Abstract
Cellular senescence is triggered by various distinct stresses and characterized by a permanent cell cycle arrest. Senescent cells secrete a variety of inflammatory factors, collectively referred to as the senescence-associated secretory phenotype (SASP). The mechanism(s) underlying the regulation of the SASP remains incompletely understood. Here we define a role for innate DNA sensing in the regulation of senescence and the SASP. We find that cyclic GMP-AMP synthase (cGAS) recognizes cytosolic chromatin fragments (CCFs) in senescent cells. The activation of cGAS, in turn triggers the production of SASP factors via Stimulator of interferon genes (STING), thereby promoting paracrine senescence. We demonstrate that diverse stimuli of cellular senescence engage the cGAS-STING pathway in vitro and we show cGAS-dependent regulation of senescence upon irradiation and oncogene activation in vivo. Our findings provide insights into the mechanisms underlying cellular senescence by establishing the cGAS-STING pathway as a crucial regulator of senescence and the SASP.
Introduction
Senescence is a cellular program characterized by a permanent growth arrest of damaged or aged cells 1 2. Cells undergoing senescence display profound phenotypic changes, which are driven by alterations in metabolism, chromatin organization and transcriptional activity. A prominent hallmark of senescent cells is the secretion of inflammatory mediators, including various cytokines, chemokines, extracellular matrix proteins and growth factors, collectively referred to as the senescence-associated secretory phenotype (SASP)3 4. Through the secretion of the SASP, senescent cells critically influence many biological processes, including wound healing5, tissue repair6, tumorigenesis7 or in vivo reprogramming8. Moreover, the inflammatory response linked to the SASP is considered to underlie many senescence-associated effects on ageing and age-related disorders 9, 10 11. Thus, understanding the regulation of the SASP is essential, both for deciphering the basis of senescent cell communication but also for discovering new targets controlling senescence effector responses. On a molecular level, several transcription factors have been implicated in the up-regulation of SASP-genes including NF-κB, C/EBP-β, p38 MAPK and GATA4 12, 13, 14, 15, 16. However, the upstream signalling pathway(s) that activate these transcriptional regulators within senescent cells remain incompletely characterized.
Inflammation is generally initiated through the activation of innate immune receptors, most mentionable various pattern recognition receptors (PRRs) 17. Despite being best known for the recognition of microbial products in the context of infection, some PRRs can also become activated by aberrant self-molecules. cGAS, which senses double-stranded DNA, represents a well-studied example of a PRR that responds equally to both microbial as well as endogenous DNA ligands 18. Upon activation, cGAS synthesizes the second messenger molecule 2`3`cyclic GMP-AMP (cGAMP), which engages STING, thereby leading to the production of inflammatory cytokines, chemokines and type I interferons (IFNs) 19 20 21 22, 23 24 25. Although the localization within the cytosol usually prevents an unintended activation of the cGAS-STING signalling pathway, conditions wherein self-DNA gains access to the cytosol can trigger – occasionally detrimental - inflammatory responses. Genetic defects that compromise endogenous DNA metabolism or instances that trigger massive, synchronized accumulation of dead cells represent examples of this phenomenon 26 27 28, 29 30. However, whether self-DNA sensing by cGAS contributes to biological processes in a more “physiological” manner remains unclear.
Here we report a function of innate DNA sensing through cGAS in the regulation of cellular senescence. Specifically, we identified that senescent cells engage the cGAS-STING pathway, thereby regulating the SASP and facilitating paracrine senescence. We demonstrate that the activation of cGAS bases on its recognition of aberrant cytosolic chromatin fragments (CCFs), which arise in senescent cells as a consequence of nuclear lamin B1 degradation 31. We found that diverse triggers of cellular senescence, including oxidative stress, oncogene signalling, irradiation and pro-senescent drugs depend upon cGAS-STING signalling to drive the production of inflammatory SASP components. Finally, we observed that cGAS-triggered senescence occurs upon irradiation and oncogene activation in vivo. Together, these findings establish endogenous DNA sensing through the cGAS-STING pathway as an important regulator of senescence and the SASP.
Deficiency of the cGAS-STING pathway impairs oxidative-stress induced senescence
While studying the function of cGAS, we repeatedly noticed that upon serial passaging under standard culture conditions mouse embryonic fibroblasts (MEFs) deficient for cGAS (cGAS KO MEFs) exhibited an accelerated proliferation rate compared to wild-type (WT) MEFs. To explore this phenomenon in further detail, we cultured MEFs from an early passage on according to the 3T3 protocol and counted cells over several weeks32. Comparing cell numbers obtained from the first few passages, we did not observe any difference in the proliferation rate of WT MEFs and cGAS KO MEFs (Fig. 1a). However, after serial passages WT MEFs proliferated less than cGAS KO MEFs (Fig. 1a). Quantifying the proliferation rate by 5-bromo-2`-deoxyuridine (BrdU) incorporation confirmed the increased replicative activity of cGAS KO MEFs compared to WT MEFs (Fig. 1b). In agreement with a senescent phenotype, WT cells became flat and enlarged and displayed high activity of senescence-associated β-galactosidase (SA-β-Gal) (Fig. 1c). In contrast, cGAS KO MEFs did not acquire these morphological changes and most of them did not become positive for SA-β-Gal (Fig. 1c). To systematically explore the altered phenotype of WT MEFs and cGAS KO MEFs, we performed gene expression profiling with RNA sequencing from cells cultured for 30 – 42 days. Notably, among the transcripts that showed most increased expression in WT MEFs we found Cdkn2a (p16Ink4a) and Cdkn2b (p15), crucial regulators of the senescent cell cycle arrest in eukaryotic cells (Fig. 1d)1, 33, 34. Moreover, several genes encoding proteins constituting the senescence-associated secretory phenotype (SASP) were overexpressed in WT MEFs relative to cGAS KO MEFs (Igfbp6, Ccl7, Vcam1, Csf1) (Fig. 1d)3. Conversely, genes with decreased expression in WT MEFs were enriched for transcripts related to mitosis and many of those are known targets of retinoblastoma (RB)-dependent E2F transcription factors (Mcm5, Top2a, Melk, Bub1b, Ccna2, Smc4, Bub1) (Supplementary Fig. 1a)35. Direct measurement of the expression of p16Ink4a and other senescence markers confirmed the results obtained by RNA sequencing (Fig. 1e, f). Similar to cGAS, deficiency of STING resulted in an accelerated proliferation rate and compromised the induction of markers related to senescence (Supplementary Fig. 1b, c, d). Together, these data suggest that the cGAS-STING signalling pathway acts as a positive regulator of senescence in MEFs.
Figure 1. Absence of cGAS attenuates the senescence response.
