SUMMARY
The spike trains of retinal ganglion cells (RGCs) are the only source of visual information to the brain. Here, we genetically identify an RGC type in mice that functions as a pixel encoder and increases firing to light increments (PixON-RGC). PixON-RGCs have medium-sized dendritic arbors and non-canonical center-surround receptive fields. From their receptive field center, PixON-RGCs receive only excitatory input, which encodes contrast and spatial information linearly. From their receptive field surround, PixON-RGCs receive only inhibitory input, which is temporally matched to the excitatory center input. As a result, the firing rate of PixON-RGCs linearly encodes local image contrast. Spatially offset (i.e., truly lateral) inhibition of PixON-RGCs arises from spiking GABAergic amacrine cells. The receptive field organization of PixON-RGCs is independent of stimulus wavelength (i.e., achromatic). PixON-RGCs project predominantly to the dorsal lateral geniculate nucleus (dLGN) of the thalamus and likely contribute to visual perception.
In Brief
Johnson et al. genetically identify a pixel-encoder retinal ganglion cell type in mice (PixON-RGCs). PixON-RGCs have spatially offset excitatory and inhibitory receptive fields and encode local image contrast approximately linearly. Their axons project to the dorsolateral geniculate nucleus of the thalamus indicating that PixON-RGCs likely contribute to visual perception.
INTRODUCTION
The output of retinal computations is conveyed to the brain through the spike trains of >30 retinal ganglion cell (RGC) types. Recent surveys in mice have highlighted the morphological and functional diversity of RGCs (Sanes and Masland, 2015; Baden et al., 2016; Helmstaedter et al., 2013), but few RGC types have been genetically identified and studied in detail. Therefore, what specific information RGCs encode, how excitatory and inhibitory circuits give rise to their characteristic light responses, and where in the brain RGCs send their information remains, for most cell types, unknown.
RGCs serve at least three broad functions: (1) to encode local contrast in the retinal image and support visual perception (Sinha et al., 2017; Schwartz et al., 2012; Pang et al., 2003; van Wyk et al., 2009); (2) to detect specific forms of motion and drive reflexive behaviors (Huang et al., 2017; Münch et al., 2009; Sabbah et al., 2017; Zhang et al., 2012); and (3) to measure overall luminance and regulate non-visual light-dependent processes (Chen et al., 2011; Lazzerini Ospri et al., 2017). Individual RGC types can contribute to more than one of these functions, which involve different subcortical targets (Dhande et al., 2015). Recent studies in mice have identified numerous RGC types that detect specific forms of motion (Zhang et al., 2012; Jacoby and Schwartz, 2017; Münch et al., 2009; Borst and Euler, 2011) and intrinsically photosensitive RGC (ipRGC) types that regulate non-visual light-dependent processes (Chen et al., 2011; Lazzerini Ospri et al., 2017). What murine RGC types contribute to visual perception, which relies on signal propagation from the retina to dorsal lateral geniculate nucleus (dLGN) and primary visual cortex, is less well understood (Kerschensteiner and Guido, 2017). In primates, midget and parasol RGCs dominate input to primary visual cortex (Wässle et al., 1989; Field and Chichilnisky, 2007). Midget RGCs encode contrast and spatial information approximately linearly (i.e., pixel encoders) and mediate high acuity pattern vision, whereas parasol RGCs integrate spatial information nonlinearly and are sensitive to motion irrespective of the precise image patterns involved (Field and Chichilnisky, 2007; Petrusca et al., 2007; Cafaro and Rieke, 2013). Alpha RGCs in mice share response properties with parasol RGCs in primates and provide motion-sensitive input to dLGN (Cruz-Martín et al., 2014; Pang et al., 2003; Rompani et al., 2017; Schwartz et al., 2012). Whether the mouse retina contains a pixel-encoder RGC type that projects to dLGN, and, if so, what circuit mechanisms give rise to its responses remains unknown.
Throughout the nervous system, excitatory and inhibitory synaptic inputs shape the computations of neurons (Isaacson and Scanziani, 2011). Many RGCs receive excitation and inhibition via one of two circuit motifs (Cafaro and Rieke, 2013; Demb and Singer, 2015; Roska et al., 2006; Roska and Werblin, 2001). In feedforward inhibition, the same bipolar cells that excite an RGC activate amacrine cells that inhibit the RGC. In crossover inhibition, bipolar cells with opposite contrast preferences (ON versus OFF) excite and inhibit—via amacrine cells—an RGC (Cafaro and Rieke, 2013; Demb and Singer, 2015; Roska et al., 2006; Roska and Werblin, 2001). In both motifs, excitatory and inhibitory receptive fields have their highest sensitivity over the dendritic field of the RGC (i.e., the receptive field center), with inhibition extending laterally beyond excitation (i.e., the receptive field surround). By contrast, inhibitory receptive fields of direction-selective ganglion cells (DSGCs) are shifted sideways relative to their excitatory receptive fields and dendrites (Fried et al., 2002; Wei et al., 2011; Yonehara et al., 2011), indicating that non-canonical arrangements of excitatory and inhibitory receptive fields help diversify RGC light responses.
Here, we genetically identify a pixel-encoder RGC type in mice (PixON-RGCs). In two-photon-guided patch-clamp recordings, we find that PixON-RGCs encode local image contrast approximately linearly and identify a novel circuit motif (i.e., truly lateral inhibition), which gives rise to their responses. Combining genetic labeling and retrograde tracing, we show that PixON-RGCs project predominantly to the dLGN and likely contribute to visual perception.
RESULTS
Genetic, Morphological, and Functional Identification of PixON-RGCs
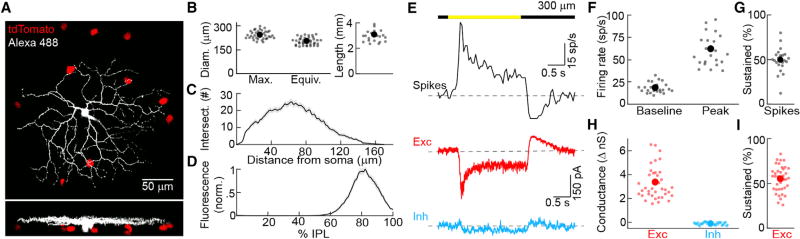
Characterizing the light responses, circuit mechanisms, and projection patterns of the >30 RGC types, which relay the output of retinal computations to the brain, is a prerequisite to understanding vision. To genetically identify unknown RGC types, we crossed a variety of Cre driver lines, including Grik4-Cre (Nakazawa et al., 2002), to the tdTomato reporter strain Ai9 (Madisen et al., 2010), and targeted fluorescent cells under two-photon guidance for patch-clamp recordings. Except where noted otherwise, we recorded cells in the ventral retina where cones express predominantly S-opsin (Baden et al., 2013; Szél et al., 1992) and presented stimuli in the UV spectrum (peak: 385 nm). In Grik4-Cre:Ai9 retinas, ON and ON-OFF DSGCs were labeled as previously reported (Ivanova et al., 2010; Martersteck et al., 2017). In addition, we frequently encountered RGCs with medium-sized, densely branched dendritic arbors stratifying close to the ganglion cell layer (Figures 1A–1D). These cells had high baseline firing rates, increased spiking in a sustained manner in response to light increments (i.e., ON stimuli), and decreased spiking during light decrements (i.e., OFF stimuli, Figures 1E–1G). Voltage-clamp recordings revealed that excitatory synaptic inputs matched the spike responses of these cells, which, remarkably, received no inhibitory input for stimuli restricted to their dendritic field (spot diameter: 300 µm, Figures 1E, 1H, 1I, and S1), and, which were not intrinsically photosensitive at light levels ranging from twilight to bright daylight (Figure S2) (Cronin et al., 2014; Johnsen et al., 2006; Milner and Do, 2017). Based on their genetic labeling, monomorphic dendrites, and consistent physiological properties, we identify these cells as a single RGC type. We refer to them as PixON-RGCs, because they appear to function as pixel encoders and increase firing to ON stimuli.
Figure 1. Morphology and Light Responses of PixON-RGCs.
(A) Orthogonal projections of a two-photon image stack through a representative PixON-RGC filled during physiological recording in Grik4-Cre:Ai9 mice.
(B) Length of the longest axis through a polygon around the PixON dendritic field (n = 38), equivalent diameter of the PixON-RGC dendritic field (n = 38), and total dendritic length (n = 23) of PixON-RGCs.
