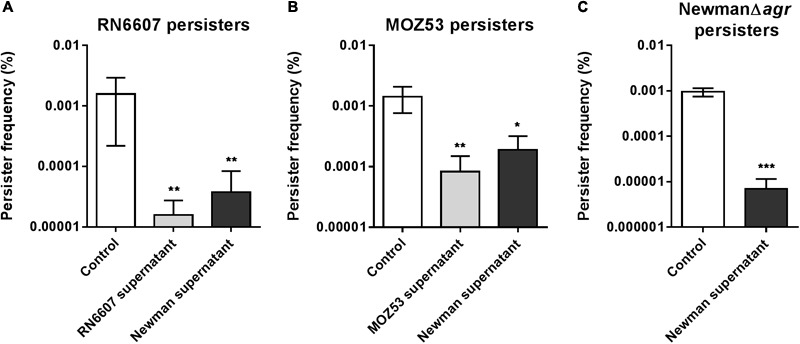
Persister reducing activity does not require induction of the agr response. Persister frequencies of S. aureus strains RN6607 (agr group II) (A) and MOZ53 (agr group III) (B) in the presence of 25% stationary phase endogenous supernatants or supernatant obtained from strain Newman (agr group I). (C) Effect of the wild type strain Newman stationary phase supernatant (25%) on persister cells isolated from strain NewmanΔagr. Persister cells were isolated using 20x MIC of ciprofloxacin and all supernatant treatments were compared to the un-supplemented control (TSB). The data represent the mean persister frequencies ± SD calculated from three biological replicates (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001).
An RNAIII Independent Activity – Persister Modulation via agr-Controlled Phenol-Soluble Modulins
The agr regulon is known to encompass a multitude of exoproteins and metabolic functions with the expression of the majority relying on the central regulatory molecule, RNAIII and a subset of targets being directly under the control of AgrA, the response regulator of the agr two-component system (Queck et al., 2008). When investigating which regulatory part of the agr regulon contributes to reduction of persister cells we found that the supernatant from RNAIII mutant cells remained equally effective as that of wild type cells in reducing the exponential phase persister frequency (Figure 6A and Supplementary Figure S7). Among the few genes under direct positive control by AgrA are those encoding the phenol-soluble modulins (PSMs) (Queck et al., 2008). They are a group of small amphipatic peptides expressed by S. aureus and other staphylococci divided into different classes with the α-class being ∼20 amino acids long and the β-class ∼44 amino acids as well as the δ-toxin that is encoded within RNAIII. In particular the α-class peptides are cytotoxic to many cell types, and work by non-specific membrane damage, while the β-class seems to lack cytotoxicity (Wang et al., 2007; Cheung et al., 2014). Interestingly, the PSMs are capable of associating into high molecular weight aggregates (Mehlin et al., 1999; Otto et al., 2004) and even form ordered amyloid structures that may stabilize biofilm structures (Schwartz et al., 2012). To further approach the hypothesis that the PSMs could be the factor influencing persister cell formation, we undertook supernatant fractionation by size-exclusion column chromatography. We found that while the active component(s) is indeed localized to the characteristic high molecular weight fractions of a wild type supernatant (i.e., they eluted early), and thus being consistent with the spin-column experiment (Figure 3B), a large proportion of the protein content within these fractions is in fact small peptides when analyzed by reducing SDS-PAGE (Supplementary Figure S8). Further, we observed that a supernatant withdrawn from a mutant unable to express PSM peptides (PSMα, PSMβ, and PSMδ families) had lost its ability to decrease the frequency of exponential phase persisters (Figure 6B) confirming that the PSMs are responsible for the reduction in exponential phase persister cell formation. Using synthetic PSMα3 we tried to recapitulate the anti-persister activity in a range of relevant concentrations without success (Supplementary Figure S9). It may be that the activity is related to one of the other peptides, a combination of several, or that the activity is confined to a processed form while at the same time being present as a high molecular weight entity.
FIGURE 6.
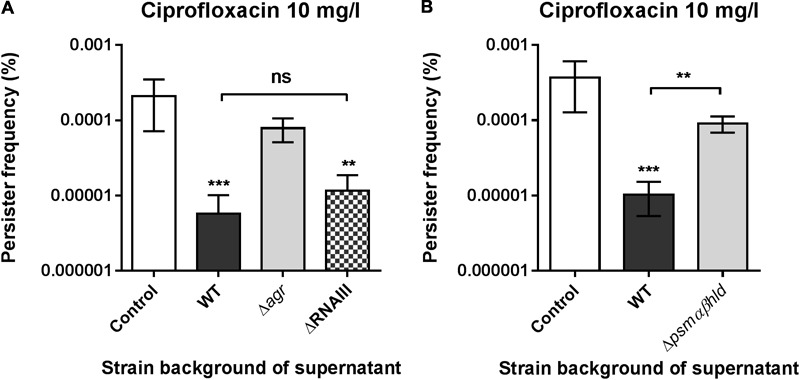
Phenol soluble modulins (PSMs) negatively affects exponential phase persister levels. (A) S. aureus strain Newman persister frequencies obtained when supplemented with 25% stationary phase endogenous supernatants from either strain Newman (WT), NewmanΔagr (Δagr) or NewmanΔRNAIII (ΔRNAIII). (B) Persister frequencies obtained when supplemented with 25% stationary phase supernatants from either strain LAC or LACΔpsmαβhld. Persister cells were selected using 20x MIC of ciprofloxacin and all supernatant treatments were compared to the un-supplemented control (TSB). The data represent the mean persister frequencies ± SD calculated from three biological replicates (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001).