(a) Proliferation curve of primary wild-type (WT) and cGAS knockout (cGAS KO) mouse embryonic fibroblasts (MEFs) cultured under 20% O2. (b) Percentages of BrdU positive WT or cGAS KO MEFs after 27 days in culture are shown. (c) Left: Images of SA-β-Gal stained WT or cGAS KO MEFs at 27 days in culture. Scale bar: 200 µm. Right: Mean and s.d. of the percentages of SA-β-Gal-positive cells are indicated. (d) Heat maps of RNA-seq analysis of WT MEFs and cGAS KO MEFs (For each genotype one sample per day; MEFs collected at Day 33, 36, 42). Genes in WT MEFs exhibiting statistically significant (n=3 biological replicates; student t-test; P < 0.05), twofold or greater increases relative to cGAS KO MEFs are shown. Senescence markers are highlighted in green. (e, f) WT MEFs or cGAS KO MEFs were harvested after 3 weeks of culture and expression of depicted genes was measured via RT-qPCR or protein expression was analysed by immunoblotting. One representative experiment out of three (a, c (left)) or mean and s.d. of n=3 independent experiments (b, c (right)) or n=3 independent biological replicates (e) are shown. P values were calculated by unpaired t-test (* P < 0.05, *** P < 0.001, **** P < 0.0001). Unprocessed original blots are shown in Supplementary Fig. 7. Source data are available in Supplementary Table 1.
Oxidative stress has been implicated as a cause of premature senescence of MEFs in vitro36. The above findings led us to investigate whether cGAS contributes to oxidative stress-induced senescence. To accelerate the induction of oxidative stress-induced senescence, we incubated MEFs under 40% O2 and monitored senescence markers after 7 days. Consistent with the pro-senescent activity of oxidative stress, we found that WT MEFs rapidly entered senescence as indicated by increased activity of SA-β-Gal, elevated expression of p16Ink4a and decreased proliferative activity (Fig. 2a, b, c, d). Conversely, cGAS KO MEFs and STING KO MEFs exhibited a compromised senescence phenotype (Fig. 2a, b, c, d). To determine whether cGAS also influences oxidative stress-induced senescence in human cells, we depleted cGAS in WI-38 cells and examined markers of senescence (Fig. 2e). Indeed, upon oxidative stress exposure, knockdown of cGAS prevented SA-β-Gal activity and p16 Ink4a expression compared to cells treated with a non-targeting control siRNA (Fig. 2f, g, h). Together, these findings suggest that cGAS and STING regulate oxidative-stress induced senescence.
Figure 2. cGAS facilitates oxidative stress-induced senescence.
(a, b, c, d) WT MEFs, cGAS KO MEFs or STING KO MEFs were cultured under hyperoxic conditions (40% O2) for 7 days and subsequently stained for SA-β-Gal activity (a), examined for percentages of EdU+ cells by immunofluorescence (b) or assessed for expression of Cdkn2a by RT-qPCR (c) or p16Ink4a by immunoblotting (d). (e) WI-38 cells were exposed to 40% O2 treatment for 9 days. On day 7 cells were transfected with a non-targeting control siRNA (si Control) or with siRNAs against cGAS (si cGAS #1, si cGAS #2). Expression of cGAS was assessed by immunoblotting on day 9. (f, g, h) WI-38 cells were treated as in (e). On day 9 SA-β-Gal activity was assessed by FACS (f) and expression of CDKN2A was examined by RT-qPCR (g) or p16Ink4a was assessed by immunoblotting (h). Numbers in (f) indicate percentages of SA-β-Gal positive cells. Mean and s.d. of n=5 independent experiments (a) or n=3 biological replicates (b, c, g) or one representative experiment out of 3 (f) or 2 (d, e, h) independent experiments are shown. P values were calculated by one-way (a) and two-way (b, c, g) ANOVA (* P < 0.05, ** P < 0.01, ns = not significant). Source data are available in Supplementary Table 1. Unprocessed original blots are shown in Supplementary Fig. 7.
The cGAS-STING pathway promotes senescence in a paracrine manner
We next sought to determine how cGAS controls senescence. In the context of innate immunity the central function of the cGAS-STING pathway is the secretion of antiviral and inflammatory cytokines and chemokines. We therefore suspected that cGAS and STING might promote senescence indirectly through regulating the SASP 37 38. Indeed, we found that conditioned medium (CM) from WT MEFs exposed to oxidative stress promoted a senescence response (as indicated by decreased BrdU incorporation and increased SA- β-Gal activity) within both WT MEFs as well as cGAS KO MEFs (Fig. 3a, b and Supplementary Fig. 2a, b). In contrast, CM from cGAS KO MEFs cultured under the same conditions failed to arrest MEFs (Fig. 3a, b and Supplementary Fig. 2a, b). To better characterize the SASP components, which are controlled by cGAS, we analysed the CM from both WT MEFs and cGAS KO MEFs using a cytokine array. Remarkably, the secretion of several distinct SASP factors including IL-6, TNF-α and several chemokines depended on cGAS (Fig. 3c). Moreover, the production of Cxcl-10, a well-known cGAS-dependent interferon-stimulated gene (ISG), was reduced in cGAS KO MEFs relative to WT MEFs (Fig. 3c). Similarly, depletion of cGAS in WI-38 cells decreased the secretion of SASP components, most prominently IL-6 (Supplementary Fig. 2c). We confirmed that the cGAS-STING signalling pathway is required for the expression of SASP factors (IL-6) and for the induction of ISGs (Ifi44) from both MEFs as well as WI-38 cells undergoing oxidative stress-induced senescence (Fig. 3d, e and Supplementary Fig. 2d, e).
Figure 3. cGAS regulates the senescence-associated secretory phenotype.
(a, b) Conditioned medium (CM) was collected from WT MEFs or cGAS KO MEFs exposed to 40% O2 for 7 days. Percentages of proliferating cells (WT MEFs) were assessed by BrdU incorporation assay (a) or induction of SA-β-Gal activity was determined by microscopy (b). (c) Cytokine profile of the CM from WT MEFs of cGAS KO MEFs exposed to 40% O2 for 7 days. Some SASP factors are highlighted in red. (d) WT MEFs, cGAS KO MEFs or STING KO MEFs were incubated in 40% O2 for 7 days and IL-6 was quantified by ELISA. (e) WI-38 cells were exposed to 40% O2 treatment for 9 days. On day 7 cells were transfected with a non-targeting control siRNA (si Control) or with siRNAs against cGAS (si cGAS #1, si cGAS #2). Expression of IL-6 was determined by RT-qPCR. (f, g) WT MEFs and cGAS KO MEFs were cultured in the presence of recombinant IFN-β as indicated for 14 days. Cells were stained for SA-β-Gal (f) or cell cycle activity was assessed by EdU incorporation (g). (h, i, j) WT MEFs of IFNAR KO MEFs were cultured under 40% O2 for 7 days and stained for SA-β-Gal (h), proliferative activity (BrdU incorporation) was assessed by FACS at day 4 (i) or expression of p16Ink4a was determined by immunoblotting after 7 days (j). Mean and SEM of n=3 (a) or mean and s.d. of n=4 independent experiments are shown (h) or one representative out of 2 independent experiments is shown (c, j). Data shown in (b, d, e, f, g, i) are from n=2 independent experiments with the column representing the mean. P value was calculated by unpaired t-test (h) (ns = not significant). Source data are available in Supplementary Table 1. Unprocessed original blots are shown in Supplementary Fig. 7.