(C) Sholl analysis of traced PixON-RGC dendrites (n = 23).
(D) Stratification of PixON-RGC dendrites within the IPL (n = 18; 0%–100% border between inner plexiform layer and inner nuclear layer to border between inner plexiform layer and ganglion cell layer). In (C) and (D), lines (shaded areas) indicate the means (±SEMs) of the traced population.
(E) Representative spiking (black), and excitatory (red) and inhibitory (cyan) currents in response to presentation of a 300-µm circle (2 s ON, 2 s OFF; 1,500 R*/rod/s background) centered on the soma of the recorded cell. Dashed lines show baselines in the absence of stimulus.
(F) Spontaneous and peak firing rates (n = 23) in response to the same stimulus as in (E).
(G) Percentage of peak spike response remaining 1.5 s after stimulus onset (n = 23).
(H) Change in excitatory and inhibitory conductance (n = 38) in response to the same stimulus as in (E).
(I) Percentage of peak excitatory conductance remaining 1.5 s after stimulus onset (n = 38).
In (F)–(I), dots represent data of individual cells, whereas larger circles (error bars) indicate means (±SEMs) of the respective populations. See also Figures S1, S2, and S4.
Linear Encoding of Contrast and Spatial Information by PixON-RGCs
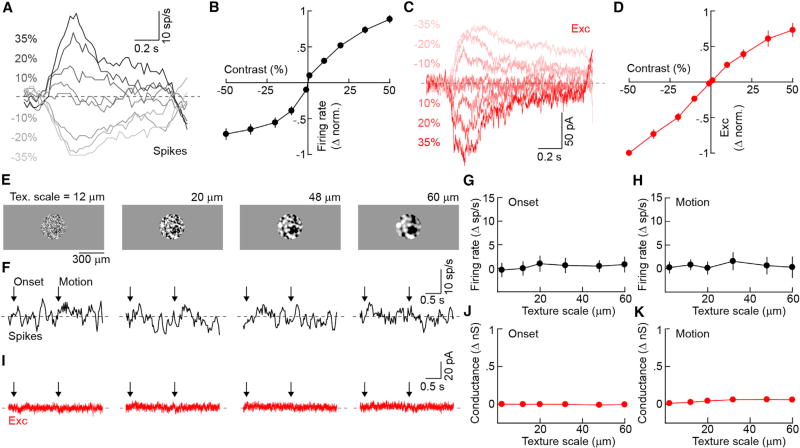
To allow the brain to infer image patterns accurately, pixel-encoder RGCs change their firing rates approximately linearly as a function of the light intensity over their dendritic fields, and do not respond to stimuli, including second-order motion (Demb et al., 2001), for which the overall light intensity in this area does not change (Sinha et al., 2017; Field and Chichilnisky, 2007). Recording spike responses to spots (diameter: 300 µm) of varying intensity, we found that PixON-RGCs increased and decreased their firing rates approximately linearly to positive and negative contrast steps, respectively (Figures 2A and 2B). These responses matched excitatory synaptic inputs to PixON-RGCs, which increased and decreased linearly from high tonic levels (1.3 ± 0.2 nano-Siemens [nS], mean ± SEM, n = 9) to light increments and decrements, respectively (Figures 2C and 2D).
Figure 2. Linear Contrast Response Functions and Linear Spatial Integration of PixON-RGCs.
(A and C) Representative spike (A) and EPSC (C) responses to contrast steps presented in a 300-µm circle centered on the soma of the recorded cell.
(B and D) Contrast response functions of normalized spike rate (B, n = 13) and excitation (D, n = 6). Circles (error bars) indicate means (±SEMs) of the respective populations.
(E) Representative texture stimuli of different spatial scales masked in a 300-µm circle. Textures appeared from a gray background (onset) and were then translated 33 µm in either the dorsal, ventral, nasal, or temporal direction (motion). Textured regions were of the same mean luminance as the background.
(F and I) Representative spike (F) and excitatory (I) responses of a PixON-RGC to presentation of textures of the spatial scales shown in (E).
(G and H) Summary data of PixON-RGC spike responses to the onset (G) and motion (H) of texture stimuli of different spatial scales. Circles (error bars) indicate means (±SEMs) of the population (n = 13).
(J and K) Analogous to (G) and (H), respectively, for excitation (n = 3).
See also Figures S3–S5.
We next recorded the spike trains of PixON-RGCs during presentation and motion of textures of varying spatial scales. We balanced textures to keep the average light intensity above the dendritic field of each RGC constant (Figure 2E). PixON-RGCs responded neither to presentation nor motion of these textures (Figures 2F–2H), and neither elicited synaptic excitation (Figures 2I and 3K). By contrast, the ON DSGCs and ON-OFF DSGCs labeled in Grik4-Cre:Ai9 retinas responded robustly to both the presentation and motion of texture stimuli (Figure S3). Thus, PixON-RGCs encode contrast and spatial information linearly. They inherit these properties from their excitatory input and receive no synaptic inhibition from their receptive field center.
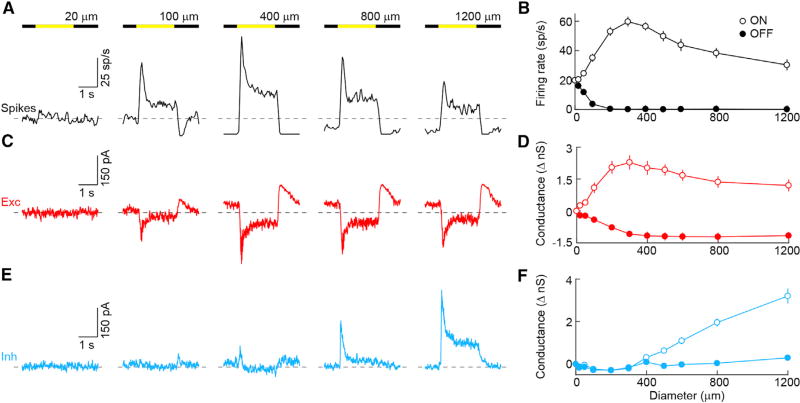
Figure 3. Spatial receptive fields of PixON-RGCs.
(A, C, and E) Representative spike (A, black), EPSC (C, red), and IPSC (E, cyan) responses to square-wave modulation (2 s ON, 2 s OFF) of circles of varying diameter. Dashed lines show baselines in the absence of stimulus.
(B, D, and F) Firing rates (B, n = 22) and changes in excitatory (D, n = 9) and inhibitory conductances (F, n = 9) during ON (open circles) and OFF (filled circles) plotted as a function of stimulus diameter. Circles (error bars) indicate means (±SEMs) of the respective populations.
See also Figure S4.
To characterize the temporal response properties of PixON-RGCs, we recorded spike trains during presentation of Gaussian white noise stimuli. Reverse correlation of responses with the stimulus revealed monophasic temporal filter kernels (Figure S4), with opposite polarity in the receptive field center and surround, in keeping with the sustained responses observed during contrast steps.
Surround Suppression of PixON-RGCs and Its Mechanisms
Many RGCs show attenuated responses to large stimuli (Kuffler, 1953; Demb and Singer, 2015). This enhances edges in their representation of the retinal image, which in turn facilitates pattern and object recognition in the brain (Egan et al., 2016; Biederman and Ju, 1988). To explore the stimulus size tuning of PixON-RGCs, we presented spots of varying diameter (20–1,200 µm) in pseudorandom sequences. ON responses increased up to a stimulus size of 300 µm and were strongly suppressed by larger stimuli (Figures 3A and 3B). Thus, PixON-RGCs fired at only 17.2% ± 8.8% (n = 22) of their maximal response rates when stimulated with 1,200-µm spots. Excitatory synaptic inputs similarly increased up to a stimulus size of 300 µm but were suppressed to a lesser degree than spike responses by larger stimuli (Figures 3C and 3D). Thus, 1,200-µm spots still activated 49.0% ± 9.4% (n = 9) of the maximal excitatory conductance of PixON-RGCs. Although spots restricted to their dendritic fields did not evoke inhibition, we found that larger stimuli gradually activated inhibitory synaptic inputs to PixON-RGCs (Figures 3E and 3F). Because PixON-RGCs received little or no tonic inhibition, only ON stimuli modulated these inputs (Figures 3E, 3F, and S5), and suppression of tonic excitation underlay the spike suppression at light OFF. Thus, PixON-RGCs encode stimulus contrast and spatial information linearly, reflecting properties of excitatory inputs from their receptive field center, and exhibit strong size selectivity (i.e., a preference for local contrast or edges) due to a combination of pre- and postsynaptic inhibition. The latter, unusually, is recruited only by stimuli that extend beyond the dendrites of PixON-RGCs into their receptive field surround.
