Abstract
Fifty fungal isolates were sampled from diseased tomato plants as result of a survey conducted in seven tomato crop areas in Algeria from 2012 to 2015. Morphological criteria and PCR-based identification, using the primers PF02 and PF03, assigned 29 out of 50 isolates to Fusarium oxysporum (Fo). The banding patterns amplified for genes SIX1, SIX3 and SIX4 served to identify races 2 and 3 of Fo f. sp. lycopersici (FOL), and Fo f. sp. radicis lycopersici (FORL) among the Algerian isolates. All FOL isolates showed pathogenicity on the susceptible tomato cv. “Super Marmande,” while nine of out 10 Algerian FORL isolates were pathogenic on tomato cv. “Rio Grande.” Inter simple sequence repeat (ISSR) fingerprints showed high genetic diversity among Algerian Fo isolates. Seventeen Algerian Trichoderma isolates were also obtained and assigned to the species T. asperellum (12 isolates), T. harzianum (four isolates) and T. ghanense (one isolate) based on ITS and tef1α gene sequences. Different in vitro tests identified the antagonistic potential of native Trichoderma isolates against FORL and FOL. Greenhouse biocontrol assays performed on “SM” tomato plants with T. ghanense T8 and T. asperellum T9 and T17, and three Fo isolates showed that isolate T8 performed well against FORL and FOL. This finding was based on an incidence reduction of crown and root rot and Fusarium wilt diseases by 53.1 and 48.3%, respectively.
Keywords: ISSR, antagonism, antibiosis, biological control, Trichoderma ghanense, Fusarium oxysporum f. sp. radicis lycopersici, Fusarium oxysporum f. sp. lycopersici
Introduction
Fusarium oxysporum Schlecht (Fo) is a free-living ascomycete fungus with no known sexual state. Fo is a complex species comprised of ubiquitous soil-borne plant pathogens, with ca. 120 formae speciales (ff. spp.) based on host specificity (Michielse and Rep, 2009; Arie, 2010). The different ff. spp. show considerable genetic diversity and have polyphyletic origin (O'Donnell et al., 1998; Nirmaladevi et al., 2016). Fo causes significant economic losses of many crops including tomato (Solanum lycopersium L.), which is one of the most worldwide cultivated vegetable crops. Fo diseases in tomato are mainly caused by f. sp. radicis-lycopersici Jarvis and Shomaker (FORL), responsible for crown and root rot, and by f. sp. lycopersici (Sacc.) Snyder and Hansen (FOL), responsible for vascular wilt disease (Edel-Hermann et al., 2012), and although both ff. spp. infect the same host plant, FOL and FORL have strict host specificity. Three physiological races of FOL (1, 2, and 3) have been differentiated depending on their ability to infect tomato cultivars carrying different resistance loci (Mes et al., 1999). The use of resistant cultivars and resistant rootstocks remains the most appropriate way to prevent Fo diseases in tomato production. Hence, determining which pathogens are emerging in the field is important in order to select the most suitable tomato cultivar. Since pathogenic strains of Fo cannot be identified morphologically, pathogenicity tests are commonly used on different tomato cultivars. However, these methods are very time-consuming and expensive (Baysal et al., 2009), and also the results of these types of biological tests can be affected by variations in temperature (Boix-Ruíz et al., 2015).
The gene sequence variability found in polygalacturonases, major enzymes involved in Fo-plant interactions (Di Pietro and Roncero, 1998), has been useful to study the genetic diversity in populations of this fungus (Kawabe et al., 2005). A PCR-based technique, using a set of primers specific to the sequences of the endo-polygalacturonase gene pg1 and the exo-polygalacturonase gene pgx4 of Fo isolates from Japan, allowed FOL and FORL, and the races of FOL (Hirano and Arie, 2006) to be differentiated. However, discrepancies have been observed in identifying some isolates collected from tomato crops in other areas of the world, such as those from the Mediterranean coast of Turkey, with pathogenicity tests and PCR carried out using this set of primers (Baysal et al., 2009; Çolak and Biçici, 2013).
FOL isolates appear to have horizontally transferred accessory chromosomes (Ma, 2014) which encode a number of putative effectors, including the set of the secreted in xylem (SIX) proteins (Houterman et al., 2009). Several SIX genes have been associated with the three races of FOL, and the molecular markers developed for these genes provide a robust PCR-based method for identifying the host specificity of FOL isolated from plant tissues (Lievens et al., 2009; Jelinski et al., 2017). In addition, the presence of SIX1 can be used to identify FOL isolates. Moreover, SIX4 allows the identification of race 1 isolates, and SIX3 variations can serve to differentiate race 2 from race 3 isolates (Lievens et al., 2009). A previous study exploring the genetic diversity of Fo and other Fusarium spp. pathogenic on tomato in different Mediterranean countries, which combine the use of the presence of SIX1, intergenic spacer (IGS) DNA typing and vegetative compatibility grouping (VCG), identified 27 out of 27 Fo Algerian isolates as FORL (Edel-Hermann et al., 2012). Unfortunately, cultivars of tomato with resistance to FORL are not yet commercially available. And, crown and root rot disease caused by FORL is widely present in most of the African and Asian Mediterranean countries, including Algeria, where tomato production is economically important, occupying second place after potato. Moreover, there are no reliable data regarding Fusarium wilt, caused by FOL races, in Algerian tomato cultivation areas.
Trichoderma species (teleomorph Hypocrea) are cosmopolitan filamentous fungi frequently found in agricultural habitats because of their ability to colonize the rhizosphere and progress in different soils (Hermosa et al., 2004; Rubio et al., 2005). The biocontrol capability of these fungi is well recognized since they are antagonists of phytopathogenic fungi, oomycetes and nematodes (Lorito et al., 2010; Medeiros et al., 2017). The biocontrol mechanisms of Trichoderma are at least based on competition for nutrients, the production of hydrolytic enzymes and/or antibiotics (Harman et al., 2004). In addition, a systemic activation of plant defense responses against biotic and abiotic damages has been observed for selected rhizosphere-competent Trichoderma strains (Hermosa et al., 2012; Ruocco et al., 2015; Rubio et al., 2017a), and the ability of Trichoderma spp. to reduce Fusarium wilt in tomato has been previously described (Cotxarrera et al., 2002; Taghdi et al., 2015).
The aim of the present study was to identify the Fo pathogenic types present in the current seven major tomato-growing areas in Algeria, to explore their genetic diversity and to propose environmentally friendly practices for controlling Fo diseases in tomato. To this end, a collection of Trichoderma spp. isolates was obtained from soils sampled in tomato plantations with low incidence of Fusarium wilt and from healthy nursery tomato plants in Algeria. These isolates were then molecularly identified and screened using both in vitro and in vivo assays to explore biocontrol potentialities against FOL and FORL. In addition, we wanted to contribute to previous studies on Trichoderma diversity in the North African Mediterranean countries in which T. atroviride and T. hamatum were prevalent in cultivated soils from Tunisia (Sadfi-Zouaoui et al., 2009), and T. asperellum, T. hamatum and T. virens were found in soil and compost samples in Morocco (Taghdi et al., 2015). It is noteworthy that race 2 and 3 of FOL are being reported for the first time in Algeria and that as far as we know T. ghanense has not been previously recorded in the Mediterranean North African countries.
Materials and methods
Collection of plant and soil samples and fungal isolation
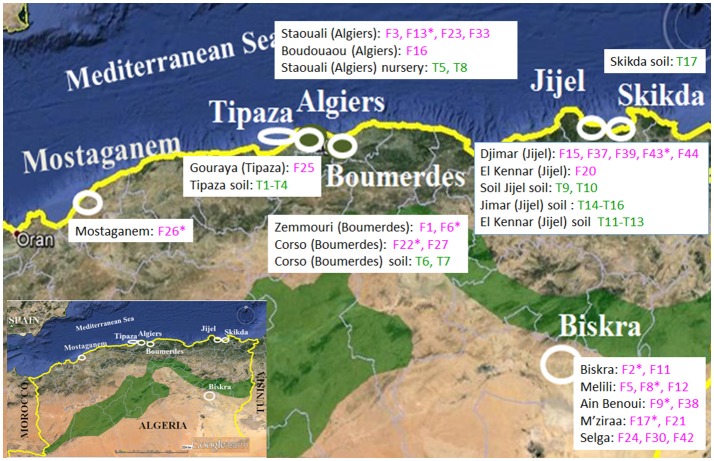
Surveys were conducted from 2012 to 2015 in seven tomato growing areas in Algeria: Mostaganem, Tipaza, Algiers, Boumerdes, Skikda, and Jijel, which are located on the coast, and Biskra in the interior of the country (Figure 1). Plants showing typical Fusarium disease symptoms (vascular tissue discoloration, leaf wilting or crown, and root rot) were collected. Tomato tissue fragments from diseased plants were surface-disinfected with 2% sodium hypochlorite for 10 min and rinsed thoroughly with sterile distilled water, and plated on potato dextrose agar (PDA, Difco Laboratories, Detroit, USA). Fifty fungal isolates recovered from these seven tomato production areas developed colonies with the typical phenotype of Fo (Table 1). Thus, monosporic cultures were obtained and the fungi preserved in the Mycological Collection of the Biotechnology Research Center (CRBt, Constatine, Algeria). In addition, two Fo isolates provided by Prof. Díaz-Mínguez (Institute CIALE, Salamanca, Spain) were used as references in this study: Forlc of FORL and 4287 of FOL (Di Pietro and Roncero, 1998). All Fo isolates were kept at 4°C on PDA plates.