The induced expression of ISGs along with the reported role of type I IFNs in senescence, prompted us to examine whether cGAS-mediated secretion of type I IFNs is involved in promoting oxidative stress-induced senescence 39, 40, 41, 42. We found that addition of recombinant IFN-β to cultures of WT MEFs and cGAS KO MEFs was sufficient to induce SA-β-Gal activity and p16Ink4a expression and to promote cell cycle arrest (Fig. 3f, g and Supplementary Fig. 2f, g). Conversely, MEFs deficient for the type I IFN receptor (IFNAR KO MEFs), displayed a compromised oxidative stress-induced senescence response (SA-β-Gal activity, BrdU incorporation, p16Ink4a expression, IL-6 production, ISG induction) compared to WT MEFs (Fig. 3h, i, j and Supplementary Fig. 2h). Collectively, these data suggest a possible role for type I IFNs in relaying cGAS-mediated senescence. However, additional cGAS-dependent SASP factors or activities most likely further enhance the senescent phenotype.
CCFs are recognized by cGAS in senescent cells
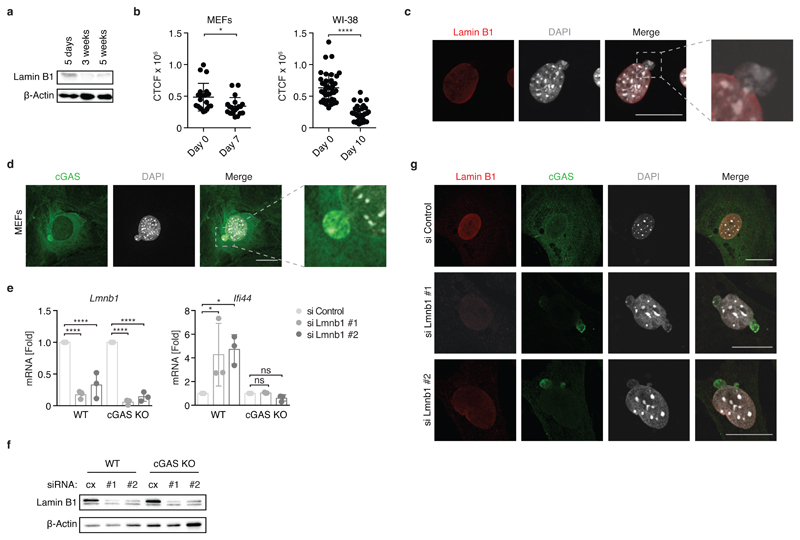
We next sought to characterize the DNA source responsible for the activation of cGAS in senescent cells. Degradation of the nuclear envelope component lamin B1 and the occurrence of cytosolic chromatin fragments (CCFs) are well-described features of senescent cells 43 31. In accordance with this, we found decreased expression of lamin B1 in senescent cells (Fig. 4a, b). Furthermore, using confocal fluorescence microscopy, we observed DNA-containing blebs budding of the nucleus as well as characteristic CCFs in cells undergoing oxidative-stress induced senescence (Fig. 4c). We thus hypothesized that the herniation of nuclear DNA into the cytosol might represent the trigger for provoking cGAS-STING signalling in senescent cells. In order to investigate this, we reconstituted cGAS KO MEFs with a doxycycline (Dox)-inducible FLAG-tagged cGAS construct (MEFs Tet-cGAS-FLAG). Remarkably, in cells undergoing oxidative stress-induced senescence, cGAS formed clusters that co-stained with DAPI-positive chromatin herniations (Fig. 4d). Likewise, endogenous cGAS co-localized with chromatin fragments at the nuclear-cytosolic border in senescent WI-38 cells (Supplementary Fig. 3). To assess whether the loss of nuclear integrity might itself be sufficient to activate cGAS-STING signalling, we knocked down lamin B1 with two distinct siRNAs in primary MEFs (Fig. 4e, f). Down-regulation of lamin B1 induced leakage of chromatin into the cytosol (Fig. 4g). More important, MEFs with reduced expression of lamin B1 displayed an elevated ISG response (Ifi44 induction), which was dependent on cGAS (Fig. 4e). Thus, it appears that the mere de-stabilization of the nuclear membrane is sufficient to engage cGAS. Consistent with this idea, we observed that cGAS enriched on chromatin containing blebs in cells knocked down for lamin B1 (Fig. 4g). Together, these results suggest that activation of cGAS in senescent cells is caused by its interaction with endogenous DNA arising from defective, senescent nuclei.
Figure 4. cGAS interacts with chromatin fragments in senescent cells.
(a) Lysates from WT MEFs and senescent MEFs were probed for the expression of lamin B1 by immunoblot. (b) MEFs (left panel) or WI-38 cells (right panel) were incubated at 40% O2 as indicated and expression of lamin B1 in individual cells was determined by confocal fluorescent microscopy (CTCF = Corrected Total Cell Fluorescence). (c) Confocal fluorescent microscopy images of WT MEFs cultured for 7 days under 40% O2 stained for lamin B1 (red) and DAPI (grey). (d) cGAS KO MEFs carrying a Dox-inducible vector expressing FLAG-tagged cGAS were cultured in the presence of Dox overnight. Cells were stained with DAPI (grey) or anti-FLAG antibody (green) and imaged by confocal microscopy. (e, f) MEFs were transfected with a non-targeting siRNA (si Control) or Lmnb1-targeting siRNAs (si Lmnb1 #1, si Lmnb1 #2). After 72 h mRNA levels of indicated genes were analysed by RT-qPCR (e) or protein expression was determined by immunoblotting after 48 h. (g) MEFs carrying a Dox-inducible vector expressing FLAG-tagged cGAS were transfected with siRNAs as indicated. 24 h later cells were treated with Dox overnight and stained for lamin B1 (red), cGAS-FLAG (green) and DAPI (grey). One representative experiment out of 3 (f, g) or 2 (a, b, c, d) is shown. Mean and s.d. of n=20 individual MEFs at Day 0; n=20 individual MEFs at Day 7; n=42 individual WI-38 cells at Day 0; n=33 individual WI-38 cells at Day 7 are shown in (b). Mean and s.d. of n=3 independent experiments are shown in (e). P values were calculated by unpaired t-test (b) or one-way ANOVA (e) (* P < 0.05, **** P < 0.0001, ns = not significant). Scale bar: 20 µm. Source data are available in Supplementary Table 1. Unprocessed original blots are shown in Supplementary Fig. 7.