Kinetics and Mechanisms of Inhibitory Synaptic Inputs to PixON-RGCs
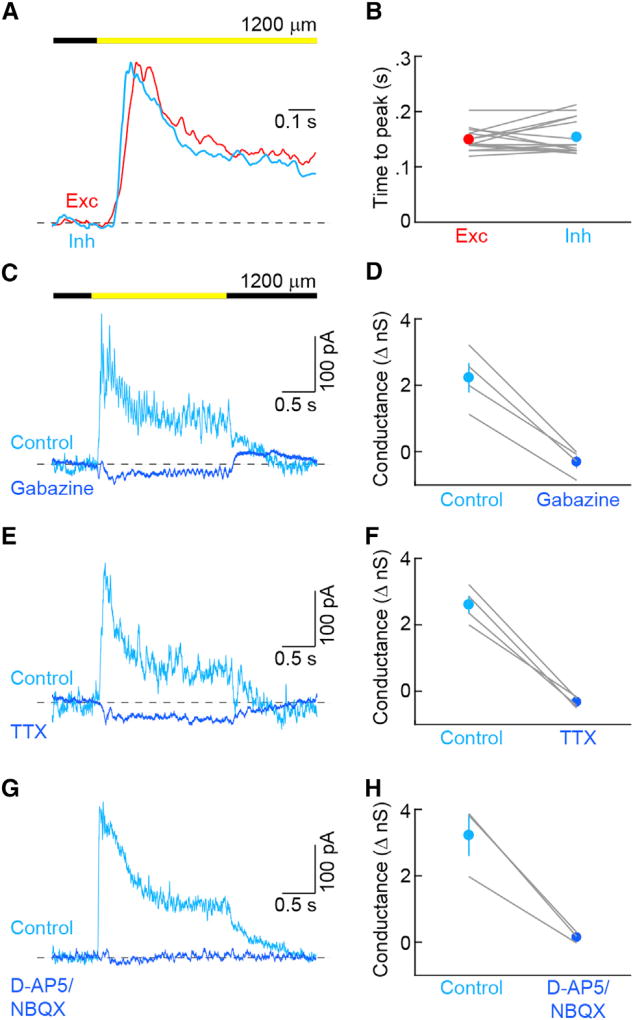
Comparing the timing of synaptic inputs elicited by large stimuli (diameter: 1,200 µm), we found that inhibition and excitation were temporally matched (Figure 4A). Thus, neither the time to peak (Figure 5B) nor the percentages of sustained input (excitation: 47.8% ± 7.0%, inhibition: 46.1% ± 2.6%, n = 15, p > 0.8) were significantly different between excitation and inhibition. Because inhibition, in spite of originating in the receptive field surround, reached PixON-RGCs simultaneously with excitation from the receptive field center, we hypothesized that spiking GABAergic wide-field amacrine cells may be its source. We tested this hypothesis pharmacologically. Both gabazine and tetrodotoxin (TTX) blocked inhibition to PixON-RGCs (Figures 4C–4F), indicating that inhibition is mediated by GABAA receptors and provided by spiking GABAergic wide-field amacrine cells. In addition to using spikes to relay signals from the surround, at least one retinal circuit accelerates inhibition by driving amacrine cells via gap-junctional rather than glutamatergic input from bipolar cells (Farrow et al., 2013). Inhibition to PixON-RGCs was blocked completely by D-AP5 and NBQX (Figures 4G and 4H), suggesting that the spiking GABAergic wide-field amacrine cells in this circuit are activated by conventional glutamatergic input from bipolar cells.
Figure 4. Kinetics and Mechanisms of PixON-RGC Inhibition.
(A) Representative normalized excitatory (red) and inhibitory (cyan) conductances in response to presentation of a 1,200-µm circle.
(B) Time from stimulus onset to peak of excitatory (n = 15) and inhibitory (n = 15) currents in response to presentation of a 1,200-µm circle (p = 0.544).
(C) Representative IPSCs in control (cyan) solution and in the presence of the GABAA receptor antagonist gabazine (blue) evoked by square-wave modulation (2 s ON, 2 s OFF) of a 1,200-µm circle.
(D) Summary data of inhibitory conductance under control conditions (cyan) or in the presence of gabazine (blue, n = 4, p < 0.004). Lines indicate results from individual cells, whereas circles (error bars) indicate the means (±SEMs) of the respective population.
(E and F) Analogous to (C) and (D), respectively, but for tetrodotoxin (TTX, n = 4, p < 0.003).
(G and H) Analogous to (C) and (D), respectively, but for NMDA (D-AP5) and AMPA (NBQX) receptor antagonists (n = 3, p < 0.03).
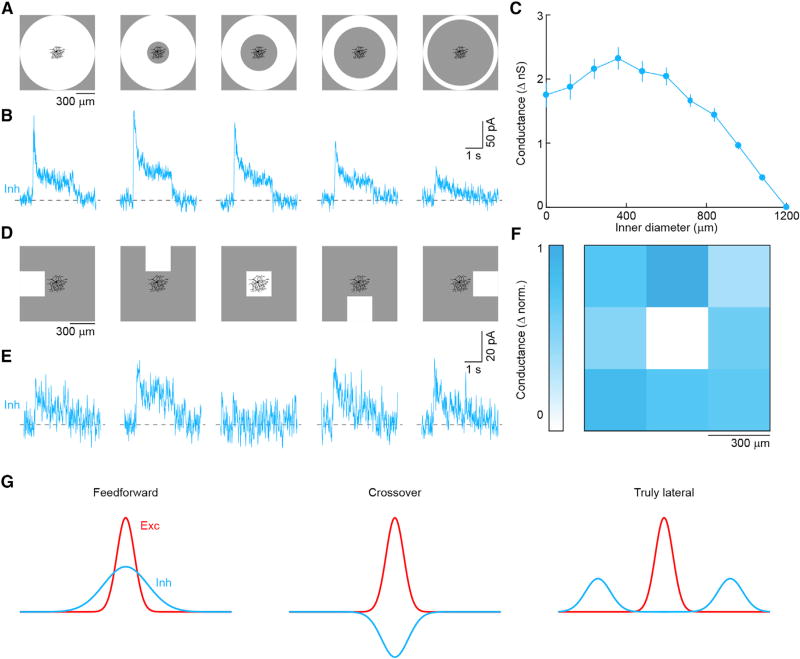
Figure 5. Spatially Offset Inhibition of PixON-RGCs.
(A) Annuli of constant 1,200-µm outer diameter and varying inner diameter were presented as 100%contrast steps (2 s ON) from a gray background (1,500 R*/rod/s).
(B) Representative IPSCs in response to annuli shown in (A).
(C) Summary data plotting inhibitory conductance as a function of the inner diameter of stimulus annuli. Circles (error bars) indicate mean (±SEM) of the recorded population (n = 7).
(D) The display was divided into a 3 × 3 grid of 300- × 300-µm squares and centered on the soma of the recorded cell. Between stimulus presentations all squares had the same luminance (1,500 R*/rod/s). During stimulus presentation, one of the nine squares increased in luminance (100% contrast, 2 s ON).
(E) Representative IPSCs in response to appearance of squares in the positions shown in (A).
(F) Map of the normalized inhibitory conductance evoked by the appearance of a square at each position (n = 4). Coloration indicates the strength of inhibition.
(G) Sensitivity profiles of excitatory and inhibitory receptive fields in feedforward inhibition, in crossover inhibition, and in truly lateral inhibition.
See also Figures S6 and S7.
