Abstract
Maintenance of homeostasis of the endoplasmic reticulum (ER) ensures the balance between loading of nascent proteins and their secretion. Certain developmental conditions or environmental stressors affect protein folding causing ER stress. The resultant ER stress is mitigated by upregulating a set of stress-responsive genes in the nucleus modulating the mechanism of the unfolded protein response (UPR). In plants, the UPR is mediated by two major pathways; by the proteolytic processing of bZIP17/28 and by the IRE1-mediated splicing of bZIP60 mRNA. Recent studies have shown the involvement of plant-specific NAC transcription factors in UPR regulation. The molecular mechanisms activating plant-UPR transducers are only recently being unveiled. This review focuses on important structural features involved in the activation of the UPR transducers like bZIP17/28/60, IRE1, BAG7, and NAC017/062/089/103. Also, we discuss the activation of the UPR pathways, including BAG7-bZIP28 and IRE1-bZIP60, in detail, together with the NAC-TFs, which adds a new paradigm to the plant UPR.
Keywords: endoplasmic reticulum, abiotic/biotic stress, UPR activation, bZIP28, bZIP60, IRE1, NAC-TFs
Introduction
In eukaryotes, the endoplasmic reticulum (ER) acts as a factory site for proper folding and maturation of secretory and membrane proteins, which comprise about one-third of the total proteome (Wallin and von Heijne, 1998). These proteins undergo post-translational modification, such as N-linked glycosylation and disulfide bond formation, in the ER lumen (Braakman and Bulleid, 2011). ER-factory is well-equipped with a protein folding machinery containing molecular chaperones, including the luminal binding protein (BiP), calnexin (CNX), and calreticulins (CRT), and folding enzymes like the protein disulfide isomerase (PDI). Molecular chaperones prevent the aggregation of denatured proteins and assist in their proper folding, while PDI catalyzes the formation of correct disulfide bridges between the cysteine residues in proteins to maintain ER-homeostasis (Vitale and Denecke, 1999; Park and Seo, 2015). In plants, abiotic stressors, including high temperature, salt, osmotic stress, drought, heavy metals, and high light intensity, or biotic agents, such as viruses, disturb protein folding (Figure 1) (Liu et al., 2007b; Gao et al., 2008; Valente et al., 2009; Liu and Howell, 2010b; Deng et al., 2011; Faria et al., 2011; Mendes et al., 2013; Korner et al., 2015; Verchot, 2016; Xi et al., 2016; Nawkar et al., 2017). As a consequence, the accumulation of unfolded or misfolded proteins exceeds the ER-protein folding capacity leading to ER stress conditions (Liu and Howell, 2010b; Deng et al., 2011). During the last decade, there have been lots of development in the field of plant-ER stress signaling and unfolded protein response (UPR) modulated by a set of stress-responsive genes in the nucleus (Liu and Howell, 2016; Wan and Jiang, 2016; Angelos et al., 2017). Primarily, to overcome ER stress, plants activate the conserved UPR mechanism (Figure 1). The UPR plays an important role in restoring the protein folding capacity of the ER membrane by increasing the levels of molecular chaperones, and reducing the protein load by enhancing ER-associated protein degradation (ERAD) (Liu and Howell, 2016). Moreover, the UPR supports vegetative root growth and plays a vital role during the development of the male reproductive parts of a plant, not only under normal but also under stress conditions (Deng et al., 2013; Bao and Howell, 2017). In particular, the UPR maintains plant fertility by regulating pollen development during stressful conditions (Fragkostefanakis et al., 2016; Zhang et al., 2017). Recently, it has been suggested that the stress-responsive, plastid originated retrograde signal, 2-C-methyl-D-erythritol-2,4-cyclopyrophosphate (MEcPP), and the plant defense hormone, salicylic acid (SA), induces UPR in plants, although the underlining mechanism is not yet completely known (Nagashima et al., 2014; Walley et al., 2015). Generally, under laboratory conditions, the UPR has been studied using several chemicals interfering with the post-translational protein modifications; examples include the inhibition of N-glycosylation by tunicamycin (Tm), hindrance to the formation of disulfide bonds by dithiothreitol (DTT), and interference in the formation of native protein structures by the proline homolog azetidine-2-carboxylic acid (AZC) (Howell, 2013).
FIGURE 1.

Activation of the unfolded protein response (UPR) in Arabidopsis in response to external stresses. Abiotic (high light intensity, temperature, salt, drought, and heavy metals) and biotic (pathogenic attack) stresses disturb protein folding in the ER lumen causing the activation of transcription factors (TFs), which bind to the ER stress response elements (ERSEs) to upregulate the UPR genes. The UPR plays an important role during vegetative root growth and reproductive development in Arabidopsis. Moreover, plant signaling compounds such as 2-C-methyl-D-erythritol-2,4-cyclopyrophosphate (MEcPP) and salicylic acid (SA) activate the UPR pathways via unknown mechanisms.
In plants, the processes of regulated intramembrane proteolysis (RIP) of basic leucine zipper (bZIP) transcription factors (TFs) like bZIP17/28, and the inositol-requiring enzyme 1 (IRE1)-mediated unconventional splicing of bZIP60 act as the primary pathways of the UPR (Iwata and Koizumi, 2012). The activation of the membrane-tethered TFs (MTFs) by the RIP and the activation of the IRE1-dependent pathway are conserved in plants, mammals, and fungi (Chakraborty et al., 2016). Recently, the plant B-cell lymphoma2 (Bcl-2)-associated athanogene 7 (BAG7) protein, belonging to an evolutionarily conserved family of co-chaperones, has been shown to be involved in the regulation of the heat-induced UPR pathway (Li et al., 2017). The protein kinase RNA-like ER kinase (PERK)-mediated translational inhibition was characterized in mammals (Ruberti and Brandizzi, 2014). Also, the plant-specific NAC [acronym derived from No apical meristem (NAM), Arabidopsis transcription activation factor (ATAF), and Cup-shaped cotyledon (CUC)]-TFs are shown to be involved in ER stress response (Sun et al., 2013; Yang et al., 2014b; Chi et al., 2017). These NAC-TFs are equivalent to the secondary UPR transducers in plants. The knowledge about the activation mechanisms of the UPR in model plants such as Arabidopsis is a prerequisite for developing stress-tolerance in the agriculturally important crops. In this review, we have summarized the structural features of individual UPR-sensors and focused on the mechanistic insights into the activation of the conserved arms of the UPR, such as the bZIP28 and IRE1-bZIP60 pathways, as well as on the plant-specific UPR transducers including NAC-TFs.