Phenol-Soluble Modulins Affect Stationary Phase Persisters
Having pointed to a role of PSMs in reducing S. aureus exponential phase persister frequency when added exogenously we aimed to assess any relevance on cells that have entered stationary growth, a condition known to sharply increase the proportion of survivors of antibiotic treatment. First, we monitored stationary phase persisters when challenged with ciprofloxacin in the presence or absence of PSM-containing supernatant simultaneously. To eliminate the effect of endogenously expressed PSMs, we assessed persister cell formation in a Δagr mutant of strain Newman. Upon addition of spent supernatant of either WT or PSM deficient cultures we observed a more than 10-fold reduction in stationary phase survivors in presence of PSMs (Figure 7A). Next, we asked if this finding translated into a direct difference in persister levels between a wild type and a psm mutant in stationary phase. Since the β-type PSMs have been reported to be expressed only in low amounts in S. aureus in vitro (Cheung et al., 2010), we focused on the α-type family. Certainly, a PSMα1-4 deficient derivative of S. aureus Newman displays well above 10-fold higher persister levels than wild type (Figure 7B) and we confirmed the PSMα-dependent phenotype by redoing the assay on specific PSMα and PSMβ mutant derivatives constructed elsewhere (Supplementary Figure S10A) (Tsompanidou et al., 2013). The generality of this paradigm was established by the significant effect obtained in reducing type I ciprofloxacin persisters of S. aureus strains SA564 and 8325-4 specifically by a PSMα-containing supernatant (Supplementary Figures S10B,C) and by confirming the high-persister phenotype of a specific psmα mutant constructed in SA564 (Supplementary Figure S11). Finally, correlating with the clear role of PSMs in reducing persister selection frequency, we note an apparent striking difference in persister frequency between wild type and agr mutant cells (compare Figure 7A and 7B). We confirmed this notion in a direct comparison and that the 10 to 100-fold difference in persister levels between the two cell types is caused by the external environment as the high-persister phenotype of the agr mutant is negated by the wild type supernatant (Figure 7C).
FIGURE 7.
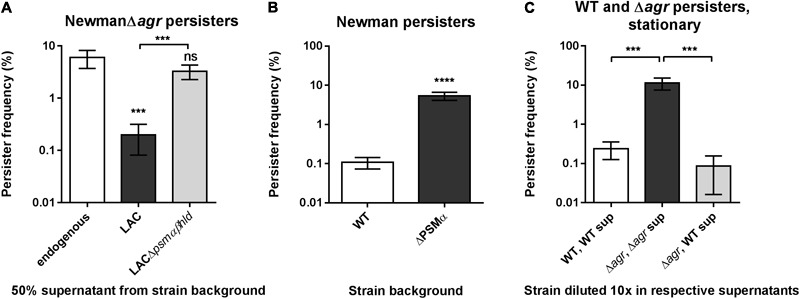
Phenol soluble modulins are strong determinants of S. aureus persister levels in stationary phase. (A) S. aureus strain NewmanΔagr stationary phase persister frequencies obtained when supplemented with 50% stationary phase supernatants from either strain LAC or LACΔpsmαβhld. Persister cells were selected using 100x MIC of ciprofloxacin and treatments were compared to the control added endogenous supernatant only. (B) Stationary phase persister frequencies of S. aureus strain Newman and the derived psmα mutant selected with 100x MIC ciprofloxacin added directly to overnight cultures. (C) Stationary phase persisters of strain Newman (WT) and NewmanΔagr (Δagr) when resuspended in 90% endogenous supernatant prior to challenge with 100x MIC ciprofloxacin in comparison to persister level obtained with NewmanΔagr in supernatant derived from WT. The data represent the mean persister frequencies ± SD calculated from three biological replicates (∗∗∗P < 0.001, ∗∗∗∗P < 0.0001).