Spatially Offset Inhibitory and Excitatory Receptive Fields of PixON-RGCs
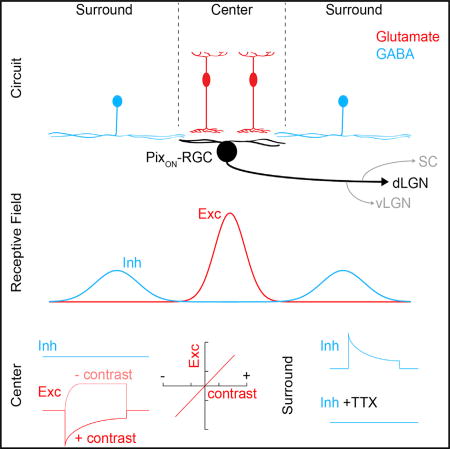
For centered spots, we found that inhibition was restricted to large stimuli, which extended beyond PixON-RGCs’ dendritic fields (Figure 3). This could either be because the wide-field amacrine cells that provide this input need to integrate bipolar cell inputs over large areas to pass spike threshold (i.e., size-thresholded inhibition) or because the wide-field amacrine cells that inhibit a given PixON-RGC receive their excitatory input outside its dendritic field (i.e., spatially offset inhibition). We designed two sets of stimuli to distinguish between these possibilities. First, we gradually eroded a large centered spot (diameter: 1,200 µm) from the inside (Figure 5A). Inhibition elicited by this stimulus declined only after the inner stimulus boundary had receded beyond the dendritic territory of PixON-RGCs (Figures 5A–5C). Second, we presented a stimulus square (side length: 300 µm) in a 3 × 3 grid centered on a PixON-RGC (Figure 5D). Consistently, the center square failed to elicit inhibitory synaptic inputs, whereas all other squares evoked robust inhibition (Figures 5D–5F). The responses of PixON-RGC inhibition differed drastically from those of ONα-RGCs (Figure S6), which receive canonical feedforward inhibition. We conclude that the inhibitory receptive fields of PixON-RGCs are spatially offset from their dendrites and their excitatory receptive fields (Figure S7) and that PixON-RGCs receive inputs from a previously unknown circuit motif, which we refer to as truly lateral inhibition (Figure 5G).
Wavelength Independence of PixON-RGC’s Receptive Field Organization
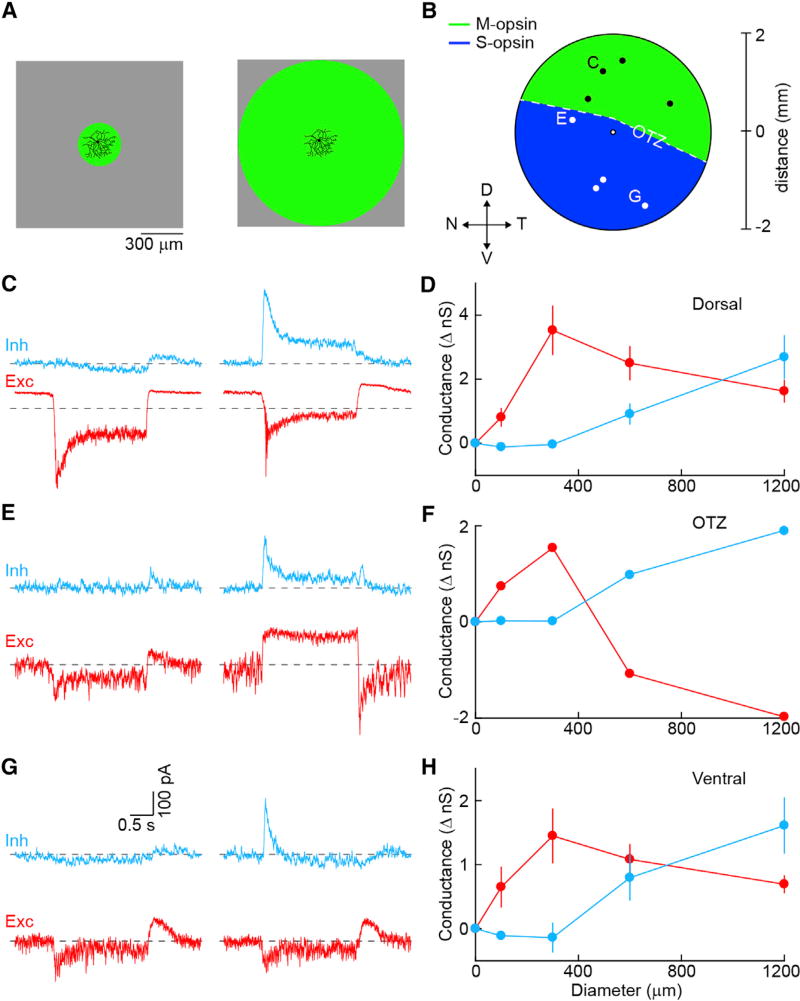
To test whether the organization of PixON-RGC receptive fields depends on stimulus wavelength and whether these cells encode chromatic information, we recorded PixON-RGCs at different points along the dorsoventral gradient of cone opsin expression (Baden et al., 2013; Szél et al., 1992) noting their position in the retina (Figures 6A and 6B). In addition, we switched from stimulating in the UV spectrum (peak: 385 nm), which favors S-opsin activation, to stimulating with green light (peak: 532 nm), favoring M-opsin activation. Excitatory and inhibitory receptive fields of PixON-RGCs probed with green stimuli in the dorsal (Figures 6C and 6D; n = 4) and ventral (Figures 6G and 6H; n = 3) retina were activated exclusively by light increments and were spatially offset, identical to our observations for UV stimuli in the ventral retina (Figure 3). In addition, with the exception of one cell in the opsin transitional zone (OTZ) (Figures 6E and 6F), the ratio of center excitation and surround inhibition was similar across the retina and stimulus wavelengths. Thus, outside of the OTZ, where color-opponent responses arise in several RGC types without cone-type selective connectivity (Chang et al., 2013), PixON-RGCs appear not to encode chromatic information and their receptive field organization is wavelength independent.
Figure 6. Spatial Receptive Field Organization of PixON-RGCs with Green Stimulus.
(A) Green visual stimulus (peak: 532 nm) was used to probe receptive field properties of PixON-RGCs.
(B) Schematic of the retina showing approximate locations of recorded cells (OTZ, opsin transition zone).
(C) Representative EPSC (red) and IPSC (cyan) responses to square-wave modulation (2 s ON, 2 s OFF) of either a 300- (left) or 1,200-µm (right) circle from a cell recorded in the dorsal retina. Dashed lines show baselines in the absence of stimulus.
(D) Summary of changes in excitatory and inhibitory conductances of cells recorded in the dorsal retina (n = 4) during the ON phase of the stimulus, plotted as a function of stimulus diameter. Circles (error bars) indicate means (±SEMs) of the respective populations.
(E–H) Analogous to (C) and (D), but for a cell recorded near the OTZ (E and F) (n = 1) or cells recorded in the ventral retina (G and H) (n = 3).
PixON-RGC Axons Project to dLGN
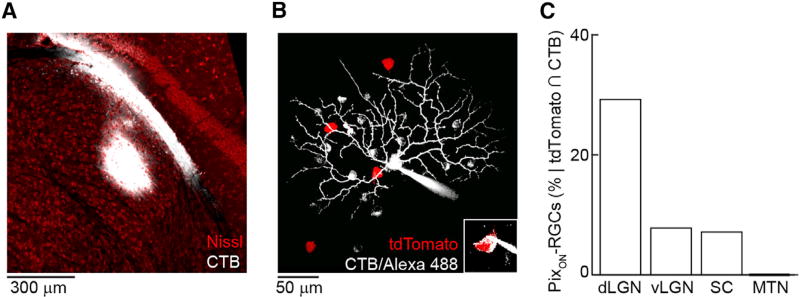
Visual perception relies on retinal input to dLGN (Saalmann and Kastner, 2011; Kerschensteiner and Guido, 2017). To determine whether PixON-RGCs could contribute to visual perception, we retrogradely traced their axonal projection patterns. We injected either cholera toxin B conjugated to Alexa 488 (CTB488) or an adeno-associated virus expressing GCaMP6f in a Credependent manner (AAV9-Syn-FLEX-GCaMP6f) into subcortical visual targets labeled in Grik4-Cre:Ai9 mice (Figure 7A). We then targeted RGCs co-labeled with tdTomato and CTB488 or GCaMP6f for patch-clamp recordings and identified PixON-RGCs by their characteristic morphology (Figure 7B) and light responses. Of the 74 PixON-RGCs included in this study, 23 were targeted in this way. PixON-RGCs accounted for approximately 1/3 of the cells co-labeled after dLGN injections. By comparison, PixON-RGCs made up smaller fractions of co-labeled cells after injections into ventral lateral geniculate nucleus (vLGN) or superior colliculus (SC), and no PixON-RGCs were labeled by injections into the medial terminal nucleus (MTN, Figure 7C). This projection pattern of PixON-RGC axons supports the notion that they contribute to visual perception.