Diverse triggers of cellular senescence engage the cGAS-STING pathway
Numerous stressors are able to induce senescence by signalling through various distinct pathways1, 2, 33. We therefore sought to determine whether the activation of cGAS is a common feature of senescent cells. To this end, we exposed WT MEFs, cGAS KO MEFs and STING KO MEFs to distinct senescence inducing stimuli and compared the expression of SASP components (IL-6, Cxcl10). We tested ionizing radiation (12 Gy) and palbociclib (PD-0332991), a CDK4 inhibitor, which mimics the effect of p16Ink4a. Of note, the production of IL-6 or the induction of Cxcl10 was largely abolished in cGAS KO MEFs or STING KO MEFs (Fig. 5a). To complement the findings with human cells, we explored the impact of cGAS on the induction of IL-6 and Cxcl10 in WI-38 cells undergoing irradiation-induced (12 Gy) or oncogene-induced (HRasV12) senescence. For both triggers and similar to MEFs, the up-regulation of IL-6 and Cxcl10 from WI-38 cells was dependent on cGAS (Fig. 5b). This shows that the cGAS signalling pathway contributes to the production of cytokines from senescent cells, whether induced by irradiation, by oncogene activation or pharmacologically. Despite severely affecting the production of SASP components, cGAS and STING only partially influenced the induction of SA-β-Gal activity in MEFs undergoing irradiation- or palbociclib-dependent senescence, suggesting differences in the extent to which MEFs rely on paracrine senescence, at least in these in vitro contexts (Supplementary Fig. 4a). In contrast, depletion of cGAS in WI-38 cells impaired the propagation of senescence following irradiation or HRasV12 signalling, as revealed by diminished SA-β-Gal activity and impaired p16Ink4a induction in cells knocked down for cGAS (Supplementary Fig. 4b, c). Using fluorescence microscopy, we determined that all treatments from above triggered the degradation of lamin B1, suggesting that the loss of nuclear envelope integrity is a common substrate for the activation of cGAS in these contexts (Fig. 5c, d). To further substantiate the direct involvement of cGAS, we visualized the distribution patterns of nuclear DNA in relation to cGAS. Remarkably, both Dox-treated Tet-cGAS-FLAG MEFs and WI-38 cells displayed an overlay between herniated chromatin and cGAS after irradiation or following treatment with palbociclib or HRasV12 signalling (Fig. 5e and Supplementary Fig. 4d). Thus, a diverse range of distinct senescence inducers engages cGAS, which is required for the induction of SASP components.
Figure 5. Engagement of cGAS is a common feature of multiple senescence triggers.
(a) MEFs with the indicated genotypes were exposed to 12 Gy ionizing irradiation or stimulated with Palbociclib. Expression levels of Cxcl10 or protein levels of IL-6 were quantified by RT-qPCR or ELISA, respectively. (b) WI-38 cells (left panel) were exposed to 12 Gy ionizing irradiation or WI-38 ER:RAS cells (right panel) were treated with 4-OHT (500 nM) for 7 days. At day 7 (irradiated cells) or at day 3 (WI-38 ER:RAS cells) were transfected with a non-targeting control siRNA (si Control) or with siRNAs against cGAS (si cGAS #1, si cGAS #2). Expression of depicted genes was determined by RT-qPCR. (c) Expression of lamin B1 in individual WT MEFs exposed to irradiation (left panel) or treated with Palbociclib (right panel) for 7 days was determined by confocal fluorescence microscopy (CTCF = Corrected Total Cell Fluorescence). (d) Expression of lamin B1 in individual WI-38 cells exposed to irradiation (left panel) or WI-38 ER:RAS cells treated with 4-OHT was assessed by confocal fluorescence microscopy (CTCF = Corrected Total Cell Fluorescence). (e) cGAS KO MEFs carrying a Dox-inducible lentiviral vector containing a FLAG-tagged murine cGAS construct were irradiated (12 Gy) or treated with Palbociclib. 2 days post treatment cells were cultured in the presence of Dox overnight and then stained for FLAG (green) and nuclei (DAPI, grey). One representative experiment out of 2 experiments is shown (e) or mean and s.d. of n=51 individual control MEFs compared with n=48 individual irradiated MEFs (c, left); n=50 individual control MEFs compared with n=58 individual Palbociclib treated MEFs (c, right); n=177 individual control WI-38 cells compared with n=158 individual irradiated WI-38 cells (d, left); n=151 individual control WI-38 cells compared with n=87 individual 4-OHT treated WI-38 cells (d, right) are shown. Data shown in (a) and (b) are from n=2 independent experiments with the column representing the mean. P values were calculated by unpaired t-test (c, d) (**** P < 0.0001, ns = not significant). Scale bar: 20 µm. Source data are available in Supplementary Table 1.
cGAS regulates cellular senescence in vivo
Finally, we explored whether the cGAS-mediated regulation of senescence also occurs in vivo. To this end, we exposed mice to total body irradiation and examined the induced expression of IL-6, Cxcl10 and Cdkn2a in the lung44. Although, at 24 h post irradiation, the abundance of ISGs was less in irradiated cGAS KO mice or STING KO mice, Cdkn1a was induced to a similar extend in WT mice, cGAS KO mice or STING KO mice, indicating that cGAS and STING do not influence the in vivo DNA damage response (Supplementary Fig. 5a). As expected, at this early time point Cdkn2a levels were not detectable. However, 16 weeks after irradiation WT mice expressed Cdkn2a in the lung (Fig. 6a). Strikingly, the up-regulation of Cdkn2a was reduced in both cGAS KO and STING KO mice. We also noted decreased expression of IL-6 and Cxcl10 in cGAS KO and STING KO mice when compared to WT mice (Fig. 6a).
Figure 6. cGAS contributes to cellular senescence in vivo.
(a) Depicted mRNA levels of lungs from WT, cGAS KO and STING KO mice 16 weeks after irradiation are shown. (b) Representative liver stainings of Nras-, p21-, and SA-β-Gal 6 days after intrahepatic delivery of NrasG12V or NrasG12V/D38A into WT or cGAS KO mice are shown. Arrows indicate positive staining. (c, d) Quantification of Nras- or p21-positive cells (c) or expression of IL-6 protein levels or Cxcl10 mRNA levels (d) from (b) are shown. Mean of n=2 (WT mice untreated, cGAS KO mice) or mean and s.d of n=3 (WT mice irradiated, STING KO mice) (a), mean and s.d. of or n=4 (cGAS KO mice) or n=5 (WT mice) (c, d) or representative images (b) are shown. P values were calculated by two-way ANOVA (comparing genotypes or treatments) (* P < 0.05, ** P < 0.01, ns = not significant). Scale bar: 100 μm. Source data are available in Supplementary Table 1.
To corroborate the effect of cGAS on the senescence response in vivo, we employed a model of oncogene-induced senescence, wherein oncogenic NrasG12V is stably delivered into mouse livers by hydrodynamic injection of transposable elements 7. Both WT mice and cGAS KO mice were injected with transposons encoding NrasG12V and markers of senescence were analysed 6 days later. Delivery of transposable elements encoding for a signalling-defective NrasG12V mutant (NrasG12V/D38A) served as a control. Interestingly, despite similar expression of NrasG12V in both genotypes (as assessed by immunostaining of Nras), livers of cGAS KO mice showed decreased levels of p21 and less activity of SA-β-Gal compared to livers of WT mice (Fig. 6b, c). Moreover, expression of several SASP factors including IL-6 and Cxcl10 was reduced in cGAS KO mice relative to WT mice (Fig. 6d). As expected, owing to immune mediated clearance, WT mice showed a reduction of Nras-positive cells after 12 days (Supplementary Fig. 5b) 7. In contrast, no significant decrease of Nras-positive cells was observed in cGAS KO mice, thus suggesting that absence of cGAS affects senescence induction and the subsequent immune cell-mediated clearance of oncogene-activated cells. Taken together, these data indicate that cGAS is important for the regulation of senescence in vivo, whether induced through irradiation or oncogenic signalling.