Conclusion
Here, we find that PSMα peptide toxins naturally produced by S. aureus and the related bacterium S. epidermidis are able to limit the S. aureus persister population. The effect is mediated both by endogenously produced peptides and by peptides added as part of spent staphylococcal supernatants, suggesting that they act exogenously. The high-persister phenotype of a stationary phase PSMα mutant upon fluoroquinolone treatment (Figure 7B) is mirrored when treating with the aminoglycoside gentamicin, but not upon challenge with wall-acting antibiotics, oxacillin and vancomycin, toward which a wild type cell remains fully tolerant also (Supplementary Figure S12). That the effect is confined only to some, yet unrelated, antibiotic classes suggests that the phenomenon is neither related to a direct killing of the persister subpopulation nor to a general resuscitation of persister cells. Further, we note that the persisters selected with oxacillin in exponential phase (Supplementary Figure S2B) were susceptible to the presence of supernatant. It may be that that effect is unrelated to PSMs or reflect a physiological difference between type I and type II persisters.
Recently, Xu et al. (2017) reported that inactivation of either psmα or psmβ genes increase persister formation over a time course of several days. They concluded that persister formation was regulated by an unclear mechanism and that the phenotype was specific to the fluoroquinolone levofloxacin. Here, we identified the activity of PSMs against persisters by a very different approach. When searching for a persister-reducing activity in spent staphylococcal supernatants we found the PSMα peptides to be responsible. The effect of the peptides was observed already within a 24 h antibiotic challenge; was evident on both exponential phase and stationary phase persisters and was seen when treated with intracellular antibiotics rather than only with fluoroquinolones. The extent to which our findings are consistent with those of Xu et al. (2017) warrants further study, including their observation that a deletion of either psmα or psmβ leads to a high-persister phenotype, which cannot be readily explained by a model that the peptides themselves reduces S. aureus persisters (i.e., PSMα is expected to be synthesized in the absence of psmβ). Importantly we observe that a PSM-containing supernatant limits the persister population not only of S. aureus but also of S. carnosus and the more distantly related Listeria monocytogenes (Supplementary Figure S6). Based on our findings we propose that PSMα peptides limit the persister population directly by interacting with the persister cells and increasing their susceptibility to antibiotics.
Originally PSMs were discovered as virulence factors that stimulate inflammatory responses and lyse human cells including leukocytes and erythrocytes with particularly the PSMα peptides being cytolytic (Wang et al., 2007; Cheung et al., 2014). However, activity against bacterial membranes have also been reported with the N-terminally processed PSMα1 and PSMα2 displaying antimicrobial activity against Streptococcus pyogenes (Joo et al., 2011). For S. aureus, PSMα peptides are found associated with the membrane where they are involved in release of lipoproteins and they promote colony spreading by exerting membrane surfactant activity (Tsompanidou et al., 2013; Kizaki et al., 2016). Strikingly, a recent report showed that particularly PSMα2 and PSMα3 can cause membrane damage in S. aureus to the extent that cellular components (proteins, nucleic acids, etc.) are released (Ebner et al., 2017). The toxicity of the PSM peptides against S. aureus explains previous observations of “non-classical” excretion of cellular proteins although the extent to which this influences viability is not clear (Ebner et al., 2017). Based on these observations we propose that the persister state of S. aureus cells is associated with changes in the bacterial membrane that reduce susceptibility to ciprofloxacin and gentamicin, however, in the presence of PSMα peptides, these changes are mitigated and susceptibility is restored. Intriguingly, Pader et al. (2016) unraveled a mechanism by which S. aureus agr-defective mutants specifically survives exposure to the membrane-active antibiotic daptomycin. While daptomycin is sequestered and inactivated by S. aureus by release of membrane phospholipids, this antibiotic-specific process is inhibited by PSMα excretion (Pader et al., 2016). It appears now that PSM-production by S. aureus may in fact alter the efficacy of multiple types of antibiotic exposure.
A surprising and interesting aspect of our findings is that they suggest that S. aureus limit the persister population under conditions where agr is induced and PSMs are expressed. Such conditions are found in acute infections where agr is considered to play an important role (Le and Otto, 2015). Persistent infections, however, are commonly reported with agr negative strains (Fowler et al., 2004; Park et al., 2013) and it appears that S. aureus can even diversify into acute and chronic subpopulations via what is termed agr bimodal switching within an otherwise clonal population (García-Betancur et al., 2017). In these cases, the PSM production will be absent and persisters are more likely to be present. Even short-term tolerance has been reported to be improved in strains lacking agr when exposed to lethal stressors such as ciprofloxacin or gentamicin (Kumar et al., 2017), however, it remains to be determined if these findings can be explained by lack of PSM production. In summary, we propose that S. aureus by producing PSMs limit the persister population under conditions of acute infection by increasing their susceptibility to antibiotics via membrane modulation whereas under chronic conditions the reduced PSM production allows the presence of persister cell with greatly reduced susceptibility to antibiotics.
Materials and Methods
Bacterial Strains and Growth Conditions
All strains used are listed in Supplementary Table S1. Strains were taken from frozen stocks (-80°C) and grown on tryptic soy agar (TSA, Oxoid) at 37°C. Liquid cultures were grown from single colonies in tryptic soy broth (TSB, Oxoid) at 37°C and were shaken at 200 rpm.