Figure 7. Retrograde Tracing of PixON-RGCs’ Axonal Projections.
(A) Representative cholera toxin subunit B (CTB) injection primarily in the core of dLGN.
(B) Morphology of a tdTomato and CTB positive (inset) cell targeted, recorded, and filled following injection of CTB into dLGN in a Grik4-Cre:Ai9 mouse.
(C) Percentage of recorded cells that were both tdTomato and CTB positive and had physiological and morphological properties of PixON-RGCs following CTB injection into different brain regions (dLGN: 19/65 cells; vLGN: 3/38 cells; SC: 1/14 cells; MTN: 0/10 cells).
DISCUSSION
In this study, we discover and characterize a pixel-encoder RGC type in the mouse retina (PixON-RGCs). The dendrites of PixON-RGCs, which morphologically resemble cluster 9n cells in the Eyewire museum (Bae et al., 2017), stratify near the boundary between the inner plexiform and ganglion cell layer, and receive sustained excitatory input from ON bipolar cells (Figure 1) (Euler et al., 2014; Franke et al., 2017). Tonic excitation likely accounts for the high baseline firing rates of PixON-RGCs (Figure 1), and bidirectional changes in excitatory input underlie their approximately linear contrast encoding (Figure 2). Similarly high baseline firing rates and linear contrast response functions have been recorded in ONα-RGCs (Grimes et al., 2014; Schwartz et al., 2012; Tien et al., 2017). However, unlike ONα-RGCs (Grimes et al., 2014; Schwartz et al., 2012; Tien et al., 2017), PixON-RGCs integrate spatial information linearly and do not respond to stimuli that do not change the average light intensity in their dendritic fields (Figure 2). Linear spatial integration and linear contrast response functions enable PixON-RGCs to encode relatively faithfully the retinal image. These properties of PixON-RGCs are reminiscent of X/beta RGCs in cats, rabbits, and guinea pigs (Enroth-Cugell and Robson, 1966; Demb et al., 2001; Cleland and Levick, 1974; Roska et al., 2006), and to midget RGCs in primates (Crook et al., 2011; Sinha et al., 2017). Compared to these pixel-encoder cell types, dendritic fields of PixON-RGCs span larger areas of visual space (7.1° ± 1.1°, n = 38). Whether other pixel-encoder RGCs with smaller dendrites exist in the mouse retina and how PixON-RGCs influence visual acuity remain to be explored (Prusky et al., 2000; Burgess et al., 2017).
PixON-RGCs receive only excitatory synaptic input from their receptive field center and receive only inhibitory synaptic input from their receptive field surround (Figures 5 and S7). We refer to this circuit motif as truly lateral inhibition. Because inhibition is excluded from the receptive field center, PixON-RGC spike responses are coupled with high gain to excitatory input elicited by small stimuli (diameter 300 µm: 17.5 ± 2.8 spikes sp/s/nS). For larger stimuli, spatially offset inhibition reduces the gain of the excitation-spike coupling (diameter 1,200 µm: 8.5 ± 3.6 sp/s/nS), which, together with presynaptic inhibition, accounts for the nearly complete surround suppression. Thus, truly lateral inhibition allows for a combination of high gain in the receptive field center and strong surround suppression. This enables PixON-RGCs to encode relatively faithfully the retinal image, deemphasizing regions of uniform intensity. Because excitation and inhibition are temporally matched (Figure 4), stimulation of the surround suppresses the amplitude of the center response without changing its dynamics. This further simplifies the apparent image encoding of PixON-RGCs.
We find that spiking GABAergic amacrine cells provide the inhibitory input to PixON-RGCs. The most parsimonious explanation for the spatial offset of inhibitory receptive fields is that these amacrine cells receive input and provide output in separate arbors (dendrite and axon, respectively) or separate regions of the same arbor. Polyaxonal amacrine cells are an appealing candidate for the spatially offset inhibitory receptive fields of PixON-RGCs (Famiglietti, 1992; Völgyi et al., 2001), which unlike the spatially offset inhibitory receptive fields of DSGCs (Briggman et al., 2011; Fried et al., 2002; Wei et al., 2011; Yonehara et al., 2011), surround the excitatory receptive fields symmetrically. The cellular composition of the circuit that mediates truly lateral inhibition of PixON-RGCs remains to be explored experimentally, which may reveal more complicated wiring schemes.
The receptive field organization of PixON-RGCs was indistinguishable between the dorsal and the ventral retina and between stimuli that preferentially activity S- or M-opsin, indicating that outside the OTZ where color-opponency can arise in several RGC types without cone-type selective wiring (Chang et al., 2013), PixON-RGCs do not encode chromatic information. A recent paper described an RGC type, referred to as M5 (Stabio et al., 2018), which morphologically resembles PixON-RGCs and which likewise receives sustained excitatory inputs. However, M5 RGCs were suggested to consistently encode chromatic information and to exhibit intrinsic photocurrents. PixON-RGCs did not stain for melanopsin and did not exhibit intrinsic photocurrents at light levels equivalent to bright daylight (Figure S2) (Cronin et al., 2014; Johnsen et al., 2006; Milner and Do, 2017), which elicited large intrinsic photocurrents in M1 RGCs (data not shown). Thus, is seems that despite their morphologic similarity PixON-RGCs and M5 RGCs may be distinct cell types.
Combining retrograde tracing and genetic labeling, we found that PixON-RGC axons project predominantly to dLGN (Figure 7). To what extent information from PixON-RGCs is preserved in dLGN or whether it is recombined with other retinal inputs before being sent to primary visual cortex remains to be determined. Interestingly, sustained ON responses that linearly encode contrast and spatial information have been recorded in dLGN (Piscopo et al., 2013), and RGCs with morphologies similar to PixON-RGCs have been shown to contribute to relay mode innervation of dLGN neurons (Rompani et al., 2017). These findings suggest that information of PixON-RGC may be preserved in dLGN and relayed faithfully to primary visual cortex. Future studies will have to test this experimentally and probe the contribution of PixON-RGCs to visual perception.
EXPERIMENTAL PROCEDURES
Animals
Throughout this study, we used Grik4-Cre mice (Nakazawa et al., 2002) (Jackson Laboratories stock #006474) crossed to the tdTomato reporter strain Ai9 (Madisen et al., 2010) (Jackson Laboratories stock #007909) to label PixON-RGCs. We isolated retinas from young adult mice (postnatal day P21–P40) of both sexes. All experiments in this study were approved by the Institutional Animal Care and Use Committee of Washington University School of Medicine and were performed in compliance with the NIH Guide for the Care and Use of Laboratory Animals.
Tissue Preparation
Mice were deeply anesthetized with CO2, killed by cervical dislocation, and enucleated. For patch-clamp recordings, mice were dark adapted overnight before their retinas were isolated under infrared illumination (>900 nm) in oxygenated mACSFNaHCO3 containing (in mM) 125 NaCl, 2.5 KCl, 1 MgCl2, 1.25 NaH2PO4, 2 CaCl2, 20 glucose, 26 NaHCO3, and 0.5 L-glutamine equilibrated with 95% O2/5% CO2. For confocal imaging, retinas were isolated in oxygenated mouse artificial cerebrospinal fluid (mACSFHEPES) containing (in mM): 119 NaCl, 2.5 KCl, 1 NaH2PO4, 2.5 CaCl2, 1.3 MgCl2, 20 HEPES, and 11 glucose (pH adjusted to 7.37 using NaOH), mounted flat on filter paper and fixed for 30 min in 4% paraformaldehyde in mACSFHEPES.
Immunohistochemistry
Flat-mount preparations were cryoprotected (1 hr 10% sucrose in PBS at room temperature [RT], 1 hr 20% sucrose in PBS at RT, and overnight 30% sucrose in PBS at 4°C), frozen and thawed three times, and blocked with 10% normal donkey serum in PBS for 2 hr before incubation with primary antibodies for 5 days at 4°C. Flat mounts were washed in PBS (3 × 1 hr) at RT, incubated with secondary antibodies for 1 day at 4°C, and washed in PBS (3 × 1 hr) at RT. The following primary antibodies were used in this study: rabbit anti-Melanopsin (1:5,000, Abcam, RRID:AB_444842) and mouse anti-RFP (1:1,000, Abcam, RRID:AB_945213). Secondary antibodies were Alexa 488 and Alexa 568 conjugates (1:1,000, Invitrogen, RRID:AB_2556546 and RRID:AB_2534013).