The Mechanism of Activation of bZIP28
To mitigate ER stress in plants, the RIP-mediated activation of the bZIP28 is the first-hand response. The molecular structure of bZIP28, a type II membrane protein, shows that it contains a single transmembrane domain (TMD), a cytoplasmic DNA-binding bZIP domain at its N-terminus, and a luminal domain at its C-terminus (Figure 2) (Liu et al., 2007a). During the past decade, accumulating evidence suggested the step-wise activation of a bZIP28 in response to ER stress (Figure 3). Under an unstressed condition, the master regulator BiP binds to an intrinsically disordered region of the luminal domain of bZIP28, while it dissociates from the bZIP28 in response to ER stress (Srivastava et al., 2013). Once released from BiP, bZIP28 interacts with Sar1a through its dibasic motifs (KK 311, 320) present in its cytosolic domain, to initiate vesicle formation with coat protein complex II (COPII) (Srivastava et al., 2012). Under ER stress conditions, the transcriptionally induced COPII components, Sar1a/Sec23a, promote the trafficking of bZIP28 from the ER to the Golgi complex (Song et al., 2015; Zeng et al., 2015). Site-1 protease (S1P) is not involved in the processing of bZIP28 in the Golgi body, despite the presence of the consensus S1P recognition motif (RRIL) at its luminal domain. Rather, as-yet unidentified protease(s) first cleaves the bZIP28 at its TMD making it available as a substrate for the site-2 protease (S2P) (Iwata et al., 2017). S2P recognizes the putative helix-breaking residue (G329) in the TMD of bZIP28 for proteolytic processing, and the active cytosolic bZIP28 translocates to the nucleus (Srivastava et al., 2012). The nuclear-localized bZIP28 forms a transcriptional complex with the nuclear factor-Y (NF-Y) TFs, and binds specifically to the ER stress response element (ERSE) that is located in the promoter regions of the UPR genes (Liu and Howell, 2010a).
FIGURE 2.
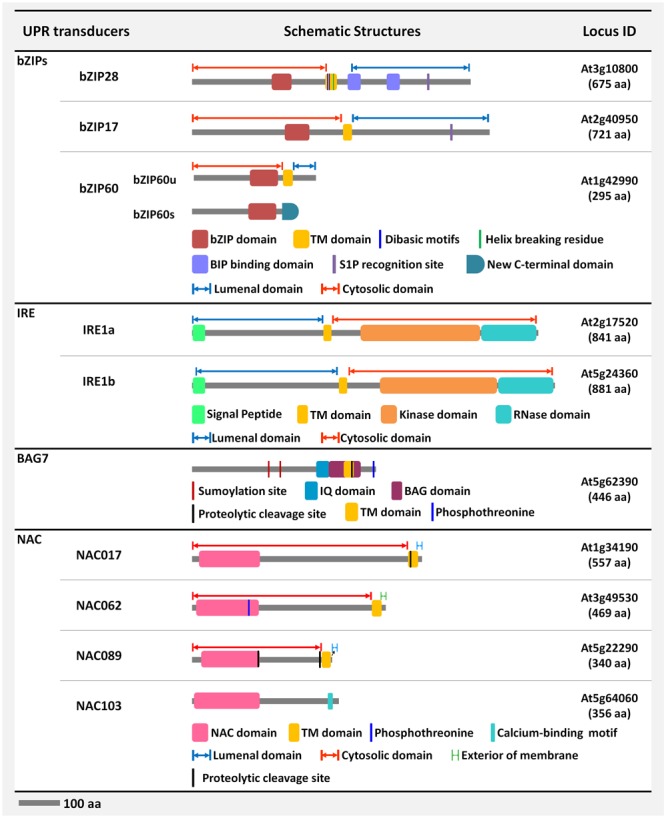
Schematic representation of the structural features of the signal transducers of the unfolded protein response (UPR) in Arabidopsis. The molecular structures of the Arabidopsis signal transducing elements in the UPR pathway are schematically represented. The structural features of the bZIP/NAC-TFs, coregulators-BAG7, and dual-functioning enzymes-IRE1 are briefly depicted.
FIGURE 3.

ER stress induced activation of bZIP28 and BAG7 in plant UPR signaling. Under normal conditions, the bZIP28 and BAG7 proteins are anchored to the ER membrane by interactions with the Binding protein (BiP). However, in response to ER stress, BiP assists the proper folding of the unfolded proteins. Then the released bZIP28 traffics to the Golgi through the Coat Protein II (COPII) vesicles. After the proteolytic cleaving of the bZIP28 by an unknown protease (X?) and by the site-2 protease (S2P), the truncated active form of bZIP28 translocates into the nucleus. The nuclear bZIP28 outcompetes HY5 for binding to the ERSE element and forms a transcriptional complex with bZIP60 and NF-Y-TFs to activate the UPR gene expression. The 26S-proteasomal degradation system eventually degrades the released HY5. Additionally, BAG7 is also released from the ER membrane by an unknown protease (X?). Subsequently, BAG7 is sumoylated and enters the nucleus. The nuclear BAG7 interacts with WRKY29 and regulates the expression of BAG7 and other chaperone proteins to mitigate ER stress.