Discussion
The secretion of the SASP is a key feature of senescent cells. Through the auto- and paracrine effects of the SASP, senescent cells on the one hand modulate their own cellular behaviour, while on the other hand they also influence their microenvironment. Thus, the SASP is considered as a pivotal effector branch of senescent cells in several physiological and pathological contexts. Here we provide evidence for a contribution of the innate DNA sensor cGAS and its downstream adaptor STING in the regulation of the SASP and show that signalling through this pathway acts to reinforce cellular senescence in a non-cell autonomous manner (Supplementary Fig. 6).
Previous work has demonstrated the down-regulation of lamin B1 and the subsequent partial loss of nuclear integrity with the occurrence of CCFs represents a characteristic feature of senescent cells 15, 31. Our data showing that these changes in the intracellular compartmentalization are connected - via cGAS/STING – to the induction of an inflammatory response, illustrate its functional consequences for cellular senescence. Interestingly, despite the immediate activation of multiple stress pathways, the emergence of the SASP is considered as a relatively late event in the development of senescence – usually requiring multiple days to arise 45. Our finding that stimulation of the cGAS/STING pathway occurs through the recognition of CCFs in cells, which are already on the “senescence track”, provides one possible explanation of how a senescence-specific inflammatory SASP-response may be initiated.
It is well established that the SASP functions as a strong amplifier of senescence. The findings presented here suggest that the cGAS-STING pathway regulates cellular senescence predominantly through the secretion of soluble factors and by involving auto- and paracrine signalling. First, unbiased examination of the cGAS-controlled secretome showed that many inflammatory cytokines and chemokines associated with promoting senescence depended on cGAS. Second, experiments using conditioned medium, best revealed that the presence of cGAS was crucial to render a senescent cell`s secretome “pro-senescent”. Conversely, loss of cGAS did not influence the responsiveness of recipient cells in committing to paracrine senescence. Consistent with a SASP-mediated regulation of senescence, we found that stimulation of cells with palbociclib, which directly engages the p16 pathway, triggered SA-β-Gal activity largely in a cGAS/STING-independent fashion, yet still depended on the latter genes for the production of inflammatory cytokines and chemokines.
We also explored specific factors that might mediate paracrine senescence. Consistent with previous reports, we found a contribution of type I IFNs in facilitating senescence 39, 40, 41 42. However, at least in vitro, type I IFNs - on their own - were less potent in triggering senescence compared to the more complex SASP present in the secretome of senescent cells. Thus, rather than a single factor, it is the joint action of several mediators that synergize to promote cGAS-dependent senescence.
We also found that loss of cGAS-STING-signalling compromised the senescence response in two distinct models of cellular senescence in vivo. Given that the decreased expression of senescence markers (SA-β-Gal or Cdkn2a) paralleled with reduced levels of inflammatory mediators, it is likely that the absence of cGAS/STING-dependent SASP factors accounts for the reduced in vivo senescence response.
In conclusion, our study reveals a key role of innate DNA sensing in controlling cellular senescence. Based on our work, we propose that beyond acting as a system to defend against infection, the cGAS-STING pathway may also serve as a cell-intrinsic surveillance system that protects an organism against the deleterious consequences of (pre-) neoplastic cells. Indeed, senescence is well described to function as a potent anticancer mechanism: First, by imposing a permanent cell cycle arrest, cellular senescence prevents the proliferation of damaged and potentially mutated cells. Second, by secreting cytokines and chemokines, senescent cells recruit immune cells, thereby facilitating their own elimination 7, 46. Given its role in the immune cell-mediated clearance of pre-malignant cells, as indicated by our study, we speculate that the cGAS-STING pathway may function to promote cell extrinsic antitumor effects.
In addition to these beneficial effects, however, cellular senescence is also implicated in ageing45 and various age-related pathological conditions, including as recently reported atherosclerosis47 or osteoarthritis48. Because of its prominent role in stimulating an inflammatory response, cGAS and STING may participate in these and other adverse effects of senescence in vivo, a point, which will need to be addressed by future studies.
Methods
Cell culture
MEFs were generated by standard methods from wild type, STING-deficient or cGAS-deficient mice. Detailed laboratory protocols are available upon request. All mice were on the C57Bl/6 background and obtained from both genders. Tmem173-/- (STING-deficient) mice were a gift from J Cambier 49. Mb21d1-/- (cGAS knockout first) mice were described in 50. Ifnar1-/- were a gift from C Reis e Sousa and were originally described in 51. This work was performed in accordance with the UK Animals (Scientific Procedures) Act 1986 and institutional guidelines for animal care. This work was approved by a project license granted by the UK Home Office (PPL No. 40/3583) and was also approved by the Institutional Animal Ethics Committee Review Board at the University of Oxford. Cells were cultured under 5% CO2 and 5% O2 at 37°C in Dulbecco’s modified Eagle medium (DMEM) (Life Technologies) containing 10% (v/v) fetal calf serum (FCS), 1% (v/v) penicillin (100 IU/ml)/streptomycin (100 μg/ml), 4.5 g/l D-glucose and 2 mM L-Glutamine. In some experiments MEFs were incubated at ambient O2 or 40% O2. Cells were repeatedly tested for mycoplasma using specific primers. WI-38 cells were purchased from ATCC CCL-75™ and cultured in DMEM supplemented with 10% (v/v) FCS and 1% (v/v) penicillin (100 IU/ml)/streptomycin (100 μg/ml). No method of cell line authentication was used.
Proliferation curves and SA-β-Gal assays
Populations of primary MEFs (2 x 105 per 10 cm dish) were counted by trypan blue exclusion every third day. SA-β-Gal staining was performed according to the manufactures instructions (Cell Signaling, SA-β-Gal staining kit (#9860)) and images were acquired with a stereomicroscope (Leica MZ 16 1FA). DAPI staining was used to identify nuclei.
For FACS-based determination of senescence, lysosomal alkalinization was induced by treating cells with 100 nM bafilomycin A1 for 1 h in fresh cell culture medium (2 ml per 35 mm dish) at 37 °C. 33 μl of 2 mM C12FDG (Thermofisher, ref. D2893) working solution was added to the cell culture medium to obtain a final concentration of 33 μM. After 1h of incubation, cells were washed, trypsinised and collected on a Gallios flow cytometer (Beckman Coulter) using Kaluza Acquisition software. Data were analysed with FlowJo software.
Generation of conditioned medium
The indicated cells (1×106) were seeded in a 10cm dish and incubated for 7 days in DMEM with 5% FCS in a 40% O2 incubator. After incubation, the conditioned medium (CM) was collected, centrifuged at 5,000g and filtered through a 0.2μm pore filter. CM was mixed with DMEM 40% FBS in a proportion of 3 to 1 to generate CM containing 10% FBS.
Antibodies
Detailed information on the antibodies used including their resources (company names, catalog numbers) and dilutions are provided in Supplementary Table 2.