Figure 1.
Location of the tomato production fields where the Fusarium (F) and Trichoderma (T) isolates were collected in Algeria. Illustration source: www.google.com/earth.
Table 1.
Algerian F. oxysporum (Fo) isolates studied.
| Code | Origin | SIX1a | f. sp | Symptoms severity on RGb | Symptoms severity on SMb |
|---|---|---|---|---|---|
| F1 | Zemmouri, Boumerdes | + | FOL | 0.0 e | 1.3 ± 0.5 e |
| F2 | Biskra | − | FORL | 3.6 ± 0.5 a | NDd |
| F3 | Staouali, Algiers | + | FOL | 0.0 e | 1.3 ± 0.5 e |
| F5 | Melili, Biskra | + | FOL | 0.0 e | 2.0 ± 0.7 bcde |
| F6 | Zemmouri, Boumerdes | − | FORL | 3.5 ± 0.7 a | ND |
| F8 | Melili, Biskra | − | FORL | 1.3 ± 0.5 d | ND |
| F9 | Ain Benoui, Biskra | − | FORL | 1.3 ± 0.5 d | ND |
| F11 | Biskra | + | FOL | 0.0 e | 3.1 ± 0.6 ab |
| F12 | Melili, Biskra | + | FOL | 0.0 e | 2.8 ± 0.9 abc |
| F13 | Staouali, Algiers | − | FORL | 1.2 ± 0.5 d | ND |
| F15 | Djimar, Jijel | + | FOL | 0.0 e | 3.6 ± 0.5 a |
| F16 | Boudouaou, Algiers | + | FOL | 0.0 e | 1.4 ± 0.5 e |
| F17 | M'ziraa, Biskra | − | FORL | 2.5 ± 0.5 bc | ND |
| F20 | El Kennar, Jijel | + | FOL | 0.0 e | 1.3 ± 0.5 e |
| F21 | M'ziraa, Biskra | + | FOL | 0.0 e | 1.3 ± 0.5 e |
| F22 | Corso, Boumerdes | − | FORL | 1.6 ± 0.5 cd | ND |
| F23 | Staouali, Algiers | + | FOL | 0.0 e | 1.6 ± 0.5 cde |
| F24 | Selga, Biskra | + | FOL | 0.0 e | 1.3 ± 0.5 e |
| F25 | Gouraya, Tipaza | + | FOL | 0.0 e | 2.0 ± 0.5 bcde |
| F26 | Mostagenem | − | FORL | 1.9 ± 0.3 cd | ND |
| F27 | Corso, Boumerdes | + | FOL | 0.0 e | 1.3 ± 0.5 e |
| F30 | Selga, Biskra | + | FOL | 0.0 e | 3.7 ± 0.5 a |
| F33 | Staouali, Algiers | + | FOL | 0.0 e | 2.1 ± 0.6 bcde |
| F37 | Djimar, Jijel | + | FOL | 0.0 e | 2.7 ± 0.7 abcd |
| F38 | Ain Bennoui, Biskra | + | FOL | 0.0 e | 1.3 ± 0.5 e |
| F39 | Djimar, Jijel | + | FOL | 0.0 e | 1.5 ± 0.5 e |
| F42 | Selga, Biskra | + | FOL | 0.0 e | 2.5 ± 0.5 abcde |
| F43 | Djimar, Jijel | − | FORL | 1.9 ± 0.3 cd | ND |
| F44 | Djimar, Jijel | − | Fo | 0.0 e | 0.0 f |
| 4287c | Córdoba, Spain | + | FOL | 0.0 e | 2.8 ± 0.4 abc |
| Forlcc | Gaza | − | FORL | 3.4 ± 0.7 ab | ND |
Information corresponding to the code, isolation location, isolate classification as ff. spp. lycopersici (FOL) or radicis lycopersici (FORL) based on the amplification of the SIX1 gene, and the severity of symptoms developed on tomato “Rio Grande” (RG) and/or “Super Marmande” (SM) varieties for all 29 Fo isolates collected from diseased tomato plants in Algeria.
Presence (+) or absence (−) of 992 bp PCR fragment using the primers P12-F2R and P12-R1 on the target gene SIX1.
Ten plants were used to test each Fo isolate, 4 weeks after inoculation in 14-day-old tomato plants, and the symptom severity was registered using a scale of 0–4. For each column, the values (mean±SD) with different letters denote the statistical significance determined by ANOVA followed by Tukey test (P < 0.05).
Isolates 4287 and Forlc were included as references of FOL and FOLR, respectively.
ND, not determined.
Soil samples were taken in tomato crop fields with a low incidence of Fusarium wilt from four Algerian littoral areas (Tipaza, Boumerdes, Jijel, and Skika), and nursery tomato plants samples were also collected from the greenhouses at the Industrial and Vegetable Crops Institute (ITCMI, Staouali, Algiers). The soil samples were taken at a depth of 10 cm at each site, placed in sterile polyethylene bags and transported to the laboratory. Ten grams of soil from each sample were mixed with 90 mL of sterile water by magnetic shaking for 1 h. An aliquot of each sample was sown on Trichoderma-selective medium (Elad et al., 1981) and then incubated at 25°C for 5 days. In the case of the nursery tomato plants, roots were disinfected with 2% sodium hypochlorite and washed with sterile water and plated on PDA medium containing 50 mg/L streptomycin. Monoconidial cultures of Trichoderma spp. isolated from the soil and plant samples were obtained.
Plant material
Two susceptible commercial tomato cultivars were used in the pathogenicity tests of the Fo isolates (Hibar et al., 2005): “Super Marmande” (SM) and “Rio Grande” (RG) (Thompson & Morgan, Ipswich, UK) for FOL and FORL, respectively. Tomato seeds were superficially disinfected as previously described (Rubio et al., 2017b).
Genomic DNA extraction
Genomic DNA from Fusarium and Trichoderma isolates was extracted after growing in potato dextrose broth (PDB, Difco) medium at 25°C and 120 rpm for 5 and 2 days, respectively. Fungal mycelia for DNA extraction were obtained as previously described (Hermosa et al., 2004). Fungal genomic DNA was isolated following the protocol described in Lee et al. (1988).
PCR reactions with Fusarium DNA
Identification of FOL races and FORL
In order to support the identification based on morphological characteristics of the 50 Fusarium isolates obtained from diseased tomato plants, a PCR reaction using primers PFO2 and PFO3 (Table 2), previously described as specific for Fo (Edel et al., 2000), was carried out. A total volume of 25 μL containing 20 ng of Fo genomic DNA, 10 pmol of each of the primers indicated above and one unit of Taq DNA polymerase (Biotools B&M Lab., Madrid, Spain) was used to perform the PCR. The amplification program included an initial denaturalization cycle of 5 min at 95°C, followed by 30 cycles of 30 s at 95°C, 30 s at 62°C, 30 s at 72°C, and a final extension step of 3 min at 72°C in a Applied Biosystems GeneAmp PCR System 9700 thermal cycler. Following amplification, the PCR products were observed after electrophoresis on 2% agarose gels.
Table 2.
Primers used in this study and size of amplicons.
| Primer | Sequence (5′-3′) | Amplicon (in bp) | Target | References |
|---|---|---|---|---|
| PFO2 | CCCAGGGTATTACACGGT | 70 | rDNA-Fo | Edel et al., 2000 |
| PFO3 | CGGGGGATAAAGGCGG | Edel et al., 2000 | ||
| P12-F2B | TATCCCTCCGGATTTTGAGC | 992 | SIX1 | van der Does et al., 2008 |
| P12-R1 | AATAGAGCCTGCAAAGCATG | Rep et al., 2004 | ||
| SIX3-F1 | CCAGCCAGAAGGCCAGTTT | 608 | SIX3 | van der Does et al., 2008 |
| SIX3-R2 | GGCAATTAACCACTCTGCC | van der Does et al., 2008 | ||
| SIX4-F1 | TCAGGCTTCACTTAGCATAC | 967 | SIX4 | van der Does et al., 2008 |
| SIX4-R1 | GCCGACCGAAAAACCCTAA | van der Does et al., 2008 | ||
| SIX3-G121AF2a | ACGGGGTAACCCATATTGCA | 429 | SIX3 | Lievens et al., 2009 |
| SIX3-G134AF2a | TTGCGTGTTTCCCGGCCA | 414 | SIX3 | Lievens et al., 2009 |
| SIX3-G137CF2a | GCGTGTTTCCCGGCCGCCC | 412 | SIX3 | Lievens et al., 2009 |
| ITS1b | AATAGAGCCTGCAAAGCATG | 580–600 | ITS | White et al., 1990 |
| ITS4 b | GCCATCCTTGGAGACCAGC | White et al., 1990 | ||
| EF1-728Fb | GCCATCCTTGGAGACCAGC | 520–565 | tef1alpha | Chaverri et al., 2003 |
| tef1-rev b | GCCATCCTTGGAGACCAGC | Kullnig-Gradinger et al., 2002 |
When used with SIX3-R2, a fragment of 429, 414 or 412 bp is generated for FOL race 3 isolates by PCR.