In addition to the RIP-induced activation of the bZIP28 pathway of plant UPR, different layers of regulation cascade in the bZIP28-mediated UPR activation have been identified. Firstly, the retention of bZIP28 by the master regulator BiP under unstressed conditions is assisted by the co-chaperone function of the BAG7 protein located in the ER membrane. The BAG7 protein facilitates the direct interaction of bZIP28 with BiP under unstressed conditions. However, similar to bZIP28, ER stress also triggers a proteolytic release of the BAG7 protein, which is then sumoylated and translocated to the nucleus where it interacts with TF WRKY29 that regulates the stress-responsive genes (Williams et al., 2010; Li et al., 2017). Secondly, the activity of bZIP28 as a TF for the UPR is inhibited by another bZIP-TF, elongated hypocotyl 5 (HY5) under unstressed conditions, since they compete for binding to the G-box element (CACGTG) present at the ERSE motifs in the promoters of the UPR genes. Under ER stress conditions, the negative regulator of UPR, HY5, undergoes proteasomal degradation. After that, HY5 is out-competed by the nuclear-localized bZIP28 for binding to the ERSE motif, and activates the UPR (Nawkar et al., 2017). Moreover, the interaction of bZIP28/60 with the core components of the COMPASS-like complex mediates chromatin remodeling of the active promoters of the UPR genes through the sequence-specific histone H3K4 trimethylation (H3K4me3). This chromatin modification is critically essential for the up-regulation of the UPR genes (Song et al., 2015).
Activation of the IRE1-bZIP60 Pathway
In many eukaryotes, activation of the IRE1-mediated unconventional splicing of mRNA is the most conserved arm of the UPR (Chen and Brandizzi, 2013; Ruberti et al., 2015). In Arabidopsis, there are two isoforms of IRE1-IRE1a and IRE1b (Koizumi et al., 2001). Both the isoforms, IRE1a and IRE1b, are classified as type I single-pass transmembrane proteins. They consist of multi-functional domains such as an N-terminal signal-peptide, an ER-stress sensing domain facing the ER lumen, a protein kinase domain, and a C-terminal ribonuclease domain facing the cytosol (Figure 2). In response to biotic and abiotic stresses, IRE1a and IRE1b specifically activate the unconventional splicing of the bZIP60 mRNA (Deng et al., 2011; Moreno et al., 2012). The plant IRE1a and IRE1b can form homo/heterodimers to enable the activation of the IRE1-dependent UPR signaling pathway, similar to the activation of the IRE pathway in yeast and mammals (Credle et al., 2005; Zhou et al., 2006; Zhang et al., 2015). The yeast IRE1p activation is a stepwise mechanism that is fine-tuned by the binding of BiP to the luminal domain of IRE1p (Pincus et al., 2010). Importantly, the luminal IRE1a/1b sensor domains of plants functionally complement the yeast IRE1p sensor domain and rescue the ER stress-sensitive phenotype of the Δire1 yeast mutant (Koizumi et al., 2001). Under ER stress conditions, the luminal domain of the yeast IRE1p forms a composite groove structure by dimerization, where unfolded proteins bind and trigger IRE1p oligomerization and clustering (Korennykh et al., 2009; Gardner and Walter, 2011). Although the exact mechanism of IRE1 activation in plants has not been elucidated yet, the Arabidopsis IRE1b, but not IRE1a, showed the oligomerization structure and clustering when expressed heterologously in the yeast cells. It indicates that the first step of IRE1 activation is conserved not only in yeast, and mammals but also in Arabidopsis (Zhou et al., 2006; Zhang et al., 2016). Proper clustering and activation of Arabidopsis IRE1 requires the function of the ER-shaping GTPase Root Hair Defective 3 (RHD3), of which the underlining mechanism of action is unknown (Lai et al., 2014). Once activated, the yeast IRE1p aligns its cytosolic kinase domains in such a way that they trans-autophosphorylate each other resulting in conformational changes. The altered confirmation leads to the activation of the RNase domain of the yeast IRE1p enzyme (Shamu and Walter, 1996; Lee et al., 2008; Korennykh et al., 2011). Based on the in vitro kinase activity of the plant IRE1 and the presence of a higher degree of conserved cytosolic kinase and RNase domains among the eukaryotes, it is believed that the plant IRE1 also gets activated by trans-autophosphorylation similar to the yeast IRE1p (Koizumi et al., 2001; Zhang et al., 2016).
The yeast IRE1p clusters mediates the selective recruitment of a bipartite element in the 3′-untranslated region of the HAC1 (homologous to ATF/CREB 1) mRNA. Then the positive charge of a cytosolic linker in the yeast IRE1p mediates mRNA docking, which is an essential step for the unconventional splicing of the HAC1 mRNA (Aragon et al., 2009). In Arabidopsis, the selective targeting and docking of the unspliced bZIP60 mRNA toward the IRE1 clusters have not been reported. However, in the case of plants, it is postulated that the bZIP60 protein with a hydrophobic membrane-anchoring domain mediates the pre-recruitment of mRNA to the ER membrane. Furthermore, the conserved basic linker motif of the IRE1 homologs in plants serves as an mRNA docking site, which needs more experimental validations (van Anken et al., 2014). Similar to that of the yeast IRE1p, the activated RNase function of the Arabidopsis IRE1a/b mediates the unconventional splicing of the bZIP60 mRNA at two conserved sites (CXG| XXG) present in the kissing stem–loop structure. It results in the release of a single intron, and the spliced-fragments of the bZIP60 mRNA are ligated by the tRNA ligase RLG1 in the cytoplasm (Figure 4) (Sidrauski et al., 1996; Sidrauski and Walter, 1997; Deng et al., 2011; Nagashima et al., 2011, 2016). Upon translation of the spliced mRNA, the active form of bZIP60 without TMD (i.e., bZIP60s) translocates into the nucleus and binds to the promoter region of the UPR genes to up-regulate the UPR genes (Walter and Ron, 2011; Iwata and Koizumi, 2012). Apart from the unspliced bZIP60 mRNA, the Arabidopsis IRE1 also targets the degradation of the mRNAs encoding the proteins functioning in a secretory pathway designated as the Regulated IRE1-Dependent Decay (RIDD) of mRNAs pathway (Mishiba et al., 2013). To mediate the degradation of the mRNAs, the mammalian IREα recognizes the consensus mRNA sequence (CUGCAG) accompanied by a stem–loop structure (Oikawa et al., 2010). It will be interesting to identify such a structural specificity for the Arabidopsis IRE1-RIDD targets.