Strain Constructions
The isaA and sceD mutations (Stapleton et al., 2007) were moved from the SH1000 background into strain Newman and the Δagr::tetM mutation from RN6911 (Novick et al., 1993) was transferred into 8325-4; all by phage transduction (Φ11) and selection for integrated antibiotic resistance markers. Unmarked clean deletions of the psmα1-4 locus were constructed in strain Newman and SA564 via temperature-sensitive allelic exchange using the shuttle vector pBASE6 (Geiger et al., 2012). Approximately, 1 kb chromosomal regions surrounding the psmα operon from the two strains were PCR amplified using primer pairs 5′-GATACAGAGCTCTTCCTGCATGCATAATTGCC-3′/5′-GAATTTTAAGTATTCAATTCGCTTAAATAAGATTACCTCCTTTGCTTATGAG-3′ and 5′-TTTAAGCGAATTGAATACTTAAAATTC-3′/5′-GATACAAGATCTCGAGTCAGCAGGATGGATC-3′, respectively. These products were subsequently joined in a spliced overlap extension PCR combining the forward and reverse primers from the previous reactions, thus generating strain specific deletion fragments that were cloned into pBASE6 via BglII/SacI. The resulting plasmids were purified from E. coli IM08B (Monk et al., 2015) and transformed directly into the wild type strains at 30°C followed by chromosomal integration by plating on TSA (10 μg/ml chloramphenicol) at 43°C. Plasmid cross-out was performed by passage at 30°C followed by plating on TSA (500 ng/ml anhydrotetracycline). Colonies were replica-plated to select for sensitivity toward chloramphenicol and successful allelic exchange were screened for by PCR amplification using primers 5′-TAAGACAACAAATTCTGAAGTAG-3′/5′-AGTTAGAATAACACCACCTGC-3′ positioned outside the chromosomal region used for homologous recombination.
Preparation of Bacterial Supernatants
All supernatants were derived from TSB cultures grown from single colonies in shaking flasks at 37°C/200 rpm. Unless otherwise stated, supernatants were withdrawn after 24 h of growth. Cells were separated from the medium by centrifugation (10,000 g for 10 min) and the supernatant was passed through a 0.2 μm sterile filter (Minisart, Sartorius). Heat treated supernatant was prepared by heating in a water bath at 95°C for 20 min followed by an additional filtration step. Protease treatment was performed by addition of a Streptomyces griseus protease mixture (Sigma-Aldrich) at 1 mg/ml and incubation at 37°C for 30 min followed by heat inactivation (80°C for 20 min). Molecular weight fractionation was performed by passing a full supernatant through Vivaspin 6 ultrafiltration columns (Sartorius) with different molecular weight cut-offs (MWCOs) according to the manufacturer’s instruction. Size-exclusion chromatography was performed using an Äkta Pure 25 L1 and a Superdex 200 Increase 10/300 GL size exclusion column (both GE Healthcare Life Sciences). Supernatants were up-concentrated approximately 20-fold by use of Amicon® Ultra 15 mL spin Filters (Millipore) with a 10 kDa cut-off and 500 μl of each sample were loaded onto the size-exclusion column and fractionated using 1x PBS as mobile phase and a flow-rate of 0.75 ml/min. All supernatants and fractions were freshly prepared or kept at -20°C until use.
SDS-PAGE of Supernatant Proteins
Supernatant proteins were precipitated with ethanol (50% v/v) at 4°C overnight and resuspended in TE-buffer. Samples were analyzed on a NuPAGE® Novex 4-12% Bis-Tris gel under reducing conditions using the XCell SureLock® Mini-Cell system and stained with SimplyBlueTM SafeStain according to the manufacturer (Thermo Fischer Scientific). Samples from the size-exclusion chromatography were mixed 1:1 with reducing (50 mM DTT) 2x Laemli buffer and run immediately using the same SDS-PAGE system.
Determination of Minimum Inhibitory Concentrations
Minimum inhibitory concentrations of ciprofloxacin, gentamicin, oxacillin, and vancomycin (all Sigma-Aldrich) in TSB were estimated by standard twofold broth dilution. S. aureus strain Newman was inoculated in TSB (or TSB + 25% bacterial supernatant) and dispensed into microtiter trays added a twofold serial dilution of respective antibiotics reaching an initial cell count of approximately 5 × 105 CFU/ml. Following 24 h of incubation at 37°C, MIC values were recorded as the lowest concentration of each antibiotic inhibiting visual growth.