Primer pairs used to amplify fragments and sequencing from Trichoderma isolates.
All Fo isolates were subjected to PCR analysis using primer sets P12-F2B/P12-R1 (SIX1 gene), SIX3-F1/SIX3-R2, SIX4-F1/SIX4-R1, SIX3-G121A-F2/SIX3-R2, SIX3-G134A-F2/SIX3-R2, and/or SIX3-G137C-F1/SIX3-R2 (Table 2). Reaction mixtures and Thermal conditions were as previously described (van der Does et al., 2008; Lievens et al., 2009). PCR products were electrophoresed as described above, and the results of this PCR analysis were scored as previously described (Lievens et al., 2009).
Inter simple sequence repeat (ISSR) PCR analysis
The genetic diversity existing among 31 Fo isolates (the 29 isolated in Algeria and the two Spanish included as references) was analyzed by PCR with the primers (GA)9C and (GA)9T and the Taq polymerase system (Biotools). The PCR amplification was carried out in a 25 μL reaction volume with 2.5 μL of 10X PCR buffer, 0.2 mM dNTPs, 10 pmol of primer, 1.25 units of Taq DNA polymerase and 50 ng genomic DNA as the template. For each primer-isolate combination, amplifications were repeated two times to assure reproducibility. The PCR program was as previously described (Nirmaladevi et al., 2016). PCR products were electrophoresed on 1.5% agarose gel as described above, and photographs were taken when visualized in a UV transilluminator.
For data analysis, the presence or absence of a given band was scored as 1 and 0, respectively. The application of the Jaccard's coefficient to the binary matrix served to calculate the pairwise distance among the isolates, which were clustered by UPGMA (Unweighted Pair Group method using arithmetic means). Genetic diversity for each primer was calculated as previously described (Owen et al., 1998) using the following formula (H = 1–Σxi2), where H is the expected heterozygosity and xi is the frequency of allele i, including “nulls” as a separate band.
PCR amplification, sequencing and DNA analysis for Trichoderma isolates
The amplification and sequencing of the ITS regions and a fragment of ca. 0.56 kb of the tef1α (translation elongation factor 1-alpha) gene from 17 Trichoderma isolates were performed using the primers ITS1/ITS4 and EF1-728F/tef1-rev (Table 2), respectively, as described previously (Hermosa et al., 2004; Sadfi-Zouaoui et al., 2009). The Trichoderma ITS and tef1α sequences obtained in this work were analyzed by comparison with sequences of Trichoderma strains deposited in the NCBI GenBank database.
Pathogenicity test for Fo isolates
Tomato seeds of “SM” and “RG” varieties were disinfected as indicated above, and sowed in a commercial substrate Projar Professional-Comercial (Projar SA, Valencia, Spain), previously autoclaved for 1 h at 121°C on two successive days and deposited in multi-cell growing trays. Seedlings were maintained under greenhouse conditions and inoculated with Fo when they were 14 days old as previously described (Rubio et al., 2017b), except that 3 L pots containing a sterilized mixture of sand:peat:vermiculite (1:1:1) were used in the present study. Five pots per condition and two seedlings per pot were used for each Fo. Results were expressed for “SM” and “RG” as symptoms severity index, using the following symptoms scale (0–4): 0, healthy plant; 1, 2, and 3, slight, moderate and severe chlorosis, wilting or stunting “SM” plant, respectively; 1, 2, and 3, slight, moderate and severe dark brown lesions on crown and roots of “RG” plant, respectively; and 4, dead plant.
In vitro antifungal assays
Isolates T5, T8, T9, and T17 of Trichoderma were in vitro tested for their antifungal potential against the Fo isolates F17, Forlc, 4287, and F42 using direct confrontation and membrane assays as previously described (Rubio et al., 2009, 2017b), except that the Fo colony diameters were measured after 6 and 5 days, respectively. Each Trichoderma-Fo combination was tested in triplicate. Results were expressed as the percentage of growth inhibition of each Fo by each Trichoderma isolate with respect to the mean colony diameters of each Fo grown alone (direct confrontation assay) or Fo control culture (membrane assays).
In vivo biocontrol assays in tomato plants
The ability of three Trichoderma isolates (T8, T9, and T17) to control diseases caused by three Fo isolates (Forlc, 4287, and F42) in tomato cv. “SM” was evaluated in in vivo assays. Surface-sterilized tomato seeds were coated with Trichoderma conidia (1 mL of an aqueous suspension containing 2 × 107 conidia per mL was used to coat 30 seeds) as previously described (Pérez et al., 2015). Trichoderma-treated tomato seeds were sown in the commercial substrate indicated above in multi-cell growing trays maintained in a greenhouse. Untreated tomato seeds were used as control. Fo inoculation was performed as previously described (Rubio et al., 2017b). Ten pots containing one seedling per pot were used in each Trichoderma-Fo combination tested. The experiment lasted 3 weeks and was conducted twice with similar results. The severity of the symptoms was recorded using the index range (0–4) already mentioned. The percentage of the incidence of disease (ID) and the decrease in ID (DID) due to the treatment with Trichoderma were determined as previously described (Song et al., 2004).
Statistical analysis
Data from in vitro and in vivo assays were subjected to analysis of variance (ANOVA) using STATISTIC v10.0 (NH Analytical Software, Roseville, USA) and means compared by Tukey's test (P < 0.05).
Results
Identification of algerian Fo isolates
The amplification of a 70 bp PCR fragment with the primer pair PFO2–PFO3 served to identify 29 out of 50 fungal isolates collected from diseased tomato plants in Algeria as Fo (Table 1, Figure 1). In order to discriminate between FORL and FOL isolates, PCR amplifications of the SIX1 gene, which encodes a FOL virulence factor toward tomato, were performed using the P12-F2R and P12-R1 primers, and the DNA from two reference isolates were also included as controls. As a result, 20 isolates, including the Spanish isolate 4287 used as a FOL reference, exhibited the 992-bp PCR fragment indicative of the f. sp. FOL (Table 1).
All of the 31 (29 from Algeria and two control) isolates of Fo were utilized in the pathogenicity tests performed on tomato plants with the aim to confirm the identification of each isolate as being either FOL and FORL as shown by the SIX1 PCR amplifications. The results of pathogenicity tests are presented in Table 1. Ten (nine collected in Algeria and the reference isolate Forlc) out of 11 FORL isolates presented virulence on tomato cv. “RG”. Plants inoculated with isolates F2 and F6 gave the highest symptom severity indexes, these values being significantly different from those obtained for the seven other Algerian FORL isolates, although no differences were detected with that of Forlc. Then, a pathogenicity test was performed on tomato plants cv. “SM” for the remaining 20 FOL isolates, plus the Fo isolate F44, that were non-virulent on “RG.” All FOL isolates (19 from Algeria and the reference isolate 4287) caused the typical symptoms of wilt disease, which were observed 4 weeks after the plants were inoculated. The symptoms severity values obtained for FOL-inoculated plants were diverse, ranging from 1.3 (for seven isolates) to 3.7 for F30. Isolates F15 and F30 gave the highest values but did not differ significantly from the values recorded for the plants inoculated with isolate 4287. Since the Fo isolate F44 did not cause disease symptoms on either “RG” or “SM” tomato plants, it was recorded as being a tomato non-pathogenic Fo isolate.
Additional PCR amplifications were carried out with SIX3 and SIX4 pair primers (Table 2) to discriminate the FOL races. The three different amplification patterns obtained with the 31 Fo isolates showed the presence of FOL races 2 and 3 and FORL among the Algerian ones (Table 3). A 608-bp fragment of SIX3 was amplified with SIX3-F1 and SIX3-R2 primers for 20 isolates, but no amplification was observed with SIX4-F1 and SIX4-R1 primers, indicating that they were not FOL race 1. This result is in concordance with the amplification of the 992-bp PCR fragment of the SIX1 gene seen in these 20 isolates, and supports the results obtained from the pathogenicity test. The presence of the 412-bp fragment PCR detected for isolates F12 and F20 with the SIX3-G137C-F1 and SIX3-R2 pair primers, and the absence of an amplicon when SIX3-G121AF2 or SIX3-G134AF2 primer was combined with the primer SIX3-R2, indicated that these isolates were FOL race 3. No amplification bands for genes SIX3 and SIX4 were observed for the 10 isolates, including the FORL reference isolate, scored previously as SIX1 negative and exhibiting virulence on tomato cv. “RG”, confirming that they were FORL.