FIGURE 4.
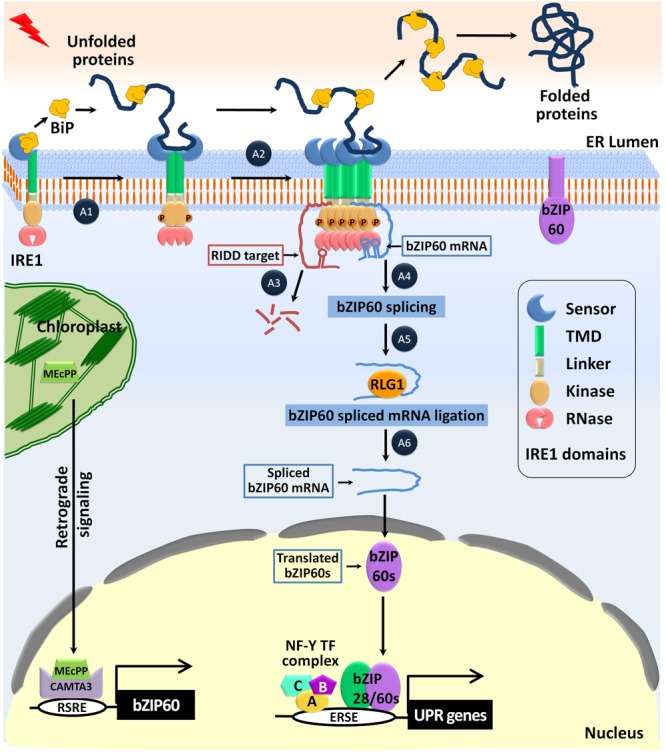
Activation of the IRE1-bZIP60 mediated UPR pathway against ER stress. Under normal conditions, the sensor domain of IRE1 binds to the ER-luminal Binding protein (BiP) and the full-length bZIP60 is anchored in the ER membrane. In response to ER-stress, 2-C-methyl-D-erythritol-2,4-cyclopyrophosphate (MEcPP) is accumulated in the chloroplast and activates the Calmodulin-Binding Transcription Activator 3 (CAMTA3)-TF, which binds to the rapid stress response element (RSRE) and up-regulates bZIP60 expression. A1–A6 indicates the step-wise activation process of IRE1. (A1) In response to ER stress, BiP dissociates from IRE1 to assist the proper folding of the accumulated unfolded proteins. The released IRE1 is dimerized to align its cytosolic kinase domains in such a way that they trans-autophosphorylate each other to activate the RNase function. (A2) The luminal domain of IRE1 binds to the hydrophobic domain of the unfolded proteins and triggers oligomerization and clustering. (A3) The mRNAs encoding the secretory pathway proteins are promiscuously cleaved by the proteins of the regulated IRE1-dependent decay (RIDD) pathway. (A4) The basic linker region of IRE1 mediates specific recruitment and docking of the unspliced bZIP60 mRNA to IRE1 clusters where it is cleaved explicitly at the two conserved sites (CXG| XXG) present in the kissing stem–loop structure. (A5) The tRNA ligase RLG1 catalyzes the ligation of the resulting fragments of the bZIP60 mRNA. (A6) Translation of the spliced bZIP60 mRNA results in the production of the active bZIP60 protein (bZIP60s). It translocates to the nucleus forming the bZIP28-NF-Y-TFs complex. These TF-complexes bind to the ERSE element of the UPR genes and positively regulate the UPR-related downstream gene expression.
In the beginning, Arabidopsis bZIP60 was characterized as a Tm-induced bZIP-TF, which regulates the UPR. Moreover, bZIP60 induces its transcription through an ERSE-like sequence present in the promoter region (Iwata and Koizumi, 2005). Recently, MEcPP, a metabolite functioning in stress-specific retrograde signaling, has also been shown to be involved in the regulation of bZIP60 at the transcript level (Benn et al., 2016). MEcPP is a stress-induced isoprenoid intermediate produced by the methylerythritol phosphate (MEP) pathway (Xiao et al., 2012). MEcPP positively regulates the general stress response (GSR) by activating the Calmodulin-Binding Transcription Activator 3 (CAMTA3)-TF, which binds to a rapid stress response element (RSRE). MEcPP-CAMTA3 induces the expression of IRE1 and bZIP60 through RSRE, following which the complex initiates the GSR, an essential process for restoration of ER-homeostasis (Figure 4) (Walley et al., 2015; Benn et al., 2016).
Activation of the NAC-TFs Pathway
The plant-specific NAC proteins belong to one of the largest gene families of TFs (Jensen et al., 2010). In the Arabidopsis genome, about 117 NAC genes have been identified, out of which 13 are predicted to be type II membrane-associated proteins (Seo et al., 2008; Puranik et al., 2012). These NAC-TFs are involved in various developmental pathways and phytohormonal interplay, and are upregulated at the transcription level by different abiotic stresses (He et al., 2005; Jiang and Deyholos, 2006; Kim et al., 2007; Tran et al., 2007). The NAC-MTFs are proteolytically processed in response to different stress stimuli such as cold, drought, high salinity, osmotic stress, or hormonal stimuli, including abscisic acid (ABA), gibberellic acid (GA), and cytokinin. Regulated activation of the MTFs is essential for rapid stress-response and plant development (Kim et al., 2008; Yoon et al., 2008; Seo et al., 2010b).