Immunoblots
Cell pellets were lysed in a lysis buffer containing 0.5% Triton X-100, 20 mM HEPES (pH 7.4), 150 mM NaCl, 1.5 mM MgCl2, 2 mM EGTA, protease and phosphatase inhibitors (Protease Inhibitor Cocktail I, Animal-Free CALBIOCHEM). Protein concentration was measured using the BCA Pierce Protein assay kit and normalized to the lowest concentration. Samples were diluted with a loading buffer (100 mM Tris HCl (pH 6.8), 10% (v/v) SDS, 20% (v/v) glycerol, bromophenolblue and 4% β-mercaptoethanol). The primary antibody was incubated in 0.1% NFDM for 16 h at 4°C. The secondary antibodies anti-mouse or anti-rabbit HRP-conjugated antibodies were incubated for 1 h at room temperature. Proteins were visualized with the enhanced chemiluminescence substrate ECL (Pierce, ThermoScientific) and imaged using the ChemiDoc XRS Biorad Imager and Image Lab Software. Imaging was performed in two channels: Chemiluminescence and colorimetry, merged together in Supplementary Figure 7 for uncropped images.
Immunofluorescence
Cells were seeded onto coverslips at a density of 75’000 cells per coverslip and fixed with 2% (v/v) paraformaldehyde for 10 min, permeabilized for 5 min in 0.1% (v/v) Triton X-100 and blocked with 2% BSA in PBS for 20 min at room temperature. Cells were incubated with the primary antibody in PBS containing 2% (v/v) BSA for 1 h at room temperature in a humid chamber. After washing cells were incubated with the secondary antibody in PBS containing 5 μg/ml 4,6-diamino-2-phenylindole (DAPI) for 1 h at room temperature. Coverslips were mounted with Dako (fluorescent mounting medium). Images were acquired by a wide field fluorescence microscope (Zeiss Axioplan) and processed in ImageJ software. Confocal sections were obtained with a confocal laser scanning microscope (Zeiss LSM700).
Corrected total cell fluorescence
Ten images from lamin B1 stained cells per condition were acquired using a Zeiss LSM 700 confocal with a 63x lens. The corrected total cell fluorescence was determined using imageJ following the methods of 52. Briefly, the integrated density of lamin B1, area and mean grey value from nuclei were measured by imageJ. The corrected total cell fluorescence was calculated using the following formula: CTCF = Integrated Density – (Area of selected cell x Mean fluorescence of background readings).
BrdU incorporation assay
Cells plated in 10 cm dishes were incubated with 10 μM BrdU (Sigma-Aldrich) for 3h. Cells were fixed with 70% (v/v) cold ethanol overnight. After washing cells were resuspended in 2 M HCl for 30 min and then incubated in PBS-Tween 0.1% containing anti-BrdU antibodies conjugated to FITC (BD Biosciences, Clone B44) for 1 h. Samples were measured using a Gallios Flow Cytometer (Beckman Coulter). Data were analysed by the Kaluza software (Beckman Coulter).
Click-iT EdU incorporation assay
The Click-iT EdU reaction was performed following the instructions of the Click-iT imaging kit (molecular probes by Life Technologies). Ten images per condition were acquired with a 40x magnification objective. The number of EdU positive cells and DAPI positive cells was determined using imageJ.
Senescence induction
Palbociclib (PD-0332991 HCl, Selleckchem) was added to the culture medium at a final concentration of 4 μM. Cells (MEFs, WI-38 cells) were irradiated with 12 Gy (RS-2000 X-Ray Irradiator). To induce senescence via oxidative stress MEFs were incubated at 20% and 40% O2 for the indicated times. Recombinant IFN-β was obtained form Biolegend and used at the indicated concentrations. Cells were treated with IFN-β every day for two weeks.
WI-38 cells were transduced with a neomycin selectable retroviral vector containing pLNCX2 ER:RAS (addgene, Plasmid #67844) using polybrene at a concentration of 5 μg/ml. Transduced and neomycin selected WI-38 cells (WI-38 ER:RAS cells) were treated with 500 nM of 4’-hydroxy-tamoxifen (SIGMA) every day. Four days after the experimental start cells were treated with the indicated siRNAs for two days and then processed for analysis. WI-38 cells were incubated at 40% O2 or irradiated with 12 Gy, cultured for seven days and then transfected with the indicated siRNAs.
Lentiviral vector production and transduction
HEK 293T cells were transfected with pCMVDR8.74, pMD2.G plasmids and the puromycin selectable lentiviral vector pTRIPZ containing the open reading frame of cGAS-FLAG by the calcium phosphate precipitation method. The supernatant containing lentiviral particles was harvested at 48 and 72 h, pooled and concentrated by ultracentrifugation. WT MEFs were transduced with the lentiviral vectors by directly adding 10 μl to the culture medium.
Transfection
Cells were transfected with siRNAs using Lipofectamin 2000 reagent (Thermo Fisher Scientific) according to the manufacurer’s instruction. Silencer select predesigned siRNAs were purchased from Life Technologies. Sequence of mmLmnB1 siRNA Nr. 2: sense: 5’ CAUCAGUCAGUUACAAAUATT 3’; antisense: 5’ UAUUUGUAACUGACUGAUGTG 3’. mmLmnB1 siRNA Nr. 1: sense: 5’ GGAAGUUUAUUCGCUUGAAtt 3’; antisense: 5’ UUCAAGCGAAUAAACUUCCca 3’. Human MB21D1 (cGAS) siRNAs were purchased from ThermoFisher Scientific (Cat: AM16708, sicGAS 1 ID: 129125, sicGAS 2 ID: 129126).
Quantitative real-time PCR
Cells were lysed with the RLT buffer (Qiagen). RNA was extracted according to the manufacture’s protocol (QIAGEN RNeasy Mini kit) and treated with RNase-free DNase (Thermo Scientific). 500 ng of RNA were reverse transcribed (RevertAid, Thermo Scientific) and analysed by RT-qPCR in duplicates using the Maxima SYBR Green/ROX qPCR Master Mix (Life Technologies). The qPCR reactions were run on a 7900HT Fast qPCR instrument or a QuantStudio 5 Real-Time PCR system (Thermo Fisher Scientific). GAPDH was used as a housekeeping gene for normalization
Primer sequences for RT-qPCR:
mm Cdkn2a forward 5’ GATTCAGGTGATGATGATGGGC 3’;
mm Cdkn2a reverse 5’ TGCACCGTAGTTGAGCAGAAG 3’;
mm GAPDH forward 5’ GTCATCCCAGAGCTGAACG 3’;
mm GAPDH reverse 5’ TCATACTTGGCAGGTTTCTCC 3’;
mm p21 forward 5’ CTGGTGATGTCCGACCTGTT 3’;
mm p21 reverse 5’ TCAAAGTTCCACCGTTCTCG 3’;
mm IFI44 forward 5’ TGCACTCTTCTGAGCTGGTG 3’;
mm IFI44 reverse 5’ CCAGCTTGGACTTCACAGGA 3’;
mm Cdkn2b forward 5’ ACGGAGCAGAACCCAACTG 3’;
mm Cdkn2b reverse 5’ ATACCTCGCAATGTCACGGT 3’;
mm Ccl7 forward 5’ GAGGATCTCTGCCACGCT TC 3’;
mm Ccl7 reverse 5’ ACACCGACTACTGGTGATCC 3’;
mm Ccl2 forward 5’ CCTGCTGCTACTCATTCACCA 3’;
mm Ccl2 reverse 5’ ATTCCTTCTTGGGGTCAGCA 3’;
mm Igfbp7 forward 5’ GCGAGCAAGGGTCTCTGATA 3’;
mm Igfbp7 reverse 5’ AAGAACACCTTGGCACCAGT 3’
mm Igfbp6 forward 5’ GCAGAAGAATCCACGGACCT 3’;
mm Igfbp6 reverse 5’ GGAACGACACTGCTGCTTTC 3’;
mm Cxcl10 forward 5’ AAGTGCTGCCGTCATTTTCT 3’;
mm Cxcl10 reverse 5’ GTGGCAATGATCTCAACACG 3’;
hs GAPDH forward 5’ GAGTCAACGGATTTGGTCGT 3’;
hs GAPDH reverse 5’ GACAAGCTTCCCGTTCTCAG 3’;
hs CDKN2A forward 5’ TCCCTCAGACATCCCCGATTG 3’;
hs CDKN2A reverse 5’ AAATGAAAACTACGAAAGCGG 3’;
hs IL6 forward 5’ CCCCTGACCCAACCACAAAT 3’;
hs IL6 reverse 5’ ATTTGCCGAAGAGCCCTCAG 3’;
hs CXCL10 forward 5’ AGTGGCATTCAAGGAGTACC 3’;
hs CXCL10 rev 5’ TGATGGCCTTCGATTCTGGA 3’;
hs IFI44 forward 5’ GATGTGAGCCTGTGAGGTCC 3’;
hs IFI44 reverse 5’ CTTTACAGGGTCCAGCTCCC 3’;
ELISA
Supernatants were harvested and centrifuged at 1’000g to remove cell debris and dead cells. ELISA was performed following the instructions of the Mouse IL-6 ELISA Set from BD Biosciences (BD OpetEIA, Cat. No. 555240).