Confocal Imaging
Image stacks of fixed tissue were acquired through a 20 × 0.85 numerical aperture (NA) oil immersion objective (Olympus) on an upright laser scanning confocal microscope (FV1000, Olympus) and processed with Fiji (Schindelin et al., 2012).
Electrophysiology
Cell-attached and whole-cell patch-clamp recordings were obtained primarily from the ventral halves of dark-adapted retinas, flat-mounted on transparent membrane discs (13 mm Whatman Anodisc) superfused (~7 mL/min) with warm (30°C–33°C) mACSFNaHCO3. Fluorescent RGCs were targeted under two-photon guidance (excitation wavelength: 940 nm) in Grik4-Cre:Ai9 retinas. The intracellular solution for current-clamp recordings contained (in mM) 125 K-gluconate, 10 NaCl, 1 MgCl2, 10 EGTA, 5 HEPES, 5 ATP-Na2, and 0.1 GTP-Na (pH adjusted to 7.2 with KOH). The intracellular solution for voltage-clamp recordings contained (in mM) 120 Cs-gluconate, 1 CaCl2, 1 MgCl2, 10 Na-HEPES, 11 EGTA, 10 TEA-Cl, 2 Qx314, ATP-Na2, and 0.1 GTP-Na (pH adjusted to 7.2 with CsOH). Patch pipettes had resistances of 3–6 MΩ (borosilicate glass). Signals were amplified with a Multiclamp 700B amplifier (Molecular Devices), filtered at 3 kHz (8-pole Bessel low-pass), and sampled at 10 kHz (Digidata 1550, Molecular Devices). For voltage-clamp recordings, series resistance (<15 MΩ) was compensated electronically by ~60%. All reported voltages were corrected for a liquid junction potential of ~−15 mV. Excitatory postsynaptic currents (EPSCs) were measured near the reversal potential of inhibitory conductances (−70 mV) and inhibitory postsynaptic currents (IPSCs) were measured near the reversal potential of excitatory conductances (0 mV). In some experiments (Figure 5), the following pharmacological agents were individually added to mACSFNaHCO3 and bath-applied: gabazine (5 µM, Tocris), D-AP5 (30 µM, Tocris), NBQX (10 µM, Tocris), or TTX (1 µM, Sigma). To measure intrinsic photosensitivity (Figure S2), the following agents were applied simultaneously: L-AP4 (100 µM, Tocris), NBQX (10 µM, Tocris), gabazine (5 µM, Tocris), strychnine (2 µM, Sigma-Aldrich), and D-AP5 (30 µM, Tocris).
Retrograde Labeling
Mice were anesthetized with ketamine (1 mg/10 g) and Cholera toxin B conjugated to Alexa 488 (1 g/L; 150 nL, Thermo Fisher Scientific) or AAV9-Syn-FLEX-GCaMP6f (University of Pennsylvania Vector Core) were injected into different brain regions using a NanojectII (Drummond). Approximately 72 hr after injection, retinas were prepared for physiological recordings as described above, while brains were removed and placed in 4% paraformaldehyde overnight. The following day, brains were sectioned, stained with Neuro-Trace (Thermo Fisher Scientific), and mounted for confocal imaging. Brain slices were imaged on a confocal microscope to verify injection accuracy.
Visual Stimulation
All visual stimuli were written using the Cogent Graphics toolbox (John Romaya, Laboratory of Neurobiology at the Wellcome Department of Imaging Neuroscience, University College London) in MATLAB (MathWorks). A UV E4500 MKII PLUS II projector illuminated by a 385-nm light emitting diode (LED) (EKB Technologies) was used for stimulus presentation, except for Figures 6 and S2 in which the green (peak: 532 nm) and blue (peak: 452 nm) LEDs, respectively, of a DLP LightCrafter 4500 (Texas Instruments) were used. Stimuli were focused onto the photoreceptors via a substage condenser of an upright two-photon microscope (Scientifica). All stimuli were centered on the soma of the recorded cell. A background luminance of 1,500 rhodopsin isomerizations/rod/s (R*) was used for all visual stimuli unless otherwise noted. In Figures 1 and 4, a circular region (Figure 1 diameter: 300 µm, Figure 4 diameter: 1,200 µm) was square-wave modulated at a frequency of 0.25 Hz (Michelson contrast: 100%). In Figures 2 and S5, contrast sensitivity was tested by 1-s luminance steps within a circular region (Figure 2 diameter: 300 µm, Figure S5 diameter: 1,200 µm) every 3 s. To test spatiotemporal filtering (Figure S4), the receptive field center was stimulated by a 300-µm circle, and the surround was stimulated by a series of annuli of equal area to the center circle, but of varying inner and outer diameters. The intensities of these regions were chosen randomly from a normal distribution (root-mean-square [RMS] contrast: 40%) every 16.7 ms (refresh rate: 60 Hz) for 18–24 min. In another series of experiments, only the center was stimulated. In Figures 2, S3, and S5, textures of varying scale (2, 12, 20, 32, 48, 60 µm) were generated by convolving random binary maps with 2D Gaussian filters. Textures were masked within a 300- (Figures 2 and S3) or 1,200-µm (Figure S5) circle. The mean luminance of the textured region was equal to that of the background. In Figure 3, circles of varying diameters were presented in a pseudo random sequence and square-wave modulated at 0.25 Hz (Michelson contrast: 100%). In Figure S2, a full-field light pulse from a black background was presented for 20 s. The intensity of these light pulses (peak: 452 nm) ranged from ~1,000 R* (~2 * 1011 photons/cm2/s) to ~107 R* (~2 * 1015 photons/cm2/s), the equivalent of twilight and bright daylight, respectively (Cronin et al., 2014; Johnsen et al., 2006; Milner and Do, 2017). In Figures 5 and S6, annuli with outer diameters of 1,200 µm and inner diameters ranging from 0 µm (i.e., a 1,200-µm circle) to 1,200 µm (i.e., uniform background) were presented in a pseudorandom order as +100% contrast steps from background for 2 s. Also in Figures 5 and S6, the display was divided into a 3 × 3 grid of squares (300-µm side lengths), with the center square centered on the recorded RGC soma. With each stimulus presentation, 1 of the 9 squares increased in luminance (+100% contrast) for 2 s. The visual stimulus in Figure 6 was analogous to the visual stimulus in Figure 3, but green light (peak: 532 nm) was used instead of UV (peak: 385 nm). In Figure S7, annuli of a constant width (60 µm) and varying inner and outer diameters were presented in pseudorandom sequences as +100% contrast steps from background for 2 s.
Electrophysiology Analysis
Average spike rates or baseline-subtracted average conductances were measured during 100- to 200-ms time windows. The percentages of responses that were sustained were calculated by dividing the response 1.5 s after stimulus onset by the peak response. Excitation preference (Figure S1) was calculated as the difference between the peak excitatory and inhibitory conductances evoked by presentation of a 300-µm circle divided by the sum of these conductances. A value of 1 indicates pure excitation, and a value of −1 indicates pure inhibition. Following white noise stimulation (Figure S4), spike-triggered stimulus averages (STAs) were calculated by reverse correlation and used to map spatiotemporal receptive fields (Johnson et al., 2017). All analyses were performed using custom scripts written in MATLAB.
Morphological Analysis
To calculate dendritic field diameters, maximum intensity projections of Alexa 488 filled cells were made in Fiji. Using custom software written in MATLAB, a polygon was drawn around the edges of a cell’s dendrites, and the dendritic field diameter was calculated as the longest distance across the polygon. The area of the polygon was also calculated, and the equivalent diameter was calculated as the diameter of a circle with them same area as the polygon ( , where A is the area of the polygon). Neurite length was calculated by tracing z stack images of Alexa 488 filled cells using the Simple Neurite Tracer plugin (Longair et al., 2011) in Fiji. Dendritic branching patterns were analyzed using the Sholl Analysis plugin (Ferreira et al., 2014) in Fiji. To calculate inner plexiform layer (IPL) depth, IPL borders were detected from transmitted light images. Z stack images of filled cells were registered by their relative position within the inner plexiform layer (0%–100% from its border with inner nuclear layer to its border with the ganglion cell layer). Fluorescence intensity at each depth was measured using custom scripts written in MATLAB.