Difference in the stresses affecting protein folding leads to variations in the ER stress. The induction of stress-specific NAC-TFs in plants is needed to rapidly up-regulate the UPR downstream genes to withstand these stress conditions. For example, the NAC-TFs NAC017, NAC062, NAC089, and NAC103 are activated in response to the canonical ER stress-inducing agents such as Tm and DTT (Figure 5) (Sun et al., 2013; Yang et al., 2014b; Chi et al., 2017). The mitochondrial retrograde signaling component NAC017, which has a predicted TMD, is localized in the connections and junctions of the ER and F-actin. It acts as a high-level transcriptional regulator of the H2O2-mediated primary stress response in plants. The presence of a consensus rhomboid protease cleavage site (LSIVGA) just before the TMD of the NAC017-TF regulates its activation under stress conditions (Ng et al., 2013). Furthermore, the active NAC017 without TMD imparts ER stress tolerance (Chi et al., 2017). The expression of NAC062 and NAC103 is induced by bZIP60 through direct binding to UPRE III (TCATCG) present in their cis-elements (Sun et al., 2013; Yang et al., 2014a). NAC062 is localized to the plasma membrane and activated under cold/pathogen stresses to regulate the expression of the PR1 genes under stress conditions (Seo et al., 2010a). Although the exact mechanism for the proteolytic activation of NAC062 is poorly understood, it has been suggested that changes in the lipid composition of the plasma membrane due to ER stress or cold stress triggers the proteolytic cleavage of NAC062 by unknown zinc-dependent proteases (Seo et al., 2010b). The overexpression of NAC062 lacking the TMD and the nuclear-localized NAC103 involved in the up-regulation of the UPR genes positively mediate ER stress tolerance (Sun et al., 2013; Yang et al., 2014a). Interestingly, the overexpression of the other ER-localized protein NAC089 without TMD, results in programmed cell death (PCD), while the knock-down of its coding gene imparts ER stress tolerance (Yang et al., 2014b). Both bZIP28 and bZIP60 are involved in the upregulation of the NAC089 transcript under ER stress condition, but the protease enzyme involved in the activation of NAC089 is still unknown. Two cleavage sites in the NAC089 sequence at the amino acid position 163 (VVCRVRR| NK), a particular Arg/Lys-specific site, and a second site at position 297 (RPSQKKK| GK) have been predicted. An active NAC089 regulates the genes involved in PCD such as NAC094, metacaspase 5 (MC5), and BAG6 (Yang et al., 2014b). Moreover, the redox-dependent NAC089 functions as a negative regulator of the stromal ascorbate peroxidase (sAPX) gene expression, which is involved in the chloroplast antioxidant defense system (Klein et al., 2012; Yang et al., 2014b).
FIGURE 5.
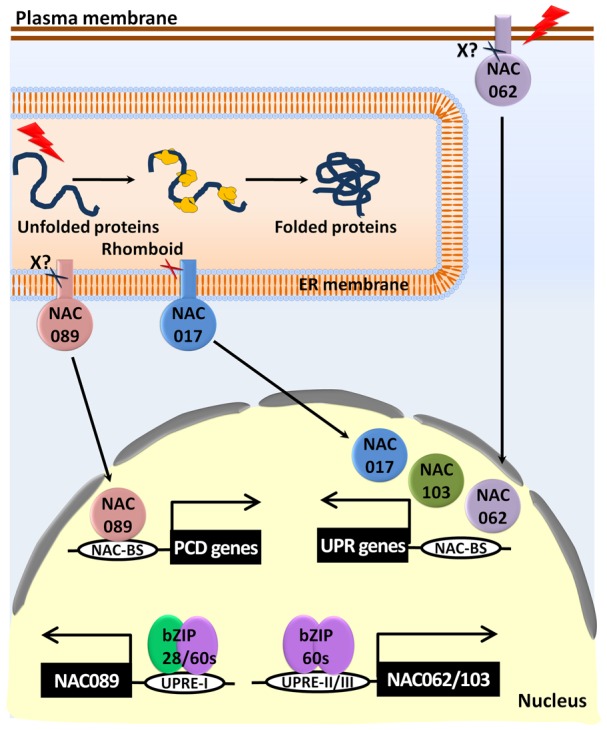
Activation of the NAC-TFs mediated UPR signaling pathways in response to ER stress. In response to ER stress, bZIP60s binds to the UPR element-II/III (UPRE-II/III) to transcriptionally upregulate the expression of NAC062 and NAC103. An unknown protease activates the plasma membrane-localized NAC062. A rhomboid protease is involved in the activation of the ER membrane-localized NAC017. The nuclear localized-NAC103 along with the activated NAC017/062 binds to the NAC-binding sites and activates the UPR gene expressions. Under chronic ER stress conditions, bZIP28/60s binds to the UPRE-I element of the NAC089 promoter and activates its gene expression. Moreover, NAC089 is processed by an unknown protease and translocates to the nucleus to upregulate the genes involved in programmed cell death (PCD).
Concluding Remarks
In Arabidopsis, all of the UPR transducers possess a TMD and are localized in the ER membrane, except NAC103 and NAC062. NAC103 is a constitutively nuclear-localized protein lacking a TMD, while NAC062 is a plasma membrane-localized protein. The lengths of the luminal domains vary among the sensor/transducer proteins, which results in the complexity in understanding the mechanism of UPR activation. For example, the interaction of the master regulator BiP with a luminal domain of bZIP28, BAG7, and IRE1 acts as a switch in the bZIP28-BAG7 and IRE1-bZIP60 pathways, respectively. Contrarily, the NAC-TFs have a shorter luminal domain and their interaction with BiP is not yet reported. Therefore, information about the key regulators in the NAC-TF-mediated pathway is still missing. For a rapid response, the MTFs are regulated at two levels; first at the post-translation level, second at the transcript level. The MTFs are located in the membrane and activated upon the onset of stress. The regulated activation of the MTFs depends on various proteases that are mostly unknown. Identification of the proteases processing specific MTFs under certain physiological conditions will help to understand the cross-talk between the activation pathways of the UPR and signaling by hormones including auxin, brassinosteroids, and SA. The advancement in the understanding of the relationship between proteases and MTFs will be helpful in plant biotechnology to improve the stress-tolerance of crop varieties and in molecular pharming.
Author Contributions
GN, CHK, and SYL conceived the idea and designed the outlines of the article. GN, ESL, RS, JHP, and SWR wrote the article. GN, RS, CHK, and SYL revised the article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by a grant from the Next-Generation BioGreen 21 Program (SSAC, PJ01317301), RDA to SHL, and by a grant from Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (2016R1D1A1B01016551) to CHK, South Korea.