Persister Assay
The frequency of S. aureus persister cells in growing cultures (type II persisters) was determined by bactericidal antibiotic killing of the susceptible population and enumeration of surviving cells. Cells from an overnight culture was diluted 1/1000 in 2 ml fresh TSB and incubated in 15 ml test tubes at 37°C and 200 rpm for 2.5 h to reach exponential growth and a cell density of approximately 1–4 × 107 CFU/ml. An aliquot was taken to allow determination of the actual CFUs (by serial dilutions on TSA plates) and the culture was supplemented with a lethal concentration (20x MIC) of antibiotic. After 24 h of incubation, 1 ml of culture was withdrawn and centrifuged (12,000 g for 5 min), washed with 0.9% NaCl to remove the antibiotic, and plated on TSA. Plates were incubated at 37°C for 24 h and the persister frequency was calculated as the plate count relative to the CFUs obtained at time 0 h. The persister level was arbitrarily set to 0.5 CFU/ml if a sample had less than 1 CFU/ml, i.e., below the detection limit. Stationary phase persister cells (type I persisters) were evaluated by transfer of 24 h cultures to new test tubes followed by addition of 100x MIC of antibiotic and comparing the CFUs before and after another 24 h of incubation. For some assays stationary phase cells were supplemented with exogenous supernatants prior to antibiotic challenge. A minimum of three biological replicates (overnight cultures originating from individual colonies) were included for each assay and experimental condition. All experimental results are representative of repeated independent experiments. Representative colonies from the persister assay were isolated to verify the non-inherited nature of persistence by confirmation of their susceptibility toward the antibiotic with which they were isolated and an unaltered persister frequency in subsequent assays.
Statistical Analysis
Persister frequencies within individual experiments were compared by two-tailed Student’s t-test or One-way ANOVA followed by Dunnett’s or Tukey’s (where applicable) multiple comparisons test using GraphPad Prism version 6.00 for Windows, GraphPad Software, La Jolla, CA, United States1. Persister frequencies were log transformed prior to analysis to normalize the variance. Regular and multiplicity adjusted values of P < 0.05 were considered statistically significant.
Author Contributions
MB and HI conceived the project and designed the work. MB and SL performed the experiments. MB, MV, and HI analyzed the data and drafted the manuscript. All authors approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Szabolcs Semsey for helpful input on the manuscript, to Dr. Simon J. Foster for providing the SH1000ΔisaA and SH1000ΔsceD strains, to Dr. Michael Otto for sharing the LAC and LACpsmα/β/hld strains, and to Dr. Jan Maarten van Dijl for sharing the Newman psm mutant strains.
Funding. This work was supported by grants from the Danish Council for Independent Research – Technology and Production (1337-00129 and 1335-00772) to MB and by the Danish National Research Foundation (DNRF120) to HI.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00255/full#supplementary-material
References
- Amato S. M., Orman M. A., Brynildsen M. P. (2013). Metabolic control of persister formation in Escherichia coli. Mol. Cell. 50 475–487. 10.1016/j.molcel.2013.04.002 [DOI] [PubMed] [Google Scholar]
- Balaban N. Q., Merrin J., Chait R., Kowalik L., Leibler S. (2004). Bacterial persistence as a phenotypic switch. Science 305 1622–1625. 10.1126/science.1099390 [DOI] [PubMed] [Google Scholar]
- Bæk K. T., Bowman L., Millership C., Søgaard M. D., Kaever V., Siljamäki P., et al. (2016). The cell wall polymer lipoteichoic acid becomes nonessential in Staphylococcus aureus cells lacking the ClpX chaperone. mBio 7:e01228-16. 10.1128/mBio.01228-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung G. Y., Joo H.-S., Chatterjee S. S., Otto M. (2014). Phenol-soluble modulins–critical determinants of staphylococcal virulence. FEMS Microbiol. Rev. 38 698–719. 10.1111/1574-6976.12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung G. Y., Rigby K., Wang R., Queck S. Y., Braughton K. R., Whitney A. R., et al. (2010). Staphylococcus epidermidis strategies to avoid killing by human neutrophils. PLoS Pathog. 6:e1001133. 10.1371/journal.ppat.1001133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen N. R., Lobritz M. A., Collins J. J. (2013). Microbial persistence and the road to drug resistance. Cell Host Microbe 13 632–642. 10.1016/j.chom.2013.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon B. P. (2014). Staphylococcus aureus chronic and relapsing infections: evidence of a role for persister cells. Bioessays 36 991–996. 10.1002/bies.201400080 [DOI] [PubMed] [Google Scholar]
- Conlon B. P., Nakayasu E. S., Fleck L. E., LaFleur M. D., Isabella V. M., Coleman K., et al. (2013). Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503 365–370. 10.1038/nature12790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon B. P., Rowe S. E., Gandt A. B., Nuxoll A. S., Donegan N. P., Zalis E. A., et al. (2016). Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 1:16051. 10.1038/nmicrobiol.2016.51 [DOI] [PubMed] [Google Scholar]