Table 3.
Molecular identification of Algerian F. oxysporum (Fo) isolates based on patterns of SIX3 and SIX4 gene amplifications (+, amplicon was present; −, amplicon was absent).
| Isolates | Primer pairs | Fo identify | ||
|---|---|---|---|---|
| SIX3-F1/SIX3-R2 | SIX4-F1/SIX4-R1 | SIX3-G137C-F1/SIX3-R2 | ||
| (608 bp) | (967 bp) | (412 bp) | ||
| F1, F3, F5, F11, F15, F16, F21, F23-F25, F27, F30, F33, F37-F39, F42, 4287a | + | − | − | Race 2 FOL |
| F12, F20 | + | − | + | Race 3 FOL |
| F2, F6, F8, F9, F13, F17, F22, F26, F43, F44 | ||||
| Forlcb | − | − | − | FORL |
Isolate 4287 was included as reference for f. sp. lycopersici (FOL) race 2.
Isolate Forlc was included as reference for f. sp. radicis lycopersici (FORL).
Genetic diversity among algerian Fo isolates by ISSR fingerprinting
Two primers were analyzed by their capacity to produce polymorphism amplicons on the set of 31 Fo isolates, which included the 29 from Algeria and the reference isolates Forlc and 4287. The (GC)9C or (GT)9T primers resulted in reproducible banding patterns for 26 out of 31 isolates (Table 4). The two primers (GA)9C and (GA)9T produced 20 and 17 bands among 22 and 26 Fo isolates, respectively. The (GA)9C primer produced 1–8 bands ranging from approximately 400–1,900 bp and (GA)9T produced 1–7 bands ranging from 300 to 2,000 bp. The average Nei's gene diversity was 0.22, with the highest value recorded for primer (GA)9T (0.26).
Table 4.
ISSR typing using two primers to analyze the genetic diversity among 31 isolates of F. oxysporum, 29 collected in Algeria and 4287 and Folrc included as references, with the Nei's gene diversity (h).
| ISSR primer | Number of loci | Number of isolates with null band | Nei's gene diversity (h) |
|---|---|---|---|
| (GA)9C | 20 | 9 | 0.19 |
| (GA)9T | 17 | 5 | 0.26 |
The drawn UPGMA dendrogram (Figure 2), based on the band patterns observed with both primers and the Jaccard similarity coefficient, showed high diversity among the 26 Fo isolates. A similarity index of 1 was only recorded for the FOL race 2 isolate 4287 and FOL isolate F33. The typology of this dendrogram clustered the 26 Fo isolates into seven major groups, identified as I-VII, using a similarity index limit of 0.26. In general, FOL and FORL isolates were distributed across the dendrogram, and their clusters did not show a correlation with either the severity of the symptoms found in the pathogenicity tests or the geographical locations in which the isolates were collected. Nevertheless, five out of 10 isolates collected in the Biskra area were located within group IV, but this cluster also included four out of the 10 FORL isolates and it was not supported with bootstrap stability.
Figure 2.
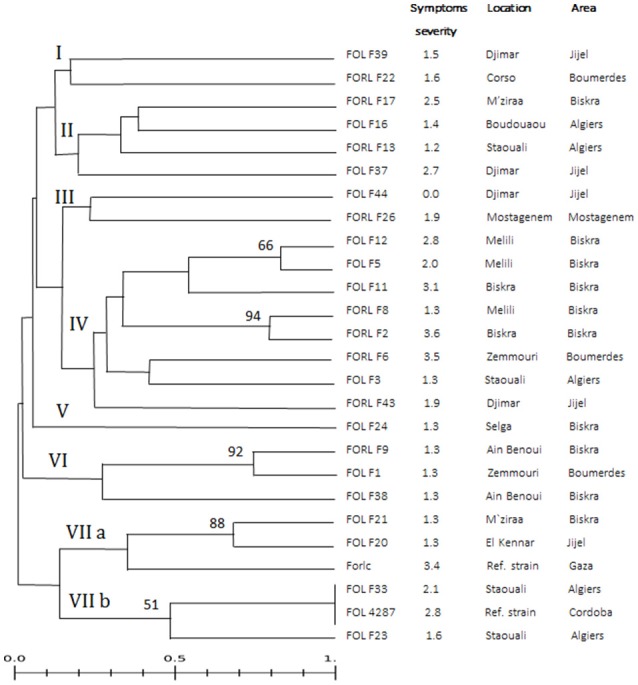
Grouping of Algerian F. oxysporum (Fo) isolates based on genetic similarity. Unweighted pair-group method algorithm dendrogram of relative genetic similarity among 26 Fo isolates, calculated in the ISSR analysis with (GA)9C and (GA)9T primers and using Jaccard's coefficient. Relative support for particular clades in the UPGMA dendrogram was estimated by using 1,000 replications of the bootstrap procedure.
Identification of Trichoderma isolates and potential biocontrol activity against Fo
Seventeen Trichoderma isolates were collected, 15 from cultivated tomato soils without symptoms of disease and two of nursery tomato plants (Figure 1). Their identity was confirmed at the species level by analyzing the sequences of two molecular markers (Table 5). The sequences containing the ITS1, ITS2, and 5.8S gene were 584–600 bp in length, and their alignment served to identify three different sequence types. A ca 600 bp fragment of the tef1α gene fragment also served to identify three different types of sequences. Both ITS and tef1α sequences gave 100% identity with sequences from reference strains deposited in the databases, and identified four isolates as T. harzianum, 12 as T. asperellum and one as T. ghanense.
Table 5.
Source and identification of Algerian Trichoderma isolates analyzed in this study.
| Isolate code | Source/geographical origin | Identified as | 100% Identity to strain/accession numbera |
|---|---|---|---|
| T1 | Soil/Tipaza | T. harzianum | GJS 99-5/FJ442236 |
| T2 | Soil/Tipaza | T. asperellum | CBS 433.97/AF456907 |
| T3 | Soil/Tipaza | T. asperellum | CBS 433.97/AF456907 |
| T4 | Soil/Tipaza | T. asperellum | CBS 433.97/AF456907 |
| T5 | ITCMIb nursery/Staouali Algiers | T. harzianum | GJS 99-5/FJ442236 |
| T6 | Soil/Corso, Boumerdes | T. harzianum | GJS 99-5/FJ442236 |
| T7 | Soil/Corso, Boumerdes | T. harzianum | GJS 99-5/FJ442236 |
| T8 | ITCMIb nursery/Staouali Algiers | T. ghanense | DAOM 220800/EU280100 |
| T9 | Soil/Jijel | T. asperellum | CBS 433.97/AF456907 |
| T10 | Soil/Jijel | T. asperellum | CBS 433.97/AF456907 |
| T11 | Soil/El Kennar, Jijel | T. asperellum | CBS 433.97/AF456907 |
| T12 | Soil/El Kennar, Jijel | T. asperellum | CBS 433.97/AF456907 |
| T13 | Soil/El Kennar, Jijel | T. asperellum | CBS 433.97/AF456907 |
| T14 | Soil/Jimar, Jijel | T. asperellum | CBS 433.97/AF456907 |
| T15 | Soil/Djimar, Jijel | T. asperellum | CBS 433.97/AF456907 |
| T16 | Soil/Djimar, Jijel | T. asperellum | CBS 433.97/AF456907 |
| T17 | Soil/Skikda | T. asperellum | CBS 433.97/AF456907 |
The classification of Trichoderma species was based on the sequences obtained from ITS1-ITS4 region of the rDNA and a fragment of tef1alpha gene.
100% identity of ITS and tef1alpha-fragment sequences with database (NCBI GenBank) sequences from reference Trichoderma strains.
ITCMI: Industrial and Vegetable Crops Institute, Staouali Algiers, Algeria. Strains are available for research proposals in the Mycological Collection of the Biotechnology Research Center (CRBt, Constatine, Algeria) (www.crbt.dz).
Two Algerian Fo isolates, representing FOL and FORL, the reference isolates FOL 4287 and FORL Forlc, and four Trichoderma isolates (T5, T8, T9, and T17), representing the species diversity found within the Algerian samples (Figure 1), were included in in vitro antifungal assays to test the biocontrol potential of Trichoderma against Fo (Table 6). When confronted in dual cultures, the four Trichoderma isolates were able to overgrow the colonies of the four Fo isolates and sporulate. Differences were only detected in the growth inhibition of the dual cultures containing T5 against Forlc or 4287 and T8 against F17.
Table 6.