References
- Angelos E., Ruberti C., Kim S. J., Brandizzi F. (2017). Maintaining the factory: the roles of the unfolded protein response in cellular homeostasis in plants. Plant J. 90 671–682. 10.1111/tpj.13449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon T., Van Anken E., Pincus D., Serafimova I. M., Korennykh A. V., Rubio C. A., et al. (2009). Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature 457 736–740. 10.1038/nature07641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y., Howell S. H. (2017). The unfolded protein response supports plant development and defense as well as responses to abiotic stress. Front. Plant Sci. 8:344. 10.3389/fpls.2017.00344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn G., Bjornson M., Ke H., De Souza A., Balmond E. I., Shaw J. T., et al. (2016). Plastidial metabolite MEcPP induces a transcriptionally centered stress-response hub via the transcription factor CAMTA3. Proc. Natl. Acad. Sci. U.S.A. 113 8855–8860. 10.1073/pnas.1602582113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I., Bulleid N. J. (2011). Protein folding and modification in the mammalian endoplasmic reticulum. Annu. Rev. Biochem. 80 71–99. 10.1146/annurev-biochem-062209-093836 [DOI] [PubMed] [Google Scholar]
- Chakraborty R., Baek J. H., Bae E. Y., Kim W.-Y., Lee S. Y., Kim M. G. (2016). Comparison and contrast of plant, yeast, and mammalian ER stress and UPR. Appl. Biol. Chem. 59 337–347. 10.1007/s13765-016-0167-6 [DOI] [Google Scholar]
- Chen Y., Brandizzi F. (2013). IRE1: ER stress sensor and cell fate executor. Trends Cell Biol. 23 547–555. 10.1016/j.tcb.2013.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y. H., Melencion S. M. B., Alinapon C. V., Kim M. J., Lee E. S., Paeng S. K., et al. (2017). The membrane-tethered NAC transcription factor, AtNTL7, contributes to ER-stress resistance in Arabidopsis. Biochem. Biophys. Res. Commun. 488 641–647. 10.1016/j.bbrc.2017.01.047 [DOI] [PubMed] [Google Scholar]
- Credle J. J., Finer-Moore J. S., Papa F. R., Stroud R. M., Walter P. (2005). On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 102 18773–18784. 10.1073/pnas.0509487102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Humbert S., Liu J. X., Srivastava R., Rothstein S. J., Howell S. H. (2011). Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 7247–7252. 10.1073/pnas.1102117108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Srivastava R., Howell S. H. (2013). Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110 19633–19638. 10.1073/pnas.1314749110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria J. A., Reis P. A., Reis M. T., Rosado G. L., Pinheiro G. L., Mendes G. C., et al. (2011). The NAC domain-containing protein, GmNAC6, is a downstream component of the ER stress- and osmotic stress-induced NRP-mediated cell-death signaling pathway. BMC Plant Biol. 11:129. 10.1186/1471-2229-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkostefanakis S., Mesihovic A., Hu Y., Schleiff E. (2016). Unfolded protein response in pollen development and heat stress tolerance. Plant Reprod. 29 81–91. 10.1007/s00497-016-0276-8 [DOI] [PubMed] [Google Scholar]
- Gao H., Brandizzi F., Benning C., Larkin R. M. (2008). A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105 16398–16403. 10.1073/pnas.0808463105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner B. M., Walter P. (2011). Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science 333 1891–1894. 10.1126/science.1209126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X. J., Mu R. L., Cao W. H., Zhang Z. G., Zhang J. S., Chen S. Y. (2005). AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44 903–916. 10.1111/j.1365-313X.2005.02575.x [DOI] [PubMed] [Google Scholar]
- Howell S. H. (2013). Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 64 477–499. 10.1146/annurev-arplant-050312-120053 [DOI] [PubMed] [Google Scholar]
- Iwata Y., Ashida M., Hasegawa C., Tabara K., Mishiba K. I., Koizumi N. (2017). Activation of the Arabidopsis membrane-bound transcription factor bZIP28 is mediated by site-2 protease, but not site-1 protease. Plant J. 91 408–415. 10.1111/tpj.13572 [DOI] [PubMed] [Google Scholar]
- Iwata Y., Koizumi N. (2005). An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. U.S.A. 102 5280–5285. 10.1073/pnas.0408941102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y., Koizumi N. (2012). Plant transducers of the endoplasmic reticulum unfolded protein response. Trends Plant Sci. 17 720–727. 10.1016/j.tplants.2012.06.014 [DOI] [PubMed] [Google Scholar]
- Jensen M. K., Kjaersgaard T., Nielsen M. M., Galberg P., Petersen K., O’shea C., et al. (2010). The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426 183–196. 10.1042/BJ20091234 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Deyholos M. K. (2006). Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol. 6:25. 10.1186/1471-2229-6-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. G., Lee A. K., Yoon H. K., Park C. M. (2008). A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J. 55 77–88. 10.1111/j.1365-313X.2008.03493.x [DOI] [PubMed] [Google Scholar]
- Kim S. Y., Kim S. G., Kim Y. S., Seo P. J., Bae M., Yoon H. K., et al. (2007). Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res. 35 203–213. 10.1093/nar/gkl1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P., Seidel T., Stocker B., Dietz K. J. (2012). The membrane-tethered transcription factor ANAC089 serves as redox-dependent suppressor of stromal ascorbate peroxidase gene expression. Front. Plant Sci. 3:247. 10.3389/fpls.2012.00247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi N., Martinez I. M., Kimata Y., Kohno K., Sano H., Chrispeels M. J. (2001). Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol. 127 949–962. 10.1104/pp.010636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh A. V., Egea P. F., Korostelev A. A., Finer-Moore J., Zhang C., Shokat K. M., et al. (2009). The unfolded protein response signals through high-order assembly of Ire1. Nature 457 687–693. 10.1038/nature07661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh A. V., Korostelev A. A., Egea P. F., Finer-Moore J., Stroud R. M., Zhang C., et al. (2011). Structural and functional basis for RNA cleavage by Ire1. BMC Biol. 9:47. 