Cytokine array
Supernatants were harvested from indicated cells and centrifuged at 1’000g to remove dead cells. Cytokines from human cells were analysed following the instructions on Human Cytokine Antibody Array (abcam, ab133997). Cytokines from MEFs were analysed following the instructions of the ProteomeProfiler Array from R&D SYSTEMS. Dot blots were analysed using the plugin MicroArray Profile (OptiNav, Inc.) on Fiji software.
RNA sequencing
RNA was isolated from the WT, cGAS KO MEFs exposed to 20% O2 after 33, 36 and 42 days in culture using the RNeasy Mini kit (Qiagen). RNA was further processed for sequencing by the Gene Expression Core Facility GECF (EPFL, Switzerland). mRNA-seq libraries were prepared using the TruSeq mRNA stranded LT (Illumina kit). Samples were sequenced by the NextSeq 500 system sequencing with 1x75 cycle (“single read”), “high output” mode (=expected minimal 400mio reads), chemistry “v2”. Sequencing data were processed using HTSstation online software from the Bioinformatics and Biostatistics Core Facility (EPFL, Switzerland). Heat maps were produced from normalized expression data using Cluster 3.0 for computation and JTreeview for visualization.
In vivo total body irradiation and oncogene-induced senescence
WT, cGAS-/- or STING-/- mice (8-14 weeks of age) were mock-irradiated or exposed to IR (4.25 Gy) and lung tissue was obtained after 24 h or after 16 weeks. Mice from both gender were randomly allocated to different groups and experiments were conducted non-blinded. Mice were maintained under specific pathogen-free (SPF) conditions at Ecole Polytechnique Federale de Lausanne (EPFL), Switzerland. These animal experiments were approved by the Service de la consommation et des affaires vétérinaires (1066 Épalinges, Canton of Vaud, Switzerland). The transposon system encoding for oncogenic NrasG12V or an effector loop mutant incapable of downstream signalling (NrasG12V/D38A) were previously described 53. For intrahepatic delivery of the transposon system, mice were hydrodynamically injected with a 5:1 molar ratio of transposon to transposase-encoding plasmid (30 μg total DNA) via the tail vein as previously described. Livers were obtained on day 6 and on day 12 after injection. No statistical method was used to predetermine sample size. The investigators were not blinded to allocation during experiments and outcome assessment. Mice were maintained under specific pathogen-free (SPF) conditions at the University Hospital Tübingen. These animal experiments were approved by the local German authorities of the state of Baden-Wuerttemberg (Regierungspräsidium Tübingen).
Immunohistochemistry
Immunohistochemical stainings for Nras (clone F155, 1:50, Santa Cruz) and p21 (clone SXM30, 1:100, BD Pharmingen), were performed on paraffin embedded liver sections. SA-β Gal staining on frozen liver sections was performed at pH = 5.5 as previously described 54.
Statistics and Reproducibility
All experiments were performed three or more times independently under similar conditions, except experiments shown in Figs 1f, 2d, 2e, 2h, 3b-j, 4a-d, 5 and Supplementary Figs 1b-d, 2a-d, 2f-h, 3, 4, 5a which were performed twice and Fig 1d, Fig 6 and Supplementary Fig 1a and Fig 5b, which were performed once. Statistical significance was calculated by unpaired t-test (Figs 1b, c, e, 3h, 4b, 5c, d) or one-way (Fig 2a, 4e) and two-way (Fig 2b, c, g, 6a, c, d, Supplementary Fig 2e, 5a, b) ANOVA as described in the figure legends. Prism 6 software was used to generate graphs and to perform statistical analysis. P values of statistical significance are represented as **** P < 0.0001, *** P < 0.001, ** P < 0.01, * P < 0.05.
Data availability
RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) under the accession code (GSE100102). Source data from Fig. 1-6 and Supplementary Fig.1, 2, 4 and 5 are shown in Supplementary Table 1 or available from the corresponding author on request.
Supplementary Material
Acknowledgements
We thank Nathalie Jordan for the preparation of viral particles. This research was supported by grants from the SNF (BSSGI0-155984, 31003A_159836), the Gebert Rüf Foundation (GRS-059_14) and the Else Kröner-Fresenius Stiftung (2014_A250) to A.A.. In addition, this work was supported by the German Research Foundation (DFG; grants FOR2314 (L.Z.) and SFB685 (L.Z.)), the Gottfried Wilhelm Leibniz Program (L.Z.)), the European Research Council (projects ‘CholangioConcept’ (L.Z.),‘Heptromic’(L.Z.)), the German Ministry for Education and Research (BMBF) (eMed (Multiscale HCC)) (L.Z.), the German Universities Excellence Initiative (third funding line: ‘future concept’) (L.Z.), the German Center for Translational Cancer Research (DKTK) (L.Z.), and the German-Israeli Cooperation in Cancer Research (DKFZ-MOST) (L.Z.). B.G. is supported by a long-term EMBO fellowship (ALTF 203-2016). J.R. is supported by grants from the UK Medical Research Council [MRC core funding of the MRC Human Immunology Unit] and the Wellcome Trust (grant number 100954).