Antagonism of Algerian Trichoderma spp. isolates against F. oxysporum (Fo).
| Trichoderma-Fo combination | Dual culture | Cellophane | Dialysis membrane |
|---|---|---|---|
| T5-FORL F17 | 64.6 ± 1.1 ab | 17.0 ± 1.0 de | 47.6 ± 1.4 de |
| T5-FORL Forlc | 69.8 ± 0.0 a | 12.5 ± 2.9 e | 29.8 ± 2.2 fg |
| T5-FOL 4287 | 70.6 ± 1.1 a | 35.3 ± 2.1 bcd | 27.5 ± 1.0 fg |
| T5-FOL F42 | 69.5 ± 0.0 ab | 35.1 ± 2.4 bcd | 22.6 ± 2.5 g |
| T8-FORL F17 | 59.1 ± 1.1 b | 35.8 ± 1.3 bcd | 57.7 ± 1.5 bcd |
| T8-FORL Forlc | 62.8 ± 0.0 ab | 63.5 ± 1.8 a | 72.5 ± 1.5 ab |
| T8-FOL 4287 | 63.0 ± 1.0 ab | 47.1 ± 1.5 abc | 53.7 ± 1.0 cd |
| T8-FOL F42 | 68.0 ± 1.1 ab | 51.2 ± 1.4 ab | 56.9 ± 1.3 bcd |
| T9-FORL F17 | 62.4 ± 1.5 ab | 37.0 ± 1.8 bc | 41.8 ± 2.6 def |
| T9-FORL Forlc | 63.6 ± 1.1 ab | 62.7 ± 0.6 a | 78.5 ± 1.0 a |
| T9-FOL 4287 | 63.0 ± 0.0 ab | 46.2 ± 1.0 abc | 68.3 ± 2.9 abc |
| T9-FOL F42 | 66.4 ± 1.1 ab | 26.5 ± 1.5 cde | 30.5 ± 1.91 efg |
| T17-F17 | 64.6 ± 1.1 ab | 28.5 ± 1.7 cde | 49.2 ± 1.4 d |
| T17-FORL Forlc | 63.6 ± 1.1 ab | 64.4 ± 2.2 a | 82.1 ± 2.6 a |
| T17-FOL 4287 | 63.0 ± 0.0 ab | 52.1 ± 1.9 ab | 68.9 ± 2.3 abc |
| T17-FOL F42 | 68.7 ± 1.1 ab | 29.9 ± 1.9 cde | 31.3 ± 2.9 efg |
Colony growth inhibition of Fo f. sp radicis lycopersici F17 and Forlc, and Fo f. sp. lycopersici 4287 and F42 isolates by T. harzianum T5, T. ghanense T8 and T. asperellum T9 and T17, after dual culture growth for 6 days and by metabolites from 2-day cultures of Trichoderma strains on cellophane or a 14-kDa cut-off dialysis membrane for 5 daysa. aResults are expressed as the percentage of inhibition of each Fo isolate by each Trichoderma with respect to the mean colony diameter of each fungus grown alone. Values are means of three replicates with the corresponding standard deviation. Values in the same row with different letters are significantly different according to Tukey's test (P < 0.05).
When Trichoderma isolates were tested on cellophane and dialysis (cut-off 14 kDa) membranes, the subsequent Fo cultivation always led to a reduced colony size. The effects of metabolites and hydrolytic enzymes (cellophane) or only metabolites (dialysis) secreted by the four Trichoderma isolates on the growth of the four Fo isolates are reported as Fo colony growth inhibition percentages in Table 6. No significant differences were found among the four Trichoderma isolates against FOL 4287 in the cellophane assay, but differences were observed for the inhibition of FORL F17 between T9 and T5 in that same assay. The isolate T5 also displayed a significantly worse performance against Forlc. In the case of F42, significant growth inhibition differences were detected between the best isolate (T8: 51.2%) and the worst (T9: 26.5%).
In dialysis membrane tests (Table 6), no significant differences were observed in the antifungal activities of the four Trichoderma isolates against FORL F17. However, T5 showed the significantly lowest inhibition values against Forlc and 4287. Isolates T8 and T9 also showed the significant highest and lowest inhibition records against FOL F42, respectively. These results indicate that Fo is sensitive to metabolites released by T8, T9, and T17 and, at least in in vitro assays, these isolates had the highest antagonistic potential against Fo. These three Trichoderma isolates were selected for succesive in planta tests.
Biocontrol of Trichoderma against Fo
The effect of treatment with T. ghanense T8 and T. asperellum T9 and T17 on diseases caused by FOL F42, and the reference isolates FOL 4287 and FORL Forlc, in tomato cv. “SM” is shown in Table 7. No symptoms were detected on the non-inoculated plants and those grown from T8-, T9-, or T17-treated seeds (data not shown). The ID caused by Forlc ranged from 80 to 97.5%, while the percentages of the ID in F42- and 4287-inoculated plants were lower. Furthermore, biocontrol activity against Fo in tomato was detected for the three Trichoderma strains tested. The highest percentages of DID were recorded for isolate T8, which were 53.1 and 48.3%, against Forlc and F42, respectively. Isolate T9 gave a higher DID against FOL 4287 than T8 and T17. Based on these results, T. ghanense T8 seemed to be the best isolate for controlling tomato diseases caused by FORL and FOL.
Table 7.
Incidence of the disease (ID) caused by F. oxysporum f. sp. radicis lycopersici Forlc and F. oxysporum f. sp. lycopersici F42 and 4287 in “Super Marmande” tomato plants single treated with T. ghanense T8 and T. asperellum T9 and T17a.
| Fusarium treatments | ID (%) | Trichoderma-Fusarium treatments | ID (%) | Decrease of disease incidence (DID)b |
|---|---|---|---|---|
| FORL Forlc | 80 ± 10.5 | T8-Forlc | 37.5 ± 13.2 a | 53.1 |
| FORL Forlc | 97.5 ± 7.9 | T9-Forlc | 77.5 ± 7.9 b | 20.5 |
| Forlc | 90 ± 12.9 | T17-Forlc | 80 ± 10.5 b | 11.1 |
| FOL 4287 | 77.5 ± 7.9 | T8-4287 | 57.5 ± 12.1 ab | 25.8 |
| FOL 4287 | 62.5 ± 13.2 | T9-4287 | 37.5 ± 13.2 a | 40.0 |
| FOL 4287 | 82.5 ± 12.1 | T17-4287 | 67.5 ± 12.1 b | 18.2 |
| FOL F42 | 72.5 ± 7.9 | T8-F42 | 37.5 ± 13.2 a | 48.3 |
| FOL F42 | 55 ± 10.5 | T9-F42 | 30 ± 10.5 a | 45.4 |
| FOL F42 | 42.5 ± 12.1 | T17-F42 | 30 ± 10.5 a | 29.4 |
Plants were obtained from untreated or Trichoderma-treated seeds. Fusarium isolates were applied when tomato seedlings were 14 days old, and the incidence of disease was evaluated 3 weeks after Fusarium inoculation. Data are the mean of the results obtained from 10 plants per single Fusarium or combined Trichoderma-Fusarium treatments with the corresponding standard deviation. For each Trichoderma-Fo combination tested, ID values followed by different letters are significantly different according to Tukey's test (P < 0.05).
DID was calculated as the difference between DI in the Fo treatment and in the Fo-Trichoderma treatment divided by DI in the Fo treatment expressed as a percentage.
Discussion
The Mediterranean climate with warm to hot, dry and long summers and very mild winters favors tomato crop field production, which can be accompanied by soil-borne diseases, as is the case of those produced by Fo (Muñoz et al., 2008). The use of cultivars resistant to FOL and grafted material to prevent FORL (only tolerant varieties are available against this last f. sp.) are common strategies employed in many countries that have led to a decrease in Fusarium diseases in tomato crops (Hibar et al., 2007; Baysal et al., 2009). However, Fo diseases continue to be a major sanitary problem in tomato production areas of countries like Turkey, Malta, Morocco, Tunisia, Algeria (Porta-Puglia and Mifsud, 2005; Baysal et al., 2009; Edel-Hermann et al., 2012; Taghdi et al., 2015), or India (Nirmaladevi et al., 2016). In many cases, in addition to the lack of knowledge concerning the pathogenic forms present in a given cultivation area, farmers produce their own seeds from old tomato cultivars, which can lead to the spread of FOL and FORL pathogens through seeds, transplants and contaminated soil (Edel-Hermann et al., 2012).