10.1186/1741-7007-9-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner C. J., Du X., Vollmer M. E., Pajerowska-Mukhtar K. M. (2015). Endoplasmic reticulum stress signaling in plant immunity–at the crossroad of life and death. Int. J. Mol. Sci. 16 26582–26598. 10.3390/ijms161125964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y. S., Stefano G., Brandizzi F. (2014). ER stress signaling requires RHD3, a functionally conserved ER-shaping GTPase. J. Cell Sci. 127 3227–3232. 10.1242/jcs.147447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. P., Dey M., Neculai D., Cao C., Dever T. E., Sicheri F. (2008). Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell 132 89–100. 10.1016/j.cell.2007.10.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Williams B., Dickman M. (2017). Arabidopsis B-cell lymphoma2 (Bcl-2)-associated athanogene 7 (BAG7)-mediated heat tolerance requires translocation, sumoylation and binding to WRKY29. New Phytol. 214 695–705. 10.1111/nph.14388 [DOI] [PubMed] [Google Scholar]
- Liu J. X., Howell S. H. (2010a). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22 782–796. 10.1105/tpc.109.072173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. X., Howell S. H. (2010b). Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22 2930–2942. 10.1105/tpc.110.078154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. X., Howell S. H. (2016). Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol. 211 418–428. 10.1111/nph.13915 [DOI] [PubMed] [Google Scholar]
- Liu J. X., Srivastava R., Che P., Howell S. H. (2007a). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19 4111–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. X., Srivastava R., Che P., Howell S. H. (2007b). Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 51 897–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes G. C., Reis P. A., Calil I. P., Carvalho H. H., Aragao F. J., Fontes E. P. (2013). GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc. Natl. Acad. Sci. U.S.A. 110 19627–19632. 10.1073/pnas.1311729110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishiba K., Nagashima Y., Suzuki E., Hayashi N., Ogata Y., Shimada Y., et al. (2013). Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 110 5713–5718. 10.1073/pnas.1219047110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno A. A., Mukhtar M. S., Blanco F., Boatwright J. L., Moreno I., Jordan M. R., et al. (2012). IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS One 7:e31944. 10.1371/journal.pone.0031944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima Y., Iwata Y., Ashida M., Mishiba K., Koizumi N. (2014). Exogenous salicylic acid activates two signaling arms of the unfolded protein response in Arabidopsis. Plant Cell Physiol. 55 1772–1778. 10.1093/pcp/pcu108 [DOI] [PubMed] [Google Scholar]
- Nagashima Y., Iwata Y., Mishiba K., Koizumi N. (2016). Arabidopsis tRNA ligase completes the cytoplasmic splicing of bZIP60 mRNA in the unfolded protein response. Biochem. Biophys. Res. Commun. 470 941–946. 10.1016/j.bbrc.2016.01.145 [DOI] [PubMed] [Google Scholar]
- Nagashima Y., Mishiba K., Suzuki E., Shimada Y., Iwata Y., Koizumi N. (2011). Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci. Rep. 1:29. 10.1038/srep00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawkar G. M., Kang C. H., Maibam P., Park J. H., Jung Y. J., Chae H. B., et al. (2017). HY5, a positive regulator of light signaling, negatively controls the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114 2084–2089. 10.1073/pnas.1609844114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S., Ivanova A., Duncan O., Law S. R., Van Aken O., De Clercq I., et al. (2013). A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25 3450–3471. 10.1105/tpc.113.113985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa D., Tokuda M., Hosoda A., Iwawaki T. (2010). Identification of a consensus element recognized and cleaved by IRE1 alpha. Nucleic Acids Res. 38 6265–6273. 10.1093/nar/gkq452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C. J., Seo Y. S. (2015). Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathol. J. 31 323–333. 10.5423/PPJ.RW.08.2015.0150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus D., Chevalier M. W., Aragon T., Van Anken E., Vidal S. E., El-Samad H., et al. (2010). BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. 8:e1000415. 10.1371/journal.pbio.1000415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S., Sahu P. P., Srivastava P. S., Prasad M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17 369–381. 10.1016/j.tplants.2012.02.004 [DOI] [PubMed] [Google Scholar]
- Ruberti C., Brandizzi F. (2014). Conserved and plant-unique strategies for overcoming endoplasmic reticulum stress. Front. Plant Sci. 5:69. 10.3389/fpls.2014.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruberti C., Kim S. J., Stefano G., Brandizzi F. (2015). Unfolded protein response in plants: one master, many questions. Curr. Opin. Plant Biol. 27 59–66. 10.1016/j.pbi.2015.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P. J., Kim M. J., Park J. Y., Kim S. Y., Jeon J., Lee Y. H., et al. (2010a). Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 61 661–671. 10.1111/j.1365-313X.2009.04091.x [DOI] [PubMed] [Google Scholar]
- Seo P. J., Kim M. J., Song J. S., Kim Y. S., Kim H. J., Park C. M. (2010b). Proteolytic processing of an Arabidopsis membrane-bound NAC transcription factor is triggered by cold-induced changes in membrane fluidity. Biochem. J. 427 359–367. 10.1042/BJ20091762 [DOI] [PubMed] [Google Scholar]
- Seo P. J., Kim S. G., Park C. M. (2008). Membrane-bound transcription factors in plants. Trends Plant Sci. 13 550–556. 10.1016/j.tplants.2008.06.008 [DOI] [PubMed] [Google Scholar]
- Shamu C. E., Walter P. (1996). Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 15 3028–3039. [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C., Cox J. S., Walter P. (1996). tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 87 405–413. 10.1016/S0092-8674(00)81361-6 [DOI] [PubMed] [Google Scholar]