- Conlon B. P., Rowe S. E., Lewis K. (2015). “Persister cells in biofilm associated infections,” in Biofilm-Based Healthcare-Associated Infections. Advances in Experimental Medicine and Biology ed. Donelli G. (Cham: Springer; ) 1–9. [DOI] [PubMed] [Google Scholar]
- DeLeo F. R., Otto M., Kreiswirth B. N., Chambers H. F. (2010). Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375 1557–1568. 10.1016/S0140-6736(09)61999-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner P., Luqman A., Reichert S., Hauf K., Popella P., Forchhammer K., et al. (2017). Non-classical protein excretion is boosted by PSMα-induced cell leakage. Cell Rep. 20 1278–1286. 10.1016/j.celrep.2017.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Halfawy O. M., Klett J., Ingram R. J., Loutet S. A., Murphy M. E., Martín-Santamaría S., et al. (2017). Antibiotic capture by bacterial lipocalins uncovers an extracellular mechanism of intrinsic antibiotic resistance. mBio 8:e00225-17. 10.1128/mBio.00225-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., Gollan B., Helaine S. (2017). Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 15 453–464. 10.1038/nrmicro.2017.42 [DOI] [PubMed] [Google Scholar]
- Fowler V. G., Jr., Sakoulas G., McIntyre L. M., Meka V. G., Arbeit R. D., Cabell C. H., et al. (2004). Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J. Infect. Dis. 190 1140–1149. 10.1086/423145 [DOI] [PubMed] [Google Scholar]
- Frees D., Qazi S. N., Hill P. J., Ingmer H. (2003). Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 48 1565–1578. 10.1046/j.1365-2958.2003.03524.x [DOI] [PubMed] [Google Scholar]
- Fujimoto D. F., Bayles K. W. (1998). Opposing roles of the Staphylococcus aureus virulence regulators, Agr and Sar, in Triton X-100-and penicillin-induced autolysis. J. Bacteriol. 180 3724–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Betancur J.-C., Goñi-Moreno A., Horger T., Schott M., Sharan M., Eikmeier J., et al. (2017). Cell differentiation defines acute and chronic infection cell types in Staphylococcus aureus. Elife 6:e28023. 10.7554/eLife.28023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T., Francois P., Liebeke M., Fraunholz M., Goerke C., Krismer B., et al. (2012). The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog. 8:e1003016. 10.1371/journal.ppat.1003016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger E., Isberg R. R. (2015). Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog. 11:e1004691. 10.1371/journal.ppat.1004691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaber J., Cohn M. T., Frees D., Andersen T. J., Ingmer H. (2012). Planktonic aggregates of Staphylococcus aureus protect against common antibiotics. PLoS One 7:e41075. 10.1371/journal.pone.0041075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaber J., Friberg C., McCreary M., Lin R., Cohen S. N., Ingmer H. (2015). Reversible antibiotic tolerance induced in Staphylococcus aureus by concurrent drug exposure. mBio 6:e02268-14. 10.1128/mBio.02268-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms A., Maisonneuve E., Gerdes K. (2016). Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354:aaf4268. 10.1126/science.aaf4268 [DOI] [PubMed] [Google Scholar]
- Helaine S., Cheverton A. M., Watson K. G., Faure L. M., Matthews S. A., Holden D. W. (2014). Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343 204–208. 10.1126/science.1244705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. H., Wang X., O’Connor H. F., Benedik M. J., Wood T. K. (2012). Bacterial persistence increases as environmental fitness decreases. Microbial. Biotechnol. 5 509–522. 10.1111/j.1751-7915.2011.00327.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito R., Mustapha M. M., Tomich A. D., Callaghan J. D., McElheny C. L., Mettus R. T., et al. (2017). Widespread fosfomycin resistance in Gram-negative bacteria attributable to the chromosomal fosA gene. mBio 8:e00749-17. 10.1128/mBio.00749-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji G., Beavis R., Novick R. P. (1997). Bacterial interference caused by autoinducing peptide variants. Science 276 2027–2030. 10.1126/science.276.5321.2027 [DOI] [PubMed] [Google Scholar]
- Johnson P. J., Levin B. R. (2013). Pharmacodynamics, population dynamics, and the evolution of persistence in Staphylococcus aureus. PLoS Genet. 9:e1003123. 10.1371/journal.pgen.1003123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo H.-S., Cheung G. Y., Otto M. (2011). Antimicrobial activity of community-associated methicillin-resistant Staphylococcus aureus is caused by phenol-soluble modulin derivatives. J. Biol. Chem. 286 8933–8940. 10.1074/jbc.M111.221382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I., Kaldalu N., Spoering A., Wang Y., Lewis K. (2004). Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230 13–18. 10.1016/S0378-1097(03)00856-5 [DOI] [PubMed] [Google Scholar]