Footnotes
Author Contributions
S.G., B.G., M.F.G., K.W., T.-W.K., L.Z. and A.A. designed experiments and analysed the data. S.G., B.G., M.F.G., K.W., T.-W.K. and A.A. performed experiments. N.A.S. assisted in the establishment of methods. A.B. and J.R. provided reagents and cells. S.G., B.G. and A.A. wrote the manuscript with help from all authors. A.A. supervised the project.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes & development. 2010;24(22):2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campisi J. Aging, cellular senescence, and cancer. Annual review of physiology. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annual review of pathology. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tchkonia T, Zhu Y, van Deursen J, Campisi J, Kirkland JL. Cellular senescence and the senescent secretory phenotype: therapeutic opportunities. The Journal of clinical investigation. 2013;123(3):966–972. doi: 10.1172/JCI64098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demaria M, Ohtani N, Youssef SA, Rodier F, Toussaint W, Mitchell JR, et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Developmental cell. 2014;31(6):722–733. doi: 10.1016/j.devcel.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134(4):657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang TW, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479(7374):547–551. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- 8.Mosteiro L, Pantoja C, Alcazar N, Marion RM, Chondronasiou D, Rovira M, et al. Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science. 2016;354(6315) doi: 10.1126/science.aaf4445. [DOI] [PubMed] [Google Scholar]
- 9.Baker DJ, Childs BG, Durik M, Wijers ME, Sieben CJ, Zhong J, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530(7589):184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Childs BG, Durik M, Baker DJ, van Deursen JM. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nature medicine. 2015;21(12):1424–1435. doi: 10.1038/nm.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133(6):1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 13.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133(6):1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 14.Chien Y, Scuoppo C, Wang X, Fang X, Balgley B, Bolden JE, et al. Control of the senescence-associated secretory phenotype by NF-kappaB promotes senescence and enhances chemosensitivity. Genes & development. 2011;25(20):2125–2136. doi: 10.1101/gad.17276711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freund A, Patil CK, Campisi J. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. The EMBO journal. 2011;30(8):1536–1548. doi: 10.1038/emboj.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang C, Xu Q, Martin TD, Li MZ, Demaria M, Aron L, et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science. 2015;349(6255):aaa5612. doi: 10.1126/science.aaa5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 18.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell reports. 2013;3(5):1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, et al. cGAS produces a 2'-5'-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498(7454):380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Molecular cell. 2013;51(2):226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, et al. Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153(5):1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ablasser A, Gulen MF. The role of cGAS in innate immunity and beyond. Journal of molecular medicine. 2016 doi: 10.1007/s00109-016-1423-2. [DOI] [PubMed] [Google Scholar]
- 24.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nature immunology. 2016;17(10):1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 25.Barber GN. STING: infection, inflammation and cancer. Nature reviews Immunology. 2015;15(12):760–770. doi: 10.1038/nri3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao D, Li T, Li XD, Chen X, Li QZ, Wight-Carter M, et al. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(42):E5699–5705. doi: 10.1073/pnas.1516465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray EE, Treuting PM, Woodward JJ, Stetson DB. Cutting Edge: cGAS Is Required for Lethal Autoimmune Disease in the Trex1-Deficient Mouse Model of Aicardi-Goutieres Syndrome. Journal of immunology. 2015;195(5):1939–1943. doi: 10.4049/jimmunol.1500969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ablasser A, Hemmerling I, Schmid-Burgk JL, Behrendt R, Roers A, Hornung V. TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. Journal of immunology. 2014;192(12):5993–5997. doi: 10.4049/jimmunol.1400737. [DOI] [PubMed] [Google Scholar]
- 29.Crow YJ, Manel N. Aicardi-Goutieres syndrome and the type I interferonopathies. Nature reviews Immunology. 2015;15(7):429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 30.Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity. 2014;41(5):843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanov A, Pawlikowski J, Manoharan I, van Tuyn J, Nelson DM, Rai TS, et al. Lysosome-mediated processing of chromatin in senescence. The Journal of cell biology. 2013;202(1):129–143. doi: 10.1083/jcb.201212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Current protocols in molecular biology / edited by Frederick M Ausubel [et al] 2005 doi: 10.1002/0471142727.mb2801s70. Chapter 28: Unit 28 21. [DOI] [PubMed] [Google Scholar]
- 33.Munoz-Espin D, Serrano M. Cellular senescence: from physiology to pathology. Nature reviews Molecular cell biology. 2014;15(7):482–496. doi: 10.1038/nrm3823. [DOI] [PubMed] [Google Scholar]
- 34.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130(2):223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends in biochemical sciences. 2004;29(8):409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nature cell biology. 2003;5(8):741–747. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS biology. 2008;6(12):2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acosta JC, Banito A, Wuestefeld T, Georgilis A, Janich P, Morton JP, et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nature cell biology. 2013;15(8):978–990. doi: 10.1038/ncb2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moiseeva O, Mallette FA, Mukhopadhyay UK, Moores A, Ferbeyre G. DNA damage signaling and p53-dependent senescence after prolonged beta-interferon stimulation. Molecular biology of the cell. 2006;17(4):1583–1592. doi: 10.1091/mbc.E05-09-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu Q, Katlinskaya YV, Carbone CJ, Zhao B, Katlinski KV, Zheng H, et al. DNA-damage-induced type I interferon promotes senescence and inhibits stem cell function. Cell reports. 2015;11(5):785–797. doi: 10.1016/j.celrep.2015.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katlinskaya YV, Katlinski KV, Yu Q, Ortiz A, Beiting DP, Brice A, et al. Suppression of Type I Interferon Signaling Overcomes Oncogene-Induced Senescence and Mediates Melanoma Development and Progression. Cell reports. 2016;15(1):171–180. doi: 10.1016/j.celrep.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Braumuller H, Wieder T, Brenner E, Assmann S, Hahn M, Alkhaled M, et al. T-helper-1-cell cytokines drive cancer into senescence. Nature. 2013;494(7437):361–365. doi: 10.1038/nature11824. [DOI] [PubMed] [Google Scholar]
- 43.Freund A, Laberge RM, Demaria M, Campisi J. Lamin B1 loss is a senescence-associated biomarker. Molecular biology of the cell. 2012;23(11):2066–2075. doi: 10.1091/mbc.E11-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le ON, Rodier F, Fontaine F, Coppe JP, Campisi J, DeGregori J, et al. Ionizing radiation-induced long-term expression of senescence markers in mice is independent of p53 and immune status. Aging cell. 2010;9(3):398–409. doi: 10.1111/j.1474-9726.2010.00567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Deursen JM. The role of senescent cells in ageing. Nature. 2014;509(7501):439–446. doi: 10.1038/nature13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445(7128):656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Childs BG, Baker DJ, Wijshake T, Conover CA, Campisi J, van Deursen JM. Senescent intimal foam cells are deleterious at all stages of atherosclerosis. Science. 2016;354(6311):472–477. doi: 10.1126/science.aaf6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeon OH, Kim C, Laberge RM, Demaria M, Rathod S, Vasserot AP, et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nature medicine. 2017 doi: 10.1038/nm.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin L, Hill KK, Filak H, Mogan J, Knowles H, Zhang B, et al. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J Immunol. 2011;187(5):2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bridgeman A, Maelfait J, Davenne T, Partridge T, Peng Y, Mayer A, et al. Viruses transfer the antiviral second messenger cGAMP between cells. Science. 2015;349(6253):1228–1232. doi: 10.1126/science.aab3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264(5167):1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 52.McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A. Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell cycle. 2014;13(9):1400–1412. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang TW, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479(7374):547–551. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- 54.Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134(4):657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) under the accession code (GSE100102). Source data from Fig. 1-6 and Supplementary Fig.1, 2, 4 and 5 are shown in Supplementary Table 1 or available from the corresponding author on request.