Nineteen out of the 29 Algerian isolates of Fo analyzed in our study, collected between 2012 and 2015, have been classified as FOL, according to the pathogenicity tests and the amplification of the SIX genes. Previous studies have highlighted the usefulness of primers designed to SIX genes for the rapid discrimination by PCR-based methods of FOL races among the Fusarium species (van der Does et al., 2008; Lievens et al., 2009). Our results indicate that SIX genes were useful to discriminate between FOL and FORL isolates, since the results obtained from the amplification of the SIX genes and the pathogenicity tests showed a high level of concordance in 30 out of the 31 Fo isolates analyzed. Isolate F44 did not cause symptoms when used to inoculate the “RG” tomato variety. Moreover, it is not uncommon to find non-pathogenic Fo strains among isolates originally obtained from collections (Cai et al., 2003), monosporic-origin inocula (Boix-Ruíz et al., 2015), or soil samples (Jelinski et al., 2017). FOL race 2 was overrepresented among the Algerian isolates tested, although our results show that race 3 (isolates F12 and F20) is also present in this country. Our results agree with those reported for Fo isolates collected from the Mediterranean coast of Turkey (Baysal et al., 2009), where race 2 and 3 of FOL, and FORL were detected. While FOL race 3 was overrepresented in Turkish samples, in the present study its prevalence only reached 6.9%. The existence of race 3 in a North African country such as Algeria reinforces the fact that this FOL race is distributed worldwide.
Since the use of grafted material is not a common practice in this North African region, it could be expected that FORL is often present in tomato growing areas (Hibar et al., 2007; Edel-Hermann et al., 2012). Nine out of the 29 Algerian isolates analyzed in our study were classified as FORL, representing only 34.5% of total number of isolates. A previous study carried out with Fo isolates from eight Mediterranean countries, showed that the Algerian isolates, all of them identified as FORL, were genetically diverse from those of the other countries (Edel-Hermann et al., 2012). Our results show the presence of FORL and FOL, and even different FOL races in a same location, i.e., isolates F5, F8, and F12 in Melili (Biskra). Thus, the 19 Algerian isolates identified as FOL in the present study represent the first formal report of the presence of FOL race 2 and 3 in Algeria.
Correlation between FOL races and field location has been proposed for isolates sampled in Turkey (Baysal et al., 2009). For instance, race 3 and FORL were associated with coastal fields, whereas race 2 was more frequent in inland crops. However, our study, covering the seven major tomato production areas in Algeria and including isolates from 13 geographical locations, did not identify a predictable dissemination pattern for Fo (Figure 1). FORL and the races 2 and 3 of FOL were detected in inland fields and at a single tomato growing area in Melili (Biskra). In addition, seven out of the 17 FOL race 2 isolates were also sampled in the Biskra region.
Molecular markers based on PCR, such as ISSR and RAPD, are useful to analyze genetic variation within and between fungal species. Several studies have used the ISSR analysis to assess genetic variability in Fo from different food crops (Dubey and Shio, 2008; Baysal et al., 2009, 2010; Edel-Hermann et al., 2012; Nirmaladevi et al., 2016). Because of the use of longer primers, normally 18–24 bp long, ISSR markers are referred as more reproducible than those generated by RAPD ones (Godwin et al., 1997). In this work, we have examined the genetic diversity among 29 Algerian Fo isolates by ISSR analysis, using two primers and including two reference isolates for comparative purposes. Despite the fact that five isolates did not produce amplified bands, a high divergence among isolates was still observed (83.9%). In addition, the polymorphism detected as a whole with these two primers suggests a large genetic variation within FOL and FORL. This is in accordance with the high degree of genetic diversity detected among Indian FOL isolates using the same primers to carry out the ISSR analysis (Nirmaladevi et al., 2016). By contrast, low genetic diversity was reported for race 2 isolates in Taiwan, which was the predominant race in that particular study (Sheu and Wang, 2006). RAPD-based genetic diversity within and among VCGs of FORL has also been described (Balmas et al., 2005). Moreover, it has been reported that European Fo isolates are less diverse than those from North Africa, and that FORL populations in Algeria, which comprised numerous IGS types and VCGs, presented differences when compared to other populations originating from other Mediterranean countries (Edel-Hermann et al., 2012). This appreciation is supported by the UPGMA tree generated in our study, where most of the Algerian isolates were separately clustered from the two reference isolates of FOL and FORL.
In our ISSR analysis, 26 Fo isolates were distributed in seven UPGMA groups, but this clustering did not correspond to any of the field sampling locations, ff. spp. or the level of symptoms of disease in tomato. For instance, as indicated above, FOL races 2 and 3 and FORL occurred in the same location. Some studies have reported correlation between the groupings of Fo isolates and geographical origins at various levels, while others have not. Baysal et al. (2009) reported an association between UPGMA groupings and climatic conditions for FOL race 3 isolates coming from Turkey. However, occurrences of more than one race of the Fo f. sp. ciceris in the same place of India have also been reported (Dubey and Shio, 2008).
Fo diseases have become a major threat to tomato production in Algeria because resistant hosts are not always available or used, the chemical treatments usually fail to control FOL and FORL in the field and, as indicated above, farmers themselves are contributing to the spread of these pathogens (Baysal et al., 2009). In addition, it has been proposed that “new pathogenic forms of FORL could have evolved from non-pathogenic local populations in Algeria” (Edel-Hermann et al., 2012). Previous studies have demonstrated the ability of biocontrol strains of Trichoderma against Fusarium wilt in different crops, such as tomato (Cotxarrera et al., 2002; Taghdi et al., 2015) and melon (Martínez-Medina et al., 2014). Recently, it has been reported that two T. harzianum isolates collected in Western Algeria displayed biocontrol activity against the tomato crown and root rot disease caused by Algerian FORL isolates (Kerroum et al., 2015). It is well established that biocontrol agents isolated from a given crop would have better adaptation to that crop, providing better control than those isolated from other plants (Cook, 1993). Considering the high genetic variability of the FORL and FOL isolates sampled in the different Algerian regions, we performed a survey in nursery tomato plants and tomato field soils with low incidence of Fusarium diseases to collect native Trichoderma isolates following a similar approach to that used by Taghdi et al. (2015) in Morocco. The most frequently isolated species in our study was T. asperellum. This finding agrees with previous studies that described the presence of this species in tomato cultivation fields in other North African countries like Egypt (El Komy et al., 2015) and Morocco (Taghdi et al., 2015). T. ghanense, a member of the Trichoderma section Longibrachiatum, has a worldwide distribution (Druzhinina et al., 2012) and has scarcely been cited as being a biocontrol agent. However, in addition to having plant growth promoting abilities, some T. ghanense isolates have shown capacity to control Fusarium wilt in melon (Martínez-Medina et al., 2014) and the root disease caused by Pythium arrhenomanes in rice plants (Banaay et al., 2012). In our study, an isolate sampled from a tomato plant from the ITCMI nursery (Staouali, Algiers) was identified as T. ghanense.
It is clear that the screening procedures for the selection of biological control agents must include in vitro and in vivo tests because of the influence of environmental and edaphic factors in the mechanisms of action displayed by the antagonist. Based on the species diversity and abundance of the Trichoderma isolates found in Algeria, the isolates T5, T8, T9, and T17 were selected to carry out laboratory tests against two FORL and two FOL isolates in order to explore any antagonistic activity. Results show that the Trichoderma isolates T5, T8, T9, and T17 were effective against FORL and FOL via different mechanisms. All of them were able to overgrow FORL and FOL isolates and inhibit their growth. We have previously reported that T. asperellum isolates sampled in Morocco were not able to overgrow the colonies of FOL (Taghdi et al., 2015). Thus, the ability to do so, as observed in T9 and T17 against FOL and FORL, must not to be considered a common trait of T. asperellum but rather a particular feature of these two isolates. Antibiosis is a major biocontrol mechanism in Trichoderma that has been proven to be efficient against FORL and FOL, as indicated by the results obtained in membrane assays. The lowest inhibition values, obtained for most of the T5-Fo combinations in the two different membrane assays, are indicate that T. harzianum T5 has a lower antifungal activity against FORL and FOL isolates. However, higher Fo growth inhibition values in the dialysis membrane assay for eight out of the 16 Trichoderma-Fo combinations tested was detected. These results show that both FORL and FOL are sensitive to the small compounds secreted by Trichoderma. This is in agreement with previous studies showing that metabolites secreted by Trichoderma spp. are a major contributor to biocontrol of this fungus against different plant pathogenic fungi (Lorito et al., 2010; Taghdi et al., 2015). No differences were noted in the biocontrol potential of T8, T9, and T17 when representative FORL and FOL isolates were used as targets. Therefore, these three Trichoderma isolates were further tested to assess their efficiency suppressing crown and root rot and Fusarium wilt in tomato caused by isolates of FORL and FOL, respectively.
Greenhouse assays performed on susceptible “SM” plants with three Fo isolates, representing FORL and FOL and the two reference isolates, showed high ID values in Trichoderma untreated plants. The highest reduction in ID corresponded to T. ghanense T8 and T. asperellum T9, indicating these species could satisfactorily protect tomato against FORL and FOL. The differences in the biocontrol potential observed between T. asperellum T9 and T17 show how two isolates of the same species can display similar antagonistic activity against different Fo isolates in in vitro assays but show different biocontrol behavior in in planta tests.