- Kizaki H., Omae Y., Tabuchi F., Saito Y., Sekimizu K., Kaito C. (2016). Cell-surface phenol soluble modulins regulate Staphylococcus aureus colony spreading. PLoS One 11:e0164523. 10.1371/journal.pone.0164523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K., Chen J., Drlica K., Shopsin B. (2017). Tuning of the lethal response to multiple stressors with a single-site mutation during clinical infection by Staphylococcus aureus. mBio 8:e01476-17. 10.1128/mBio.01476-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le K. Y., Otto M. (2015). Quorum-sensing regulation in staphylococci—an overview. Front. Microbiol. 6:1174 10.3389/fmicb.2015.01174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner S., Lewis K., Bertram R. (2012). Staphylococcus aureus persisters tolerant to bactericidal antibiotics. J. Mol. Microbiol. Biotechnol. 22 235–244. 10.1159/000342449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung V., Lévesque C. M. (2012). A stress-inducible quorum-sensing peptide mediates the formation of persister cells with noninherited multidrug tolerance. J. Bacteriol. 194 2265–2274. 10.1128/JB.06707-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin-Reisman I., Ronin I., Gefen O., Braniss I., Shoresh N., Balaban N. Q. (2017). Antibiotic tolerance facilitates the evolution of resistance. Science 355 826–830. 10.1126/science.aaj2191 [DOI] [PubMed] [Google Scholar]
- Lewis K. (2010). Persister cells. Annu. Rev. Microbiol. 64 357–372. 10.1146/annurev.micro.112408.134306 [DOI] [PubMed] [Google Scholar]
- Lowy F. D. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339 520–532. 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- Maisonneuve E., Castro-Camargo M., Gerdes K. (2013). (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154 1140–1150. 10.1016/j.cell.2013.07.048 [DOI] [PubMed] [Google Scholar]
- Maisonneuve E., Gerdes K. (2014). Molecular mechanisms underlying bacterial persisters. Cell 157 539–548. 10.1016/j.cell.2014.02.050 [DOI] [PubMed] [Google Scholar]
- Mechler L., Herbig A., Paprotka K., Fraunholz M., Nieselt K., Bertram R. (2015). A novel point mutation promotes growth phase-dependent daptomycin tolerance in Staphylococcus aureus. Antimicrob. Agents Chemother. 59 5366–5376. 10.1128/AAC.00643-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlin C., Headley C. M., Klebanoff S. J. (1999). An inflammatory polypeptide complex from Staphylococcus epidermidis: isolation and characterization. J. Exp. Med. 189 907–918. 10.1084/jem.189.6.907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. B., Bassler B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55 165–199. 10.1146/annurev.micro.55.1.165 [DOI] [PubMed] [Google Scholar]
- Möker N., Dean C. R., Tao J. (2010). Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum-sensing signaling molecules. J. Bacteriol. 192 1946–1955. 10.1128/JB.01231-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk I. R., Tree J. J., Howden B. P., Stinear T. P., Foster T. J. (2015). Complete bypass of restriction systems for major Staphylococcus aureus lineages. mBio 6:e00308-15. 10.1128/mBio.00308-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Gámez S., Sorg R. A., Domenech A., Kjos M., Weissing F. J., van Doorn G. S., et al. (2017). Quorum sensing integrates environmental cues, cell density and cell history to control bacterial competence. Nat. Commun. 8:854. 10.1038/s41467-017-00903-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris R. P., Nguyen L., Gatfield J., Visconti K., Nguyen K., Schnappinger D., et al. (2005). Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 102 12200–12205. 10.1073/pnas.0505446102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy L. R., Burns J. L., Lory S., Lewis K. (2010). Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 192 6191–6199. 10.1128/JB.01651-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P. (2003). Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48 1429–1449. 10.1046/j.1365-2958.2003.03526.x [DOI] [PubMed] [Google Scholar]
- Novick R. P., Ross H., Projan S., Kornblum J., Kreiswirth B., Moghazeh S. (1993). Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12 3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orman M. A., Brynildsen M. P. (2015). Inhibition of stationary phase respiration impairs persister formation in E. coli. Nat. Commun. 6:7983. 10.1038/ncomms8983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M., O’mahoney D. S., Guina T., Klebanoff S. J. (2004). Activity of Staphylococcus epidermidis phenol-soluble modulin peptides expressed in Staphylococcus carnosus. J. Infect. Dis. 190 748–755. 10.1086/422157 [DOI] [PubMed] [Google Scholar]
- Pader V., Hakim S., Painter K. L., Wigneshweraraj S., Clarke T. B., Edwards A. M. (2016). Staphylococcus aureus inactivates daptomycin by releasing membrane phospholipids. Nat. Microbiol. 2:16194. 10.1038/nmicrobiol.2016.194 [DOI] [PubMed] [Google Scholar]
- Park S.-Y., Chong Y., Park H., Park K.-H., Moon S., Jeong J.-Y., et al. (2013). agr dysfunction and persistent methicillin-resistant Staphylococcus aureus bacteremia in patients with removed eradicable foci. Infection 41 111–119. 10.1007/s15010-012-0348-0 [DOI] [PubMed] [Google Scholar]