Statistics
Paired t tests and ANOVA were used to assess the statistical significance of observed differences. Unless otherwise noted, population data are reported as mean ± SEM, and n represents the numbers of cells analyzed.
Supplementary Material
Highlights.
A pixel-encoder retinal ganglion cell type (PixON-RGC) is labeled in Grik4-Cre mice
PixON-RGCs have spatially offset excitatory and inhibitory receptive fields
PixON-RGCs encode local image contrast approximately linearly
PixON-RGCs project to the dLGN and likely contribute to visual perception
Acknowledgments
We thank members of the Kerschensteiner lab for helpful comments and suggestions throughout this study. This work was supported by the NIH (EY023341, EY026978, and EY027411 to D.K., GM008151-32 to K.P.J., and the Vision Core Grant EY0268) and by an unrestricted grant from the Research to Prevent Blindness Foundation to the Department of Ophthalmology and Visual Sciences at Washington University.
Footnotes
Supplemental Information includes seven figures and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.01.037.
AUTHOR CONTRIBUTIONS
The study was conceived and designed by K.P.J. and D.K.; data were acquired by K.P.J. and L.Z.; data were analyzed and interpreted by K.P.J. and D.K.; and the manuscript was written by K.P.J. and D.K.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Baden T, Schubert T, Chang L, Wei T, Zaichuk M, Wissinger B, Euler T. A tale of two retinal domains: Near-optimal sampling of achromatic contrasts in natural scenes through asymmetric photoreceptor distribution. Neuron. 2013;80:1206–1217. doi: 10.1016/j.neuron.2013.09.030. [DOI] [PubMed] [Google Scholar]
- Baden T, Berens P, Franke K, Román Rosón M, Bethge M, Euler T. The functional diversity of retinal ganglion cells in the mouse. Nature. 2016;529:345–350. doi: 10.1038/nature16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae JA, Mu S, Kim JS, Turner NL, Tartavull I, Kemnitz N, Jordan CS, Norton AD, Silversmith WM, Prentki R, et al. Structural and functional diversity of a dense sample of retinal ganglion cells. bioRxiv. 2017 https://doi.org/10.1101/182758.
- Biederman I, Ju G. Surface versus edge-based determinants of visual recognition. Cognit. Psychol. 1988;20:38–64. doi: 10.1016/0010-0285(88)90024-2. [DOI] [PubMed] [Google Scholar]
- Borst A, Euler T. Seeing things in motion: Models, circuits, and mechanisms. Neuron. 2011;71:974–994. doi: 10.1016/j.neuron.2011.08.031. [DOI] [PubMed] [Google Scholar]
- Briggman KL, Helmstaedter M, Denk W. Wiring specificity in the direction-selectivity circuit of the retina. Nature. 2011;471:183–188. doi: 10.1038/nature09818. [DOI] [PubMed] [Google Scholar]
- Burgess CP, Lak A, Steinmetz NA, Zatka-Haas P, Bai Reddy C, Jacobs EAK, Linden JF, Paton JJ, Ranson A, Schröder S, et al. High-yield methods for accurate two-alternative visual psychophysics in head-fixed mice. Cell Rep. 2017;20:2513–2524. doi: 10.1016/j.celrep.2017.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafaro J, Rieke F. Regulation of spatial selectivity by crossover inhibition. J. Neurosci. 2013;33:6310–6320. doi: 10.1523/JNEUROSCI.4964-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Breuninger T, Euler T. Chromatic coding from cone-type unselective circuits in the mouse retina. Neuron. 2013;77:559–571. doi: 10.1016/j.neuron.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Chen SK, Badea TC, Hattar S. Photoentrainment and pupillary light reflex are mediated by distinct populations of ipRGCs. Nature. 2011;476:92–95. doi: 10.1038/nature10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland BG, Levick WR. Brisk and sluggish concentrically organized ganglion cells in the cat’s retina. J. Physiol. 1974;240:421–456. doi: 10.1113/jphysiol.1974.sp010617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin TW, Johnson S, Marshall NJ, Warrant EJ. Visual Ecology. Princeton University Press; 2014. [Google Scholar]
- Crook JD, Manookin MB, Packer OS, Dacey DM. Horizontal cell feedback without cone type-selective inhibition mediates “red-green” color opponency in midget ganglion cells of the primate retina. J. Neurosci. 2011;31:1762–1772. doi: 10.1523/JNEUROSCI.4385-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Martín A, El-Danaf RN, Osakada F, Sriram B, Dhande OS, Nguyen PL, Callaway EM, Ghosh A, Huberman AD. A dedicated circuit links direction-selective retinal ganglion cells to the primary visual cortex. Nature. 2014;507:358–361. doi: 10.1038/nature12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demb JB, Singer JH. Functional circuitry of the retina. Annu. Rev. Vis. Sci. 2015;1:263–289. doi: 10.1146/annurev-vision-082114-035334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demb JB, Zaghloul K, Sterling P. Cellular basis for the response to second-order motion cues in Y retinal ganglion cells. Neuron. 2001;32:711–721. doi: 10.1016/s0896-6273(01)00484-6. [DOI] [PubMed] [Google Scholar]
- Dhande OS, Stafford BK, Lim JA, Huberman AD. Contributions of retinal ganglion cells to subcortical visual processing and behaviors. Annu. Rev. Vis. Sci. 2015;1:291–328. doi: 10.1146/annurev-vision-082114-035502. [DOI] [PubMed] [Google Scholar]
- Egan J, Sharman RJ, Scott-Brown KC, Lovell PG. Edge enhancement improves disruptive camouflage by emphasising false edges and creating pictorial relief. Sci. Rep. 2016;6:38274. doi: 10.1038/srep38274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enroth-Cugell C, Robson JG. The contrast sensitivity of retinal ganglion cells of the cat. J. Physiol. 1966;187:517–552. doi: 10.1113/jphysiol.1966.sp008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euler T, Haverkamp S, Schubert T, Baden T. Retinal bipolar cells: Elementary building blocks of vision. Nat. Rev. Neurosci. 2014;15:507–519. doi: 10.1038/nrn3783. [DOI] [PubMed] [Google Scholar]
- Famiglietti EV. Polyaxonal amacrine cells of rabbit retina: PA2, PA3, and PA4 cells. Light and electron microscopic studies with a functional interpretation. J. Comp. Neurol. 1992;316:422–446. doi: 10.1002/cne.903160404. [DOI] [PubMed] [Google Scholar]
- Farrow K, Teixeira M, Szikra T, Viney TJ, Balint K, Yonehara K, Roska B. Ambient illumination toggles a neuronal circuit switch in the retina and visual perception at cone threshold. Neuron. 2013;78:325–338. doi: 10.1016/j.neuron.2013.02.014. [DOI] [PubMed] [Google Scholar]
- Ferreira TA, Blackman AV, Oyrer J, Jayabal S, Chung AJ, Watt AJ, Sjöström PJ, van Meyel DJ. Neuronal morphometry directly from bitmap images. Nat. Methods. 2014;11:982–984. doi: 10.1038/nmeth.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field GD, Chichilnisky EJ. Information processing in the primate retina: Circuitry and coding. Annu. Rev. Neurosci. 2007;30:1–30. doi: 10.1146/annurev.neuro.30.051606.094252. [DOI] [PubMed] [Google Scholar]
- Franke K, Berens P, Schubert T, Bethge M, Euler T, Baden T. Inhibition decorrelates visual feature representations in the inner retina. Nature. 2017;542:439–444. doi: 10.1038/nature21394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried SI, Münch TA, Werblin FS. Mechanisms and circuitry underlying directional selectivity in the retina. Nature. 2002;420:411–414. doi: 10.1038/nature01179. [DOI] [PubMed] [Google Scholar]
- Grimes WN, Schwartz GW, Rieke F. The synaptic and circuit mechanisms underlying a change in spatial encoding in the retina. Neuron. 2014;82:460–473. doi: 10.1016/j.neuron.2014.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstaedter M, Briggman KL, Turaga SC, Jain V, Seung HS, Denk W. Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature. 2013;500:168–174. doi: 10.1038/nature12346. [DOI] [PubMed] [Google Scholar]