- Sidrauski C., Walter P. (1997). The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90 1031–1039. 10.1016/S0092-8674(00)80369-4 [DOI] [PubMed] [Google Scholar]
- Song Z. T., Sun L., Lu S. J., Tian Y., Ding Y., Liu J. X. (2015). Transcription factor interaction with COMPASS-like complex regulates histone H3K4 trimethylation for specific gene expression in plants. Proc. Natl. Acad. Sci. U.S.A. 112 2900–2905. 10.1073/pnas.1419703112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R., Chen Y., Deng Y., Brandizzi F., Howell S. H. (2012). Elements proximal to and within the transmembrane domain mediate the organelle-to-organelle movement of bZIP28 under ER stress conditions. Plant J. 70 1033–1042. 10.1111/j.1365-313X.2012.04943.x [DOI] [PubMed] [Google Scholar]
- Srivastava R., Deng Y., Shah S., Rao A. G., Howell S. H. (2013). BINDING PROTEIN is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in Arabidopsis. Plant Cell 25 1416–1429. 10.1105/tpc.113.110684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Yang Z. T., Song Z. T., Wang M. J., Sun L., Lu S. J., et al. (2013). The plant-specific transcription factor gene NAC103 is induced by bZIP60 through a new cis-regulatory element to modulate the unfolded protein response in Arabidopsis. Plant J. 76 274–286. 10.1111/tpj.12287 [DOI] [PubMed] [Google Scholar]
- Tran L. S., Nakashima K., Sakuma Y., Osakabe Y., Qin F., Simpson S. D., et al. (2007). Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 49 46–63. 10.1111/j.1365-313X.2006.02932.x [DOI] [PubMed] [Google Scholar]
- Valente M. A., Faria J. A., Soares-Ramos J. R., Reis P. A., Pinheiro G. L., Piovesan N. D., et al. (2009). The ER luminal binding protein (BiP) mediates an increase in drought tolerance in soybean and delays drought-induced leaf senescence in soybean and tobacco. J. Exp. Bot. 60 533–546. 10.1093/jxb/ern296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Anken E., Pincus D., Coyle S., Aragon T., Osman C., Lari F., et al. (2014). Specificity in endoplasmic reticulum-stress signaling in yeast entails a step-wise engagement of HAC1 mRNA to clusters of the stress sensor Ire1. Elife 3:e05031. 10.7554/eLife.05031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot J. (2016). How does the stressed out ER find relief during virus infection? Curr. Opin. Virol. 17 74–79. 10.1016/j.coviro.2016.01.018 [DOI] [PubMed] [Google Scholar]
- Vitale A., Denecke J. (1999). The endoplasmic reticulum-gateway of the secretory pathway. Plant Cell 11 615–628. 10.2307/3870888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley J., Xiao Y., Wang J. Z., Baidoo E. E., Keasling J. D., Shen Z., et al. (2015). Plastid-produced interorgannellar stress signal MEcPP potentiates induction of the unfolded protein response in endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 112 6212–6217. 10.1073/pnas.1504828112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin E., von Heijne G. (1998). Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci 7 1029–1038. 10.1002/pro.5560070420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Ron D. (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334 1081–1086. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Wan S., Jiang L. (2016). Endoplasmic reticulum (ER) stress and the unfolded protein response (UPR) in plants. Protoplasma 253 753–764. 10.1007/s00709-015-0842-1 [DOI] [PubMed] [Google Scholar]
- Williams B., Kabbage M., Britt R., Dickman M. B. (2010). AtBAG7, an Arabidopsis Bcl-2-associated athanogene, resides in the endoplasmic reticulum and is involved in the unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 107 6088–6093. 10.1073/pnas.0912670107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H., Xu H., Xu W., He Z., Xu W., Ma M. (2016). A SAL1 loss-of-function Arabidopsis mutant exhibits enhanced cadmium tolerance in association with alleviation of endoplasmic reticulum stress. Plant Cell Physiol. 57 1210–1219. 10.1093/pcp/pcw069 [DOI] [PubMed] [Google Scholar]
- Xiao Y., Savchenko T., Baidoo E. E., Chehab W. E., Hayden D. M., Tolstikov V., et al. (2012). Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149 1525–1535. 10.1016/j.cell.2012.04.038 [DOI] [PubMed] [Google Scholar]
- Yang Z. T., Lu S. J., Wang M. J., Bi D. L., Sun L., Zhou S. F., et al. (2014a). A plasma membrane-tethered transcription factor, NAC062/ANAC062/NTL6, mediates the unfolded protein response in Arabidopsis. Plant J. 79 1033–1043. 10.1111/tpj.12604 [DOI] [PubMed] [Google Scholar]
- Yang Z. T., Wang M. J., Sun L., Lu S. J., Bi D. L., Sun L., et al. (2014b). The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet. 10:e1004243. 10.1371/journal.pgen.1004243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H. K., Kim S. G., Kim S. Y., Park C. M. (2008). Regulation of leaf senescence by NTL9-mediated osmotic stress signaling in Arabidopsis. Mol. Cells 25 438–445. [PubMed] [Google Scholar]
- Zeng Y., Chung K. P., Li B., Lai C. M., Lam S. K., Wang X., et al. (2015). Unique COPII component AtSar1a/AtSec23a pair is required for the distinct function of protein ER export in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 112 14360–14365. 10.1073/pnas.1519333112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen H., Brandizzi F., Verchot J., Wang A. (2015). The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genet. 11:e1005164. 10.1371/journal.pgen.1005164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhang C., Wang A. (2016). Divergence and conservation of the major UPR branch IRE1-bZIP signaling pathway across eukaryotes. Sci. Rep. 6:27362. 10.1038/srep27362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. S., Yang H., Ding L., Song Z. T., Ma H., Chang F., et al. (2017). Tissue-specific transcriptomics reveals an important role of the unfolded protein response in maintaining fertility upon heat stress in Arabidopsis. Plant Cell 29 1007–1023. 10.1105/tpc.16.00916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Liu C. Y., Back S. H., Clark R. L., Peisach D., Xu Z., et al. (2006). The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 103 14343–14348. 10.1073/pnas.0606480103 [DOI] [PMC free article] [PubMed] [Google Scholar]