In conclusion, the results of the present study have shown that FORL and races 2 and 3 of FOL are present in Algeria, and that they show high genetic diversity. Also, the results support the hypothesis that native species display better biocontrol, where two isolates of T. ghanense and T. asperellum can protect against crown and root rot and Fusarium wilt in tomato, in cases of severe disease. Therefore, it can be said that Trichoderma biocontrol is a suitable means for the integrated management of these two widespread tomato diseases when used in combination with resistant host plants.
Author contributions
AD and HB carried out the collection of plant and soil samples and fungal isolation. AD carried out PCR analysis and in vitro and greenhouse assays. RH prepared tables and figures. RH, EM, and AD wrote the manuscript. RH designed and led the study. All authors have read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research project was funded by the Spanish Ministry of Economy and Competitiveness (Project no. AGL2015-70671-C2). Ali Debbi received a residential fellowship 2015–2016 supported by the PNE Program funding from the Ministry of the Higher Education and the Scientific Research (MESRS), Algeria. The authors wish to thank Prof. J.M. Díaz-Mínguez (Genetics Group, CIALE, University of Salamanca) for providing the 4287 and Forlc isolates.
References
- Arie T. (2010). Phylogeny and phytopathogenicity mechanisms of soilborne Fusarium oxysporum. J. Gen. Plant Pathol. 76, 403–405. 10.1007/s10327-010-0264-z [DOI] [Google Scholar]
- Balmas V., Scherm B., Di Primo P., Rau D., Marcello A., Migheli Q. (2005). Molecular characterisation of vegetative compatibility groups in Fusarium oxysporum f. sp. radicis-lycopersici and f. sp. lycopersici by random amplification of polymorphic DNA and microsatellite-primed PCR. Euro. J. Plant Pathol. 111, 1–8. 10.1007/s10658-004-1602-9 [DOI] [Google Scholar]
- Banaay C. G. B., Cuevas V. C. C., Vera-Cruz C. M. (2012). Trichoderma ghanense promotes plant growth and controls disease caused by Pythium arrhenomanes in seedlings of aerobic rice variety apo. Philipp. Agric. Scientist 95, 175–184. [Google Scholar]
- Baysal Ö., Siragusa M., Gümrükcü E., Zengin S., Carimi F., Sajeva M., et al. (2010). Molecular characterization of Fusarium oxysporum f. melongenae virulence by ISSR and RAPD markers on eggplant. Biochem. Genet. 48, 524–537. 10.1007/s10528-010-9336-1 [DOI] [PubMed] [Google Scholar]
- Baysal Ö., Siragusa M., Ikten H., Polat I., Gümrükcü E., Yigit F., et al. (2009). Fusarium oxysporum f. sp. lycopersici races and their genetic discrimination by molecular markers in West Mediterranean region of Turkey. Physiol. Mol. Plant Pathol. 74, 68–75. 10.1016/j.pmpp.2009.09.008 [DOI] [Google Scholar]
- Boix-Ruíz A., Gálvez-Patón L., de Cara-García M., Palmero-Llamas D., Camacho-Ferre F., Tello-Marquina J. C. (2015). Comparison of analytical techniques used to identify tomato-pathogenic strains of Fusarium oxysporum. Phytoparasitica 43, 471–483. 10.1007/s12600-014-0444-z [DOI] [Google Scholar]
- Cai G., Gale L. R., Schneider R. W., Kistler H. C., Davis R. M., Elias K. S., et al. (2003). Origin of race 3 of Fusarium oxysporum f. sp. lycopersici at a single site in California. Phytopathology 93, 1014–1022. 10.1094/PHYTO.2003.93.8.1014 [DOI] [PubMed] [Google Scholar]
- Chaverri P., Castlebury L. A., Samuels G. J., Geiser D. M. (2003). Multilocus phylogenetic structure within the Trichoderma harzianum/Hypocrea lixii complex. Mol. Phylogenet. Evol. 27, 302–313. 10.1016/S1055-7903(02)00400-1 [DOI] [PubMed] [Google Scholar]
- Cook R. J. (1993). Making greater use of introduced microorganisms for biological control of plant pathogens. Ann. Rev. Phytopathol. 31, 53–80. 10.1146/annurev.py.31.090193.000413 [DOI] [PubMed] [Google Scholar]
- Çolak A., Biçici M. (2013). PCR detection of Fusarium oxysporum f. sp. radicis-lycopersici and races of F. oxysporum f. sp. lycopersici of tomato in protected tomato-growing areas of the eastern Mediterranean region of Turkey. Turk. J. Agric. For. 37, 457–467. 10.3906/tar-1203-71 [DOI] [Google Scholar]
- Cotxarrera L., Trillas-Gay M. I., Steinberg C., Alabouvette C. (2002). Use of sewage sludge compost and Trichoderma asperellum isolates to suppress Fusarium wilt of tomato. Soil Biol. Biochem. 34, 467–476. 10.1016/S0038-0717(01)00205-X [DOI] [Google Scholar]
- Di Pietro A., Roncero M. I. G. (1998). Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum. Mol. Plant Microbe Interact. 11, 91–98. 10.1094/MPMI.1998.11.2.91 [DOI] [PubMed] [Google Scholar]
- Druzhinina I. S., Komon-Zelazowska M., Ismaiel A., Jaklitsch W., Mullaw T., Samuels G. J., et al. (2012). Molecular phylogeny and species delimitation in the section Longibrachiatum of Trichoderma. Fungal Gen. Biol. 49, 358–368. 10.1016/j.fgb.2012.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey S. C., Shio R. S. (2008). Virulence analysis and oligonucleotide fingerprinting to detect genetic diversity among Indian isolates of Fusarium oxysporum f. sp. ciceris causing chickpea wilt. Mycopathologia 165, 389–406. 10.1007/s11046-008-9090-7 [DOI] [PubMed] [Google Scholar]
- Edel V., Steinberg C., Gautheron N., Alabouvette C. (2000). Ribosomal DNA-targeted oligonucleotide probe and PCR assay specific for Fusarium oxysporum. Mycol. Res. 104, 518–526. 10.1017/S0953756299001896 [DOI] [Google Scholar]
- Edel-Hermann V., Gautheron N., Steinberg C. (2012). Genetic diversity of Fusarium oxysporum and related species pathogenic on tomato in Algeria and other mediterranean countries. Plant Pathol. 61, 787–800. 10.1111/j.1365-3059.2011.02551.x [DOI] [Google Scholar]
- Elad Y., Chet I., Henis Y. (1981). A selective medium for improving quantitative isolation of Trichoderma spp. from soil. Phytoparasitica 9, 59–67. 10.1007/BF03158330 [DOI] [Google Scholar]
- El Komy M. H., Saleh A. A., Eranthodi A., Molan Y. Y. (2015). Characterization of novel Trichoderma asperellum isolates to select effective biocontrol agents against tomato Fusarium wilt. Plant Pathol. J. 31, 50–60. 10.5423/PPJ.OA.09.2014.0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin I. D., Aitken A. B., Smith L. W. (1997). Application of inter simple sequence repeat (ISSR) markers to plant genetics. Electrophoresis 18, 1524–1528. 10.1002/elps.1150180906 [DOI] [PubMed] [Google Scholar]
- Harman G. E., Howell C. R., Viterbo A., Chet I., Lorito M. (2004). Trichoderma species-opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56. 10.1038/nrmicro797 [DOI] [PubMed] [Google Scholar]
- Hermosa M. R., Keck E., Chamorro I., Rubio B., Sanz L., Vizcaíno J. A., et al. (2004). Genetic diversity shown in Trichoderma biocontrol isolates. Mycol. Res. 108, 897–906. 10.1017/S0953756204000358 [DOI] [PubMed] [Google Scholar]
- Hermosa R., Viterbo A., Chet I., Monte E. (2012). Plant-beneficial effects of Trichoderma and of its genes. Microbiology 158, 17–25. 10.1099/mic.0.052274-0 [DOI] [PubMed] [Google Scholar]
- Hibar K., Daami-Remadi M., Ayed F., El-Mahjoub M. (2007). Fusarium crown and root rot of tomato and its chemical control. Int. J. Agric. Res. 2, 687–695. 10.3923/ijar.2007.687.695 [DOI] [Google Scholar]
- Hibar K., Daami-Remadi M., Khiareddine H., El Mahjoub M. (2005). Effet inhibiteur in vitro et in vivo du Trichoderma harzianum sur Fusarium oxysporum f. sp. radicis-lycopersici. Biotechnol. Agron. Soc. Environ. 9, 163–171. [Google Scholar]
- Hirano Y., Arie T. (2006). PCR-based differentiation of Fusarium oxysporum f. sp. lycopersici and radicis- lycopersici and races of F. oxysporum f. sp. lycopersici. J. Gen. Plant Pathol. 72, 273–283. 10.1007/s10327-006-0287-7 [DOI] [Google Scholar]