- Pascoe B., Dams L., Wilkinson T. S., Harris L. G., Bodger O., Mack D., et al. (2014). Dormant cells of Staphylococcus aureus are resuscitated by spent culture supernatant. PLoS One 9:e85998. 10.1371/journal.pone.0085998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquaroli S., Zandri G., Vignaroli C., Vuotto C., Donelli G., Biavasco F. (2013). Antibiotic pressure can induce the viable but non-culturable state in Staphylococcus aureus growing in biofilms. J. Antimicrob. Chemother. 68 1812–1817. 10.1093/jac/dkt086 [DOI] [PubMed] [Google Scholar]
- Prax M., Bertram R. (2014). Metabolic aspects of bacterial persisters. Front. Cell. Infect. Microbiol. 4:148 10.3389/fcimb.2014.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que Y.-A., Hazan R., Strobel B., Maura D., He J., Kesarwani M., et al. (2013). A quorum sensing small volatile molecule promotes antibiotic tolerance in bacteria. PLoS One 8:e80140. 10.1371/journal.pone.0080140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queck S. Y., Jameson-Lee M., Villaruz A. E., Bach T.-H. L., Khan B. A., Sturdevant D. E., et al. (2008). RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell. 32 150–158. 10.1016/j.molcel.2008.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzikowski J. L., Vedelaar S., Siegel D., Ortega ÁD., Schmidt A., Heinemann M. (2016). Bacterial persistence is an active σ S stress response to metabolic flux limitation. Mol. Syst. Biol. 12:882. 10.15252/msb.20166998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstein R., Nerz C., Biswas L., Resch A., Raddatz G., Schuster S. C., et al. (2009). Genome analysis of the meat starter culture bacterium Staphylococcus carnosus TM300. Appl. Environ. Microbiol. 75 811–822. 10.1128/AEM.01982-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoulas G., Eliopoulos G. M., Moellering R. C., Jr., Novick R. P., Venkataraman L., Wennersten C., et al. (2003). Staphylococcus aureus accessory gene regulator (agr) group II: is there a relationship to the development of intermediate-level glycopeptide resistance? J. Infect. Dis. 187 929–938. 10.1086/368128 [DOI] [PubMed] [Google Scholar]
- Schumacher M. A., Balani P., Min J., Chinnam N. B., Hansen S., Vulić M., et al. (2015). HipBA-promoter structures reveal the basis of heritable multidrug tolerance. Nature 524 59–64. 10.1038/nature14662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz K., Syed A. K., Stephenson R. E., Rickard A. H., Boles B. R. (2012). Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog. 8:e1002744. 10.1371/journal.ppat.1002744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian J., Swaminath S., Nair R. R., Jakkala K., Pradhan A., Ajitkumar P. (2017). De novo emergence of genetically resistant mutants of Mycobacterium tuberculosis from the persistence phase cells formed against antituberculosis drugs in vitro. Antimicrob. Agents Chemother. 61:e01343-16. 10.1128/AAC.01343-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton M. R., Horsburgh M. J., Hayhurst E. J., Wright L., Jonsson M., Tarkowski A., et al. (2007). Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus. J. Bacteriol. 189 7316–7325. 10.1128/JB.00734-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsompanidou E., Denham E. L., Becher D., de Jong A., Buist G., van Oosten M., et al. (2013). Distinct roles of phenol-soluble modulins in spreading of Staphylococcus aureus on wet surfaces. Appl. Environ. Microbiol. 79 886–895. 10.1128/AEM.03157-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bergh B., Fauvart M., Michiels J. (2017). Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol. Rev. 41 219–251. 10.1093/femsre/fux001 [DOI] [PubMed] [Google Scholar]
- Van den Bergh B., Michiels J. E., Wenseleers T., Windels E. M., Boer P. V., Kestemont D., et al. (2016). Frequency of antibiotic application drives rapid evolutionary adaptation of Escherichia coli persistence. Nat. Microbiol. 1:16020. 10.1038/nmicrobiol.2016.20 [DOI] [PubMed] [Google Scholar]
- Verstraeten N., Knapen W. J., Kint C. I., Liebens V., Van den Bergh B., Dewachter L., et al. (2015). Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol. Cell. 59 9–21. 10.1016/j.molcel.2015.05.011 [DOI] [PubMed] [Google Scholar]
- Vestergaard M., Leng B., Haaber J., Bojer M. S., Vegge C. S., Ingmer H. (2016). Genome-wide identification of antimicrobial intrinsic resistance determinants in Staphylococcus aureus. Front. Microbiol. 7:2018. 10.3389/fmicb.2016.02018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Braughton K. R., Kretschmer D., Bach T.-H. L., Queck S. Y., Li M., et al. (2007). Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13 1510–1514. 10.1038/nm1656 [DOI] [PubMed] [Google Scholar]
- Waters E. M., Rowe S. E., O’Gara J. P., Conlon B. P. (2016). Convergence of Staphylococcus aureus persister and biofilm research: can biofilms be defined as communities of adherent persister cells? PLoS Pathog. 12:e1006012. 10.1371/journal.ppat.1006012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Wang X.-Y., Cui P., Zhang Y.-M., Zhang W.-H., Zhang Y. (2017). The agr quorum sensing system represses persister formation through regulation of phenol soluble modulins in Staphylococcus aureus. Front. Microbiol. 8:2189. 10.3389/fmicb.2017.02189 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