- Huang L, Yuan T, Tan M, Xi Y, Hu Y, Tao Q, Zhao Z, Zheng J, Han Y, Xu F, et al. A retinoraphe projection regulates serotonergic activity and looming-evoked defensive behaviour. Nat. Commun. 2017;8:14908. doi: 10.1038/ncomms14908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson JS, Scanziani M. How inhibition shapes cortical activity. Neuron. 2011;72:231–243. doi: 10.1016/j.neuron.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova E, Hwang GS, Pan ZH. Characterization of transgenic mouse lines expressing Cre recombinase in the retina. Neuroscience. 2010;165:233–243. doi: 10.1016/j.neuroscience.2009.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby J, Schwartz GW. Three small-receptive-field ganglion cells in the mouse retina are distinctly tuned to size, speed, and object motion. J. Neurosci. 2017;37:610–625. doi: 10.1523/JNEUROSCI.2804-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen S, Kelber A, Warrant E, Sweeney AM, Widder EA, Lee RL, Jr, Hernández-Andrés J. Crepuscular and nocturnal illumination and its effects on color perception by the nocturnal hawkmoth Deilephila elpenor. J. Exp. Biol. 2006;209:789–800. doi: 10.1242/jeb.02053. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Tien NW, Shen N, Pearson JT, Soto F, Kerschensteiner D. Homeostatic plasticity shapes the visual system’s first synapse. Nat. Commun. 2017;8:1220. doi: 10.1038/s41467-017-01332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschensteiner D, Guido W. Organization of the dorsal lateral geniculate nucleus in the mouse. Vis. Neurosci. 2017;34:E008. doi: 10.1017/S0952523817000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuffler SW. Discharge patterns and functional organization of mammalian retina. J. Neurophysiol. 1953;16:37–68. doi: 10.1152/jn.1953.16.1.37. [DOI] [PubMed] [Google Scholar]
- Lazzerini Ospri L, Prusky G, Hattar S. Mood, the circadian system, and melanopsin retinal ganglion cells. Annu. Rev. Neurosci. 2017;40:539–556. doi: 10.1146/annurev-neuro-072116-031324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longair MH, Baker DA, Armstrong JD. Simple neurite tracer: Open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics. 2011;27:2453–2454. doi: 10.1093/bioinformatics/btr390. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martersteck EM, Hirokawa KE, Evarts M, Bernard A, Duan X, Li Y, Ng L, Oh SW, Ouellette B, Royall JJ, et al. Diverse central projection patterns of retinal ganglion cells. Cell Rep. 2017;18:2058–2072. doi: 10.1016/j.celrep.2017.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner ES, Do MTH. A population representation of absolute light intensity in the mammalian retina. Cell. 2017;171:865–876. doi: 10.1016/j.cell.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch TA, da Silveira RA, Siegert S, Viney TJ, Awatramani GB, Roska B. Approach sensitivity in the retina processed by a multifunctional neural circuit. Nat. Neurosci. 2009;12:1308–1316. doi: 10.1038/nn.2389. [DOI] [PubMed] [Google Scholar]
- Nakazawa K, Quirk MC, Chitwood RA, Watanabe M, Yeckel MF, Sun LD, Kato A, Carr CA, Johnston D, Wilson MA, Tonegawa S. Requirement for hippocampal CA3 NMDA receptors in associative memory recall. Science. 2002;297:211–218. doi: 10.1126/science.1071795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang JJ, Gao F, Wu SM. Light-evoked excitatory and inhibitory synaptic inputs to ON and OFF alpha ganglion cells in the mouse retina. J. Neurosci. 2003;23:6063–6073. doi: 10.1523/JNEUROSCI.23-14-06063.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrusca D, Grivich MI, Sher A, Field GD, Gauthier JL, Greschner M, Shlens J, Chichilnisky EJ, Litke AM. Identification and characterization of a Y-like primate retinal ganglion cell type. J. Neurosci. 2007;27:11019–11027. doi: 10.1523/JNEUROSCI.2836-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piscopo DM, El-Danaf RN, Huberman AD, Niell CM. Diverse visual features encoded in mouse lateral geniculate nucleus. J. Neurosci. 2013;33:4642–4656. doi: 10.1523/JNEUROSCI.5187-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusky GT, West PW, Douglas RM. Behavioral assessment of visual acuity in mice and rats. Vision Res. 2000;40:2201–2209. doi: 10.1016/s0042-6989(00)00081-x. [DOI] [PubMed] [Google Scholar]
- Rompani SB, Müllner FE, Wanner A, Zhang C, Roth CN, Yonehara K, Roska B. Different modes of visual integration in the lateral geniculate nucleus revealed by single-cell-initiated transsynaptic tracing. Neuron. 2017;93:1519. doi: 10.1016/j.neuron.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roska B, Werblin F. Vertical interactions across ten parallel, stacked representations in the mammalian retina. Nature. 2001;410:583–587. doi: 10.1038/35069068. [DOI] [PubMed] [Google Scholar]
- Roska B, Molnar A, Werblin FS. Parallel processing in retinal ganglion cells: How integration of space-time patterns of excitation and inhibition form the spiking output. J. Neurophysiol. 2006;95:3810–3822. doi: 10.1152/jn.00113.2006. [DOI] [PubMed] [Google Scholar]
- Saalmann YB, Kastner S. Cognitive and perceptual functions of the visual thalamus. Neuron. 2011;71:209–223. doi: 10.1016/j.neuron.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbah S, Gemmer JA, Bhatia-Lin A, Manoff G, Castro G, Siegel JK, Jeffery N, Berson DM. A retinal code for motion along the gravitational and body axes. Nature. 2017;546:492–497. doi: 10.1038/nature22818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Masland RH. The types of retinal ganglion cells: Current status and implications for neuronal classification. Annu. Rev. Neurosci. 2015;38:221–246. doi: 10.1146/annurev-neuro-071714-034120. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz GW, Okawa H, Dunn FA, Morgan JL, Kerschensteiner D, Wong RO, Rieke F. The spatial structure of a nonlinear receptive field. Nat. Neurosci. 2012;15:1572–1580. doi: 10.1038/nn.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R, Hoon M, Baudin J, Okawa H, Wong RO, Rieke F. Cellular and circuit mechanisms shaping the perceptual properties of the primate fovea. Cell. 2017;168:413–426. doi: 10.1016/j.cell.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabio ME, Sabbah S, Quattrochi LE, Ilardi MC, Fogerson PM, Leyrer ML, Renna JM, Kim MT, Kim I, Schiel M, Briggman KL, Berson DM. The M5 cell: A color-opponent intrinsically photosensitive retinal ganglion cell. Neuron. 2018;97:150–163. doi: 10.1016/j.neuron.2017.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szél A, Röhlich P, Caffé AR, Juliusson B, Aguirre G, Van Veen T. Unique topographic separation of two spectral classes of cones in the mouse retina. J. Comp. Neurol. 1992;325:327–342. doi: 10.1002/cne.903250302. [DOI] [PubMed] [Google Scholar]
- Tien NW, Soto F, Kerschensteiner D. Homeostatic plasticity shapes cell-type-specific wiring in the retina. Neuron. 2017;94:656–665. doi: 10.1016/j.neuron.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wyk M, Wässle H, Taylor WR. Receptive field properties of ON- and OFF-ganglion cells in the mouse retina. Vis. Neurosci. 2009;26:297–308. doi: 10.1017/S0952523809990137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völgyi B, Xin D, Amarillo Y, Bloomfield SA. Morphology and physiology of the polyaxonal amacrine cells in the rabbit retina. J. Comp. Neurol. 2001;440:109–125. doi: 10.1002/cne.1373. [DOI] [PubMed] [Google Scholar]
- Wässle H, Grünert U, Röhrenbeck J, Boycott BB. Cortical magnification factor and the ganglion cell density of the primate retina. Nature. 1989;341:643–646. doi: 10.1038/341643a0. [DOI] [PubMed] [Google Scholar]
- Wei W, Hamby AM, Zhou K, Feller MB. Development of asymmetric inhibition underlying direction selectivity in the retina. Nature. 2011;469:402–406. doi: 10.1038/nature09600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonehara K, Balint K, Noda M, Nagel G, Bamberg E, Roska B. Spatially asymmetric reorganization of inhibition establishes a motion- sensitive circuit. Nature. 2011;469:407–410. doi: 10.1038/nature09711. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kim IJ, Sanes JR, Meister M. The most numerous ganglion cell type of the mouse retina is a selective feature detector. Proc. Natl. Acad. Sci. USA. 2012;109:E2391–E2398. doi: 10.1073/pnas.1211547109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.