- Houterman P. M., Cornelissen B. J., Rep M. (2009). Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog. 4:e1000061. 10.1371/journal.ppat.1000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinski N. A., Broz K., Jonkers W., Ma L. J., Kistler H. C. (2017). Effector gene suites in some soil isolates of Fusarium oxysporum are not sufficient predictors of vascular wilt in tomato. Phytopathology 107, 842–851. 10.1094/PHYTO-12-16-0437-R [DOI] [PubMed] [Google Scholar]
- Kawabe M., Kobayashi Y., Okada G., Yamaguchi I., Teraoka T., Arie T. (2005). Three evolutionary lineages of tomato wilt pathogen, Fusarium oxysporum f. sp. lycopersici, based on sequences of IGS, MAT1, and pg1, are each composed of isolates of a single mating type and a single or closely related vegetative compatibility group. J. Gen. Plant Pathol. 71, 263–272. 10.1007/s10327-005-0203-6 [DOI] [Google Scholar]
- Kerroum F., Noureddine K., Eddine H. J., Mebrouk K. (2015). Biological control of Fusarium crown and root rot disease of tomato by Trichoderma harzianum in the west of Algeria. Int. J. Sci. Nat. 6, 141–146. [Google Scholar]
- Kullnig-Gradinger C. M., Szakacs G., Kubicek C. P. (2002). Phylogenetic and evolution of the genus Trichoderma: a multigene approach. Mycol. Res. 106, 757–767. 10.1017/S0953756202006172 [DOI] [Google Scholar]
- Lee S. B., Milgroom M. G., Taylor J. W. (1988). A rapid, high yield mini-prep method for isolation of total genomic DNA from fungi. Fungal Genet. Newsl. 35, 23–24. 10.4148/1941-4765.1531 [DOI] [Google Scholar]
- Lievens B., Houterman P. M., Rep M. (2009). Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp lycopersici races and discrimination from other formae speciales. FEMS Microbiol. Lett. 300, 201–215. 10.1111/j.1574-6968.2009.01783.x [DOI] [PubMed] [Google Scholar]
- Lorito M., Woo S. L., Harman G. E., Monte E. (2010). Translational research on Trichoderma: from 'omics to the field. Annu. Rev. Phytopathol. 48, 395–417. 10.1146/annurev-phyto-073009-114314 [DOI] [PubMed] [Google Scholar]
- Ma L. J. (2014). Horizontal chromosome transfer and rational strategies to manage Fusarium vascular wilt diseases. Mol. Plant Pathol. 15, 763–766. 10.1111/mpp.12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Medina A., Del Mar Alguacil A., Pascual J. A., Van Wees S. C. (2014). Phytohormone profiles induced by Trichoderma isolates correspond with their biocontrol and plant growth-promoting activity on melon plants. J. Chem. Ecol. 40, 804–815. 10.1007/s10886-014-0478-1 [DOI] [PubMed] [Google Scholar]
- Medeiros H. A., Araújo Filho J. V., Grassi de Freitas L., Castillo P., Rubio M. B., Hermosa R., et al. (2017). Tomato progeny inherit resistance to the nematode Meloidogynejavanica linked to plant growth induced by the biocontrol fungus Trichoderma atroviride. Sci. Rep. 7:40216. 10.1038/srep40216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mes J. J., Weststeijn E. A., Herlaar F., Lambalk J. J., Wijbrandi J., Haring M. A., et al. (1999). Biological and molecular characterization of Fusarium oxysporum f. sp. lycopersici divides race 1 isolates into separate virulence groups. Phytopathology 89, 156–160. 10.1094/PHYTO.1999.89.2.156 [DOI] [PubMed] [Google Scholar]
- Michielse C. B., Rep M. (2009). Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 10, 311–324. 10.1111/j.1364-3703.2009.00538.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz P., Antón A., Nu-ez M., Paranjpe A., Ari-o J., Castells X., et al. (2008). Comparing the environmental impacts of greenhouse versus open-field tomato production in the Mediterranean region. Acta Hortic. 801, 1591–1596. 10.17660/ActaHortic.2008.801.197 [DOI] [Google Scholar]
- Nirmaladevi D., Venkataramana M., Srivastava R. K., Uppalapati S. R., Gupta V. K., Yli-Mattila T., et al. (2016). Molecular phylogeny, pathogenicity and toxigenicity of Fusarium oxysporum f. sp. lycopersici. Sci. Rep. 6:21367. 10.1038/srep21367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell K., Kistler H. C., Cigelnik E., Ploetz R. C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. U.S.A. 95, 2044–2049. 10.1073/pnas.95.5.2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen P. G., Pei M., Karp A., Royle D. J., Edwards K. J. (1998). Isolation and characterization of microsatellite loci in the wheat pathogen Mycosphaerella graminicola. Mol. Ecol. 7, 1611–1612. [DOI] [PubMed] [Google Scholar]
- Pérez E., Rubio M. B., Cardoza R. E., Gutiérrez S., Monte E., Bettiol W., et al. (2015). The importance of chorismate mutase in the biocontrol potential of Trichoderma parareesei. Front. Microbiol. 6:1181. 10.3389/fmicb.2015.01181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta-Puglia A., Mifsud D. (2005). First record of Fusarium oxysporum f. sp. radicis-lycopersici in Malta. J. Plant Pathol. 87, 150. [Google Scholar]
- Rep M., van der Does H. C., Meijer M., van Wijk R., Houterman P. M., Dekker H. L., et al. (2004). A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 53, 1373–1383. 10.1111/j.1365-2958.2004.04177.x [DOI] [PubMed] [Google Scholar]
- Rubio M. B., Hermosa M. R., Keck E., Monte E. (2005). Specific PCR assays for the detection and quantification of DNA from the biocontrol strain Trichoderma harzianum 2413 in soil. Microb. Ecol. 49, 25–33. 10.1007/s00248-003-0171-3 [DOI] [PubMed] [Google Scholar]
- Rubio M. B., Hermosa R., Reino J. L., Collado I. G., Monte E. (2009). Thctf1 transcription factor of Trichoderma harzianum is involved in 6-pentyl-2H-pyran-2-one production and antifungal activity. Fungal Genet. Biol. 46, 17–27. 10.1016/j.fgb.2008.10.008 [DOI] [PubMed] [Google Scholar]
- Rubio M. B., Hermosa R., Vicente R., Gómez-Acosta F. A., Morcuende R., Monte E., et al. (2017a). The combination of Trichoderma harzianum and chemical fertilization leads to the deregulation of phytohormone networking, preventing the adaptative responses of tomato plants to salt stress. Front. Plant Sci. 8:294 10.3389/fpls.2017.00294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio M. B., Pardal A. J., Cardoza R. E., Gutiérrez S., Monte E., Hermosa R. (2017b). Involvement of the transcriptional coactivator ThMBF1 in the biocontrol activity of Trichoderma harzianum. Front. Microbiol. 8:2273. 10.3389/fmicb.2017.02273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruocco M., Lanzuise S., Lombardi N., Woo S. L., Vinale F., Marra R., et al. (2015). Multiple roles and effects of a novel Trichoderma hydrophobin. Mol. Plant Microbe Interact. 28, 167–179. 10.1094/MPMI-07-14-0194-R [DOI] [PubMed] [Google Scholar]
- Sadfi-Zouaoui N., Hannachi I., Rouaissi M., Hajlaoui M. R., Rubio M. B., Monte E., et al. (2009). Biodiversity of Trichoderma strains in Tunisia. Can. J. Microbiol. 55, 154–162. 10.1139/W08-101 [DOI] [PubMed] [Google Scholar]
- Sheu Z. M., Wang T. C. (2006). First report of race 2 of Fusarium oxysporum f sp. lycopersici, the causal agent of Fusarium wilt on tomato in Taiwan. Plant Dis. 90:111 10.1094/PD-90-0111C [DOI] [PubMed] [Google Scholar]
- Song W., Zhou L., Yang C., Cao X., Zhang L., Liu X. (2004). Tomato Fusarium wilt and its chemical control strategies in a hydroponic system. Crop Protect. 23, 243–247. 10.1016/j.cropro.2003.08.007 [DOI] [Google Scholar]
- Taghdi Y., Hermosa R., Domínguez S., Rubio M. B., Essalmani H., Nicolas C., et al. (2015). Effectiveness of composts and Trichoderma strains for control of Fusarium wilt of tomato. Phytopathol. Mediterr. 54, 232–240. 10.14601/Phytopathol_Mediterr-15226 [DOI] [Google Scholar]
- van der Does H. C., Lievens B., Claes L., Houterman P. M., Cornelissen B. J., Rep M. (2008). The presence of a virulence locus discriminates Fusarium oxysporum isolates causing tomato wilt from other isolates. Environ. Microbiol. 10, 1475–1485. 10.1111/j.1462-2920.2007.01561.x [DOI] [PubMed] [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, in PCR Protocols: A Guide to Methods and Applications, eds Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (San Diego, CA: Academic Press; ), 315–322. [Google Scholar]